ABSTRACT
Giant viruses able to replicate in Acanthamoeba castellanii penetrate their host through phagocytosis. After capsid opening, a fusion between the internal membranes of the virion and the phagocytic vacuole triggers the transfer in the cytoplasm of the viral DNA together with the DNA repair enzymes and the transcription machinery present in the particles. In addition, the proteome analysis of purified mimivirus virions revealed the presence of many enzymes meant to resist oxidative stress and conserved in the Mimiviridae. Megavirus chilensis encodes a predicted copper, zinc superoxide dismutase (Cu,Zn-SOD), an enzyme known to detoxify reactive oxygen species released in the course of host defense reactions. While it was thought that the metal ions are required for the formation of the active-site lid and dimer stabilization, megavirus chilensis SOD forms a very stable metal-free dimer. We used electron paramagnetic resonance (EPR) analysis and activity measurements to show that the supplementation of the bacterial culture with copper and zinc during the recombinant expression of Mg277 is sufficient to restore a fully active holoenzyme. These results demonstrate that the viral enzyme's activation is independent of a chaperone both for disulfide bridge formation and for copper incorporation and suggest that its assembly may not be as regulated as that of its cellular counterparts. A SOD protein is encoded by a variety of DNA viruses but is absent from mimivirus. As in poxviruses, the enzyme might be dispensable when the virus infects Acanthamoeba cells but may allow megavirus chilensis to infect a broad range of eukaryotic hosts.
IMPORTANCE Mimiviridae are giant viruses encoding more than 1,000 proteins. The virion particles are loaded with proteins used by the virus to resist the vacuole's oxidative stress. The megavirus chilensis virion contains a predicted copper, zinc superoxide dismutase (Cu,Zn-SOD). The corresponding gene is present in some megavirus chilensis relatives but is absent from mimivirus. This first crystallographic structure of a viral Cu,Zn-SOD highlights the features that it has in common with and its differences from cellular SODs. It corresponds to a very stable dimer of the apo form of the enzyme. We demonstrate that upon supplementation of the growth medium with Cu and Zn, the recombinant protein is fully active, suggesting that the virus's SOD activation is independent of a copper chaperone for SOD generally used by eukaryotic SODs.
INTRODUCTION
The nucleocytoplasmic large DNA viruses (NCLDV) are a monophyletic group of viruses that infect eukaryotes (1, 2). Mimiviridae are among its largest members, with particle sizes that make them visible by light microscopy and genome complexities overlapping those of the cellular world. They encode proteins thought to be the trademarks of cells, such as elements of the translation machinery as well as original metabolic pathways (3–7). Megavirus chilensis was isolated from an environmental sample from the coast of central Chile by cocultivation with Acanthamoeba castellanii (8). Like mimivirus (9), megavirus chilensis is a giant virus from the Mimiviridae family, with a 1.259-Mb double-stranded DNA genome encoding 1,120 genes. The virion consists of an icosahedral capsid of 400 nm in diameter enclosed in a layer of fibrils. It shares ∼600 genes with the single species of the genus Mimivirus, with an average of 50% of residues being identical at the protein sequence level (8, 10). Mimivirus penetrates A. castellanii cells by phagocytosis. After the capsid opens (11), the virion's internal membrane fuses with the vacuole membrane, allowing the nucleocore containing the viral DNA and associated proteins to be unloaded into the host cytoplasm. The replication cycle takes place in a transitory organelle common to all cytoplasmic viruses, the virion factory (12–14). A proteomic study of purified mimivirus particles revealed the presence of a virally encoded transcription machinery, several DNA repair enzymes, and many proteins meant to resist oxidative stress (12, 15). The corresponding genes are well conserved among the Mimiviridae (16–19). Megavirus chilensis encodes a predicted superoxide dismutase (SOD) which is found in some members of the Mimiviridae but is absent from mimivirus (20). The enzyme is present in the purified virion particles.
