ABSTRACT
Nervous necrosis virus (NNV) is a devastating pathogen of cultured marine fish and has affected more than 40 fish species. NNV belongs to the betanodaviruses of Nodaviridae and is a nonenveloped icosahedral particle with 2 single-stranded positive-sense RNAs. To date, knowledge regarding NNV entry into the host cell remains limited, and no NNV-specific receptor protein has been published. Using grouper fin cell line GF-1 and purified NNV capsid protein in a virus overlay protein binding assay (VOPBA), grouper heat shock cognate protein 70 (GHSC70) and grouper voltage-dependent anion selective channel protein 2 (GVDAC2) were investigated as NNV receptor protein candidates. We cloned and sequenced the genes for GHSC70 and GVDAC2 and expressed them in Escherichia coli for antiserum preparation. Knockdown of the expression of GHSC70 and GVDAC2 genes with specific short interfering RNAs (siRNAs) significantly downregulated viral RNA expression in NNV-infected GF-1 cells. By performing an immunoprecipitation assay, we confirmed that GHSC70 interacted with NNV capsid protein, while VDAC2 did not. Immunofluorescence staining and flow cytometry analysis revealed the presence of the GHSC70 protein on the cell surface. After a blocking assay, we detected the NNV RNA2 levels after 1 h of adsorption to GF-1 cells; the level was significantly lower in the cells pretreated with the GHSC70 antiserum than in nontreated cells. Therefore, we suggest that GHSC70 participates in the NNV entry of GF-1 cells, likely functioning as an NNV receptor or coreceptor protein.
IMPORTANCE Fish nodavirus has caused mass mortality of more than 40 fish species worldwide and resulted in huge economic losses in the past 20 years. Among the four genotypes of fish nodaviruses, the red-spotted grouper nervous necrosis virus (RGNNV) genotype exhibits the widest host range. In our previous study, we developed monoclonal antibodies with high neutralizing efficiency against grouper NNV in GF-1 cells, indicating that NNV-specific receptor(s) may exist on the GF-1 cell membrane. However, no NNV receptor protein has been published. In this study, we found GHSC70 to be an NNV receptor (or coreceptor) candidate through VOBPA and provided several lines of evidence demonstrating that GHSC70 protein has a role in the NNV entry step of GF-1 cells. To the best of our knowledge, this is the first report identifying grouper HSC70 and its role in NNV entry into GF-1 cells.
INTRODUCTION
The viral life cycle can be divided into the following stages: entry, genome replication, protein expression, assembly, and release from host cells. Different viruses employ various strategies to enter host cells, such as clathrin-mediated endocytosis, caveolar endocytosis, clathrin- and caveolin-independent pathways, and macropinocytosis. Enveloped viruses use membrane fusion or interact with cellular receptors to cross the cell membrane, while most nonenveloped viruses use receptor-mediated endocytosis to enter the host cells (1). Cellular receptors determine the host range of viruses. Numerous viral receptors have been identified in mammalian cells, such as the IgG superfamily for poliovirus, integrin for echovirus, and coxsackie-adenovirus receptor (CAR) protein for adenovirus (2–4). However, only a few viral receptors of aquatic viruses have been reported, including Penaeus monodon RAB7 (PmRab7) protein and β-integrin (5, 6) for white spot syndrome virus (WSSV) in shrimp cells and several receptors for infectious pancreatic necrosis virus (IPNV) (7).
Nervous necrosis virus (NNV) is the causative agent of viral nervous necrosis (VNN) disease, also known as viral encephalopathy and retinopathy (VER), and has resulted in high mortality of cultured marine fish, especially at the larval stage (8, 9). NNV has a wide host range and can infect fish from at least 5 orders, totaling 16 families of fish species (9). The target organ of NNV is the central nervous system, and infection results in the vacuolation of the brain and retina (10). The NNV genome consists of 2 single-stranded positive-sense RNAs. RNA1 encodes RNA-dependent RNA polymerase (RdRp), and RNA2 encodes viral capsid protein (11). RNA3, a subgenome of RNA1, encodes the B1 and B2 proteins (12). The B1 protein is expressed at the early stage of infection, exhibiting an antinecrotic cell death function by reducing mitochondrial membrane potential (MMP) loss and, thus, enhancing cell viability (13). The B2 protein binds to NNV intermediate double-stranded RNA (dsRNA) and prevents host RNA interference (RNAi)-mediated cleavage (14).
The NNV coat protein is the only structural protein of the virion and has been proved to determine the host range (15). NNV-specific monoclonal antibodies with high neutralizing titers were developed (16), suggesting that NNV-specific receptors exist on the host cell membrane. NNV has been reported to infect cells through receptor-mediated endocytosis and macropinocytosis, and the sialic acid residue is the binding target for NNV during viral binding in susceptible SSN-1 cells (17). However, no NNV-specific receptor protein has been reported. The objective of this study was to identify candidate proteins involved in the NNV entry of GF-1 cells through virus overlay protein binding assay (VOPBA), with heat shock cognate protein 70 (HSC70) and voltage-dependent anion selective channel protein 2 (VADC2) as targets. We cloned HSC70 and VDAC2 and conducted functional assays of these proteins in NNV-infected cells. Finally, we concluded that HSC70 has a role in NNV entry.
MATERIALS AND METHODS
Cells, virus, and NNV-specific antibodies.
The GF-1 cell line, which is highly permissive to NNV, was derived from the fin tissue of grouper (Epinephelus coioides) (18) and was subcultured in Leibovitz's L-15 medium (Gibco) with 5% fetal bovine serum (FBS) (Gibco). NNV isolated from grouper larvae with VNN disease (19) was used in this study. NNV was proliferated in GF-1 cells with a multiplicity of infection (MOI) of 1 and was incubated at 28°C in the L-15 medium with 1% FBS. After complete cytopathic effect (CPE) was apparent, the cultured supernatant was collected by centrifugation at 12,000 × g for 10 min and then titrated in GF-1 cells. The NNV particles were purified using the procedure described by Chi et al. (19). The specific polyclonal antibodies (serum) against NNV were prepared as in our previous work (20).
VOPBA.
The membrane-associated proteins of GF-1 cells were extracted using a Mem-PER protein purification kit (Thermo), following the manufacturer's instructions. The detergent used to extract membrane fractions was removed using a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample preparation kit (Thermo). Total membrane-associated proteins were separated using 10% SDS-PAGE and then transferred to polyvinylidene fluoride (PVDF) membranes (Millipore) as described previously (21). The membrane was blocked with Tris-buffered saline (TBS) buffer containing 5% skim milk and 3% bovine serum albumin (BSA) overnight at 4°C, transferred to protein binding buffer (50 mM Tris-HCl, 5 mM CaCl2, 10 mM MgCl2, pH 6.5), and incubated with 10 μg of purified NNV overnight at 4°C. The membrane was washed 3 times with TBS buffer containing 0.1% Tween 20 (TBST) and then incubated with an NNV-specific rabbit antiserum (1:10,000) diluted in blocking buffer at room temperature for 1 h. After 3 washes with TBST, the membrane was reacted with peroxidase-conjugated goat anti-rabbit serum (KPL) (1:10,000) diluted in TBST at room temperature for 1 h. After extensive washing with TBST, the antigen signal was developed by adding LumiGLO chemiluminescent substrate (KPL) and was visualized through autoradiography.
MS analysis.
