ABSTRACT
Until the recent emergence of two human-pathogenic tick-borne phleboviruses (TBPVs) (severe fever with thrombocytopenia syndrome virus [SFTSV] and Heartland virus), TBPVs have been neglected as causative agents of human disease. In particular, no studies have addressed the global distribution of TBPVs, and consequently, our understanding of the mechanism(s) underlying their evolution and emergence remains poor. In order to provide a useful tool for the ecological and epidemiological study of TBPVs, we have established a simple system that can detect all known TBPVs, based on conventional reverse transcription-PCR (RT-PCR) with degenerate primer sets targeting conserved regions of the viral L genome segment. Using this system, we have determined that several viruses that had been isolated from ticks decades ago but had not been taxonomically identified are novel TBPVs. Full-genome sequencing of these viruses revealed a novel fourth TBPV cluster distinct from the three known TBPV clusters (i.e., the SFTS, Bhanja, and Uukuniemi groups) and from the mosquito/sandfly-borne phleboviruses. Furthermore, by using tick samples collected in Zambia, we confirmed that our system had enough sensitivity to detect a new TBPV in a single tick homogenate. This virus, tentatively designated Shibuyunji virus after the region of tick collection, grouped into a novel fourth TBPV cluster. These results indicate that our system can be used as a first-line screening approach for TBPVs and that this kind of work will undoubtedly lead to the discovery of additional novel tick viruses and will expand our knowledge of the evolution and epidemiology of TBPVs.
IMPORTANCE Tick-borne phleboviruses (TBPVs) have been largely neglected until the recent emergence of two virulent viruses, severe fever with thrombocytopenia syndrome virus and Heartland virus. Little is known about the global distribution of TBPVs or how these viruses evolved and emerged. A major hurdle to study the distribution of TBPVs is the lack of tools to detect these genetically divergent phleboviruses. In order to address this issue, we have developed a simple, rapid, and cheap RT-PCR system that can detect all known TBPVs and which led to the identification of several novel phleboviruses from previously uncharacterized tick-associated virus isolates. Our system can detect virus in a single tick sample and novel TBPVs that are genetically distinct from any of the known TBPVs. These results indicate that our system will be a useful tool for the surveillance of TBPVs and will facilitate understanding of the ecology of TBPVs.
INTRODUCTION
Until the recent emergence of two highly virulent human-pathogenic tick-borne phleboviruses (TBPVs), severe fever with thrombocytopenia syndrome virus (SFTSV) (1) and Heartland virus (HRTV) (2), the TBPVs were largely neglected as causative agents of human disease, whereas the mosquito- or sandfly-borne phleboviruses, such as Rift Valley fever virus or Toscana virus, have been well studied. At present, there are three distinct genetic groups of TBPVs within the genus Phlebovirus, family Bunyaviridae (the SFTS group, the Bhanja group, and the Uukuniemi group) (3, 4). The global distribution of these viruses is poorly understood, except for the local distributions of SFTSV and HRTV in their respective geographic areas where they are endemic (5–8). Despite the worldwide distribution of ticks and their occurrence in diverse ecologic zones (9), the evolutionary mechanism(s) underlying the emergence of pathogenic TBPVs, such as SFTSV and HRTV, is unknown.
Except for SFTSV and HRTV, the potential virulence of other TBPVs for humans and/or animals is still unclear. Bhanja virus (BHAV), a representative of the Bhanja group, is known to have caused febrile illness in a few patients following both natural and laboratory infections (10, 11). Uukuniemi virus (UUKV), a representative of the Uukuniemi group, has also been documented as a suspected causative agent of a febrile illness in three patients (12). Serological studies of TBPVs from both the Bhanja and Uukuniemi groups indicate that humans and several other animal species can be infected with these viruses (12–15). Despite the reports of isolated human cases and seropositivity in some populations, no outbreaks associated with Bhanja or Uukuniemi group virus infection have been recognized. After the initial isolation and identification of SFTSV, additional SFTS cases were identified retrospectively using stored tissue samples from patients who died of a febrile illness of unknown origin (16–19). This suggests that the diseases caused by TBPVs may be difficult to detect and to identify clinically due to their limited geographic distributions and atypical symptoms. A similar scenario occurred following the initial isolation and description of other tick-borne bacterial or parasitic agents (Borrelia burgdorferi, Ehrlichia chaffeensis, and Babesia microti [9]). Indeed, SFTS was initially diagnosed as anaplasmosis (1, 20).
Another reason why the TBPVs have been neglected could be the genetic divergence among different groups of TBPVs; for example, nucleotide sequence identities are 40 to 45% between the SFTS and Bhanja groups and only about 35% between the Uukuniemi group and the two other groups (3). This has hampered our ability to develop a single diagnostic tool for all TBPVs and has made it difficult to detect novel TBPVs using the existing tools. In order to identify the potential impact of TBPV infections on public health, a diagnostic tool that could also be used in field studies and that targeted a wide range of TBPVs was needed. Recent advancements and increased availability of next-generation sequencing (NGS) technologies, which can read nucleotide sequences using sequence-independent amplification, allowed us to easily determine the genome sequences of several novel TBPVs. While all the complete genome sequences of TBPVs reported since the initial description of SFTSV in 2011 (1), HRTV (2), BHAV (3, 21), Lone Star virus (22), Malsoor virus (23), Hunter Island virus (24), Khasan virus (25), Komandory virus (26), American dog tick phlebovirus, and blacklegged tick phlebovirus (27) were done using NGS, it would be technically difficult to apply this approach for large-scale screening and field material, because of the cost and labor needed to produce sufficient reads of unknown pathogen sequences. Therefore, a detection system based on conventional reverse transcription-PCR (RT-PCR) that targets a wide range of TBPVs would be required for initial screening of samples prior to NGS.
Currently, approximately 40 viruses are listed as bunyaviruses but remain taxonomically unassigned to a species/genus due to the lack of genome sequence information and/or serological cross-reactivity with other known bunyaviruses (28). Recent work on the retrospective identification of uncharacterized bunya-like viruses by us and others has identified the tick-borne BHAV and Lone Star virus as novel species in the genus Phlebovirus that are genetically related to the SFTS/Heartland group (3, 21, 22). Intriguingly, several other taxonomically unassigned bunyaviruses, such as Kaisodi virus (29), Lanjan virus (30), and Silverwater virus (31), and a serological member of the Bhanja group, Kismayo virus (32), were also isolated from ticks, suggesting that NGS of such taxonomically unassigned tick-borne bunyaviruses may help us to gain further knowledge about the taxonomy, evolution, and epidemiology of TBPVs, and thus help us to better assess the zoonotic disease potential of these viruses.
In the present study, we report the establishment of an RT-PCR system that can detect all known TBPVs, based on degenerate primers that bind to conserved regions of the L segment RNA of TBPVs. Moreover, we applied our RT-PCR practically to both the retrospective identification of Kaisodi, Lanjan, Silverwater, and Kismayo viruses and to the discovery of a novel TBPV from field tick samples collected in Zambia where the distribution of TBPVs is totally unknown. Further characterization of these latter novel TBPVs was also done.
MATERIALS AND METHODS
Viruses and viral RNAs.
