ABSTRACT
Human cytomegalovirus (HCMV) is an important, ubiquitous pathogen that causes severe clinical disease in immunocompromised individuals, such as organ transplant recipients and infants infected in utero. The envelope glycoprotein B (gB) of HCMV is a major antigen for the induction of virus-neutralizing antibodies. We have begun to define target structures within gB that are recognized by virus-neutralizing antibodies. Antigenic domain 5 (AD-5) of gB has been identified as an important target for neutralizing antibodies in studies using human monoclonal antibodies (MAbs). Anti-AD-5 MAbs share a target site on gB, despite originating from different, healthy, HCMV-infected donors. Mutational analysis of AD-5 identified tyrosine 280 in combination with other surface-exposed residues (the YNND epitope) as critical for antibody binding. The YNND epitope is strictly conserved among different HCMV strains. Recombinant viruses carrying YNND mutations in AD-5 were resistant to virus-neutralizing MAbs. Competition enzyme-linked immunosorbent assays (ELISAs) with human HCMV-convalescent-phase sera from unselected donors confirmed the conserved antibody response for the YNND epitope in HCMV-infected individuals and, because a significant fraction of the gB AD-5 response was directed against the YNND epitope, further argued that this epitope is a major target of anti-AD-5 antibody responses. In addition, affinity-purified polyclonal anti-AD-5 antibodies prepared from individual sera showed reactivity to AD-5 and neutralization activity toward gB mutant viruses that were similar to those of AD-5-specific MAbs. Taken together, our data indicate that the YNND epitope represents an important target for anti-gB antibody responses as well as for anti-AD-5 virus-neutralizing antibodies.
IMPORTANCE HCMV is a major global health concern, and a vaccine to prevent HCMV disease is a widely recognized medical need. Glycoprotein B of HCMV is an important target for neutralizing antibodies and hence an interesting molecule for intervention strategies, e.g., vaccination. Mapping the target structures of neutralizing antibodies induced by naturally occurring HCMV infection can aid in defining the properties required for a protective capacity of vaccine antigens. The data presented here extend our knowledge of neutralizing epitopes within gB to include AD-5. Collectively, our data will contribute to optimal vaccine design and development of antibody-based therapies.
INTRODUCTION
Human cytomegalovirus (HCMV) is an important, ubiquitously distributed human pathogen that is found throughout all geographic locations and socioeconomic groups. Initial infection with HCMV is followed by a lifelong persistence characterized by periodic reactivation episodes. While most infections occur subclinically in the immunocompetent host, HCMV can cause severe disease and death in immunocompromised patients and newborns infected in utero. HCMV is the most frequent viral cause of congenital infection and affects 0.5 to 2% of all live births worldwide (1). It is the leading infectious cause of childhood sensorineural hearing loss and an important cause of mental retardation (2, 3). Few drugs are available for treatment of HCMV infection, and their use is limited by toxicity and the development of drug-resistant virus isolates. Therefore, prophylactic vaccination has been argued to be the preferred approach for prevention of HCMV infection of pregnant women and subsequent transmission to the fetus, and the development of an HCMV vaccine has been given highest priority by governmental agencies (4). However, no vaccine has been licensed to date, and a preventive vaccine remains elusive.
As a result of long coevolution with its only host, humans, HCMV is readily controlled by a multilayered and partially redundant immune response in the immunocompetent host. The natural immunity to HCMV is largely protective against horizontal and vertical transmission (5). However, control is not complete, as seropositive subjects can be reinfected or reactivate endogenous virus (6, 7). Natural immunity following infection includes innate and adaptive immune effector functions. Different studies in humans have documented reduced maternal infection or maternal-fetal transmission in women with preexisting HCMV immunity (8, 9). Antiviral antibodies are considered to be an important component of the maternal immune response in prevention of intrauterine transmission of HCMV. This protective role of antiviral antibodies is further supported by the observation that pregnant women who develop high titers of neutralizing antibodies are less likely to transmit the infection to the fetus (10). The protective activity of antibodies has also been demonstrated in different animal model systems. For example, in the guinea pig animal model, preconception infection protected against in utero transmission of guinea pig CMV (GPCMV), most likely due to the generation of neutralizing antibodies in the mother (11, 12). Additionally, complete protection from infection was achieved in immunodeficient mice infected with murine CMV by prophylactic or therapeutic administration of polyclonal antibody preparations (13).
HCMV is a complex virus that can express more than 165 viral antigens during its replication cycle (14, 15). The viral envelope contains glycoproteins which elicit neutralizing antibodies, and the predominant targets are glycoprotein B (gB), gM/gN, and the gH/gL-containing complexes. While the function of gM/gN in the entry process remains largely unknown, gB and gH/gL constitute the core fusion machinery, and the different gH/gL complexes determine target cell tropism (16). The gH/gL/gO complex is required for infection of fibroblasts via membrane fusion, while gH/gL, together with the UL128, UL130, and UL131A proteins, which form the gH/gL pentamer complex, are essential for epithelial/endothelial cell tropism via an endocytic pathway (17–19). gB is the actual fusion mediator, and its fusogenic activity presumably must be triggered via interaction with gH/gL (20, 21). In addition, gB is an important immunogen, as gB-specific antibodies can be detected in all naturally infected individuals (22). A major fraction of neutralizing antibodies in sera from HCMV-seropositive donors is directed against gB, and the overall neutralizing capacity correlates with the anti-gB antibody titer when tested on fibroblasts (23, 24). While in the past gB has been considered the dominant protein for the induction of neutralizing antibodies, components of the gH/gL pentameric complex have recently received increasing attention as major targets of virus-neutralizing antibodies. Gerna et al. reported that the neutralizing activity of sera was underestimated because serum neutralizing titers were mostly explored on fibroblasts, not on endothelial/epithelial cells (25). The increase in neutralizing activity on endothelial/epithelial cells presumably results from the presence of extremely potent neutralizing antibodies against the UL128-UL131A complex in human sera. Examples of such monoclonal antibodies (MAbs) were isolated by Macagno et al. (26). However, the neutralizing activity of this type of antibody is limited to endothelial, epithelial, and dendritic cells, since these MAbs have no activity on fibroblasts or trophoblasts, cells which are crucial HCMV targets in vertical transmission. Thus, the protective capacity of anti-gH/gL-pentamer antibodies in vivo remains to be evaluated. Anti-gB and anti-gH/gL antibodies, in contrast, show potent and robust neutralization in the nanomolar range, independent of the target cell type (26–28). More importantly, studies with the guinea pig model of maternal and congenital CMV infection determined that passive immunization with polyclonal anti-GPCMV antibodies or polyclonal anti-gB antibodies protected animals from infection or transmission (29, 30). In addition, in the model of murine CMV-induced encephalitis/neuropathology, significant protection from brain pathology in infected newborn mice by passive administration of a gB-specific MAb was reported (31).
In any case, gB remains an attractive target for inclusion in a human vaccine and has been a major focus of experimental vaccination strategies (5, 32, 33). In fact, an efficacy study of the gB/MF59 vaccine in postpartum, HCMV-seronegative women provided approximately 50% protection from acquisition of HCMV infection (34). Another phase II study, using solid organ transplant recipients, showed 50% efficacy in controlling viremia in high-risk patients (35). In addition, vaccination studies with gB in rhesus macaques and subsequent rhesus CMV (RhCMV) challenge showed significantly reduced RhCMV DNA in plasma (36, 37). Finally, a number of studies using the guinea pig model demonstrated that congenital infection and mortality in pups were reduced following gB DNA or recombinant protein subunit vaccination strategies (38).
In conventional views of vaccine development, vaccine-induced immunity should be comparable to that in humans undergoing natural infection. But the exact correlate of gB-mediated protection in terms of antibody response remains poorly defined for both natural infection and vaccination (5). Despite the fact that gB has been used as a vaccine antigen in humans, our knowledge about the anti-gB antibody response in terms of fine specificity and antiviral activity remains incomplete. Five antigenic domains (AD) have been described for gB, among which four can induce neutralizing antibodies during infection (27). All neutralizing anti-gB MAbs have similar neutralizing activities, and all can neutralize 50% of input virus at concentrations in the nanomolar range. The major targets of neutralizing antibodies are located within AD-4 and AD-5 of gB, and antibodies to both regions have comparable activities on fibroblasts and epithelial/endothelial cells (27). Further characterization of critical binding residues within AD-4 recently identified an important epitope for gB-directed neutralization (39). In contrast, information on neutralizing epitopes within AD-5 is limited.
AD-5 is broadly immunogenic, and antibodies to AD-5 can be detected in most cases of HCMV infection. Importantly, AD-5 contains the two internal fusion loops that are required for the fusogenic activity of herpesviral gB (40). In herpes simplex virus 1 (HSV-1) gB, this part of the molecule is referred to as the fusion domain (41). To better understand the target structures on gB that are important for neutralization of free virus, we started to characterize the epitopes of neutralizing human MAbs within AD-5. We found that three MAbs originating from different, healthy donors bound to a common antigenic site in AD-5 and that mutation of this antigenic site rendered the virus resistant to neutralization by the human MAbs. Importantly, AD-5-specific polyclonal antibodies prepared from individual human sera revealed that this antigenic site, in some cases, is the exclusive target of all neutralizing activity directed toward AD-5.
MATERIALS AND METHODS
Cells and viruses.