Three GF-1 cell membrane-associated proteins (GFMPs) of interest, including GFMP-70 and GFMP-28, were excised from the gels, reduced with DTE buffer (50 mM dithioerythreitol and 25 mM ammonium bicarbonate) at 37°C for 1 h, alkylated with IAA buffer (100 mM iodoacetamide and 25 mM ammonium bicarbonate) at room temperature for 1 h in the dark, washed twice with 25 mM ammonium bicarbonate buffer (pH 8.5) in 50% acetonitrile for 15 min each time, dehydrated with 100% acetonitrile for 5 min, vacuum dried, and rehydrated with 0.0225 μg of trypsin in 25 mM ammonium bicarbonate (pH 8.5) at 37°C for 16 h. The tryptic peptides were extracted twice with 5% trifluoroacetic acid (TFA) in 50% acetonitrile for 1.5 min with sonication. The peptide samples were pooled and dried using speed vacuum, dissolved in 0.1% formic acid and 50% acetonitrile, and analyzed using liquid chromatography-tandem mass spectrometry (LC-MS/MS). The coding region of each protein was identified by blasting against the United States National Library of Medicine (NLM) National Center for Biotechnology Information (NCBI) protein database and expressed sequence tag (EST) sequences by using Mascot Server (Matrix Science); GFMP-70 exhibited 42% coverage with HSC70, whereas GFMP-28 showed 20% coverage with voltage-dependent anion-selective channel protein 2 (VDAC2).
Cloning of GFMP fragment.
The GF-1 cells were scraped from a 25-cm2 tissue culture flask and spun down in a 1.5-ml microtube. Total RNAs were extracted using TRIzol (Invitrogen) reagent according to the manufacturer's instructions and dissolved in diethyl pyrocarbonate (DEPC)-treated water. For reverse transcription (RT), 2 μg of extracted RNA was incubated at 42°C for 1 h in a reaction buffer comprised of 40 μM deoxynucleoside triphosphates (dNTPs), 50 mM Tris-HCl buffer, 30 nM oligo(dT)20 primer, 11.7 mM dithiothreitol (DTT), 40 U of RNasin (Promega), and 60 U of Moloney murine leukemia virus (MMLV) reverse transcriptase (Promega). For PCR, an aliquot (5 μl) of cDNA was amplified in 25 μl of PCR buffer containing 0.4 μM each degenerate primer set (deHSC702-F/R or deVDAC22-F/R), 0.25 mM dNTPs, and 1 U of Taq DNA polymerase (New England BioLabs). PCR was conducted in a GeneAmp 2700 PCR system (Applied Biosystems) with a denaturing step of 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 44°C (for deHSC702-F/R primer set) or 50°C (for deVDAC22-F/R primer set) for 30 s, and 72°C for 1 min, with a final extension of 72°C for 5 min. The PCR products, including the 938-bp deHSC702-F/R primer set and the 377-bp deVDAC22-F/R primer set, were cloned into pGEM-T easy vectors (Promega) and sequenced by Mission Biotech Company (Taiwan).
RACE cloning of full-length GFMP cDNAs.
Rapid amplification of cDNA ends (RACE) was conducted using GeneRacer primers (Invitrogen), including 5′ abridged anchor primer (AAP), 3′ adapter primer (3′ AP), and abridged universal amplification primer (AUAP). The gene-specific primers (GSP) used in RACE were designed from the 938-bp-fragment sequence of grouper heat shock cognate protein 70 (GHSC70) and the 377-bp-fragment sequence of GVDAC2, which were obtained as described above, using primers including HSC70RA934-R1, HSC70RA532-R2, HSC70RA64-F12, HSC70RA312-F22, VDAC2RA03-F12, VDAC2RA69-F22, VDAC2RA299-R1, and VDAC2RA241-R2 (Table 1).
TABLE 1.
Primers and siRNAs used in this study
| Primer | Sequence (5′–3′)a |
|---|---|
| deHSC702-F | AAGATGAARGARATTGCWGARGCMTACCT |
| deHSC702-R | ACACCRGGYTGRTTGTCWGARTARGTGGT |
| deVDAC22-F | AGAAGTGGAAYACYGAYAAYACCYTGG |
| deVDAC22-R | CACYTTYTGRTAGATRGARCCWCCAAAYTC |
| HSC70RA934-R1 | CGGGCTGGTTGTCAGAATAGGTGGTGAAGG |
| HSC70RA532-R2 | CGAAGCGAGCCCTGGTGATGGAGGTGTA |
| HSC70RA64-F12 | CCAGCTTACTTCAACGATTC |
| HSC70RA312-F22 | AGGAGAGGACTTCGATAACC |
| VDAC2RA03-F12 | AAGTGGAATACTGACAATACCC |
| VDAC2RA69-F22 | AAGCTGACCTTTGACACATC |
| VDAC2RA299-R1 | GGGCGAAGTTGTTCTGCGCCAGTTTGGA |
| VDAC2RA241-R2 | GGCCAGCCAGCCCTCGTATCCCAGCAC |
| AAP | GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGII |
| AUAP | GGCCACGCGTCGACTAGTAC |
| 3′ AP | GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT |
| GHSC70_BamHI_F | CGCGGATCCCTGATCAAGCGCAACACCACCATTCCAA |
| GHSC70_HindIII_R | CCCAAGCTTGGGGTTGCAGACCTTCTCCACCTCTTTCT |
| GVDAC2_BamHI_F | CGCGGATCCATGGCTGTTCCTCCTGCGTA |
| GVDAC2_HindIII_R | CCCAAGCTTCTACGCCTCCAGCTCGAACCC |
| NNVR3 | CGAGTCAACACGGGTGAAGA |
| GHSC70_RT235_F | AACTGTACCAGCTTACTTCAACGAT |
| GHSC70_RT235_R | AGTGGACTTCACTTCAAAGATTCC |
| GVDAC2_RT229_F | AAACTGGCGCAGAACAACTT |
| GVDAC2_RT229_R | TGTTCACTTTGGCAGACAGC |
| Gactin235_RT_F | GGCCGCGACCTCACAGACTACCTC |
| Gactin235_RT_R | CCTCTGGGCAACGGAACCTCTCAT |
| NNVRNA2_RT_F | CAGTCCGACCTCAGTACAC |
| NNVRNA2_RT_R | AACACTCCAGCGACACAG |
| GHSC70-siRNA | r(CGGUGUUCCUCAGAUUGAA)dTdT |
| GVDAC2-siRNA | r(GAAGUGGAACACUGACAAC)dTdT |
| Nonsilencing control siRNA | r(UUCUUCGAACGUGUCACGU)dTdT |
R = A or G, W = A or T, M = A or C, and Y = C or T.
For 5′-RACE, the total RNAs of GF-1 were reverse transcribed into cDNAs by using GSP HSC70RA934-R1 and VDAC2RA299-R1, respectively. The cDNAs were purified using the QIAquick gel extraction kit (Qiagen) and eluted with buffer EB from the kit. Poly(C) tails were synthesized at the ends of cDNAs with 5 U of terminal transferase (New England BioLabs) in reaction buffer (1× NE Buffer 4, 0.25 mM CoCl2, 0.2 mM dCTP) and incubated at 37°C for 30 min. The reaction was stopped by heating at 95°C for 5 min. Poly(C)-tailed cDNAs were amplified with PCR using AAP and the first 5′-RACE GSP (HSC70RA934-R1 for GHSC70 and VDAC2RA299-R1 for GVDAC2). The PCR program consisted of 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 1 min, with a final extension of 72°C for 10 min. Nested PCR was performed by using AUAP and the second 5′-RACE GSP (HSC70RA532-R2 for GHSC70 or VDAC2RA241-R2 for GVDAC2) with the same PCR program as in the first 5′-RACE amplification. PCR products were cloned into pGEM-T easy vectors and sequenced.
For 3′-RACE PCR, total RNAs were reverse transcribed to cDNA with the 3′ AP primer. The cDNA was amplified with a 3′-RACE GSP (HSC70RA64-F12 for GHSC70 and VDAC2RA03-F12 for GVDAC2). The PCR program consisted of 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min, with a final extension of 72°C for 10 min. Nested PCR was performed by using AUAP and the second 3′-RACE GSP (HSC70RA312-F22 for GHSC70 and VDAC2RA69-F22 for GVDAC2) with the same PCR program as in the first 3′-RACE PCR amplification. PCR products were cloned into pGEM-T easy vectors and sequenced. The 5′- and 3′-RACE sequences overlapped by 220 bp with GHSC70 and 172 bp with GVDAC2, respectively.