The virus strains used in this study were kindly provided by the Division of Vector-Borne Diseases (DVBD), arbovirus reference collection, Centers for Disease Control and Prevention (CDC), the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) arthropod-borne virus reference collection at the University of Texas Medical Branch (UTMB) and by Mifang Liang at the Chinese Center for Disease Control and Prevention (Chinese CDC). The information regarding their origin is summarized in Table S1 in the supplemental material. Viral RNAs used in the present study were extracted from the original stock material provided by the resources named above or supernatants of cell culture using the QIAamp viral RNA Mini Kit (Qiagen).
RNA extraction from ticks.
Ticks attached to cattle were removed, and each individual tick was homogenized with 300 μl of Dulbecco's modified Eagle medium (DMEM; Sigma-Aldrich) using a homogenizer (TOMY SEIKO) run twice at 3,000 rpm. Then, 140 μl of the homogenized sample was mixed with 500 μl of TRIzol LS reagent (Invitrogen), and total RNA was extracted following the manufacturer's protocol. Experiments with these tick samples were performed at the Hokudai Center for Zoonosis Control in Zambia.
Cells and virus titration.
The Huh-7 (human hepatocellular carcinoma) cell line kindly provided by Yoshiharu Matsuura (Osaka University) was grown in DMEM supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 50 U/ml penicillin, and 50 μg/ml streptomycin (Life Technologies). SFTSV strain SD4, HRTV, and BHAV strain R-1819 were passaged in Huh-7 cells and titrated by immunostaining with mouse antibodies raised against each virus. Immunostaining was performed as follows. Huh-7 cells were infected with serial dilutions of each virus and fixed with 10% formaldehyde in phosphate-buffered saline (PBS) 3 to 5 days after infection. Then, the cells were permeabilized with 0.5% Triton X-100 in PBS for 10 min followed by blocking with 3% BSA–PBS (PBS containing 3% bovine serum albumin) for 1 h. After the cells were rinsed with PBS, they were incubated with the mouse antibodies (diluted 1:100 to 1,000 in PBS) for 1 h and rinsed three times. Virus-infected cells were visualized using horseradish peroxidase (HRP)-conjugated donkey anti-mouse IgG (H+L) (Jackson ImmunoResearch) and Sigmafast 3,3′-diaminobenzidine tablets (Sigma-Aldrich).
Primers and one-step RT-PCR.
The initial screening of the primers (the primer sequences available upon request) was performed with RNAs of SFTSV HB29 and SD4, HRTV Mo4, BHAV R-1819, and UUKV S23 with the PrimeScript one-step RT-PCR kit, version 2 (Dye Plus) (TaKaRa) using 1 μl of viral RNA and 4 pmol of each primer in 10 μl of reaction solution with the following incubation program: (i) 50°C for 30 min; (ii) 94°C for 2 min; (iii) 40 cycles with 1 cycle consisting of 94°C for 30 s, 55°C/58°C/60°C for 30 s, and 72°C for 1 min/1 kb; and (iv) 72°C for 5 min. Then, two primer sets were selected. The ppL1 set consisted of primers TBPVL2759F (F stands for forward) and TBPVL3267R, and the ppL2 set consisted of primers HRT-GL2759F and HRT-GL3276R (Table 1). One-step RT-PCR was performed with the PrimeScript one-step RT-PCR kit version 2 (Dye Plus) using 1 μl of viral RNA and 4 pmol of each primer in 10 μl of reaction solution with the following incubation program: (i) 50°C for 30 min; (ii) 94°C for 2 min; (iii) 40 cycles with 1 cycle consisting of 94°C for 30 s, 55°C for 30 s, and 72° for 30 s; and (iv) 72°C for 5 min. RT-PCR products were separated by electrophoresis on a 1% agarose gel and visualized by ethidium bromide staining.
TABLE 1.
Primers used in the RT-PCR
| Primer set | Primer | Binding regiona | Sequenceb (5′ → 3′) |
|---|---|---|---|
| ppL1 | TBPVL2759F | 2786–2803 | CAGCATGGIGGICTIAGAGAGAT |
| TBPVL3267R | 3281–3300 | TGIAGIATSCCYTGCATCAT | |
| ppL2 | HRT-GL2759F | 2786–2814 | CAGCATGGIGGIYTIAGRGAAATYTATGT |
| HRT-GL3276R | 3281–3309 | GAWGTRWARTGCAGGATICCYTGCATCAT |
Nucleotide positions in the L segment RNA (cRNA) of Uukuniemi virus strain S23 (GenBank accession number D10759).
Nonstandard nucleotides are as follows; I, inosine; R, adenine (A) and guanine (G); S, G and cytosine (C); W, A and thymine (T); Y, T and C.
qRT-PCR and synthesized RNA standards.
Quantitative real-time RT-PCR (qRT-PCR) was performed using the PrimeScript RT Master Mix Perfect Real Time (TaKaRa) for reverse transcription following the manufacturer's protocol and using SYBR Premix Ex Taq (Tli RNase H Plus; TaKaRa) for quantitative PCR. The qPCRs was performed using 1 μl of RT product and 4 pmol of each primer in 10 μl of reaction solution with the following incubation program on a CFX96 real-time system (Bio-Rad): (i) 94°C for 2 min; (ii) 40 cycles with 1 cycle consisting of 94°C for 30 s, 55°C for 30 s, and 72° for 30 s; (iii) a melting temperature cycle. Quantification cycle (Cq) values for each RNA dilution were calculated as an average of 5 replicates.
Virus growth and NGS.
Growth of Silverwater virus and Kismayo virus was attempted in several cell lines. Briefly, viruses were grown in cells in DMEM supplemented with 2% FCS, 2 mM l-glutamine, 50 U/ml penicillin, 50 μg/ml streptomycin (Life Technologies), and 10 μg/ml MycoKill AB (GE Healthcare). Virus growth was monitored using the RT-PCR established in the present study. The culture supernatants that were positive with the RT-PCR were harvested, and RNA was extracted using TRIzol LS reagent for NGS as previously described (3). The nucleotide sequences were resequenced by conventional RT-PCR and Sanger sequencing (primer sequences available upon request).
Multiple-sequence alignment and phylogenetic analysis.
The nucleotide sequences obtained for each genome segment, or the deduced amino acid sequences of each of the open reading frames (ORFs), were aligned together with representative sequences of other known phleboviruses available from GenBank (see Table S2 in the supplemental material) using MUSCLE as implemented in MEGA, version 6 (33). Multiple-sequence alignments were modified manually. Phylogenetic trees were constructed using the maximum likelihood (ML) method. For ML analysis, the Tamura-Nei model with gamma distributed with invariant sites (G+I) built into MEGA 6 was used. The robustness of the nodes was tested by 1,000 bootstrap replications. The scale bar in each tree represents the distance resulting from one character change.
Nucleotide sequence accession numbers.
The genome sequences of all TBPVs sequenced in the present study, including partial L segment sequences of Kaisodi virus, Lanjan virus, and Shibuyunji virus, were deposited in GenBank under the following accession numbers (for S segment, M segment, and L segment): KM114246 to KM114257 and KM370974 to KM370979.
RESULTS
Establishment of an RT-PCR system to detect a wide range of TBPVs.