Human embryonic kidney cells (HEK 293T), primary human foreskin fibroblasts (HFF), and human fetal lung fibroblasts (MRC-5) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies) supplemented with 10% fetal calf serum (FCS) (Sigma-Aldrich), glutamine (100 μg/ml), and gentamicin (350 μg/ml). Spodoptera frugiperda (Sf9) cells were cultured in SF-900 II SFM (Life Technologies, Germany) in suspension at 27°C.
HCMV TB40/E and its recombinant derivatives were constructed using markerless mutagenesis of vBAC4-luc (42), a TB40-BAC4-derived BACmid in which the genes for UL5 to UL9 were replaced by a simian virus 40 (SV40) promoter-driven luciferase expression cassette. Viruses were propagated in MRC-5 cells, and viral titers were determined by titration of virus stocks in MRC-5 cells, using an indirect immunofluorescence assay with the MAb p63-27, directed against the HCMV immediate early 1 (IE1) protein (43), or by measurement of luciferase activity.
Antibodies.
The following antibodies were used: gB AD-5-specific human MAbs SM10, 2C2, and 1G2; gB AD-4-specific human MAb SM5-1 (27); gB AD-2-specific human MAb C23 (Ti23) (44); a murine anti-hemagglutinin (anti-HA) MAb (clone HA-7) from Sigma-Aldrich; and a murine anti-myc MAb (clone 2G8D5) from GenScript. AD-5-specific polyclonal antibody preparations (polyAD-5 1 to 9) were prepared from individual human sera by AD-5 immunoaffinity chromatography, and the polyclonal anti-AD-4 antibody preparation has been described previously (27). Secondary antibodies and streptavidin-horseradish peroxidase (HRP) were purchased from Southern Biotech, Jackson Immuno Research, or Thermo Scientific.
Expression plasmids.
Candidate residues for the dialanine scan were identified based on a structural model of HCMV gB (27). In the first step, residues with a solvent-exposed surface area of >50 Å2 were identified and subsequently grouped into spatially proximal pairs of residues. Initially, this procedure resulted in a total of 16 pairs (set 1) of residues selected for mutagenesis. The DNAs for expression of AD-5 and its mutant derivatives were chemically synthesized by Life Technologies and cloned into the pcUL132sigHA expression vector as described previously (27). Two types of AD-5 mutant expression constructs were utilized: gB residues I133 to T343 (set 1; AD-5 only) and residues P116 to V440 (set 2; AD-5 surrounded by AD-4 for protein stability) of HCMV strain AD169. Both constructs contained an N-terminal HA tag and a C-terminal myc tag, each connected to the protein by a 5×GS spacer. Additionally, in the second set of mutants, the hydrophobic residues (YIY155–157 and W240) of the fusion loops were changed to their less hydrophobic HSV-1 gB counterparts (GHR and A) to prevent protein aggregation. For AD-5 wild-type (wt) and mutant protein expression, HEK 293T cells were transfected by calcium phosphate precipitation, and protein-containing supernatants were harvested 4 to 6 days later. For enzyme-linked immunosorbent assay (ELISA) applications, the specific protein amount in the supernatant was adjusted by titration of the supernatants with the anti-myc or SM5-1 (anti-AD-4) control antibodies.
ELISA.
For AD-5 mutant binding ELISAs, the anti-HA MAb was diluted to 1 μg/ml in 0.05 M sodium carbonate buffer, pH 9.6, and 50 μl/well was used to coat polystyrene 96-well plates overnight at 4°C. All following reactions were performed at 37°C. Reaction wells were blocked with phosphate-buffered saline (PBS) containing 2% FCS for 1 h, washed three times with PBS plus 0.1% Tween 20, and incubated with comparable amounts of AD-5 wt or mutant proteins for 2 h. The plate was washed three times with PBS plus 0.1% Tween 20, and primary antibody (MAb or polyclonal antibody preparation) was added in serial log2 dilutions starting at 0.5 μg/ml for 1 h. Unbound antibody was removed by washing three times, and the appropriate peroxidase-conjugated secondary antibody was added for 1 h. After three washing steps, 100 μl of tetramethylbenzidine (TMB) peroxidase substrate was added for 3.5 min, diluted 1:1 in peroxidase substrate solution B (KPL). The reaction was stopped by adding 100 μl of 1 M phosphoric acid. The optical density at 450 nm (OD450) was determined using an Emax microplate reader (Eurofins MWG Operon). For each antibody, the OD450 value of AD-5 wt recognition was set to 100%, and percent recognition of mutant proteins was calculated.
For competition ELISAs, antibodies SM10, 2C2, and 1G2 were biotinylated using an EZ-Link sulfo-NHS-biotinylation kit from Thermo Scientific. The ELISA protocol explained above was modified as follows: 25 ng of soluble gB was used to coat plates and incubated with serial log2 dilutions of MAb or serum. Unbound antibody was removed in three stringent washing steps with PBS plus 0.1% Tween 20 and 0.2 M NaCl. Biotinylated antibody was added at a constant concentration below saturation and subsequently detected using streptavidin-HRP.
Preparation of AD-5 and immunoaffinity chromatography.
DNA encoding residues I133 to T343 of HCMV gB strain AD169, together with an N-terminal thrombin cleavage site and a 6×His tag, was chemically synthesized by Life Technologies and inserted into the pAcGP67 vector in frame with the gp67 signal sequence coding region, C-terminally creating the additional residues ADP in the cloning procedure. To prevent aggregation of the purified AD-5 protein, the hydrophobic residues of the fusion loops (YIY155–157 and W240) were changed to alanine. Recombinant baculovirus for expression of AD-5 was generated by using BD BaculoGold Bright technology (BD Biosciences Pharmingen) according to the manufacturer's instructions. For AD-5 protein expression, a passage 2 stock of baculovirus was used to infect Sf9 cells at 2 × 106 cells/ml, with a multiplicity of infection (MOI) of 1. Supernatant was harvested at 72 to 96 h postinfection. To this end, cells were pelleted by centrifugation at 800 rpm at 4°C for 10 min. The supernatant was collected, and residual cellular debris was removed by centrifugation at 5,000 rpm at 4°C for 30 min. The supernatant was dialyzed to binding buffer for nickel ion-affinity chromatography and subsequently filtered through a 0.45-nm filter. AD-5 protein was purified using HisTrap FF columns on an ÄKTAprime machine (GE Healthcare) according to the manufacturer's protocol.
To prepare an affinity matrix for immunoaffinity chromatography, 2 mg of purified AD-5 protein was dialyzed against coupling buffer and conjugated to AminoLink Plus coupling resin (Thermo Fisher Scientific) according to the manufacturer's instructions. Approximately 50 ml of individual HCMV-convalescent-phase serum, diluted 1:4 (vol/vol) with PBS, was passed over 2 ml of antigen-coupled beads, followed by extensive washing with PBS. Bound IgG was eluted with 0.1 M glycine-HCl, pH 3.0, in 1-ml fractions, and fractions were dialyzed against PBS. The total IgG concentration was determined by ELISA as described previously (27).
BAC mutagenesis and reconstitution of BAC-derived viruses.
A luciferase-expressing HCMV TB40/E-derived bacterial artificial chromosome (BAC) (vBAC4-luc) (42) was used to generate mutant viruses by using the mutagenesis protocol of Tischer et al. (45). Recombination of vBAC4-luc to introduce alanine exchanges in gB at the positions of interest and reconstitution of recombinant TB40/E viruses were performed as described previously (39). Viral DNA was isolated from infected cells by use of proteinase K, and the correctness of reconstituted viruses was confirmed by nucleotide sequence analysis.
Microneutralization assay.
HFF (1 × 104 cells/well) were seeded in 96-well plates. HCMV TB40/E or recombinant viruses (3,000 to 9,000 PFU) were preincubated with serial log2 dilutions of MAb or polyclonal antibody preparations for 1 h at 37°C, and the mixtures were added to the cells for 4 h. The inoculum was replaced with fresh medium, and the cells were incubated at 37°C for 48 h. Cells were lysed using 100 μl Glo lysis buffer (Promega) per well. Thirty microliters of each cell lysate was transferred to white 96-well LIA plates (Costar), and 50 μl assay buffer (15 mM KH2PO4, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 5 mM ATP, 1 mM dithiothreitol [DTT]) was added to each well. Luciferase activity was measured by injection of 50 μl d-luciferin (P.J.K.) solution (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 2 mM DTT, and 0.05 mM d-luciferin) per well, and detection of chemiluminescence was performed by use of an Orion microplate luminometer (Berthold Technologies). The data were plotted as percent neutralization versus that in control wells, to which no antibody was added, and the 50% inhibitory concentrations (IC50s) were determined.
Ethics statement.
Ethics approval for the serum sample collection was obtained from the Ethics Committee of the Medical Faculty of the Friedrich-Alexander Universität Erlangen-Nürnberg. Written informed consent was obtained from all donors.
RESULTS
AD-5-specific monoclonal antibodies share a binding structure on gB.