The amino acid sequence alignments of GHSC70 and GVDAC2 in reference to those of various animal species were performed by blasting against the NCBI protein database (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Phylogenetic trees (neighbor-joining trees) were devised using MEGA version 5 (22), with bootstrapping values extracted from 1,000 replicates.
RNAi knockdown of GHSC70 or GVDAC2.
Short interfering RNAs (siRNAs) designed by GenePharma Company (Shanhai, China) were used to knock down GHSC70 or GVDAC2 in GF-1 cells. The siRNA sequences of GHSC70, GVDAC2, and a nonsilencing siRNA control (NS-siRNA) are shown in Table 1. To transfect the siRNAs, 5 μl (1 × 10−10 mol) of siRNA and 2 μl of Lipofectamine 2000 (Invitrogen) were diluted separately with 100 μl of L-15 medium, incubated at room temperature for 5 min, and then mixed and incubated for another 15 min at room temperature. Finally, 0.2 ml of the mixture was added to GF-1 cells preseeded in a 12-well plate.
The GF-1 cells were transfected separately with GHSC70-specific siRNA, GVDAC2-specific siRNA, or NS-siRNA for 24 h and then infected with NNV (MOI of 10). After 24 h of NNV infection, the total RNAs of the cells were extracted for real-time reverse transcription (RT)-PCR analysis of the target genes, and the levels of the GHSC70 and GVDAC2 proteins were analyzed through Western blotting. The virions extracted from both sets of NNV-infected cells after 24 h postinfection (hpi) were frozen and thawed 3 times and then titrated.
Real-time RT-PCR.
Total RNAs from siRNA-transfected and NNV-infected cells were extracted and reverse transcribed into cDNA by using dT20 and NNVR3 primers. The expression levels of GHSC70, GVDAC2, NNV RNA2, and actin (as internal control) were assayed using real-time PCR. The sequences of the primer sets for GHSC70 (GHSC70_RT235_F/R), GVDAC2 (GVDAC2_RT229_F/R), NNV RNA2 (NNVRNA2_RT_F/R), and actin (Gactin235_RT_F/R) are detailed in Table 1. The cDNA was added into real-time PCR buffer, which included 0.5 mM forward and reverse primers in 1× iQ SYBR green supermix (Bio-Rad). Amplification was performed on an iCycler iQ real-time PCR detection system (Bio-Rad) with a PCR program of 94°C for 3 min, followed by 40 cycles of 94°C for 20 s, 55°C for 20 s, and 72°C for 20 s, and fluorescence detection was set at 85°C for 20 s. All samples were analyzed in triplicate. The specificity of the PCR products was confirmed by conducting melting curve analysis, and the relative gene expression levels were normalized with the expression level of the internal control (actin).
Preparation of rabbit antisera against GHSC70 and GVDAC2.
The partial sequence of GHSC70 and the complete sequence of GVDAC2 were amplified with primer sets GHSC70_BamHI_F and GHSC70_HindIII_R for GHSC70 and GVDAC2_BamHI_F and GVDAC2_HindIII_R for GVDAC2. The amplified products were then cloned into pGEM-T easy vectors. The BamHI and HindIII restriction sites were used to insert sequences into prokaryotic expression vector pQE30 (Qiagen), and the plasmid inserts were named pQE-GHSC70p and pQE-VDAC2, respectively. The 2 recombinant plasmids were transformed into Escherichia coli M15 (Qiagen) to generate 6-histidine-tagged recombinant proteins. Recombinant GHSC70p and GVDAC2 were purified through Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) to prepare the rabbit antisera.
Western blotting.
Total GF-1 proteins were extracted through sonication for 10 min, using protein lysis buffer containing 50 mM Tris-HCl (pH 8), 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1 mM DTT, and 1× protease inhibitor (Roche). The total protein lysate was centrifuged at 10,000 × g at 4°C for 10 min, the concentration was adjusted to 10 μg per well for 10% SDS-PAGE, and the proteins were transferred to a PVDF membrane. The membrane was blocked with 5% skim milk in TBST buffer at room temperature for 1 h and then incubated separately with 1:1,000-diluted rabbit preimmune serum and antiserum against GHSC70 and GVDAC2 at room temperature for 1 h. After extensive washing, the antigen signal was developed by using LumiGLO chemiluminescent substrate (KPL) and visualized with autoradiography. Protein intensities were analyzed using ImageJ software (23).
IP assay.
The GF-1 cells were infected with NNV (MOI of 100) for 24 h, and total proteins were harvested using lysis buffer. Proteins extracted from noninfected GF-1 cells and purified NNV were used as the controls. Immunoprecipitation (IP) and Western blotting (WB) assays were conducted using an ImmunoCruz IP/WB optima F system (Santa Cruz) according to the manufacturer's instructions. The GHSC70- and GVDAC2-NNV capsid protein complexes were immunoprecipitated using the rabbit antiserum against NNV and then analyzed with Western blotting by using the rabbit antisera against GHSC70 and GVDAC2 separately. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), identified with anti-GAPDH antibodies (GeneTex, Taiwan), was used as the negative control.
Immunofluorescence staining.
The GF-1 cells, preseeded on 2 cover glasses, were fixed with 10% formalin in phosphate-buffered saline (PBS) for 10 min at room temperature. One cover glass was treated with 0.5% Triton X-100 (Sigma) in PBS for membrane permeabilization, and the other was not. After 3 washes with PBS, the cells were incubated with blocking buffer (5% BSA in PBS) at room temperature for 1 h and then reacted with 1:200-diluted rabbit anti-GHSC70 for 1 h. After washing, the cells were incubated with fluorescein isothiocyanate (FITC)-labeled anti-rabbit antiserum for 1 h. The cell nuclei were stained with Hoechst 33258 (1:500 diluted in PBS) for 30 min. Finally, the cells were observed using confocal microscopy (Technology Commons, National Taiwan University).
To examine the colocalization of GHSC70 and NNV on intact GF-1 cells, the cells were infected with NNV (MOI of 1,000) on ice for 1 h of adsorption. After 3 washes with PBS, the cells were incubated with rabbit anti-GHSC70 or mouse anti-NNV antibodies. The cells were washed with PBS 3 times and separately immunostained with rhodamine-labeled anti-rabbit antibodies or FITC-conjugated anti-mouse antibodies.
Flow cytometry analysis.
The GF-1 cells, preseeded in a 25-cm2 flask, were suspended using 0.15% trypsin (Sigma). After PBS washing, the cells were fixed with 10% formalin in PBS at room temperature for 10 min and then incubated with 1:200-diluted rabbit preimmune serum or antiserum against GHSC70 at room temperature for 1 h. After PBS washing, the cells were incubated with the FITC-labeled anti-rabbit antiserum at room temperature for 1 h. After three washes with PBS, the cells were analyzed using flow cytometry (FACSCanto II; BD, USA). As a negative control, a mock-treatment group that had no interaction with primary antibodies was incubated with FITC-labeled secondary antibodies.
Blocking assay.
The GF-1 cells, preseeded on 6-well plates, were incubated separately with rabbit antiserum against GHSC70 or GVDAC2 at 28°C for 1 h. After 3 washes with the L-15 medium, the cells were infected with NNV (MOI of 10) for 1 h. Following viral adsorption, the cells were washed 3 times, and the total RNA was extracted from the cells for real-time RT-PCR detection of NNV RNA2. The control cells were infected directly with NNV without an antiserum pretreatment against GHSC70 or GVDAC2.
Amino acid sequence accession numbers.
GHSC70 was deposited in GenBank under accession number JX207115. GVDAC2 was deposited in GenBank under accession number JX207116.
RESULTS
Identification of GF-1 cell membrane proteins bound with NNV.