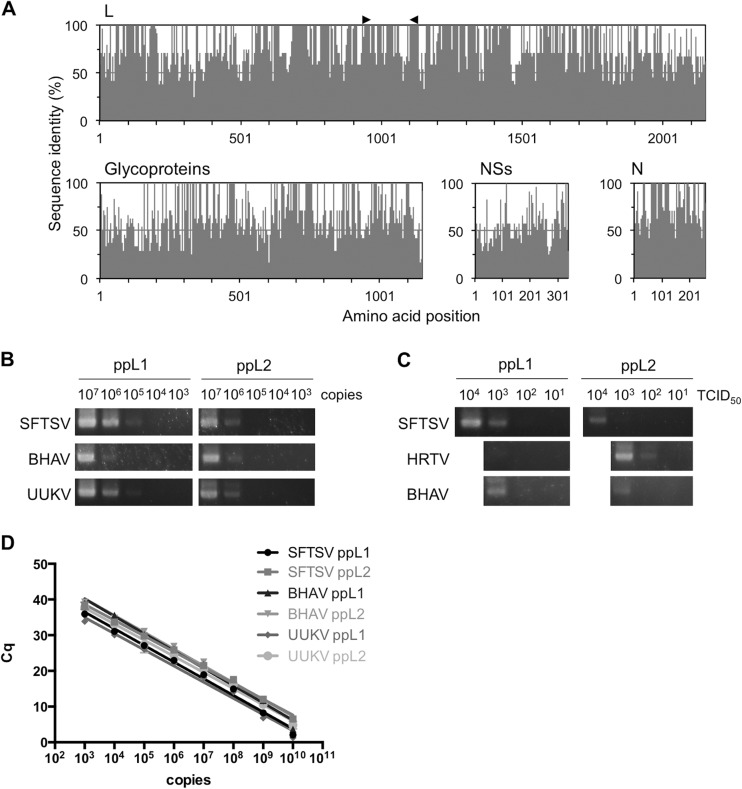
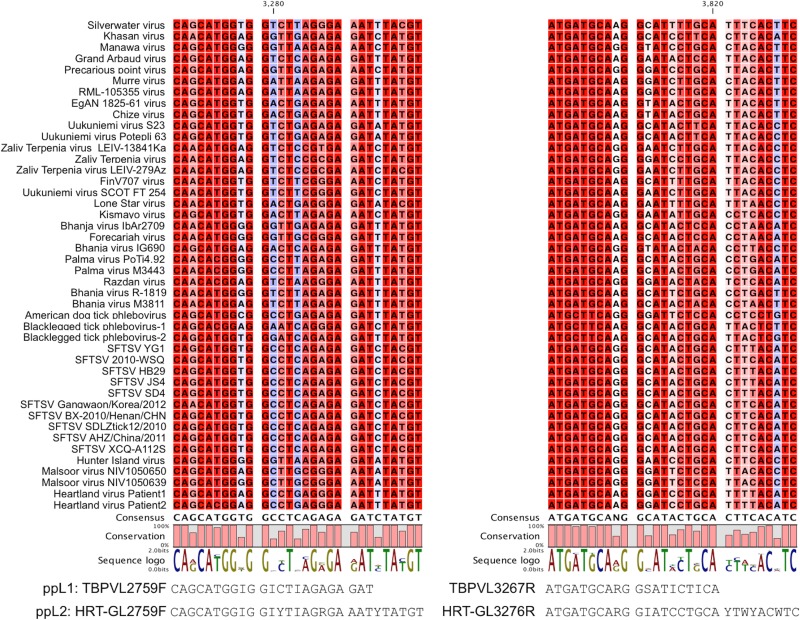
To find conserved regions of TBPV genomic RNAs minimizing degeneracy of the primers, multiple-sequence alignments were performed using the complete amino acid sequences of known TBPV proteins obtained from GenBank (Fig. 1A). Conserved regions were identified manually from the alignment, and degenerate primers were designed based on the corresponding nucleotide sequences. Initial screening of the primers was performed with one-step RT-PCR using RNAs from representative TBPVs (i.e., SFTSV HB29 and SD4, HRTV Mo4, BHAV R-1819, and UUKV S23) under different conditions. Two primer sets binding to the same regions of the L gene (ppL1 and ppL2 [Table 1]) that could detect all the viruses tested with minimal nonspecific amplifications were selected, and a single cycle protocol that maximized the sensitivity of these primer sets was established (data not shown).
FIG 1.
Sensitivity of RT-PCR using newly designed primers. (A) Conserved regions of tick-borne phlebovirus (TBPV) proteins were manually selected based on the multiple-sequence alignments. Two sets of primers (ppL1 and ppL2) were designed from the same conserved regions in the L protein ORF (indicated by black arrowheads) to minimize the degeneracy of nucleotide sequences. (B and C) The sensitivities of conventional one-step RT-PCR using these two primer sets were tested with serial dilutions of in vitro-synthesized RNAs (B) and viral RNAs (C) as the templates. The sensitivity of one-step quantitative RT-PCR (qRT-PCR) was tested using serial dilutions of in vitro-synthesized RNAs. (D) Average Cq values (five replicates) are plotted against the number of RNA copies, and trend lines calculated by semilog-line nonlinear regression analysis are also shown). Abbreviations: L, RNA-dependent RNA polymerase; NSs, nonstructural protein; N, nucleocapsid protein; SFTSV, severe fever with thrombocytopenia syndrome virus; BHAV, Bhanja virus; TCID50, 50% tissue culture infectious dose; Cq, quantification cycle.
The sensitivity of the primer sets was determined using both in vitro-transcribed RNA and viral RNA as the templates for both conventional RT-PCR and qRT-PCR. The minimum amount of synthesized RNA standard that could be detected using the conventional RT-PCR with the ppL1 primers was 105 copies of the SFTSV or UUKV standard or 106 copies of the BHAV standard. With the ppL2 primers, the detection limit for all viruses was 106 copies of the RNA standard (Fig. 1B). The detection limits for RNAs extracted from virus culture were equivalent to 102 50% tissue culture infectious doses (TCID50) of HRTV, using the ppL2 primers, and approximately 103 TCID50 equivalents for SFTSV and BHAV. With the ppL1 primers, all viruses were first detected at 103 TCID50 equivalents (Fig. 1C). The standard curves indicated that the detection limits for qRT-PCR (the lowest RNA copy number with Cq value less than 40) were also between 102 and 103 copies with all synthesized RNAs tested (Fig. 1D). The specificity of the amplification products was confirmed by Sanger sequencing.
The cross-reactivity of the primer sets to known TBPVs was tested using the available TBPV RNAs (Fig. 2). Both primer sets amplified specific fragments from all of the templates, except for FinV707 virus, which was not detected using ppL2, and Zaliv Terpenia virus, which was not detected using ppL1. Thus, we have established a one-step RT-PCR system that can detect a range of TBPVs from all three known groups, Uukuniemi, Bhanja, and SFTS/Heartland groups, using these two primer sets.
FIG 2.
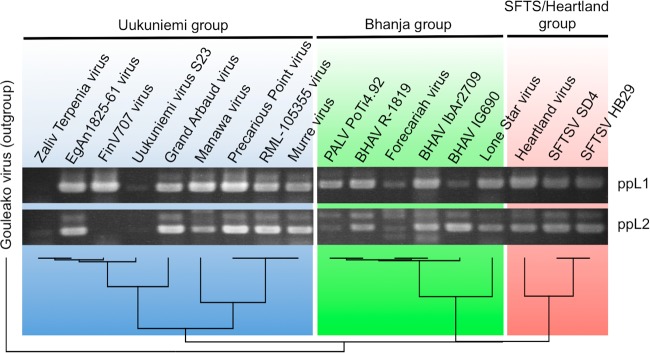
Cross-reactivity of RT-PCR using newly designed primers with known TBPVs. Cross-reactivity of the TBPV RT-PCR was tested using viral RNAs of known TBPVs. Since viral RNA concentrations were not standardized, the band intensity does not reflect the sensitivity of the system. The phylogenetic tree, which was constructed by the neighbor-joining (NJ) algorithm, at the bottom indicates the genetic relationship among the TBPVs tested. Abbreviations: SFTSV, severe fever with thrombocytopenia syndrome virus; BHAV, Bhanja virus; PALV, Palma virus.