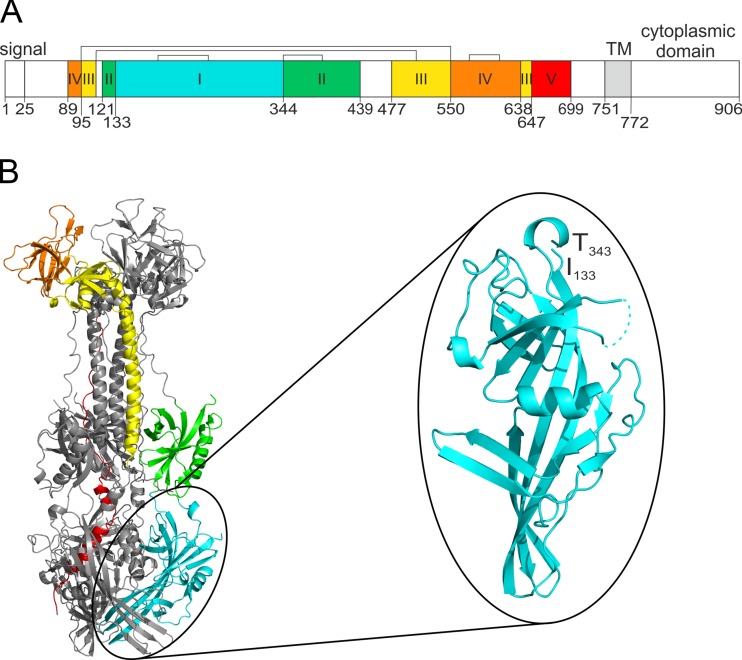
In a previous study, we identified AD-5 of HCMV gB as an antigenic determinant on gB that is targeted by neutralizing human antibodies (27). Within a structural model of gB (Fig. 1) based on the crystal structure of HSV-1 gB (46), AD-5 is formed by a continuous sequence comprising amino acids (aa) 133 to 343 of HCMV gB strain AD169. According to the three-dimensional (3D) model of HCMV gB, AD-5 corresponds to structural domain I (Dom I) and is located proximal to the viral membrane at the base of the gB trimer. Three AD-5-specific neutralizing human MAbs (SM10, 2C2, and 1G2) were isolated (27). Importantly, these antibodies were obtained from three different, healthy, HCMV-infected donors. All MAbs showed broad virus-neutralizing activity, in the nanomolar range, when assayed on fibroblasts and endothelial and epithelial cells, and this activity appeared to be independent of the inhibition of virus attachment (27). Additionally, bivalent binding of the antibodies to the antigen is not required for neutralization, as antibody Fab fragments have an IC50 comparable to that of the whole IgG molecule (see Fig. S1A in the supplemental material). To obtain additional information about the epitope(s) recognized by AD-5-specific antibodies, we initially tested their capacity to compete for binding to gB. We found that in contrast to an anti-AD-2 antibody (C23) or an anti-AD-4 antibody (SM5-1), the antibodies SM10, 2C2, and 1G2 showed strong competition for binding to gB in ELISA (see Fig. S1B). This result led us to hypothesize that the antibodies share a binding structure on AD-5, which might be of general importance for the gB-directed neutralizing antibody response, as MAbs specific for this structure were independently isolated from three donors.
FIG 1.
Domain architecture and 3D model of HCMV gB. (A) Linear representation of HCMV gB strain AD169. The individual protein domains are displayed in different colors. Brackets indicate disulfide bonds, and numbers indicate the beginnings of the domains. TM, transmembrane anchor. (B) 3D model of a gB trimer, with the structural domains of one protomer colored as in panel A. Structural domain I, corresponding to AD-5, is enlarged and rotated. Numbers in panel B indicate the beginning and end of AD-5, and residues missing from the model (aa V306 to E317) are displayed as dashed lines.
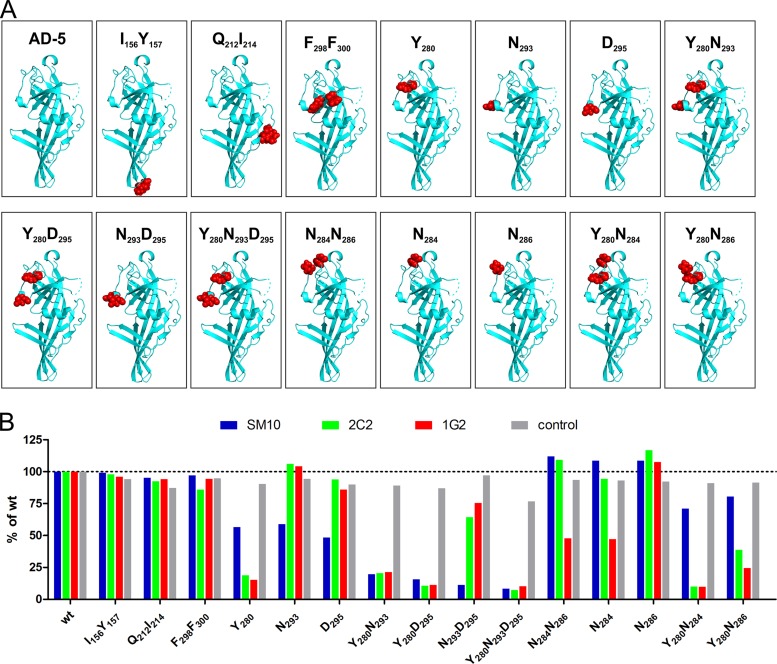
To more completely define this common binding structure, we performed dialanine scans of AD-5 to identify critical antibody binding residues. In the first set of mutants (n = 16), two predicted surface-adjacent residues per construct were selected for mutation to alanine, using the 3D model of HCMV gB, and in a second set, additional mutants (n = 12) of interest were generated based on the results for the first set. (The complete set of generated mutants is given in Fig. S2A and B in the supplemental material.) As prokaryotic expression of AD-5 resulted in a misfolded protein that was not recognized by any of the anti-AD-5 MAbs (data not shown), recombinant wt AD-5 and the respective mutants were expressed into the cell culture supernatants of transiently transfected HEK 293T cells. All constructs contained an N-terminal HA tag and a C-terminal myc tag (set 1; see Fig. S2A). A subset of constructs additionally contained AD-4 (gB aa 121 to 132 and 344 to 438) (set 2; see Fig. S2B), a discontinuous sequence surrounding and stabilizing AD-5. Additionally, in AD-5 mutants of the second set, the fusion loops were mutated to their less hydrophobic HSV-1 gB counterparts to prevent the protein aggregation that has been described to be mediated by the hydrophobic residues of the fusion loops of full-length gB (40). Following incubation for 4 to 6 days after transfection, the protein was captured from the cell culture supernatant by using an anti-HA MAb and was tested for recognition by the anti-AD-5 antibodies in ELISA. Anti-myc and anti-AD-4 (SM5-1) MAbs were used to control for comparable protein amounts and structural integrity. Representative ELISA results from sets 1 and 2 are shown in Fig. 2. For most AD-5 mutants, no significant difference in recognition was observed compared to the wt, and three representative examples (I156Y157, Q212I214, and F298F300) are shown in Fig. 2B. In examining the individual antibodies, however, we saw antibody-specific patterns of altered mutant protein recognition. For the SM10 antibody, AD-5 residues Y280, N293, and D295 were identified as critical for binding. Mutants involving combinations of Y280, N293, and/or D295 showed a complete loss of AD-5 recognition by SM10. Furthermore, mutation of Y280 also abolished binding of MAbs 2C2 and 1G2, indicating that Y280 is a critical binding residue for all three antibodies. The combinatorial mutation of N293D295 also resulted in slightly weaker AD-5 recognition by the 2C2 MAb. In addition, mutation of N284 to alanine reduced and the combined mutation of Y280 and N284 completely abolished binding of MAb 1G2 to AD-5, suggesting an involvement of both amino acids in the 1G2–AD-5 interaction. In contrast, mutation of N286 did not affect binding of any AD-5-specific antibody. Loss of binding was confirmed with selected mutants analyzed by indirect immunofluorescence of AD-5 mutant-transfected Cos7 cells (data not shown).
FIG 2.
AD-5 mutants and recognition by human antibodies. (A) Structural models of AD-5 wt and mutant proteins. AD-5 is depicted as a ribbon diagram, with the respective residues that were mutated to alanine highlighted as red spheres. (B) Representative ELISA results with AD-5 wt and mutant proteins. Recombinant AD-5 proteins were captured from the cell culture supernatants of transfected HEK 293T cells via an anti-HA MAb and tested for binding with the anti-AD-5 MAbs. Either an anti-myc antibody (I156Y157 and Q212I214 mutants) or the AD-4-specific antibody SM5-1 (all other mutants) served as a control. All assays were carried out as log2 dilution titrations to exclude antibody saturation, and data from the linear range of the assay are shown. OD450 values of wt AD-5 were set to 100% for each antibody, and percent recognition of the AD-5 mutants was calculated.
In summary, the mutational study of AD-5 revealed that binding of all three antibodies to recombinant AD-5 is sensitive to mutation of Y280 in combination with other surface-adjacent residues, i.e., Y280N293D295 for SM10 and Y280N284 for 1G2. Note that none of the AD-5-specific antibodies target the fusion loops but instead target a highly conserved region within the pleckstrin homology domain of AD-5.
Loss of binding to recombinant AD-5 correlates with loss of neutralization of gB mutant viruses.
The 3D model of gB most likely represents the postfusion conformation of the protein. However, on the infectious virion, gB would adopt a prefusion conformation, a structure that has not been defined. As there may be significant structural differences between the post- and prefusion forms of the protein, the results of antibody binding assays using recombinant AD-5 may not be equivalent to antibody binding to virion gB.