To identify the GF-1 cell membrane proteins that interacted with NNV capsid protein, we conducted VOPBA, followed by LC-MS/MS analysis. Figure 1 shows a PVDF membrane on which were detected three NNV-binding proteins from the membrane fraction of GF-1 cells, with molecular masses of 28 kDa, 40 kDa, and 70 kDa, which were named GFMP-28, GFMP-40, and GFMP-70. By conducting an analysis using the MASCOT program, we found GFMP-28, GFMP-40, and GFMP-70 to be similar to VDAC2, beta-actin, and HSC70 proteins (Table 2). Because beta-actin was reportedly located in the inner face of the cell membrane, we chose to focus on GHSC70 and GVDAC2 for the following studies.
FIG 1.
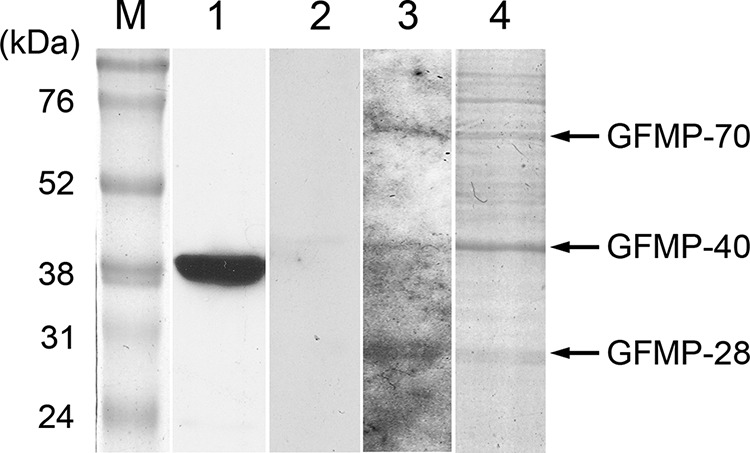
Detection of GF-1 cell membrane proteins (GFMPs) with interactivity with NNV capsid protein by VOPBA assay. Total GFMPs were resolved by 10% SDS-PAGE and transferred to PVDF membrane. Purified-NNV overlay was then performed. Bound virions were detected by rabbit anti-NNV polyclonal antibodies. Lanes: M, molecular mass marker; 1, Western blot of purified NNV using rabbit anti-NNV polyclonal antibodies; 2, VOPBA without purified NNV overlay; 3, VOPBA with NNV overlay; 4, Coomassie blue-stained profile of an electrophoresis gel similar to that used to excise the corresponding bands for LC-MS/MS analysis.
TABLE 2.
Analysis of GFMPs by LC-MS/MS and MASCOT program
| Protein | Similar protein, organism, accession no. | Size (kDa) | Score | Coverage (%) |
|---|---|---|---|---|
| GFMP-28 | Voltage-dependent anion-selective channel protein 2, Salmo salar, ACI32932 | 33 | 418 | 20 |
| GFMP-40 | Beta-actin, Monopterus albus, AAQ21403 | 41 | 2,241 | 79 |
| GFMP-70 | Heat shock cognate 70-kDa protein, Carassius gibelio, AAO43731 | 71 | 1,332 | 42 |
Cloning and characterization of the GHSC70 and GVDAC2 genes of GF-1 cells.
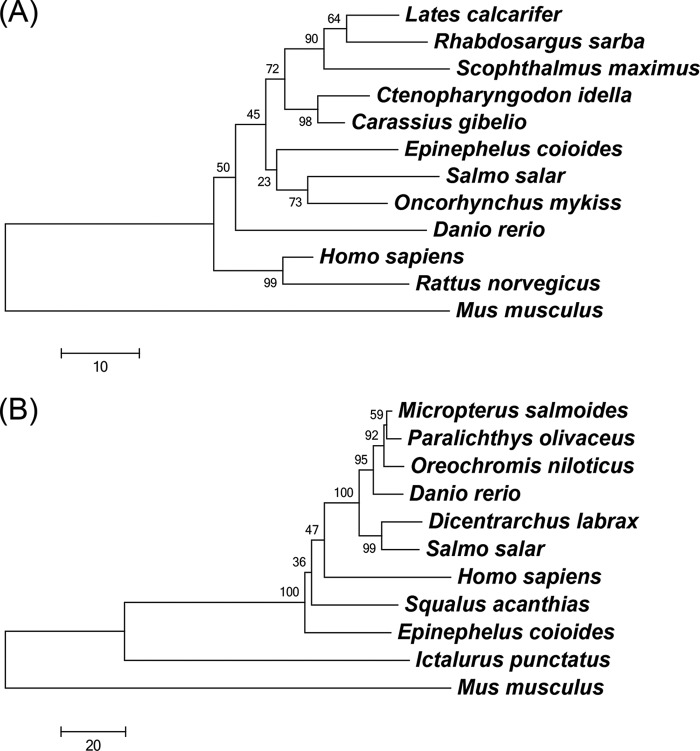
The full-length cDNA of GHSC70 contains a 72-bp 5′-untranslated region (UTR), a 1,953-bp open reading frame (ORF), and a 226-bp 3′-UTR. The encoded GHSC70 protein consists of 650 amino acids with a predicted molecular mass of 71 kDa, containing an actin-like ATPase domain at the amino terminus and a peptide-binding domain at the carboxy terminus, which are conserved domains in the HSC70 superfamily. BLAST analysis of GHSC70 revealed the amino acid sequence identities with HSC70 in other species, showing that GHSC70 shared 96% homology with HSC70 sequences of rainbow trout (Oncorhynchus mykiss), Prussian carp (Carassius gibelio), and humans (Homo sapiens) (Table 3). However, the neighbor-joining tree for comparison of the HSC70 amino acid sequences of different organisms revealed that GHSC70 had substantially greater homology with Atlantic salmon (Salmo salar) and rainbow trout (Oncorhynchus mykiss) clusters than with other fish species (Fig. 2A).
TABLE 3.
Amino acid sequence identities between GHSC70a of Epinephelus coioides and HSC70 proteins of other species
| Species | GenBank accession no. | Amino acid residues compared | % Identity |
|---|---|---|---|
| Oncorhynchus mykiss | NP001117704 | 1–612 | 96 |
| Carassius gibelio | AAO43731 | 1–614 | 96 |
| Homo sapiens | NM006597 | 1–613 | 96 |
| Salmo salar | ACN11074 | 1–614 | 95 |
| Lates calcarifer | AEH27543 | 1–614 | 95 |
| Ctenopharyngodon idella | ADX32515 | 1–650 | 95 |
| Rhabdosargus sarba | AAR97293 | 1–614 | 94 |
| Danio rerio | NP001186941 | 1–612 | 94 |
| Scophthalmus maximus | ABU50777 | 1–617 | 93 |
| Rattus norvegicus | NP077327 | 1–613 | 97 |
| Mus musculus | AAC84169 | 1–641 | 82 |
The GHSC70 GenBank accession no. is JX207115.
FIG 2.
Phylogenetic trees of HSC70 family (A) and VDAC2 family (B) proteins from different organisms using the neighbor-joining method. The trees are based on the alignment of amino acid sequences corresponding to the regions of HSC70 proteins listed in Table 3 and VDAC2 proteins listed in Table 4. Bootstrap support values are given at the branches. The scale for branch length (substitutions/site) is shown below the tree.
The complete cDNA sequence of GVDAC2 consists of a 72-bp 5′-UTR, an 852-bp ORF, a 725-bp 3′-UTR, and an ORF coding for 283 amino acids with a molecular mass of 30 kDa, containing a porin3 domain which is a conserved domain of the VDAC family. Protein BLAST analysis revealed that GVDAC2 had high identity (82%) to VDAC2 sequences of channel catfish (Ictalurus punctatus) and largemouth bass (Micropterus salmoides) (Table 4). The neighbor-joining tree analysis (Fig. 2B) revealed that GVDAC2 has higher homology to VDAC2 of spiny dogfish (Squalus acanthias) than to those of other species.
TABLE 4.