Retrospective identification of several TBPVs and assignment into novel and existing species in the genus Phlebovirus.
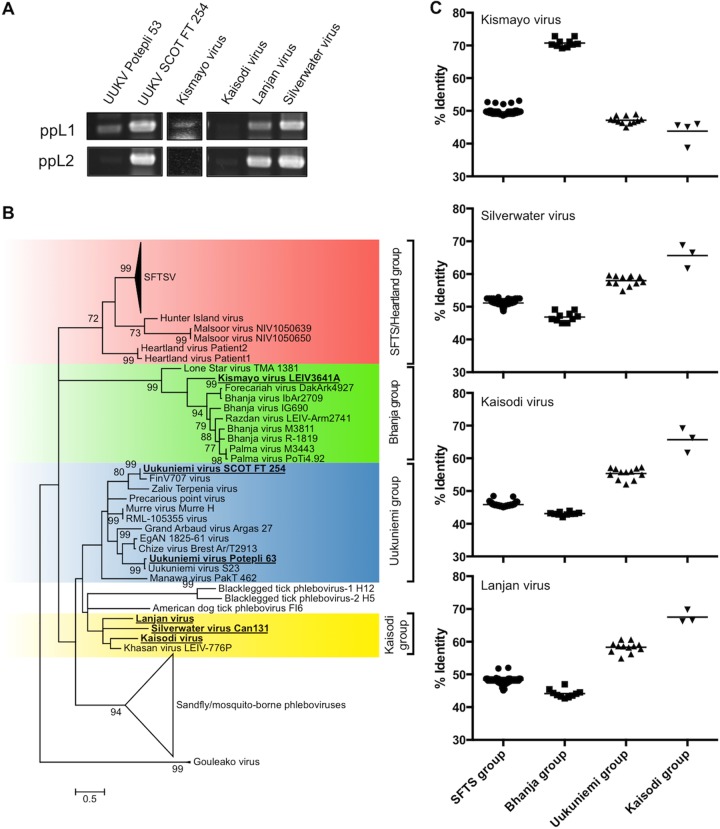
Using our RT-PCR system, we first performed a retrospective screening targeting taxonomically unassigned bunya-like viruses isolated from ticks and some genetically uncharacterized tick-associated phleboviruses. These viruses had been characterized only to a limited extent using biological and serological methods (i.e., electron microscopy, neutralization test, and complement fixation test) before the introduction of molecular biological techniques. Our RT-PCR results have confirmed that two strains of UUKV (Potepli 63 and SCOT FT 254 [34]), which were classified as UUKVs serologically, are phleboviruses. The RT-PCR results have also identified Kismayo virus, Kaisodi virus, Silverwater virus, and Lanjan virus as likely phleboviruses (Fig. 3A). All the RT-PCR products were sequenced to confirm specific amplifications from TBPV L segment RNAs. Phylogenetic analysis using the sequences obtained was employed to determine a tentative taxonomic relationship to the known TBPVs (Fig. 3B). The phylogenetic tree indicated that Kismayo virus branched between Lone Star virus and the other Bhanja group viruses; UUKV Potepli 63 and UUKV SCOT FT 254 belonged to the Uukuniemi group and were coupled with UUKV strain S23 (the prototype strain of the Uukuniemi group) and FinV707 virus, respectively. Kaisodi virus, Silverwater virus, and Lanjan virus branched from the ancestor of the Uukuniemi group together with Khasan virus to form a novel group (tentatively named the Kaisodi group, after the serological group), which was distinct from any other known groups in the Phlebovirus genus. The sequence identity between Kismayo virus and Bhanja group TBPVs was significantly higher than the values between Kismayo virus and TBPVs belonging to the other three groups (Fig. 3C). The sequence identity among Kaisodi group viruses was around 70% on average, while the identity between Kaisodi group viruses and the other three groups was less than that.
FIG 3.
Retrospective identification of TBPVs. (A) Conventional RT-PCR was performed using RNA from uncharacterized bunyaviruses obtained from multiple sources. (B) Each amplified RT-PCR fragment (around 500 bp) was sequenced, and a phylogenetic tree was constructed based on the fragment sequences and reference sequences using the maximum likelihood (ML) method with 1,000 bootstrap replicates. Bootstrap probabilities above 70% are shown near the branches. The viruses that were newly sequenced in the present study are shown in boldface type and underlined. (C) The sequence identity of the fragment between the newly identified TBPVs (i.e., Kismayo virus, Silverwater virus, Kaisodi virus, and Lanjan virus) and known TBPVs. Each symbol shows the identity value for an individual tick, and the median value for the genetic group is shown as a black bar. Abbreviations: UUKV, Uukuniemi virus; SFTSV, severe fever with thrombocytopenia syndrome virus.
Molecular characterization of retrospectively identified TBPVs.
We first tried to culture the retrospectively identified TBPVs using various cell lines to produce supernatants with less host RNA contamination than the original material (i.e., mouse brain homogenates). After two or three passages, the supernatants of these cells infected with the TBPVs were examined using our TBPV RT-PCR to verify virus replication. Kismayo virus and Silverwater virus have been confirmed to replicate in Huh-7 cells, and the genome sequences of these viruses were determined by NGS. Since the RT-PCR fragment sequences for the Potepli 63 and SCOT FT 254 strains of UUKV were close to those of previously sequenced viruses, these viruses were sequenced with RT-PCR and Sanger sequencing using primers designed based on available Uukuniemi group virus sequences (primer sequences available upon request). Due to the lack of growth in cell culture, further sequencing of Kaisodi virus and Lanjan virus was not performed.
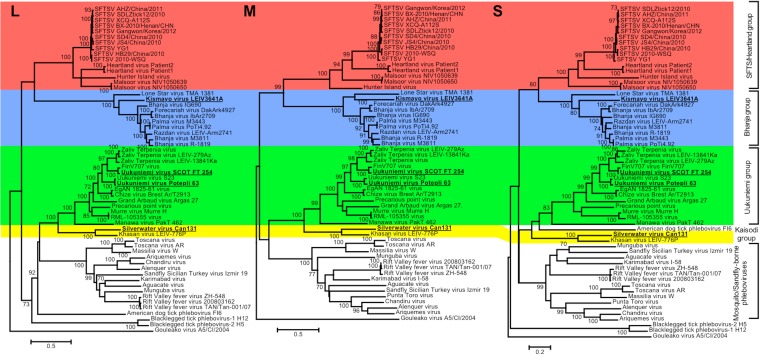
Complete sequences (except for the 5′ terminus of Silverwater virus L segment in viral RNA sense) of the new TBPVs identified genetic structures similar to those of other phleboviruses, i.e., three RNA segments that together constitute the viral genome (L, M, and S segments) and an S segment that encodes nucleocapsid (N) and nonstructural protein (NSs) genes in an ambisense orientation (Table 2). Phylogenetic analyses using these full-length sequences (Fig. 4) also demonstrated the classification of these novel TBPVs into the Uukuniemi group, the Bhanja group, or a provisional new group (Kaisodi group), as indicated by the phylogenetic tree based on the original RT-PCR fragment sequences (Fig. 3B). The topology of the trees mostly corresponds between the three genome segments; showing four distinct groups (three TBPV groups and a cluster of the other phleboviruses) and a novel group, including Silverwater virus and Khasan virus that has been identified in this study. Sequence identities of both protein or nucleotide sequences between UUKV Potepli 63 and S23 or UUKV SCOT FT 254 and FinV707 virus were above 97% for all the sequences (see Tables S3 to S6 in the supplemental material). The sequence identities between Bhanja group viruses and Kismayo virus were less than 75% but higher than those for Bhanja group viruses and Lone Star virus. The sequence identities between Silverwater virus and other viruses were all below 50%, except between Silverwater virus and Khasan virus (50 to 80% identical in amino acid sequences).