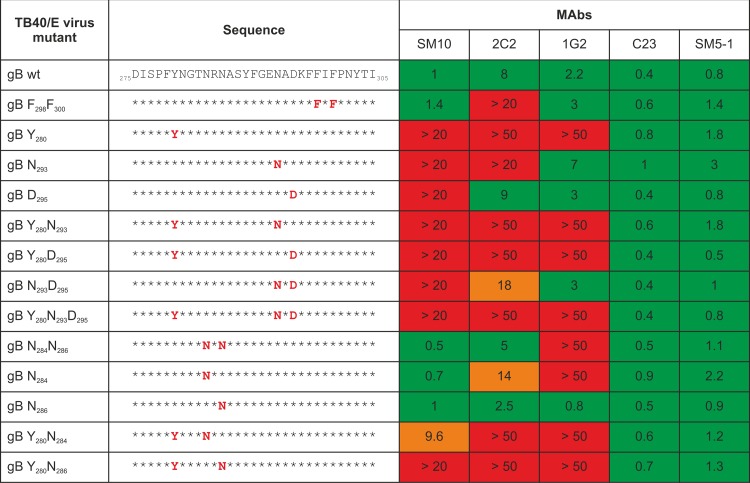
To confirm a correlation between critical residues for AD-5 binding and neutralizing activity of anti-AD-5 MAbs, we constructed recombinant viruses carrying the respective amino acid mutations to alanine in AD-5. In total, 13 recombinant viruses with single, double, or triple amino acid mutations to alanine were generated in the background of HCMV strain TB40/E (Fig. 3). All 13 recombinant viruses were replication competent, indicating that none of the changes significantly compromised gB function (see Fig. S3 in the supplemental material). The sensitivity of the recombinant viruses to neutralization by the AD-5-specific MAbs was tested in microneutralization assays on fibroblasts. Two non-AD-5-specific neutralizing MAbs, C23 (AD-2) and SM5-1 (AD-4), which should not be influenced by mutations in AD-5, were used as controls. A summary of the analysis of the recombinant viruses, with IC50 values of MAbs toward virus mutants, is provided in Fig. 3, and neutralization curves are shown in Fig. S4. As expected, the IC50 values of both control antibodies (C23 and SM5-1) were largely unaffected by mutations in AD-5 and remained in the expected range of interassay variations. In contrast, the anti-AD-5 MAbs showed distinct and antibody-specific patterns of loss of neutralizing activity toward the virus mutants that mirrored the overall result of the binding studies. However, the effects of single amino acid exchanges appeared to be more pronounced in the context of virion gB than in the mutant recombinant protein. For example, while the 1G2 MAb exhibited only a partial decrease in binding activity to the N284 mutant in ELISA, this antibody had no neutralizing activity against the N284 virus mutant. Overall, the mutation of residue Y280 alone or in combination with other surface-adjacent residues rendered the virus resistant to neutralization by any of the anti-AD-5 MAbs. The antibody 2C2 exhibited reduced neutralizing activity for the N284, N293, N293D295, and F298F300 viral mutants and complete loss of neutralizing activity toward all viruses carrying the Y280 mutation. Thus, the recognition of gB by MAb 2C2 appeared to be sensitive to local changes in the gB surface properties that surround the identified antigenic structure. In the case of MAb SM10, recombinant viruses containing mutations at N293 and/or D295 alone or in addition to Y280 were resistant to the neutralizing activity, consistent with the results from the binding assays. Interestingly, SM10 showed more potent neutralization of the N284 or N284N286 virus mutant, which MAb 1G2, in contrast, failed to neutralize. In addition, the neutralizing activity of SM10 toward Y280 was partially rescued with a supplementary mutation of N284. Thus, mutation of N284, while abolishing sensitivity to neutralization by 1G2, rendered the virus more sensitive to neutralization by SM10. The same enhancement of neutralization sensitivity was seen for 1G2 or 2C2 in viruses containing the mutation of N286 to alanine. Finally, the 1G2 antibody failed to neutralize the Y280 and N284 virus mutants and all viruses that contained these mutations. In summary, viral mutants containing Y280 in combination with additional residues in close structural proximity were resistant to the neutralizing activity of all three anti-AD-5 MAbs, suggesting that this epitope defines a common antigenic structure within AD-5. Importantly, the distinct and antibody-specific susceptibilities of the viral mutants to neutralization by the MAbs suggested that introduction of these mutations did not disrupt gB stability and function. Thus, our findings are more consistent with the interpretation that these residues in AD-5 of gB define critical contact residues for the binding of virus-neutralizing antibodies. For the sake of simplicity in describing this structure in gB, we have designated this binding determinant the YNND epitope.
FIG 3.
IC50s of anti-AD-5 antibodies for AD-5 mutant recombinant viruses. Recombinant TB40/E viruses were constructed to carry amino acid mutations to alanine at the locations indicated in red in the sequence panel. TB40/E wt and virus mutants were tested for susceptibility to neutralization by the anti-AD-5 antibodies on fibroblasts. The IC50 was determined and is given in μg/ml. Values represent medians from 2 to 10 experiments. Red, IC50 was not reached at the indicated concentration; orange, reduced neutralizing activity; green, no significant change in neutralization.
Importance of the YNND epitope for the antibody response during infection.
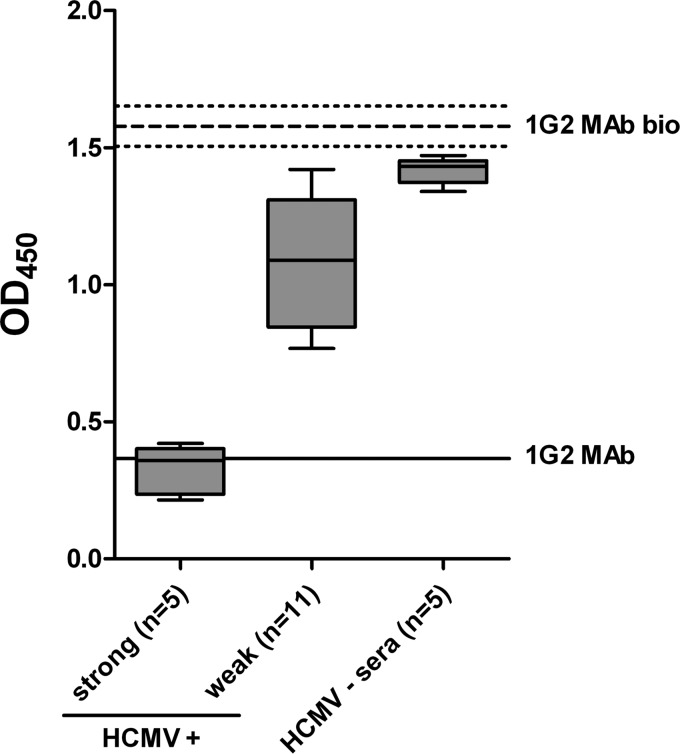
Although the MAbs utilized in this study originated from three different donors and still recognized a common structure on AD-5, it remained to be determined if these antibodies reflected the importance of this antibody binding site in the anti-AD-5 antibody response following HCMV infection. To explore this possibility, sera from 19 randomly selected, healthy, HCMV-infected individuals were collected and subsequently analyzed for reactivity with gB, AD-5, and the common antigenic structure YNND. All of the serum specimens had reactivity to recombinant gB, and 17 (90%) sera were also reactive to recombinant AD-5 (data not shown). Next, competition ELISAs were performed to determine if anti-AD-5 antibodies in individual sera could block antigen binding of anti-AD-5 MAbs. The findings from blocking experiments using MAb 1G2 are shown in Fig. 4. The results indicated that the sera could be divided into two groups: sera that almost completely blocked 1G2 binding (strong; n = 5) and those that had intermediate to weak capacities to block 1G2 binding (weak; n = 11) (Fig. 4). The serum specimens that exhibited strong blocking activity were able to reduce 1G2 binding to the same extent as that with the 1G2 MAb itself (Fig. 4). Comparable results were obtained with MAbs SM10 and 2C2 and when the binding competition assay was carried out using recombinant AD-5 instead of gB (data not shown). The capacity of sera to block anti-AD-5 MAb binding correlated with the reactivity of individual sera with AD-5, suggesting that a major fraction of polyclonal anti-AD-5 antibodies are directed against the identified antigenic structure YNND in AD-5 (data not shown).
FIG 4.
Competition for gB binding between 1G2 and individual human sera. ELISA plates were coated with 25 ng recombinant gB, followed by incubation with serial log2 dilutions of serum. After removal of residual serum components, wells were incubated with biotinylated 1G2 MAb (1G2 MAb bio) at a constant concentration of 0.1 μg/ml, which in turn was detected by HRP-conjugated streptavidin. HCMV-convalescent-phase (+) human sera were divided into strong and weak competitors, and OD450 values for an 8-fold serum dilution are given. HCMV-negative (−) sera served as a control. The thick dashed line represents the mean OD450 for 1G2 MAb bio without competitor, and thin dashed lines indicate 2 standard deviations. The solid line represents a block of 1G2 MAb bio binding by unbiotinylated 1G2 MAb at a concentration of 1.25 μg/ml.