Amino acid sequence identities between GVDAC2a of Epinephelus coioides and VDAC2 of other species.
| Species | GenBank accession no. | Amino acid residues compared | % Identity |
|---|---|---|---|
| Ictalurus punctatus | NP_001188159 | 1–281 | 82 |
| Micropterus salmoides | AEK25826 | 1–283 | 82 |
| Oreochromis niloticus | XP_003452012 | 1–283 | 81 |
| Squalus acanthias | AAF65254 | 1–283 | 81 |
| Danio rerio | AAH62525 | 1–283 | 80 |
| Paralichthys olivaceus | ABH07379 | 1–283 | 80 |
| Dicentrarchus labrax | CBN81505 | 1–283 | 77 |
| Salmo salar | ACI34280 | 1–283 | 77 |
| Mus musculus | NP_035825 | 13–295 | 75 |
| Homo sapiens | AAD54939 | 1–283 | 73 |
The GVDAC2 GenBank accession no. is JX207116.
Knockdown of GHSC70 or GVDAC2 reduced the NNV RNA2 level in GF-1 cells.
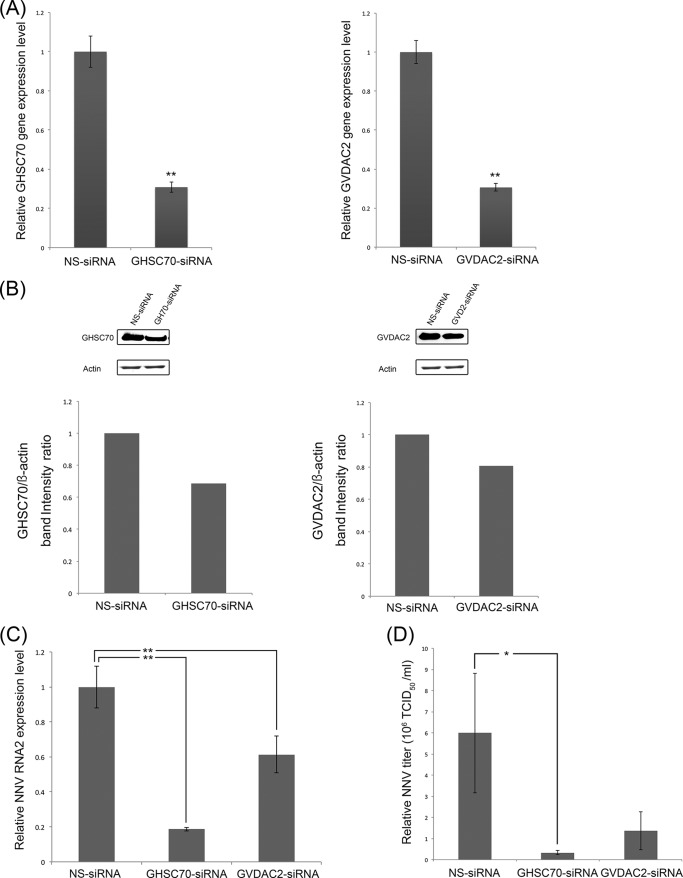
To elucidate the impact of GHSC70 and GVDAC2 on NNV infection, we transfected GHSC70-specific and GVDAC2-specific siRNAs to GF-1 cells to knock down GHSC70 and GVDAC2 gene expression. The morphology and viability of the GF-1 cells at 48 h posttransfection for both sets of siRNAs were similar to those of normal GF-1 cells. Compared with the cells transfected with NS-siRNA, the expression levels of GHSC70 and VDAC2 genes were down to 30% (Fig. 3A), and the levels of GHSC70 and GVDAC2 proteins were downregulated to 70% and 77%, respectively (Fig. 3B).
FIG 3.
siRNA downregulation of GHSC70 and GVDAC2 gene expression and its effects on NNV replication. GF-1 cells were separately transfected with GHSC70- and GVDAC2-specific siRNAs for downregulation, and nonsilencing siRNA (NS-siRNA) was used as a negative control. (A, B) The gene expression levels of GHSC70 or GVDAC2 (A) and their protein levels (B) were separately analyzed by real-time PCR and Western blotting at 24 h posttransfection. (C, D) After siRNA transfection, the cells were infected with NNV (MOI of 100), and NNV RNA2 expression levels (C) and viral titers (D) were examined at 24 h postinfection. The level in the negative control was regarded as 1. Error bars indicate standard deviations. *, P < 0.05; **, P < 0.01 (n = 3).
The transfected cells were further infected with NNV for 24 h, and we examined the levels of NNV RNA2 expression and viral titers by conducting real-time RT-PCR and titration. The relative expression levels of NNV RNA2 in the GHSC70 and GVDAC2 knockdown cells decreased by 80% and 40%, respectively (Fig. 3C). The NNV titer of the GHSC70 knockdown cells was 0.3 × 106 50% tissue culture infective dose (TCID50) ml−1, which was significantly lower than that of NS-siRNA knockdown cells (6 × 106 TCID50 ml−1) (Fig. 3D). Similarly, the viral titer of GVDAC knockdown cells decreased to 1.3 × 106 TCID50 ml−1, which was one-sixth that of the NS-siRNA knockdown cells (Fig. 3D).
GHSC70 immunoprecipitated with NNV capsid protein.
We constructed pQE-GHSC70p and pQE-VDAC2 to express the recombinant proteins for rabbit antiserum preparation, and we conducted Western blotting to confirm the specificity of the 2 antibodies by using the GF-1 cell lysate as the input (Fig. 4). The 2 antisera were used in the IP assay. The results in Fig. 5A show that GHSC70, GVDAC2, and GAPDH (as the control) existed in the cell lysate. By having them interact with NNV-specific antibodies in the IP assay, we analyzed the pulldown proteins with Western blotting by using GHSC70- and GVDAC2-specific antibodies. The results shown in Fig. 5B and C confirm that only GHSC70 could interact directly with NNV capsid protein.
FIG 4.
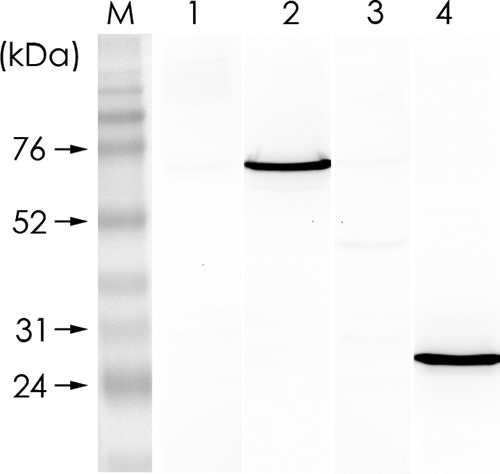
Specificities of rabbit anti-GHSC70 and anti-GVDAC2 polyclonal antibodies. GF-1 cell lysate was resolved by 10% SDS-PAGE and proteins were transferred to PVDF membrane and then incubated separately with rabbit preimmune serum for GHSC70 (lane 1), rabbit immune antiserum against GHSC70 (lane 2), rabbit preimmune serum for GVDAC2 (lane 3), and rabbit immune antiserum against GVDAC2 (lane 4) for Western blotting. M, molecular mass marker.
FIG 5.
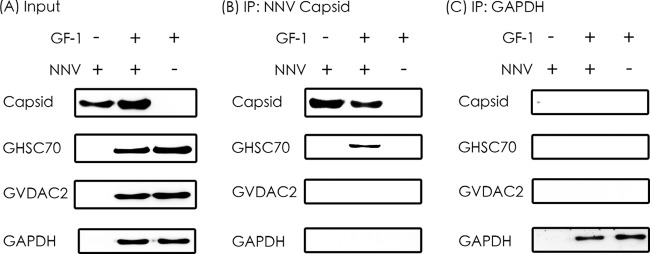
Immunoprecipitation of NNV capsid protein with GHSC70 and GVDAC2. (A) The purified NNV and cell lysates from NNV-infected and noninfected GF-1 cells were used as input for IP. (B) Western blotting of proteins immunoprecipitated (IP) by rabbit antiserum against NNV capsid protein. (C) Western blotting of proteins immunoprecipitated by rabbit anti-GAPDH antibody, which was used as the negative control.