TABLE 2.
Genome structures of newly identified tick-borne phleboviruses
| Virus | Strain | RNA segment | Length (no. of bases) | Genea | ORF positions (length)b | GenBank accession no. |
|---|---|---|---|---|---|---|
| Kismayo virus | LEIV 3641A | L | 6,341 | RdRp | 17–6,265 (6,249) | KM114252 |
| M | 3,329 | Gn and Gc | 20–3,247 (3,228) | KM114253 | ||
| S | 1,878 | N | 36–779 (744) | KM114254 | ||
| NSs | 1,857–919 (939) | |||||
| Silverwater virus | Can131 | L | >6,412c | RdRp | 17–6,379 (6,363) | KM114257 |
| M | 3,209 | Gn and Gc | 17–3,043 (3,027) | KM114255 | ||
| S | 1,732 | N | 29–793 (765) | KM114256 | ||
| NSs | 1,703–909 (795) | |||||
| Uukuniemi virus | Potepli 53 | L | 6,422 | RdRp | 17–6,325 (6,309) | KM114246 |
| M | 3,227 | Gn and Gc | 18–3,041 (3,024) | KM114247 | ||
| S | 1,720 | N | 35–796 (762) | KM114248 | ||
| NSs | 1,695–877 (819) | |||||
| SCOT FT 254 | L | 6,424 | RdRp | 17–6,325 (6,309) | KM114249 | |
| M | 3,289 | Gn and Gc | 18–3,041 (3,024) | KM114250 | ||
| S | 1,720 | N | 35–796 (762) | KM114251 | ||
| NSs | 1,695–877 (819) |
RdRp, RNA-dependent RNA polymerase; N, nucleocapsid protein; NSs, nonstructural protein. Gn and Gc are glycoproteins.
In cRNA, not including the stop codon.
The 5′ noncoding region is not sequenced fully.
FIG 4.
Phylogenetic analysis of novel TBPVs found in the present study. Phylogenetic trees were constructed using the maximum likelihood (ML) method with 1,000 bootstrap replicates based on multiple-sequence alignments of RNA sequences of the L segment, M segment, and S segment. Bootstrap probabilities above 70% are indicated near the branches. The viruses that were newly sequenced in the present study are shown in boldface type and underlined. SFTSV, severe fever with thrombocytopenia syndrome virus.
Discovery of a new TBPV from single tick samples collected in the field.
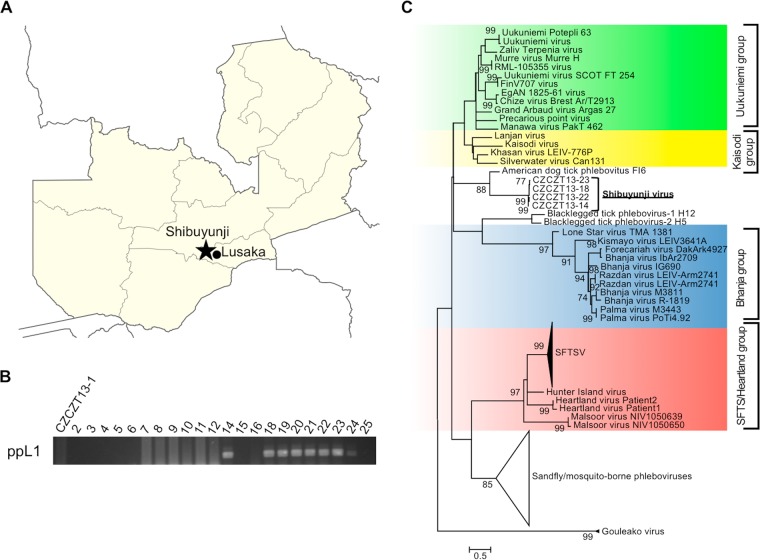
To test the usefulness of our RT-PCR system in a field situation, we gathered tick samples from cattle in the Shibuyunji District, Lusaka Province, Zambia (Fig. 5A). A total of 23 ticks, including Amblyomma spp. and Rhipicephalus spp., were collected. Total RNA extracted from each tick homogenate was tested using the RT-PCR. Eight positive samples were found (Fig. 5B), all from Rhipicephalus ticks, and specific amplification of a TBPV fragment was confirmed by sequencing 4 out of 8 positive samples (CZCZT13-14, -18, -22, and -23; the other 4 samples could not be sequenced). Phylogenetic analysis indicated that this newly identified TBPV (tentatively named Shibuyunji virus, after the area in which the ticks were collected) was genetically most closely related to American dog tick phlebovirus, which was recently found during a tick virome analysis of Dermacentor variabilis in the United States (Fig. 5C) (27).
FIG 5.
Identification of Shibuyunji virus (SHBV) from a field tick sample in Zambia. (A) The Shibuyunji district in Zambia is indicated by the black star. The Zambia location map by NordNordWest was obtained from Wikimedia Commons and modified by the authors. (B) RT-PCR was performed with field tick samples corresponding to homogenized single tick samples (CZCZT13-1 to CZCZT13-25). (C) Four out of eight RT-PCR fragments were sequenced (no sequencing reads was obtained with the other four fragments), and a phylogenetic tree was constructed based on these sequences, as well as reference sequences, using the maximum likelihood (ML) method with 1,000 bootstraps. Bootstrap probabilities above 70% are indicated near the branches. SFTSV, severe fever with thrombocytopenia syndrome virus.
DISCUSSION
With the exception of known geographic areas of SFTSV and HRTV activity where these viruses are endemic, ticks have not been extensively studied; consequently, the full geographic distributions of the various TBPVs are still largely unknown. Furthermore, in the epidemiological studies conducted for SFTSV or HRTV, the detection methods used relied on specific reactions in order to gain the highest sensitivity and specificity possible, meaning that other TBPVs were probably missed due to the high level of genetic diversity among TBPVs. Therefore, incorporating our newly established RT-PCR system, which can target a wide range of TBPVs, as part of future surveillance efforts would allow detection of both the known and yet unrecognized novel TBPVs.
The primer binding sequences selected in our system encode functional motifs (i.e., premotif A and motif B) of the L protein (RNA-dependent RNA polymerase) (35), which is the most conserved protein among phleboviruses (see Tables S3 to S5 in the supplemental material), suggesting that mutations to disrupt primer binding are likely to be infrequent. Even among the TBPVs not tested in the present study (e.g., Malsoor virus, Hunter Island virus, American dog tick phlebovirus, and blacklegged tick phlebovirus), these sequences are highly conserved, confirming the wide range of cross-reactivity of our RT-PCR system to all TBPV groups (Fig. 6), and including the detection of novel TBPVs. Although the entire amino acid sequences of the motifs selected as the targets in the present study are not conserved beyond the genus Phlebovirus (35), designing similar primer sets binding to the regions encoding these functional motifs might also be applicable for establishing an RT-PCR system that can detect a wide range of viruses in other genera.