SODs catalyze the conversion of superoxide radicals to hydrogen peroxide and dioxygen. There are four families of SODs, defined by their metal ion binding partners: Fe, Mn, Ni, or both Cu and Zn. The megavirus chilensis Mg277 gene product is homologous to the eukaryotic homodimeric copper, zinc-SOD enzymes (20). In Cu,Zn-SODs, an intramolecular disulfide bridge and the zinc atoms are essential to maintain the structural integrity of the dimeric enzyme (21), while the copper is a catalytic cofactor (22). Many other DNA viruses, including chloroviruses (23, 24), baculoviruses (25), poxviruses (26–31), and some Mimiviridae, encode a predicted Cu,Zn-SOD. In-depth studies of the poxvirus SOD protein demonstrated that it is packaged in the mature virions (30). In contrast, the enzyme was not detected in baculovirus virions (25). Surprisingly, the Amsacta moorei entomopoxvirus AMV255 (32) and the chlorovirus PBCV-1 (24) encode the only viral SODs reported to be active so far. In other poxviruses, the SOD proteins lack key residues involved in metal ion binding, required for catalytic activity. The vaccinia virus SOD-like protein (A45R) did not exhibit any enzymatic or inhibitory activity on the cellular SOD, and the disruption of the corresponding gene did not affect the virus's replication, virulence, or pathogenicity (30). Most of the enzymes lack both the zinc and the copper binding residues, except for the leporipoxvirus SOD proteins (SFV-s131R, MYX-m131R) that retained the zinc binding property (33). Thus, typical poxvirus SOD homologues might be nonfunctional. Interestingly, the restoration of the copper binding capability did not restore the activity of the Shope fibroma virus (SFV) SOD homologue (33). Many eukaryotic SODs need to be activated by a cellular copper chaperone for SOD (CCS). The C-terminal chaperone domain is homologous to the SOD and can bind to SOD monomers. It activates by inducing the critical disulfide bridge formation and inserting the copper ion into the enzyme active site. The Shope fibroma virus SOD homologue inhibits cellular SOD activity in vivo by sequestering the cellular CCS (31).
The aim of the present study was to assay and characterize the megavirus chilensis Mg277 protein's activity to validate its predicted SOD function. We determined this first viral SOD structure by X-ray crystallography and compared it to those of reference eukaryotic SODs. The metal-free enzyme forms a very stable dimer where the conserved disulfide bridge is properly formed. Combining electron paramagnetic resonance (EPR) and metal content analysis with enzymatic assay measurements, we demonstrated that the viral Cu,Zn-SOD was fully active when overexpressed in bacteria supplemented with copper and zinc.
These findings strongly suggest that the viral SOD does not need to be activated by a CCS; for the CCS-independent Cu,Zn-SOD, the viral enzyme depends only on the addition of copper and zinc ions for its activation.
MATERIALS AND METHODS
Protein production and crystallization.
The megavirus chilensis Mg277 (YP_004894328.1) recombinant protein was produced in Escherichia coli, purified, and crystallized as previously described (20). Different batches of protein were produced under different conditions to perform EPR and metal dosage studies in parallel with enzymatic activity measurements. The original Mg277 protein was expressed in E. coli Rosetta (DE3)-transformed cells grown in 2× yeast extract and tryptone (2YT) (Difco) without zinc or copper supplementation. This protein batch was concentrated to 19 g/liter in 20 mM Tris buffer (pH 8) and is referred to as Mg277b1. Another batch corresponded to the selenomethionine substitution protein produced in minimal medium as previously described (20), desalted in 20 mM Tris buffer (pH 8), and concentrated to 6.9 g/liter. This is the batch that provided the Mg277 structure and is referred to as Mg277b2. Finally, like other Cu,Zn-SOD proteins (34), the Mg277 protein was produced in E. coli Rosetta-transformed cells grown in 2YT medium, which was supplemented with copper (CuSO4, 1.2 mM) and zinc (ZnSO4, 0.4 mM) when the culture temperature was switched from 37°C to 17°C, 30 min prior to induction. Since the supplemented protein precipitated after it was desalted in the 20 mM Tris buffer (pH 8), we produced another batch of Mg277, which was desalted in 10 mM N-cyclohexyl-3-aminopropanesulfonic acid (CAPS) buffer at pH 10.5 after purification and concentrated to 6 g/liter using a Amicon Ultracel 10,000-molecular-weight-cutoff centrifugal filter device (Millipore). This batch is referred to as Mg277b3.
Structure determination.
The phase determination was performed with the Phenix AutoSol Wizard (35–39). Phases were calculated in the 35.3- to 2.2-Å-resolution range, and a single solution was found for the P43212 space group with 2 selenium atoms (for 4 methionines). The mean figure of merit for this solution was 0.323.