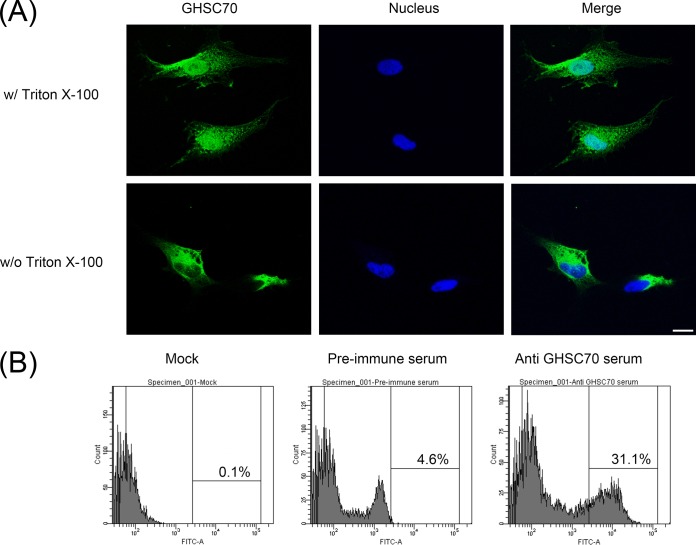
GHSC70 was detectable on the GF-1 cell surface and colocalized with NNV.
Triton X-100 is commonly used for the permeabilization of cell membranes after formalin fixation to detect intracellular antigens. Cell surface antigens can be detected if the cells are fixed with formalin without Triton X-100 treatment. In a pretest, we fixed 2 sets of GF-1 cells with formalin and permeabilized one set with Triton X-100, but not the other set. We then immunostained both sets of cells with anti-actin (an intracellular antigen) antibody, and only the permeabilized set showed positive signals.
GHSC70 is abundant in the cells. To investigate whether GHSC70 also exists on the cell surface, we fixed GF-1 cells with formalin without Triton X-100 permeabilization and then immunostained them with GHSC70-specific antibodies, and we observed immunofluorescent signals on GF-1 cells (Fig. 6A, bottom), indicating that GHSC70 existed on the GF-1 cell surface. We conducted flow cytometry to analyze nonpermeabilized cells after immunofluorescence staining with GHSC70-specific antibodies, and 31.1% of the cells showed positive signals (Fig. 6B).
FIG 6.
Detection of GHSC70 distribution of GF-1 cells by immunostaining and flow cytometry. (A) GF-1 cells were fixed with 10% formalin and then treated with (top) or without (bottom) Triton X-100. The cells were immunostained with rabbit anti-GHSC70 antiserum and then with FITC-labeled goat anti-rabbit Ig. Cell nuclei were stained with Hoechst 33258. The cells were observed under a confocal microscope. Bar = 10 μm. (B) GF-1 cells were fixed with 10% formalin without Triton X-100 permeabilization, incubated with PBS or immunostained with rabbit preimmune serum and anti-GHSC70 antiserum, stained with FITC-labeled goat anti-rabbit Ig, and finally, analyzed by flow cytometry.
To examine whether GHSC70 would colocalize with NNV particles during viral adsorption, GF-1 cells were incubated with NNV (MOI of 1,000) on ice. After 1 h of viral adsorption, the cells were fixed with formalin without Triton X-100 treatment and then immunostained with NNV-specific and GHSC70-specific antibodies. The colocalizing signal (yellow) of NNV and GHSC70 was observed (Fig. 7).
FIG 7.
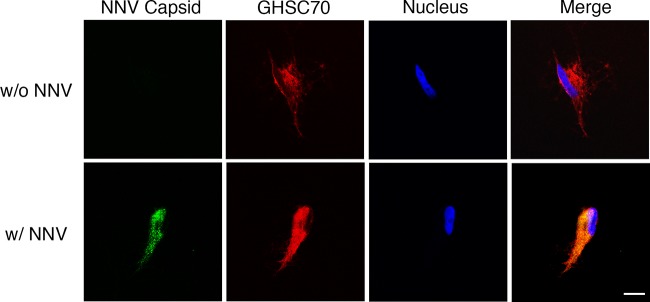
Localization of GHSC70 and NNV analyzed by immunofluorescence staining. GF-1 cells were incubated with NNV (MOI of 1,000) on ice for 1 h (bottom). Cells without NNV incubation (top) were used as a negative control. Cells were fixed with formalin without Triton X-100 permeabilization and immunostained with rabbit anti-GHSC70 and mouse anti-NNV antibodies following rhodamine-labeled and FITC-conjugated antibody staining, respectively. Cell nuclei were stained with Hoechst 33258. Samples were observed under a confocal microscope. Bar = 10 μm.
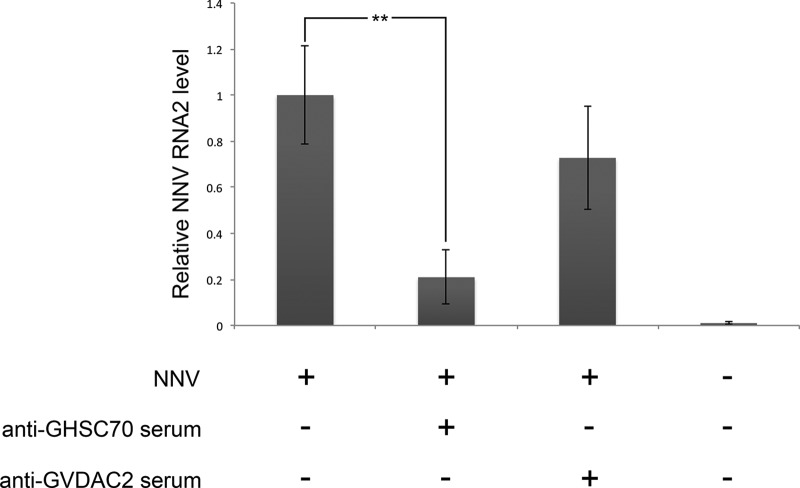
GHSC70-specific antiserum could block NNV entry into GF-1 cells.
To demonstrate the role that GHSC70 could play in NNV entry into GF-1 cells, we preincubated the cells with GHSC70- or GVDAC2-specific antibodies prior to NNV challenge. The control cells were subjected directly to NNV infection without the 2 antiserum pretreatments. The level of NNV RNA2 was analyzed by conducting real-time RT-PCR at the end of 1 h of viral adsorption. The result revealed that the NNV RNA2 level in cells pretreated with GHSC70-specific antibodies was 80% lower than that of the control group, indicating that GHSC70-specific antibodies could block NNV entry into GF-1 cells (Fig. 8).
FIG 8.
Blocking assay of NNV entry. GF-1 cells were incubated with anti-GHSC70 or anti-GVDAC2 polyclonal antibody for 1 h and then infected with NNV (MOI of 10) for 1 h. At the end of the hour of viral adsorption, total RNAs were extracted, and the NNV RNA2 level was detected by real-time RT-PCR. Error bars indicate standard deviations. **, P < 0.01 (n = 3).
DISCUSSION
The determination of viral receptor molecules on susceptible cells is pivotal for developing therapeutic agents to block viral infection. VOBPA is a useful approach and has been applied to identify the putative cellular receptors of rabbit vesivirus (24), adenovirus (25), and Japanese encephalitis virus (JEV) (26). Using VOBPA with purified NNV particles, we preliminarily assumed GHSC70 and GVDAC2 to be NNV receptor candidates.