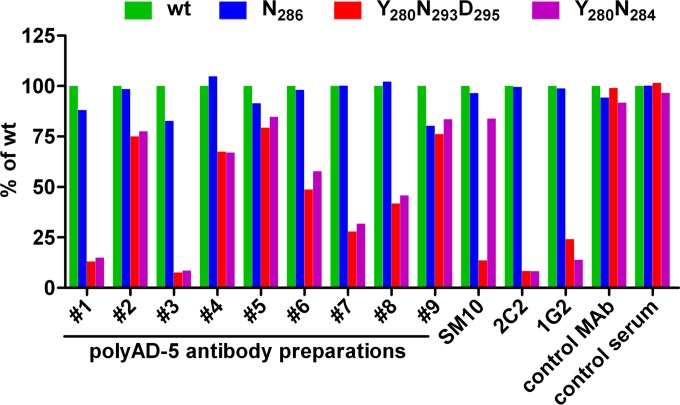
To further examine the recognition of the YNND epitope in HCMV-infected individuals, we purified polyclonal anti-AD-5 antibodies from nine individual sera by using AD-5 affinity chromatography. Sera from both the strongly and weakly competing groups were selected for purification. The specificity of the affinity purification was assessed by both ELISA and indirect immunofluorescence, and no residual reactivity with gH/gL, gM/gN, or any antigenic domain of gB, apart from AD-5, was detected in the affinity-purified antibodies (data not shown). The polyAD-5 preparations were then analyzed by ELISA for binding capacity for wt AD-5 and, for comparison, selected AD-5 mutants (Fig. 5). The polyAD-5 preparations showed different degrees of AD-5 mutant recognition, from minimal reduction to a complete loss of binding for the Y280N293D295 and Y280N284 mutants, whereas recognition of the N286 mutant remained comparable to that of the wt. Two individual specimens (preparations 1 and 3) displayed no residual reactivity to the Y280N293D295 and Y280N284 AD-5 mutants, suggesting that in the case of these sera, the YNND antigenic structure was the exclusive binding site within AD-5.
FIG 5.
Recognition of selected AD-5 mutants by polyclonal anti-AD-5 antibody preparations. Polyclonal anti-AD-5 (polyAD-5) antibodies were affinity purified from sera derived from HCMV-seropositive individuals and tested for recognition of AD-5 wt and mutant proteins. AD-5 mutant proteins were captured from cell culture supernatants of transfected HEK 293T cells by use of an anti-HA antibody and then tested for binding of polyAD-5 antibody preparations from individual sera (sera 1 to 9). The anti-AD-5 antibodies SM10, 2C2, and 1G2 were used as controls, and the anti-AD-4 antibody SM5-1 (control MAb) and a polyclonal anti-AD-4 antibody preparation (control serum) were used to confirm the specificity of the assay. OD450 values for wt AD-5 recognition were set to 100% for each antibody preparation, and the percentage of AD-5 mutant protein recognition was calculated. All experiments were performed as titrations to exclude antibody saturation, and data from the linear range of the assay are displayed.
Neutralizing activity of AD-5-specific human polyclonal antibodies.
Although our results indicated that AD-5-specific antibodies prepared from human serum recognized the YNND epitope that was the target of virus-neutralizing MAbs, an important remaining question was whether the polyclonal AD-5-specific antibodies were comparable to the MAbs in terms of their biological activity. Four polyAD-5 preparations could be tested in neutralization assays with HCMV TB40/E wt virus and viral mutants, as they contained sufficient concentrations of IgG. As shown in Fig. 6, these polyAD-5 preparations were able to effectively neutralize wt virus, with IC50 values in the range of 0.2 to 1.5 μg/ml. Interestingly, two of the four polyAD-5 preparations retained neutralizing activity toward the Y280 virus mutant and its combination with the N293D295 or N284 mutation (polyAD-5 preparations 6 and 7), while two other polyAD-5 preparations (preparations 1 and 3) exhibited no neutralizing activity against the viral mutants (Fig. 6). These results were in agreement with the binding data shown above and suggest that as yet undefined epitopes within AD-5 are targets of virus-neutralizing antibodies. However, it is important that in a significant proportion of human sera, the YNND epitope appears to be the exclusive binding site for virus-neutralizing antibodies within AD-5.
FIG 6.
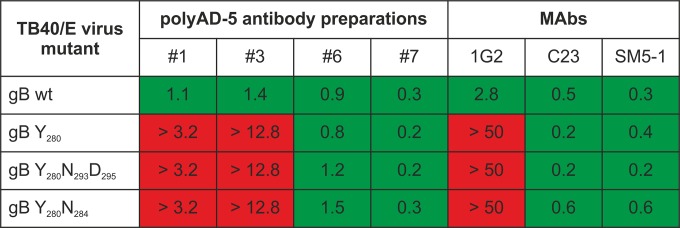
Neutralization of TB40/E wt and mutant viruses by polyAD-5 antibody preparations. TB40/E wt or recombinant viruses were incubated with antibody for 1 h at 37°C, and HFF monolayers were infected with the antibody-virus mixtures for 4 h. The extent of infection was analyzed at 48 h postinfection and calculated relative to that of the virus-only control. IC50s are given in μg/ml. Green, IC50 at the given concentration; red, IC50 was not reached at the indicated concentrations.
DISCUSSION
HCMV gB remains an important target for active and passive immune intervention strategies. However, the characteristics of a protective immune response toward gB are not well defined, and although a detailed understanding of the immune response in subjects with natural HCMV infection would appear to represent the most rational basis for the definition of parameters of a protective response, only limited information is available. Despite the fact that recombinant gB is a major component of many early-stage HCMV vaccines (32, 33), our findings clearly demonstrate that the understanding of antibody responses directed at gB during infection remains very incomplete. Developing a detailed understanding of protective B cell epitope structures on HCMV gB may allow for the generation of an antigenic map and thus provide an important guide for rational design and testing of future immune intervention strategies.
Five antigenic domains have been described on HCMV gB, among which AD-1, -2, -4, and -5 are targeted by neutralizing antibodies (16). In the present study, we analyzed the epitope(s) of three neutralizing human MAbs that target AD-5. Importantly, the antibodies were isolated from different, healthy, HCMV-infected donors and were found to recognize an overlapping, discontinuous epitope within AD-5, a domain within HCMV gB that is thought to be the fusion domain of herpesviral gB.
Critical residues and structural considerations for the AD-5 antigenic structure.
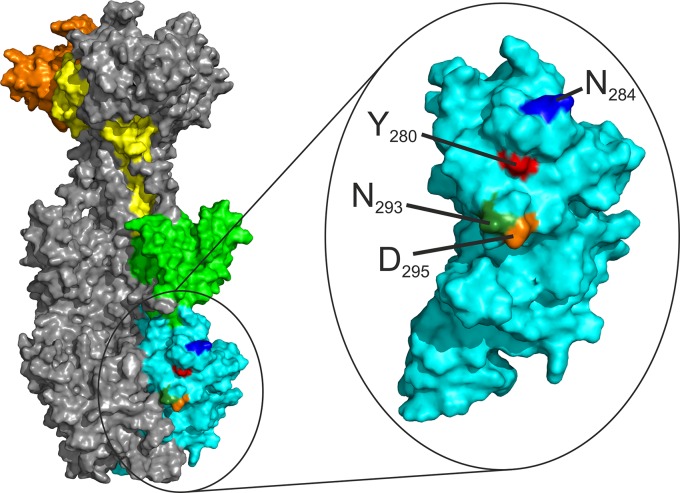
Using mutational studies based on a 3D model of HCMV gB, we identified a major antigenic structure (the YNND epitope) within AD-5 that we believe consists of residue Y280 in combination with additional surface-exposed residues in close proximity (Fig. 7). Mutation of these critical residues within the YNND epitope not only led to the reduction or loss of binding to recombinant mutant AD-5 proteins but also rendered recombinant viruses expressing these mutants resistant to neutralization by the respective antibody.
FIG 7.
The YNND epitope. The structural model shows an accessible surface area representation of the HCMV gB trimer, with one monomer highlighted as described in the legend to Fig. 1. AD-5, which corresponds to structural domain I, is displayed in an enlarged format. The critical residues for binding of human neutralizing antibodies are highlighted in different colors, and their positions in gB are given.
The current understanding of epitope-paratope interactions suggests that additional residues within AD-5 will define the complete epitope for MAb binding, but their contribution to the binding energy is likely too small to be detected in our relatively insensitive assays. It is commonly believed that the vast majority of epitopes are discontinuous, consisting of critical amino acids that are separated by stretches of amino acids of various lengths in the primary sequence but are brought together by the folding of the protein. The surface areas of epitopes can vary from <200 to >1,500 Å2, and epitopes consist of fewer than 2 to more than 34 residues (47). In the case of the AD-5-specific MAbs investigated in the current study, the space occupied by the defined critical residues covers around 560 Å2. Nevertheless, single residues have been identified as “hot spots” and as critical for antibody binding and function, since they contribute the majority of binding energy in the antibody-antigen interaction (48). Tyrosine residues have frequently been found in interaction “hot spots” (48, 49). By analogy, similar neutralizing antibody escape mutants carrying a single amino acid exchange to render a virus resistant to antibody-mediated neutralization have been described for other viral systems, such as HIV or influenza virus (50–52). Note that residues of the YNND epitope are strictly conserved between different HCMV strains (see Fig. S5 in the supplemental material), indicating that naturally occurring escape mutants may be rare or absent.