FIG 6.
Nucleotide sequence alignment of primer binding regions among TBPVs. SFTSV, severe fever with thrombocytopenia syndrome virus.
Notably, while the degeneracy of primers usually decreases the sensitivity of an RT-PCR, our RT-PCR still had sufficient sensitivity to detect Shibuyunji virus RNA in a sample from a single tick. These results indicate that our RT-PCR system has both enough cross-reactivity and enough sensitivity for use in field studies targeting ticks. However, since the titers of TBPVs in ticks may vary among viruses and tick species, and also during the life cycle of the ticks, more-specific (and more-sensitive) primers may still be required to perform further studies in the field. In the case of humans, SFTSV titers in serum varied among patients, with fatal cases showing higher viral loads than survivors (36, 37). Evidence from domestic animals infected with SFTSV suggests that our RT-PCR system would also be able to identify infection during the viremic phase (6). Therefore, the detection limit of our system appears sufficient for first-line screening in order to broadly identify a disease's causative agent, although its low specificity makes it not appropriate for use in definitive diagnosis. Indeed, with several of the viruses tested, we observed a single nonspecific band approximately 600 to 700 bp in size (Fig. 2, 3, and 5), which resulted from the misamplification of a region of L segment RNA that overlaps with the specific 500-bp product (data not shown). Designing specific primers is therefore recommended for definitive diagnosis to reduce nonspecific amplification and maximize sensitivity.
The RT-PCR system established here is simple, cheap, fast, and sensitive, allowing its use for the screening of TBPVs prior to virus isolation and/or NGS. Furthermore, due to the flexible protocols of this system (i.e., conventional RT-PCR and qRT-PCR) and the ready availability of the reagents and equipment needed, our system should be usable on-site even in resource-poor countries. The application of a novel virus discovery flow starting with our RT-PCR system, followed by classical virus isolation and NGS, as we have performed in the present study, may influence the future discovery of novel TBPVs. Moreover, comparison of phylogenetic trees based on the sequences of the 500-bp fragment (Fig. 3B) and the full-length sequences (Fig. 4) indicated that the fragments were sufficient to roughly assign a tentative taxonomic position to novel phleboviruses. In the present study, we found five provisional new TBPV species (Kismayo virus, Kaisodi virus, Lanjan virus, Silverwater virus, and Shibuyunji virus) and succeeded in growing Kismayo virus and Silverwater virus in Huh-7 cells. Because Huh-7 cells supported TBPV growth better than the other cell lines tested (data not shown), their use together with DH82 cells (used to isolate SFTSV and HRTV) may be beneficial during attempts to isolate TBPVs. Our system should be also useful in verifying virus isolation prior to NGS with TBPVs for which cytopathic effects are not obvious.
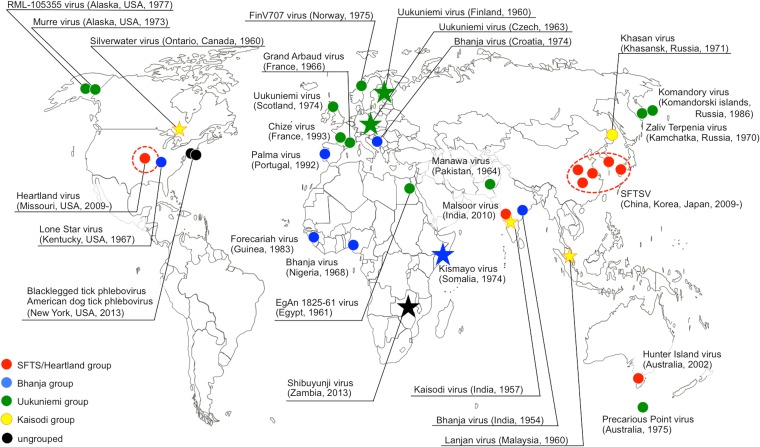
Kismayo virus, originally isolated in 1974 from Somalian brown ticks (Rhipicephalus pulchellus) (32), has been reported to cross-react partially with BHAV in serological assays (3), which was consistent with the results of our phylogenetic analysis. The sequence identity of the N protein among Bhanja group viruses (i.e., BHAVs, Palma virus, and Lone Star virus) indicated that Kismayo virus and Lone Star virus might be separate viruses, while the other BHAVs constitute a single species (see Table S5 in the supplemental material). Interestingly, while Kismayo virus has been found in East African countries, as is the case with other Bhanja group viruses, Lone Star virus has been identified only in the United States. The Kaisodi group viruses that were grouped together serologically (38) also clustered into a single genetic group in the present study. Khasan virus, which was not serologically related to the other three Kaisodi group viruses, has also been genetically classified into the Kaisodi group. Interestingly, Kaisodi virus, Silverwater virus, Lanjan virus, and Khasan virus have been found in far-flung places: India, Canada, Malaysia, and Russia (25), respectively (Fig. 7). Another interesting grouping we identified is the closely related Shibuyunji virus, found in Zambia in this study, and American dog tick virus, which was found in the United States. The sporadic discoveries of viruses that are genetically closely related to each other but lack obvious geographic and temporal relationships is characteristic throughout all known TBPV groups, suggesting that these known TBPVs represent just “the tip of the iceberg.” Our RT-PCR system will help to reveal the whole picture of TBPV genetic diversity, thus facilitating our understanding of the evolutionary mechanism(s) underlying the distinct geographic distributions of TBPVs and the emergence of pathogenic TBPVs. Furthermore, the discovery of the first TBPV in southern Africa, Shibuyunji virus, which is genetically distinct from any other known TBPVs, represents a remarkable starting point for TBPV identification in these areas. There is, however, a need for further analysis of these viruses, including virus isolation and full-length sequencing.
FIG 7.
Sporadic discoveries of tick-borne phleboviruses (TBPVs). Circles (known TBPVs) and stars (novel TBPVs identified in the present study) indicate the locations where TBPVs have been found. Representative locations of SFTSV and Heartland virus have been shown as circles, and broken-line circles designate the areas where these viruses are endemic. SFTSV, severe fever with thrombocytopenia syndrome virus.
Considering that most tick-borne diseases are zoonotic, there is a need for surveillance of both animal and tick populations to assess the risks posed by TBPVs to public health. We have demonstrated that our system can detect virus from single tick samples and could also detect TBPVs from animal tissues (i.e., suckling mouse brain homogenate). Global surveillance using this system will shed light on novel TBPVs that could potentially be pathogenic to humans and/or animals and provide us with the knowledge needed to prepare for upcoming outbreaks of disease caused by infection with novel TBPVs. SFTSV, severe fever with thrombocytopenia syndrome virus.
Supplementary Material
ACKNOWLEDGMENT
We thank the following individuals at the RML, DIR, NIAID, NIH: Sarah Anzick, Eric Dahlstrom, Daniel Bruno, Stacy Ricklefs, and Steve Porcella for performing the NGS and downstream analysis and Martha Thayer and Allison Groseth for assistance in editing the article. We also thank Brandy Russell, Division of Vector-Borne Infectious Diseases, Centers for Disease Control and Prevention, Fort Collins, CO, for her contribution to growing the viruses and Ryo Nakao and Akina Mori-Kajihara, Hokkaido University Center for Zoonosis Control, Sapporo, Japan, for their contributions to collecting ticks and performing species identification.