VDAC is a channel protein that is mainly located on the mitochondrial outer membrane (27) but can also be found on the cell membrane of lymphocytes (28), epithelial cells (29), astrocytes (30), and the postsynaptic membrane fraction of the brain (31). In MASCOT analysis, GFMP-24 performed similarly to several protein candidates, whereas VDAC2 generated the highest similarity score (data not shown). Nevertheless, the IP assay result indicated that VDAC2 did not react with NNV capsid protein, and the GVDAC2-specific antiserum failed to significantly block NNV entry. However, downregulation of GVDAC2 by siRNA could reduce RNA2 expression significantly in infected GF-1 cells, indicating that GVDAC2 still plays a vital role in NNV infection. Studies have reported that VDAC can bind adenine nucleotide translocase (ANT), forming a hydrophilic pore, and regulate the Ca2+ and ATP permeability of the mitochondria (32, 33). VDAC is also related to cell apoptosis (34). In infectious bursal disease virus (IBDV) infection, the viral nonstructural VP5 protein can interact with host VDAC2 at different time points to inhibit or promote the cell apoptotic pathway (35, 36). The role of VDAC2 during NNV infection will be studied in the future.
The heat shock protein (HSP) family consists of heat-inducible and constitutive expression proteins. The constitutive expression HSP is called heat shock cognate protein (HSC). HSC70 exhibits a chaperone function: it can bind to newly synthesized polypeptides to form correct conformations (37) and to hydrolyze ATP for degrading clathrin coated on vesicles during endocytosis (38). HSC70 has been reported to participate in the viral entry step of rotavirus, dengue virus, Japanese encephalitis virus, polyomavirus, and hepatitis C virus (HCV) (26, 38–42). Previous studies on fish HSC70 have all focused on the expression level under heat shock or stress (43–45). In this study, we demonstrated that HSC70 has a role in the NNV entry of GF-1 cells.
The downregulation of HSC70 gene expression by a specific siRNA did not affect the morphology and survival of GF-1 cells at 48 h post-siRNA transfection, but it significantly inhibited the NNV RNA2 expression level and NNV titer at 24 hpi, revealing that GHSC70 is crucial in the early stage of NNV infection. An immunoprecipitation assay revealed that GHSC70 can interact with NNV capsid protein. Through immunofluorescence staining of formalin-fixed GF-1 cells without Triton X-100 permeabilization, GHSC70-positive signals appeared on GF-1 cells, indicating the possibility of GHSC70 being located on the GF-1 cell surface. When NNV adsorption to GF-1 cells took place on ice for 1 h, followed by formalin fixation without permeabilization, immunofluorescence staining using NNV- and GHSC70-specific antibodies generated colocalization signals on the cells. Therefore, GHSC70 has the potential to be an NNV receptor or coreceptor of GF-1 cells. Finally, we presented evidence of GHSC70 being involved in the viral entry of GF-1 cells by conducting a blocking assay. The NNV RNA expression level detected after viral adsorption was significantly decreased in cells pretreated with GHSC70-specific polyclonal antibodies compared with the level in nonblocked cells. To the best of our knowledge, this is the first report identifying GHSC70, as well as its role in NNV entry into GF-1 cells of groupers.
ACKNOWLEDGMENTS
We appreciate the technical assistance and support provided by the Technology Commons, College of Life Science, National Taiwan University, with regard to confocal laser scanning microscopy, flow cytometry, and real-time PCR. We thank Lawrence Cheng and Wallace Academic Editing for English revision of the manuscript.
The work was financially supported by the Ministry of Science and Technology Taiwan under contract no. MOST 103-2313-B-002-046.
REFERENCES
- 1.Kalia M, Jameel S. 2011. Virus entry paradigms. Amino Acids 41:1147–1157. doi: 10.1007/s00726-009-0363-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mendelsohn CL, Wimmer E, Racaniello VR. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 3.Bergelson JM, Shepley MP, Chan BM, Hemler ME, Finberg RW. 1992. Identification of the integrin VLA-2 as a receptor for echovirus 1. Science 255:1718–1720. doi: 10.1126/science.1553561. [DOI] [PubMed] [Google Scholar]
- 4.Roelvink PW, Lizonova A, Lee JG, Li Y, Bergelson JM, Finberg RW, Brough DE, Kovesdi I, Wickham TJ. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J Virol 72:7909–7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sritunyalucksana K, Wannapapho W, Lo CF, Flegel TW. 2006. PmRab7 is a VP28-binding protein involved in white spot syndrome virus infection in shrimp. J Virol 80:10734–10742. doi: 10.1128/JVI.00349-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li D-F, Zhang M-C, Yang H-J, Zhu Y-B, Xu X. 2007. β-Integrin mediates WSSV infection. Virology 368:122–132. doi: 10.1016/j.virol.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 7.Ørpetveit I, Gjøen T, Sindre H, Dannevig BH. 2008. Binding of infectious pancreatic necrosis virus (IPNV) to membrane proteins from different fish cell lines. Arch Virol 153:485–493. doi: 10.1007/s00705-007-0018-1. [DOI] [PubMed] [Google Scholar]
- 8.Chi S, Shieh J, Lin S. 2003. Genetic and antigenic analysis of betanodaviruses isolated from aquatic organisms in Taiwan. Dis Aquat Org 55:221–228. doi: 10.3354/dao055221. [DOI] [PubMed] [Google Scholar]
- 9.Munday B, Kwang J, Moody N. 2002. Betanodavirus infections of teleost fish: a review. J Fish Dis 25:127–142. doi: 10.1046/j.1365-2761.2002.00350.x. [DOI] [Google Scholar]
- 10.Chi S, Lo C, Kou G, Chang P, Peng S, Chen S. 1997. Mass mortalities associated with viral nervous necrosis (VNN) disease in two species of hatchery-reared grouper, Epinephelus fuscogutatus and Epinephelus akaara (Temminck & Schlegel). J Fish Dis 20:185–193. doi: 10.1046/j.1365-2761.1997.00291.x. [DOI] [Google Scholar]
- 11.Mori K, Nakai T, Muroga K, Arimoto M, Mushiake K, Furusawa I. 1992. Properties of a new virus belonging to nodaviridae found in larval striped jack (Pseudocaranx dentex) with nervous necrosis. Virology 187:368–371. doi: 10.1016/0042-6822(92)90329-N. [DOI] [PubMed] [Google Scholar]
- 12.Sommerset I, Nerland AH. 2004. Complete sequence of RNA1 and subgenomic RNA3 of Atlantic halibut nodavirus (AHNV). Dis Aquat Org 58:117–125. doi: 10.3354/dao058117. [DOI] [PubMed] [Google Scholar]
- 13.Chen L-J, Su Y-C, Hong J-R. 2009. Betanodavirus non-structural protein B1: a novel anti-necrotic death factor that modulates cell death in early replication cycle in fish cells. Virology 385:444–454. doi: 10.1016/j.virol.2008.11.048. [DOI] [PubMed] [Google Scholar]
- 14.Fenner BJ, Thiagarajan R, Chua HK, Kwang J. 2006. Betanodavirus B2 is an RNA interference antagonist that facilitates intracellular viral RNA accumulation. J Virol 80:85–94. doi: 10.1128/JVI.80.1.85-94.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwamoto T, Okinaka Y, Mise K, Mori K-I, Arimoto M, Okuno T, Nakai T. 2004. Identification of host-specificity determinants in betanodaviruses by using reassortants between striped jack nervous necrosis virus and sevenband grouper nervous necrosis virus. J Virol 78:1256–1262. doi: 10.1128/JVI.78.3.1256-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shieh J, Chi S. 2005. Production of monoclonal antibodies against grouper nervous necrosis virus (GNNV) and development of an antigen capture ELISA. Dis Aquat Org 63:53–60. doi: 10.3354/dao063053. [DOI] [PubMed] [Google Scholar]