It may be argued that mutations that were introduced in AD-5 resulted in disruption of the AD-5 conformation. Several lines of evidence, however, suggest that the overall antigenic structure of AD-5, which contains the fusion loops of gB, remained intact in AD-5 mutants. (i) Replication-competent recombinant mutant viruses with little to no growth retardation were readily recovered following mutagenesis of the parent wt clone, suggesting that the overall function of gB was not impaired by the mutation (see Fig. S3 in the supplemental material). (ii) The MAbs recognized gB with comparable affinities (KD [equilibrium dissociation constant] range of ∼2 × 10−10) and were dependent on an intact conformation, since they were not reactive in Western blots. Thus, the differential reactivity of the MAbs for the AD-5 mutant proteins suggested that the loss of critical binding partners that followed replacement by alanine residues was the basis for this result, rather than disruption of AD-5 structural integrity. In addition, the partial rescue of loss of neutralization of the Y280 mutant virus by introduction of a second mutation (N284) further argued against structural alterations in AD-5 by the mutations (see Fig. S4). (iii) The residues constituting the YNND epitope are solvent exposed, and their side chains form only minor stabilizing interactions within AD-5. Therefore, a replacement of the respective residues by alanine was expected to have no effect on the overall structure of AD-5. (iv) The overall structural integrity of AD-5 mutant proteins was also supported by the finding that individual polyclonal anti-AD-5 antibody preparations (polyAD-5 preparations 6 and 7) completely retained their neutralizing activity against viral mutants, even though other anti-AD-5 antibody preparations exhibited a loss of neutralizing activity for the viruses expressing these mutations. Under the assumption that the vast majority of antibodies generated following infection recognized structural epitopes, these findings suggested that the overall structure of AD-5 was intact in viruses with mutations in AD-5.
Finally, epitopes defined by murine antibodies reactive with other herpesvirus epitopes have also been mapped to an AD-5-corresponding region of gB, i.e., Dom I in HSV-1 gB (see Fig. S6 in the supplemental material). Importantly, although in these cases the MAbs were derived from mice, they all had virus-neutralizing activity (53–56), indicating that this gB structural region is of general importance for Dom I-directed neutralization not only in HCMV but also in other herpesviruses.
Taken together, these data strongly argue that we have mapped a functional epitope of protective antibodies against AD-5. Ultimately, only structural analysis can verify the exact nature of the antibody-antigen interactions.
Potential mechanism of virus-neutralizing activity of anti-AD-5 antibodies.
The fact that anti-AD-5 antibodies block infection of different cell types with comparable potencies indicates that a conserved function of gB is targeted (16, 27). In light of recent attempts to understand the neutralization mechanism of other Dom I-specific antibodies (41), what could be the possible mode of action of the anti-AD-5 antibodies described in this report? Because gB is the fusion protein of HCMV and, together with gH/gL, constitutes the core fusion machinery, mutations within hydrophobic residues within the fusion loops of AD-5 could alter the fusogenic activity of gB in cell-cell fusion assays (40). To our knowledge, none of the Dom I-specific monoclonal antibodies described for HCMV or other herpesviruses so far target the hydrophobic fusion loops directly, including the antibodies described in the current study. Therefore, direct inhibition of the fusogenic residues within AD-5 seems unlikely. Nevertheless, several other mechanisms of neutralization could be envisaged, as follows.
(i) Inhibition of receptor binding.
Previous characterization of the anti-AD-5 MAbs (27) showed that they have postattachment neutralization activity, i.e., they do not prevent virus attachment to the cell surface. This is analogous to observations for other gB-specific antibodies (53). We therefore suspect that neutralization by altered virus attachment, possibly through blockade of receptor binding, is unlikely to be the mechanism of neutralization by AD-5-specific antibodies. This mode of action, however, cannot be excluded entirely, as multiple interactions of gB or other virion glycoproteins with different molecules of the host cell membrane have been described (57–59), and attachment may still occur even though gB-receptor interaction is blocked.
(ii) Restriction of conformational changes.
The crystal structures solved for gB of HSV-1 and Epstein-Barr virus (EBV) most likely represent the postfusion conformation (60, 61). Analogous to other type III viral fusion proteins, such as vesicular stomatitis virus (VSV) G or baculovirus gp64 (62), herpesviral gBs are hypothesized to undergo refolding during fusion. Experimental evidence for large-scale refolding of gB, however, has not yet been obtained (63, 64). The epitopes for the neutralizing MAbs described in this study can be mapped to the surface of the gB trimer of the postfusion model. Neutralizing antibodies, however, would be expected to recognize a prefusion conformation of gB that presumably would be present on the infectious virion. Importantly, the critical residues for antibody binding identified here are similarly accessible in a hypothetical prefusion gB model based on the crystal structure of the VSV G prefusion conformation (see Fig. S7 in the supplemental material). It can be imagined that the transition from the prefusion to the postfusion conformation requires space, which may be limited on the virion surface and which may be obstructed by bound antibody. In fact, for VSV G, for which post- and prefusion structures are available, a space-consuming conformational change seems plausible (62). Thus, a block of conformational changes in gB from a fusion-inactive to a fusion-active state by anti-AD-5 MAbs would be a mechanism of virus neutralization consistent with findings from the current study.
(iii) Block of gB-gH/gL interaction.
Another possible mechanism of virus neutralization is a block in the interaction between gB and the gH/gL complexes. In order for gB to become fusion active, it must interact with gH/gL in a specific manner to form the minimal fusion machinery. A number of studies have addressed the formation of the gB-gH/gL fusion complex. Using antibody blocking experiments, HSV-1 gB structural domains I, II, and V, but not IV, have been described as interaction partners for gH/gL (65). In contrast to these results, HCMV gB anti-AD-2 antibodies failed to block gB-gH/gL interaction (66). Using functional complementation assays for EBV gB, regions of HCMV gB corresponding to Dom III/IV/V have been postulated as interaction partners for gH/gL (67). It should be noted that interaction between gB and gH/gL on the virion surface has thus far not been demonstrated definitively. Instead, cell-cell fusion assays with individually expressed glycoproteins have been used. However, it should be noted that these assays do not take into account that additional herpesviral proteins might be involved in the modulation and regulation of the fusion process. In addition, the expression of gB as an individual protein might result in a conformation that does not precisely reflect the gB conformation that is found on the surfaces of infectious virus particles. Apparently, when the gB ectodomain is expressed as an individual molecule, it readily adopts a postfusion conformation (40), which appears to have a higher internal stability than that of a potential prefusion conformation. Thus, the exact parameters of gB interactions with gH/gL are currently undefined, and therefore the possibility of inhibition of the gB-gH/gL interaction as a mechanism of virus neutralization cannot be excluded for the activity of AD-5-specific antibodies.
(iv) Long-range conformational effects.
Lastly, long-range conformational effects that might modulate the fusogenic activity of the fusion loops may be a mode of action, as hypothesized for gB Dom I-specific neutralizing antibodies toward HSV (41).
Importance of the antigenic structure of AD-5 in infection.
The antibodies investigated in this study were obtained from three different, healthy donors. Our initial finding that these antibodies compete for gB binding strongly suggested that a shared target structure was present on gB. Alternatively, this finding may have been a coincidental result from an unrecognized bias in the B cell clone selection procedure. Thus, the question of the general relevance of this antigenic site during HCMV infection was of major interest. To address this, we first tested the ability of individual sera to compete with anti-AD-5 MAbs for binding to gB. The observation that individual sera potently blocked AD-5 MAb binding to gB or recombinant AD-5 indicated that a major fraction of anti-AD-5 antibodies that develop during infection are directed against this antigenic site. Furthermore, and supporting the binding competition results, polyclonal anti-AD-5 antibody preparations exhibited distinct patterns of AD-5 mutant protein recognition. While some polyAD-5 preparations failed to react with AD-5 after mutation of critical antigenic site residues, others showed only a slight decrease in recognition of mutant forms of AD-5, a finding that suggested the existence of additional antibody binding sites in AD-5. A multiantibody coverage of AD-5 by the antiviral antibody response seems plausible considering the size of this region. For HSV-1 gB, a neutralizing murine MAb was identified that targets a different region within structural domain I (SS55; see Fig. S6 in the supplemental material) (41).
Next, the question of the functional relevance of AD-5-specific antibodies in general and YNND epitope-specific antibodies in particular was addressed in terms of neutralizing activity toward the wt and viral mutants. In neutralization assays, the polyAD-5 preparations from individual sera showed potent neutralizing activity toward wt TB40/E virus, some even with activities comparable to those of monoclonal antibodies. When tested with the mutant viruses, however, we observed significant differences in neutralizing activity for individual polyAD-5 preparations. While some polyAD-5 preparations exhibited little difference in neutralizing activity for wt and mutant viruses, the neutralizing activities of other polyAD-5 preparations were significantly different between mutant and wt viruses. This result argued that additional virus-neutralizing antibody binding sites were present on AD-5. However, for the immune response of some HCMV-infected individuals, the identified antigenic site in AD-5 seems to be the exclusive target for virus-neutralizing antibodies directed against AD-5. Further studies are needed to define additional epitopes in AD-5 and their functional relevance for neutralization.
In summary, we have identified and characterized an antigenic site within AD-5 of HCMV gB that is targeted by neutralizing antibodies. A critical tyrosine residue at position 280 was defined in combination with other surface-adjacent amino acids (the YNND epitope). In general, this epitope appears to be recognized by a significant fraction of the anti-AD-5 antibodies that are developed following infection. In addition, it represents an important target for neutralizing activity in individual sera. Thus, induction of antibodies specific for this antigenic site should be a desirable goal following vaccination, and antibodies targeting the YNND epitope may be considered for passive immunotherapy.
Supplementary Material
ACKNOWLEDGMENTS
We thank Barbara Adler, Germany, for the kind gift of vBAC4-luc. We thank Sanofi-Pasteur, France, for the gift of recombinant gB and 4-Antibody AG, Germany, for the SM10, 2C2, and 1G2 antibody Fab fragments. Antibody C23 (TI23) was a kind gift of Teijin Pharma Limited, Japan.