The present work was funded by the Intramural Research Program of the National Institute for Allergy and Infectious Diseases and partially through the U.S.-China Biomedical Collaborative Research Program. R.B.T. was supported by NIH contract HHSN272201000040I/HHSN200004/D04.
The opinions, interpretations, conclusions, and recommendations presented here are those of the authors and are not necessarily endorsed by the NIH.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02704-14.
REFERENCES
- 1.Yu X-J, Liang M-F, Zhang S-Y, Liu Y, Li J-D, Sun Y-L, Zhang L, Zhang Q-F, Popov VL, Li C, Qu J, Li Q, Zhang Y-P, Hai R, Wu W, Wang Q, Zhan FX, Wang XJ, Kan B, Wang S-W, Wan K-L, Jing H-Q, Lu J-X, Yin W-W, Zhou H, Guan X-H, Liu J-F, Bi Z-Q, Liu G-H, Ren J, Wang H, Zhao Z, Song J-D, He J-R, Wan T, Zhang J-S, Fu X-P, Sun L-N, Dong X-P, Feng Z-J, Yang W-Z, Hong T, Zhang Y, Walker DH, Wang Y, Li DX. 2011. Fever with thrombocytopenia associated with a novel bunyavirus in China. N Engl J Med 364:1523–1532. doi: 10.1056/NEJMoa1010095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McMullan LK, Folk SM, Kelly AJ, MacNeil A, Goldsmith CS, Metcalfe MG, Batten BC, Albariño CG, Zaki SR, Rollin PE, Nicholson WL, Nichol ST. 2012. A new phlebovirus associated with severe febrile illness in Missouri. N Engl J Med 367:834–841. doi: 10.1056/NEJMoa1203378. [DOI] [PubMed] [Google Scholar]
- 3.Matsuno K, Weisend C, Travassos Da Rosa AP, Anzick SL, Dahlstrom E, Porcella SF, Dorward DW, Yu X-J, Tesh RB, Ebihara H. 2013. Characterization of the Bhanja serogroup viruses (Bunyaviridae): a novel species of the genus Phlebovirus and its relationship with other emerging tick-borne phleboviruses. J Virol 87:3719–3728. doi: 10.1128/JVI.02845-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Palacios G, Savji N, Travassos da Rosa AP, Guzman H, Yu X-J, Desai A, Rosen GE, Hutchison S, Lipkin WI, Tesh RB. 2013. Characterization of the Uukuniemi virus group (Phlebovirus: Bunyaviridae): evidence for seven distinct species. J Virol 87:3187–3195. doi: 10.1128/JVI.02719-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage HM, Godsey MS, Lambert A, Panella NA, Burkhalter KL, Harmon JR, Lash RR, Ashley DC, Nicholson WL. 2013. First detection of Heartland virus (Bunyaviridae: Phlebovirus) from field collected arthropods. Am J Trop Med Hyg 89:445–452. doi: 10.4269/ajtmh.13-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niu G, Li J, Liang M, Jiang X-L, Jiang M, Yin H, Wang Z, Li C, Zhang Q, Jin C, Wang X, Ding S, Xing Z, Wang S, Bi Z, Li D. 2013. Severe fever with thrombocytopenia syndrome virus among domesticated animals, China. Emerg Infect Dis 19:756–763. doi: 10.3201/eid1905.120245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Z, Hu J, Bao C, Li P, Qi X, Qin Y, Wang S, Tan Z, Zhu Y, Tang F, Zhou M. 2014. Seroprevalence of antibodies against SFTS virus infection in farmers and animals, Jiangsu, China. J Clin Virol 60:185–189. doi: 10.1016/j.jcv.2014.03.020. [DOI] [PubMed] [Google Scholar]
- 8.Sun JM, Zhang YJ, Gong ZY, Zhang L, Lv HK, Lin JF, Chai CL, Ling F, Liu SL, Gu SP, Zhu ZH, Zheng XH, Lan YQ, Ding F, Huang WZ, Xu JR, Chen EF, Jiang JM. 27 May 2014. Seroprevalence of severe fever with thrombocytopenia syndrome virus in southeastern China and analysis of risk factors. Epidemiol Infect doi: 10.1017/S0950268814001319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonenshine DE, Roe RM. 2013. In Sonenshine DE, Roe RM (ed), Biology of ticks, 2nd ed. Oxford University Press, New York, NY. [Google Scholar]
- 10.Calisher CH, Goodpasture HC. 1975. Human infection with Bhanja virus. Am J Trop Med Hyg 24:1040–1042. [DOI] [PubMed] [Google Scholar]
- 11.Vesenjak-Hirjan J, Calisher CH, Beus I, Marton E. 1980. First natural clinical human Bhanja virus infection, p 297–301. In Vesenjak-Hirjan J, Porterfield JS, Arslanagí c E (ed), Arboviruses in the Mediterranean countries: 6th FEMS Symposium. Fischer, Stuttgart, Germany. [Google Scholar]
- 12.Saikku P. 1973. Arboviruses in Finland. 3. Uukuniemi virus antibodies in human, cattle, and reindeer sera. Am J Trop Med Hyg 22:400–403. [DOI] [PubMed] [Google Scholar]
- 13.Hubalek Z, Juricová Z. 1984. A serological survey for Bhanja virus in Czechoslovakia. Zentralbl Bakteriol Mikrobiol Hyg A 258:540–543. [DOI] [PubMed] [Google Scholar]
- 14.Bárdos V, Hubalek Z, Mittermayer T. 1977. Bhanja virus serologic survey in Czechoslovakia. Folia Parasitol 24:381. [PubMed] [Google Scholar]
- 15.Hubalek Z, Mitterpák J, Prokopic J, Juricová Z, Kilík J. 1985. A serological survey for Bhanja and tick-borne encephalitis viruses in sheep of eastern Slovakia. Folia Parasitol 32:279–283. [PubMed] [Google Scholar]
- 16.Liu Y, Li Q, Hu W, Wu J, Wang Y, Mei L, Walker DH, Ren J, Wang Y, Yu X-J. 2012. Person-to-person transmission of severe fever with thrombocytopenia syndrome virus. Vector Borne Zoonotic Dis 12:156–160. doi: 10.1089/vbz.2011.0758. [DOI] [PubMed] [Google Scholar]
- 17.Gai Z, Liang M, Zhang Y, Zhang S, Jin C, Wang SW, Sun L, Zhou N, Zhang Q, Sun Y, Ding SJ, Li C, Gu W, Zhang F, Wang Y, Bian P, Li X, Wang Z, Song X, Wang X, Xu A, Bi Z, Chen S, Li D. 2012. Person-to-person transmission of severe fever with thrombocytopenia syndrome bunyavirus through blood contact. Clin Infect Dis 54:249–252. doi: 10.1093/cid/cir776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takahashi T, Maeda K, Suzuki T, Ishido A, Shigeoka T, Tominaga T, Kamei T, Honda M, Ninomiya D, Sakai T, Senba T, Kaneyuki S, Sakaguchi S, Satoh A, Hosokawa T, Kawabe Y, Kurihara S, Izumikawa K, Kohno S, Azuma T, Suemori K, Yasukawa M, Mizutani T, Omatsu T, Katayama Y, Miyahara M, Ijuin M, Doi K, Okuda M, Umeki K, Saito T, Fukushima K, Nakajima K, Yoshikawa T, Tani H, Fukushi S, Fukuma A, Ogata M, Shimojima M, Nakajima N, Nagata N, Katano H, Fukumoto H, Sato Y, Hasegawa H, Yamagishi T, Oishi K, Kurane I, Morikawa S, Saijo M. 2014. The first identification and retrospective study of severe fever with thrombocytopenia syndrome in Japan. J Infect Dis 209:816-827. doi: 10.1093/infdis/jit603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bao C-J, Guo X-L, Qi X, Hu J-L, Zhou M, Varma JK, Cui L-B, Yang H-T, Jiao Y-J, Klena JD, Li L-X, Tao W-Y, Li X, Chen Y, Zhu Z, Xu K, Shen A-H, Wu T, Peng H-Y, Li Z-F, Shan J, Shi Z-Y, Wang H. 2011. A family cluster of infections by a newly recognized bunyavirus in eastern China, 2007: further evidence of person-to-person transmission. Clin Infect Dis 53:1208–1214. doi: 10.1093/cid/cir732. [DOI] [PubMed] [Google Scholar]