- 17.Liu W, Hsu C-H, Hong Y-R, Wu S-C, Wang C-H, Wu Y-M, Chao C-B, Lin C-S. 2005. Early endocytosis pathways in SSN-1 cells infected by dragon grouper nervous necrosis virus. J Gen Virol 86:2553–2561. doi: 10.1099/vir.0.81021-0. [DOI] [PubMed] [Google Scholar]
- 18.Chi S, Hu W, Lo B. 1999. Establishment and characterization of a continuous cell line (GF-1) derived from grouper, Epinephelus coioides (Hamilton): a cell line susceptible to grouper nervous necrosis virus (GNNV). J Fish Dis 22:173–182. doi: 10.1046/j.1365-2761.1999.00152.x. [DOI] [Google Scholar]
- 19.Chi S, Lo B, Lin S. 2001. Characterization of grouper nervous necrosis virus (GNNV). J Fish Dis 24:3–13. doi: 10.1046/j.1365-2761.2001.00256.x. [DOI] [Google Scholar]
- 20.Wu Y-C, Lu Y-F, Chi S-C. 2010. Anti-viral mechanism of barramundi Mx against betanodavirus involves the inhibition of viral RNA synthesis through the interference of RdRp. Fish Shellfish Immun 28:467–475. doi: 10.1016/j.fsi.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gonzalez-Reyes S, Garcia-Manso A, del Barrio G, Dalton KP, Gonzalez-Molleda L, Arrojo-Fernandez J, Nicieza I, Parra F. 2009. Role of annexin A2 in cellular entry of rabbit vesivirus. J Gen Virol 90:2724–2730. doi: 10.1099/vir.0.013276-0. [DOI] [PubMed] [Google Scholar]
- 25.Li X, Bangari DS, Sharma A, Mittal SK. 2009. Bovine adenovirus serotype 3 utilizes sialic acid as a cellular receptor for virus entry. Virology 392:162–168. doi: 10.1016/j.virol.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das S, Laxminarayana SV, Chandra N, Ravi V, Desai A. 2009. Heat shock protein 70 on Neuro2a cells is a putative receptor for Japanese encephalitis virus. Virology 385:47–57. doi: 10.1016/j.virol.2008.10.025. [DOI] [PubMed] [Google Scholar]
- 27.Hoogenboom BW, Suda K, Engel A, Fotiadis D. 2007. The supramolecular assemblies of voltage-dependent anion channels in the native membrane. J Mol Biol 370:246–255. doi: 10.1016/j.jmb.2007.04.073. [DOI] [PubMed] [Google Scholar]
- 28.Cole T, Awni LA, Nyakatura E, Gotz H, Walter G, Thinnes FP, Hilschmann N. 1992. Studies on human porin VIII Expression of “Porin 31HL” channels in the plasmalemma of the acute-lymphoblastic-leukemia cell line KM3 as revealed by light- and electron-microscopy. Biol Chem Hoppe Seyler 373:891–896. doi: 10.1515/bchm3.1992.373.2.891. [DOI] [PubMed] [Google Scholar]
- 29.Puchelle E, Jacquot J, Fuchey C, Burlet H, Klossek JM, Gilain L, Triglia JM, Thinnes FP, Hilschmann N. 1993. Studies on human porin IX Immunolocalization of porin and CFTR channels in human surface respiratory epithelium. Biol Chem Hoppe Seyler 374:297–304. doi: 10.1515/bchm3.1993.374.1-6.297. [DOI] [PubMed] [Google Scholar]
- 30.Dermietzel R, Hwang TK, Buettner R, Hofer A, Dotzler E, Kremer M, Deutzmann R, Thinnes FP, Fishman GI, Spray DC. 1994. Cloning and in situ localization of a brain-derived porin that constitutes a large-conductance anion channel in astrocytic plasma membranes. Proc Natl Acad Sci U S A 91:499–503. doi: 10.1073/pnas.91.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon JI, Jung YW, Ko BH, De Pinto V, Jin I, Moon IS. 1999. Presence of a voltage-dependent anion channel 1 in the rat postsynaptic density fraction. Neuroreport 10:443–447. doi: 10.1097/00001756-199902250-00001. [DOI] [PubMed] [Google Scholar]
- 32.Gincel D, Zaid H, Shoshan-Barmatz V. 2001. Calcium binding and translocation by the voltage-dependent anion channel: a possible regulatory mechanism in mitochondrial function. Biochem J 358:147–155. doi: 10.1042/0264-6021:3580147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Regenold WT, Pratt M, Nekkalapu S, Shapiro PS, Kristian T, Fiskum G. 2012. Mitochondrial detachment of hexokinase 1 in mood and psychotic disorders: implications for brain energy metabolism and neurotrophic signaling. J Psychiatr Res 46:95–104. doi: 10.1016/j.jpsychires.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 34.Rostovtseva TK, Tan W, Colombini M. 2005. On the role of VDAC in apoptosis: fact and fiction. J Bioenerg Biomembr 37:129–142. doi: 10.1007/s10863-005-6566-8. [DOI] [PubMed] [Google Scholar]
- 35.Li Z, Wang Y, Xue Y, Li X, Cao H, Zheng SJ. 2012. Critical role for voltage-dependent anion channel 2 in infectious bursal disease virus-induced apoptosis in host cells via interaction with VP5. J Virol 86:1328–1338. doi: 10.1128/JVI.06104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu M, Vakharia VN. 2006. Nonstructural protein of infectious bursal disease virus inhibits apoptosis at the early stage of virus infection. J Virol 80:3369–3377. doi: 10.1128/JVI.80.7.3369-3377.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hightower LE. 1991. Heat shock, stress proteins, chaperones, and proteotoxicity. Cell 66:191–197. doi: 10.1016/0092-8674(91)90611-2. [DOI] [PubMed] [Google Scholar]
- 38.Goldfarb SB, Kashlan OB, Watkins JN, Suaud L, Yan W, Kleyman TR, Rubenstein RC. 2006. Differential effects of Hsc70 and Hsp70 on the intracellular trafficking and functional expression of epithelial sodium channels. Proc Natl Acad Sci U S A 103:5817–5822. doi: 10.1073/pnas.0507903103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cripe TP, Delos SE, Estes PA, Garcea RL. 1995. In vivo and in vitro association of hsc70 with polyomavirus capsid proteins. J Virol 69:7807–7813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parent R, Qu X, Petit M-A, Beretta L. 2009. The heat shock cognate protein 70 is associated with hepatitis C virus particles and modulates virus infectivity. Hepatology 49:1798–1809. doi: 10.1002/hep.22852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyes-Del Valle J, Chávez-Salinas S, Medina F, del Angel RM. 2005. Heat shock protein 90 and heat shock protein 70 are components of dengue virus receptor complex in human cells. J Virol 79:4557–4567. doi: 10.1128/JVI.79.8.4557-4567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guerrero CA, Bouyssounade D, Zárate S, Iša P, López T, Espinosa R, Romero P, Méndez E, López S, Arias CF. 2002. Heat shock cognate protein 70 is involved in rotavirus cell entry. J Virol 76:4096–4102. doi: 10.1128/JVI.76.8.4096-4102.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ojima N, Yamashita M, Watabe S. 2005. Quantitative mRNA expression profiling of heat-shock protein families in rainbow trout cells. Biochem Biopys Res Commun 329:51–57. doi: 10.1016/j.bbrc.2005.01.097. [DOI] [PubMed] [Google Scholar]
- 44.Zhang A, Zhou X, Wang X, Zhou H. 2011. Characterization of two heat shock proteins (Hsp70/Hsc70) from grass carp (Ctenopharyngodon idella): evidence for their differential gene expression, protein synthesis and secretion in LPS-challenged peripheral blood lymphocytes. Comp Biochem Physiol B Biochem Mol Biol 159:109–114. doi: 10.1016/j.cbpb.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Deane EE, Woo NYS. 2004. Differential gene expression associated with euryhalinity in sea bream (Sparus sarba). Am J Physiol Regul Integr Comp Physiol 287:R1054–63. doi: 10.1152/ajpregu.00347.2004. [DOI] [PubMed] [Google Scholar]