This work was supported by grants from the DFG (grants MA929/11-1 and GRK1071) and the NIH (grant 5R01AI089956-03).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.02393-14.
REFERENCES
- 1.Kenneson A, Cannon MJ. 2007. Review and meta-analysis of the epidemiology of congenital cytomegalovirus (CMV) infection. Rev Med Virol 17:253–276. doi: 10.1002/rmv.535. [DOI] [PubMed] [Google Scholar]
- 2.Boppana SB, Pass RF, Britt WJ, Stagno S, Alford CA. 1992. Symptomatic congenital cytomegalovirus infection: neonatal morbidity and mortality. Pediatr Infect Dis J 11:93–99. doi: 10.1097/00006454-199202000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Dahle AJ, Fowler KB, Wright JD, Boppana SB, Britt WJ, Pass RF. 2000. Longitudinal investigation of hearing disorders in children with congenital cytomegalovirus. J Am Acad Audiol 11:283–290. [PubMed] [Google Scholar]
- 4.Stratton KR, Durch JS, Lawrence RS. 2000. Vaccines for the 21st century: a tool for decisionmaking. National Academy Press, Washington, DC. [PubMed] [Google Scholar]
- 5.Fu TM, An Z, Wang D. 2014. Progress on pursuit of human cytomegalovirus vaccines for prevention of congenital infection and disease. Vaccine 32:2525–2533. doi: 10.1016/j.vaccine.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 6.Ross SA, Arora N, Novak Z, Fowler KB, Britt WJ, Boppana SB. 2010. Cytomegalovirus reinfections in healthy seroimmune women. J Infect Dis 201:386–389. doi: 10.1086/649903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reeves M, Sinclair J. 2008. Aspects of human cytomegalovirus latency and reactivation. Curr Top Microbiol Immunol 325:297–313. [DOI] [PubMed] [Google Scholar]
- 8.Fowler KB, Stagno S, Pass RF. 2003. Maternal immunity and prevention of congenital cytomegalovirus infection. JAMA 289:1008–1011. doi: 10.1001/jama.289.8.1008. [DOI] [PubMed] [Google Scholar]
- 9.Adler SP, Starr SE, Plotkin SA, Hempfling SH, Buis J, Manning ML, Best AM. 1995. Immunity induced by primary human cytomegalovirus infection protects against secondary infection among women of childbearing age. J Infect Dis 171:26–32. doi: 10.1093/infdis/171.1.26. [DOI] [PubMed] [Google Scholar]
- 10.Boppana SB, Britt WJ. 1995. Antiviral antibody responses and intrauterine transmission after primary maternal cytomegalovirus infection. J Infect Dis 171:1115–1121. doi: 10.1093/infdis/171.5.1115. [DOI] [PubMed] [Google Scholar]
- 11.Bia FJ, Griffith BP, Tarsio M, Hsiung GD. 1980. Vaccination for the prevention of maternal and fetal infection with guinea pig cytomegalovirus. J Infect Dis 142:732–738. doi: 10.1093/infdis/142.5.732. [DOI] [PubMed] [Google Scholar]
- 12.Bia FJ, Miller SA, Lucia HL, Griffith BP, Tarsio M, Hsiung GD. 1984. Vaccination against transplacental cytomegalovirus transmission: vaccine reactivation and efficacy in guinea pigs. J Infect Dis 149:355–362. doi: 10.1093/infdis/149.3.355. [DOI] [PubMed] [Google Scholar]
- 13.Klenovsek K, Weisel F, Schneider A, Appelt U, Jonjic S, Messerle M, Bradel-Tretheway B, Winkler TH, Mach M. 2007. Protection from CMV infection in immunodeficient hosts by adoptive transfer of memory B cells. Blood 110:3472–3479. doi: 10.1182/blood-2007-06-095414. [DOI] [PubMed] [Google Scholar]
- 14.Dolan A, Cunningham C, Hector RD, Hassan-Walker AF, Lee L, Addison C, Dargan DJ, McGeoch DJ, Gatherer D, Emery VC, Griffiths PD, Sinzger C, McSharry BP, Wilkinson GW, Davison AJ. 2004. Genetic content of wild-type human cytomegalovirus. J Gen Virol 85:1301–1312. doi: 10.1099/vir.0.79888-0. [DOI] [PubMed] [Google Scholar]
- 15.Van Damme E, Van Loock M. 2014. Functional annotation of human cytomegalovirus gene products: an update. Front Microbiol 5:218. doi: 10.3389/fmicb.2014.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mach M, Wiegers A, Spindler N, Winkler T. 2013. Protective humoral immunity, p 215–231. In Reddehase M. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 17.Ryckman BJ, Jarvis MA, Drummond DD, Nelson JA, Johnson DC. 2006. Human cytomegalovirus entry into epithelial and endothelial cells depends on genes UL128 to UL150 and occurs by endocytosis and low-pH fusion. J Virol 80:710–722. doi: 10.1128/JVI.80.2.710-722.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, Wagner M, Gallina A, Milanesi G, Koszinowski U, Baldanti F, Gerna G. 2004. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol 78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Shenk T. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A 102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vanarsdall AL, Ryckman BJ, Chase MC, Johnson DC. 2008. Human cytomegalovirus glycoproteins gB and gH/gL mediate epithelial cell-cell fusion when expressed either in cis or in trans. J Virol 82:11837–11850. doi: 10.1128/JVI.01623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopper M, Compton T. 2004. Coiled-coil domains in glycoproteins B and H are involved in human cytomegalovirus membrane fusion. J Virol 78:8333–8341. doi: 10.1128/JVI.78.15.8333-8341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoppel K, Kropff B, Schmidt C, Vornhagen R, Mach M. 1997. The humoral immune response against human cytomegalovirus is characterized by a delayed synthesis of glycoprotein-specific antibodies. J Infect Dis 175:533–544. doi: 10.1093/infdis/175.3.533. [DOI] [PubMed] [Google Scholar]
- 23.Marshall GS, Rabalais GP, Stout GG, Waldeyer SL. 1992. Antibodies to recombinant-derived glycoprotein B after natural human cytomegalovirus infection correlate with neutralizing activity. J Infect Dis 165:381–384. doi: 10.1093/infdis/165.2.381. [DOI] [PubMed] [Google Scholar]
- 24.Britt WJ, Vugler L, Butfiloski EJ, Stephens EB. 1990. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol 64:1079–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerna G, Sarasini A, Patrone M, Percivalle E, Fiorina L, Campanini G, Gallina A, Baldanti F, Revello MG. 2008. Human cytomegalovirus serum neutralizing antibodies block virus infection of endothelial/epithelial cells, but not fibroblasts, early during primary infection. J Gen Virol 89:853–865. doi: 10.1099/vir.0.83523-0. [DOI] [PubMed] [Google Scholar]
- 26.Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, Gerna G, Sallusto F, Lanzavecchia A. 2010. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol 84:1005–1013. doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potzsch S, Spindler N, Wiegers AK, Fisch T, Rucker P, Sticht H, Grieb N, Baroti T, Weisel F, Stamminger T, Martin-Parras L, Mach M, Winkler TH. 2011. B cell repertoire analysis identifies new antigenic domains on glycoprotein B of human cytomegalovirus which are target of neutralizing antibodies. PLoS Pathog 7:e1002172. doi: 10.1371/journal.ppat.1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zydek M, Petitt M, Fang-Hoover J, Adler B, Kauvar LM, Pereira L, Tabata T. 2014. HCMV infection of human trophoblast progenitor cells of the placenta is neutralized by a human monoclonal antibody to glycoprotein B and not by antibodies to the pentamer complex. Viruses 6:1346–1364. doi: 10.3390/v6031346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chatterjee A, Harrison CJ, Britt WJ, Bewtra C. 2001. Modification of maternal and congenital cytomegalovirus infection by anti-glycoprotein b antibody transfer in guinea pigs. J Infect Dis 183:1547–1553. doi: 10.1086/320714. [DOI] [PubMed] [Google Scholar]
- 30.Bratcher DF, Bourne N, Bravo FJ, Schleiss MR, Slaoui M, Myers MG, Bernstein DI. 1995. Effect of passive antibody on congenital cytomegalovirus infection in guinea pigs. J Infect Dis 172:944–950. doi: 10.1093/infdis/172.4.944. [DOI] [PubMed] [Google Scholar]
- 31.Cekinovic D, Golemac M, Pugel EP, Tomac J, Cicin-Sain L, Slavuljica I, Bradford R, Misch S, Winkler TH, Mach M, Britt WJ, Jonjic S. 2008. Passive immunization reduces murine cytomegalovirus-induced brain pathology in newborn mice. J Virol 82:12172–12180. doi: 10.1128/JVI.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rieder F, Steininger C. 2014. Cytomegalovirus vaccine: phase II clinical trial results. Clin Microbiol Infect 20(Suppl 5):S95–S102. doi: 10.1111/1469-0691.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plotkin SA, Plachter B. 2013. Cytomegalovirus vaccine: on the way to the future?, p 424–449. In Reddehase M. (ed), Cytomegaloviruses: from molecular pathogenesis to intervention. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 34.Pass RF, Zhang C, Evans A, Simpson T, Andrews W, Huang ML, Corey L, Hill J, Davis E, Flanigan C, Cloud G. 2009. Vaccine prevention of maternal cytomegalovirus infection. N Engl J Med 360:1191–1199. doi: 10.1056/NEJMoa0804749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffiths PD, Stanton A, McCarrell E, Smith C, Osman M, Harber M, Davenport A, Jones G, Wheeler DC, O'Beirne J, Thorburn D, Patch D, Atkinson CE, Pichon S, Sweny P, Lanzman M, Woodford E, Rothwell E, Old N, Kinyanjui R, Haque T, Atabani S, Luck S, Prideaux S, Milne RS, Emery VC, Burroughs AK. 2011. Cytomegalovirus glycoprotein-B vaccine with MF59 adjuvant in transplant recipients: a phase 2 randomised placebo-controlled trial. Lancet 377:1256–1263. doi: 10.1016/S0140-6736(11)60136-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yue Y, Wang Z, Abel K, Li J, Strelow L, Mandarino A, Eberhardt MK, Schmidt KA, Diamond DJ, Barry PA. 2008. Evaluation of recombinant modified vaccinia Ankara virus-based rhesus cytomegalovirus vaccines in rhesus macaques. Med Microbiol Immunol 197:117–123. doi: 10.1007/s00430-008-0074-5. [DOI] [PubMed] [Google Scholar]