- 20.Zhang L, Liu Y, Ni D, Li Q, Yu Y, Yu X-J, Wan K, Li D, Liang G, Jiang X, Jing H, Run J, Luan M, Fu X, Zhang J, Yang W, Wang Y, Dumler JS, Feng Z, Ren J, Xu J. 2008. Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA 300:2263–2270. doi: 10.1001/jama.2008.626. [DOI] [PubMed] [Google Scholar]
- 21.Dilcher M, Alves MJ, Finkeisen D, Hufert F, Weidmann M. 2012. Genetic characterization of Bhanja virus and Palma virus, two tick-borne phleboviruses. Virus Genes 45:311–315. doi: 10.1007/s11262-012-0785-y. [DOI] [PubMed] [Google Scholar]
- 22.Swei A, Russell BJ, Naccache SN, Kabre B, Veeraraghavan N, Pilgard MA, Johnson BJB, Chiu CY. 2013. The genome sequence of Lone Star virus, a highly divergent bunyavirus found in the Amblyomma americanum tick. PLoS One 8:e62083. doi: 10.1371/journal.pone.0062083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mourya DT, Yadav PD, Basu A, Shete A, Patil DY, Zawar D, Majumdar TD, Kokate P, Sarkale P, Raut CG, Jadhav SM. 2014. Malsoor virus, a novel bat phlebovirus, is closely related to severe fever with thrombocytopenia syndrome virus and Heartland virus. J Virol 88:3605–3609. doi: 10.1128/JVI.02617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Selleck P, Yu M, Ha W, Rootes C, Gales R, Wise T, Crameri S, Chen H, Broz I, Hyatt A, Woods R, Meehan B, McCullough S, Wang L-F. 2014. Novel phlebovirus with zoonotic potential isolated from ticks, Australia. Emerg Infect Dis 20:1040–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alkhovvskiĭ SV, Lvov DK, Shchelkanov MI, Shchetinin AM, Deriabin PG, Samokhvalov EI, Gitelman AK, Botikov AG. 2013. The taxonomy of the Khasan virus (KHAV), a new representative of Phlebovirus genera (Bunyaviridae), isolated from the ticks Haemaphysalis longicornis (Neumann, 1901) in the Maritime Territory (Russia). Vopr Virusol 58(5):15–18. (In Russian.) [PubMed] [Google Scholar]
- 26.Alkhovvskiĭ SV, Lvov DK, Shchelkanov MI, Shchetinin AM, Deryabi PG, Botikov AG, Gitel'man AK, Samokhvalov EI. 2013. Genetic characterization of new Komandory virus (KOMV; Bunyaviridae, Phlebovirus) isolated from the ticks Ixodes uriae, collected in guillemot (Uria aalgre) nesting sites on Komandorski islands, the Bering Sea. Vopr Virusol 58(6):18–22. (In Russian.) [PubMed] [Google Scholar]
- 27.Tokarz R, Williams SH, Sameroff S, Sanchez Leon M, Jain K, Lipkin WI. 2014. Virome analysis of Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks reveals novel highly divergent vertebrate and invertebrate viruses. J Virol 88:11480–11492. doi: 10.1128/JVI.01858-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Plyusnin A, Beaty BJ, Elliott RM, Goldbach R, Kormelink R, Lundkvist A, Schmaljohn CS, Tesh RB. 2012. Family Bunyaviridae, p 725–741. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ (ed), Virus taxonomy. Ninth report of the International Committee on Taxonomy of Viruses, 1st ed. Elsevier Academic Press, San Diego, CA. [Google Scholar]
- 29.Bhatt PN, Kulkarni KG, Boshell J, Rajagopalan PK, Patil AP, Goverdhan MK, Pavri KM. 1966. Kaisodi virus, a new agent isolated from Haemaphysalis spinigera in Mysore State, South India. I. Isolation of strains. Am J Trop Med Hyg 15:958–960. [DOI] [PubMed] [Google Scholar]
- 30.Tan DS, Smith CE, McMahon DA, Bowen ET. 1967. Lanjan virus, a new agent isolated from Dermacentor auratus in Malaya. Nature 214:1154–1155. doi: 10.1038/2141154a0. [DOI] [PubMed] [Google Scholar]
- 31.Hoff GL, Iversen JO, Yuill TM, Anslow RO, Jackson JO, Hanson RP. 1971. Isolations of Silverwater virus from naturally infected snowshoe hares and Haemaphysalis ticks from Alberta and Wisconsin. Am J Trop Med Hyg 20:320–325. [DOI] [PubMed] [Google Scholar]
- 32.Butenko AM, Gromashevsky VL, Lvov DK, Popov VL. 1979. Kismayo virus, a representative of the Bhanja antigenic group. Vopr Virusol 24:661–665. (In Russian.) [PubMed] [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, Filipiski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hubalek Z, Rudolf I. 2012. Tick-borne viruses in Europe. Parasitol Res 111:9–36. doi: 10.1007/s00436-012-2910-1. [DOI] [PubMed] [Google Scholar]
- 35.Müller R, Poch O, Delarue M, Bishop DH, Bouloy M. 1994. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol 75:1345–1352. doi: 10.1099/0022-1317-75-6-1345. [DOI] [PubMed] [Google Scholar]
- 36.Sun Y, Jin C, Zhan F, Wang X, Liang M, Zhang Q, Ding S, Guan X, Huo X, Li C, Qu J, Wang Q, Zhang S, Zhang Y, Wang S, Xu A, Bi Z, Li D. 2012. Host cytokine storm is associated with disease severity of severe fever with thrombocytopenia syndrome. J Infect Dis 206:1085–1094. doi: 10.1093/infdis/jis452. [DOI] [PubMed] [Google Scholar]
- 37.Gai Z-T, Zhang Y, Liang M-F, Jin C, Zhang S, Zhu C-B, Li C, Li X-Y, Zhang Q-F, Bian P-F, Zhang L-H, Wang B, Zhou N, Liu J-X, Song X-G, Xu A, Bi Z-Q, Chen S-J, Li D-X. 2012. Clinical progress and risk factors for death in severe fever with thrombocytopenia syndrome patients. J Infect Dis 206:1095–1102. doi: 10.1093/infdis/jis472. [DOI] [PubMed] [Google Scholar]
- 38.Mahy BWJ. 2009. The dictionary of virology, 4th ed. Academic Press, Burlington, MA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.