- 37.Abel K, Martinez J, Yue Y, Lacey SF, Wang Z, Strelow L, Dasgupta A, Li Z, Schmidt KA, Oxford KL, Assaf B, Longmate JA, Diamond DJ, Barry PA. 2011. Vaccine-induced control of viral shedding following rhesus cytomegalovirus challenge in rhesus macaques. J Virol 85:2878–2890. doi: 10.1128/JVI.00883-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schleiss MR. 2013. Developing a vaccine against congenital cytomegalovirus (CMV) infection: what have we learned from animal models? Where should we go next? Future Virol 8:1161–1182. doi: 10.2217/fvl.13.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spindler N, Rucker P, Potzsch S, Diestel U, Sticht H, Martin-Parras L, Winkler TH, Mach M. 2013. Characterization of a discontinuous neutralizing epitope on glycoprotein B of human cytomegalovirus. J Virol 87:8927–8939. doi: 10.1128/JVI.00434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma S, Wisner TW, Johnson DC, Heldwein EE. 2013. HCMV gB shares structural and functional properties with gB proteins from other herpesviruses. Virology 435:239–249. doi: 10.1016/j.virol.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cairns TM, Fontana J, Huang ZY, Whitbeck JC, Atanasiu D, Rao S, Shelly SS, Lou H, Ponce de Leon M, Steven AC, Eisenberg RJ, Cohen GH. 2014. Mechanism of neutralization of herpes simplex virus by antibodies directed at the fusion domain of glycoprotein B. J Virol 88:2677–2689. doi: 10.1128/JVI.03200-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scrivano L, Sinzger C, Nitschko H, Koszinowski UH, Adler B. 2011. HCMV spread and cell tropism are determined by distinct virus populations. PLoS Pathog 7:e1001256. doi: 10.1371/journal.ppat.1001256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreoni M, Faircloth M, Vugler L, Britt WJ. 1989. A rapid microneutralization assay for the measurement of neutralizing antibody reactive with human cytomegalovirus. J Virol Methods 23:157–167. doi: 10.1016/0166-0934(89)90129-8. [DOI] [PubMed] [Google Scholar]
- 44.Meyer H, Masuho Y, Mach M. 1990. The gp116 of the gp58/116 complex of human cytomegalovirus represents the amino-terminal part of the precursor molecule and contains a neutralizing epitope. J Gen Virol 71:2443–2450. doi: 10.1099/0022-1317-71-10-2443. [DOI] [PubMed] [Google Scholar]
- 45.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless red recombination system. Methods Mol Biol (Clifton, NJ) 634:421–430. doi: 10.1007/978-1-60761-652-8_30. [DOI] [PubMed] [Google Scholar]
- 46.Heldwein EE, Lou H, Bender FC, Cohen GH, Eisenberg RJ, Harrison SC. 2006. Crystal structure of glycoprotein B from herpes simplex virus 1. Science (New York, NY) 313:217–220. doi: 10.1126/science.1126548. [DOI] [PubMed] [Google Scholar]
- 47.Sharon J, Rynkiewicz MJ, Lu Z, Yang CY. 2014. Discovery of protective B-cell epitopes for development of antimicrobial vaccines and antibody therapeutics. Immunology 142:1–23. doi: 10.1111/imm.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramaraj T, Angel T, Dratz EA, Jesaitis AJ, Mumey B. 2012. Antigen-antibody interface properties: composition, residue interactions, and features of 53 non-redundant structures. Biochim Biophys Acta 1824:520–532. doi: 10.1016/j.bbapap.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogan AA, Thorn KS. 1998. Anatomy of hot spots in protein interfaces. J Mol Biol 280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 50.Kaverin NV, Rudneva IA, Govorkova EA, Timofeeva TA, Shilov AA, Kochergin-Nikitsky KS, Krylov PS, Webster RG. 2007. Epitope mapping of the hemagglutinin molecule of a highly pathogenic H5N1 influenza virus by using monoclonal antibodies. J Virol 81:12911–12917. doi: 10.1128/JVI.01522-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prabakaran M, He F, Meng T, Madhan S, Yunrui T, Jia Q, Kwang J. 2010. Neutralizing epitopes of influenza virus hemagglutinin: target for the development of a universal vaccine against H5N1 lineages. J Virol 84:11822–11830. doi: 10.1128/JVI.00891-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guo D, Shi X, Arledge KC, Song D, Jiang L, Fu L, Gong X, Zhang S, Wang X, Zhang L. 2012. A single residue within the V5 region of HIV-1 envelope facilitates viral escape from the broadly neutralizing monoclonal antibody VRC01. J Biol Chem 287:43170–43179. doi: 10.1074/jbc.M112.399402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Highlander SL, Cai WH, Person S, Levine M, Glorioso JC. 1988. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J Virol 62:1881–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kousoulas KG, Huo B, Pereira L. 1988. Antibody-resistant mutations in cross-reactive and type-specific epitopes of herpes simplex virus 1 glycoprotein B map in separate domains. Virology 166:423–431. doi: 10.1016/0042-6822(88)90513-2. [DOI] [PubMed] [Google Scholar]
- 55.Pereira L, Ali M, Kousoulas K, Huo B, Banks T. 1989. Domain structure of herpes simplex virus 1 glycoprotein B: neutralizing epitopes map in regions of continuous and discontinuous residues. Virology 172:11–24. doi: 10.1016/0042-6822(89)90102-5. [DOI] [PubMed] [Google Scholar]
- 56.Bender FC, Samanta M, Heldwein EE, de Leon MP, Bilman E, Lou H, Whitbeck JC, Eisenberg RJ, Cohen GH. 2007. Antigenic and mutational analyses of herpes simplex virus glycoprotein B reveal four functional regions. J Virol 81:3827–3841. doi: 10.1128/JVI.02710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Feire AL, Koss H, Compton T. 2004. Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc Natl Acad Sci U S A 101:15470–15475. doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soroceanu L, Akhavan A, Cobbs CS. 2008. Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature 455:391–395. doi: 10.1038/nature07209. [DOI] [PubMed] [Google Scholar]
- 59.Compton T, Nowlin DM, Cooper NR. 1993. Initiation of human cytomegalovirus infection requires initial interaction with cell surface heparan sulfate. Virology 193:834–841. doi: 10.1006/viro.1993.1192. [DOI] [PubMed] [Google Scholar]
- 60.Backovic M, Longnecker R, Jardetzky TS. 2009. Structure of a trimeric variant of the Epstein-Barr virus glycoprotein B. Proc Natl Acad Sci U S A 106:2880–2885. doi: 10.1073/pnas.0810530106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heldwein EE, Krummenacher C. 2008. Entry of herpesviruses into mammalian cells. Cell Mol Life Sci 65:1653–1668. doi: 10.1007/s00018-008-7570-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Backovic M, Jardetzky TS. 2011. Class III viral membrane fusion proteins. Adv Exp Med Biol 714:91–101. doi: 10.1007/978-94-007-0782-5_3. [DOI] [PubMed] [Google Scholar]
- 63.Vitu E, Sharma S, Stampfer SD, Heldwein EE. 2013. Extensive mutagenesis of the HSV-1 gB ectodomain reveals remarkable stability of its postfusion form. J Mol Biol 425:2056–2071. doi: 10.1016/j.jmb.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stampfer SD, Lou H, Cohen GH, Eisenberg RJ, Heldwein EE. 2010. Structural basis of local, pH-dependent conformational changes in glycoprotein B from herpes simplex virus type 1. J Virol 84:12924–12933. doi: 10.1128/JVI.01750-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Atanasiu D, Whitbeck JC, de Leon MP, Lou H, Hannah BP, Cohen GH, Eisenberg RJ. 2010. Bimolecular complementation defines functional regions of herpes simplex virus gB that are involved with gH/gL as a necessary step leading to cell fusion. J Virol 84:3825–3834. doi: 10.1128/JVI.02687-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fouts AE, Comps-Agrar L, Stengel KF, Ellerman D, Schoeffler AJ, Warming S, Eaton DL, Feierbach B. 2014. Mechanism for neutralizing activity by the anti-CMV gH/gL monoclonal antibody MSL-109. Proc Natl Acad Sci U S A 111:8209–8214. doi: 10.1073/pnas.1404653111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Plate AE, Reimer JJ, Jardetzky TS, Longnecker R. 2011. Mapping regions of Epstein-Barr virus (EBV) glycoprotein B (gB) important for fusion function with gH/gL. Virology 413:26–38. doi: 10.1016/j.virol.2010.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.