The model was refined using Phenix.Refine (39, 40) and included simulated annealing, determination of coordinates, and B-factor refinement. After a series of manual rebuilding followed by several rounds of refinement, we obtained a final Rwork of 18.6% and an Rfree of 22.6% (Table 1).
TABLE 1.
Refinement statistics
| Refinement parametera | Value(s)b |
|---|---|
| Resolution (Å) | 38.04–2.2 (2.279–2.2) |
| Wavelength (nm) | 0.9792 |
| No. of unique reflections | 15,732 (1,551) |
| Rwork | 0.1857 (0.2076) |
| Rfree | 0.2264 (0.2805) |
| No. of atoms | 2,040 |
| Protein | 1,933 |
| Water | 107 |
| Avg B factor (Å2) | 38.6 |
| Protein | 38.5 |
| Water | 40.5 |
| RMSD bond length (Å) | 0.009 |
| RMSD bond angle (°) | 1.14 |
RMSD, root mean square difference.
Statistics for the highest-resolution shell are shown in parentheses.
The structure consists of residues 6 to 158 and 4 to 158 for monomers A and B, respectively. Residues 74 to 85 and 131 to 145 of monomer A and 73 to 88 and 130 to 146 of monomer B were disordered in the crystal structure and are absent from the final model.
The quality of the model was validated using MolProbity (41). Most (96.2%) of the residues are in the most-favored regions of the Ramachandran plot, and 3.8% are in the additionally allowed regions. Refinement statistics are listed in Table 1.
Structure and sequence analysis of the predicted SOD of megavirus chilensis.
To compare megavirus chilensis Mg277 with the other viral SOD sequences and the active eukaryotic Cu,Zn-SODs, we used the Mg277 sequence and the BLASTP program (42, 43) to search against sequences in the NCBI nr database, excluding all organisms except viruses (44). We then performed a structural alignment using the 3DCoffee server (45) and included the Saccharomyces cerevisiae SOD sequence (Protein Data Bank [PDB] accession number 1SDY), human SOD sequence (GenBank accession number NP_000445.1), and viral SOD sequences (GenBank accession numbers YP_004894328.1 [megavirus chilensis], YP_007418160 [megavirus lba], AEX63072 [moumouvirus Monve], YP_007354188.1 [moumouvirus], AGF85544.1 [moumouvirus goulette], ADX05811.1 [Organic Lake phycodnavirus 1 {OLPV1}], YP_004061898.1 [micromonas virus], AGE58970.1 [Paramecium bursaria chlorella virus 1 {PBCV1}], AGE49091.1 [Acanthocystis turfacea chlorella virus 1 {ATCV1}], YP_874302.1 [Ectropis obliqua nuclear polyhedrosis virus {NPV}], AGX01251.1 [Bombyx NPV], YP_004376256.1 [Clostera granulovirus], YP_006908566.1 [Epinotia granulovirus], YP_008378623.1 [Choristoneura alphabaculovirus], YP_008004053.1 [Adoxophyes entomopoxvirus], NP_065037.1 [Amsacta entomopoxvirus], YP_227518.1 [deerpox virus], NP_659703.1 [sheeppox virus], NP_570289.1 [Shope fibroma virus {SFV} S131R, swinepox virus], AF349016_1 [cowpox virus], A45R [vaccinia virus], and NP_042201.1 [variola virus]). Both the Mg277 and Cu,Zn-SOD spinach structures (PDB accession number 1SRD) (46) were used as references for the structural alignment.
Metal content analysis.
Fluorescence scans were obtained with a multichannel fluorescence detector (MCA Rontec) implemented on the BM30 beamline at the European Synchrotron Radiation Facility (ESRF), exposing the crystal that provided the structure to an X-ray beam at a wavelength of 1.040 Å.
The metal contents of the megavirus chilensis SOD proteins obtained under various conditions were measured by using inductively coupled plasma mass spectrometry (ICP-MS) analysis (HP 4500 spectrometer, calibrated using an external standard).
EPR experiments.
The EPR spectra were recorded on a Bruker Elexsys E500 spectrometer fitted with an Oxford Instruments ESR 900 helium flow cryostat. Quantifications were made by using an external standard solution of freshly prepared 1 mM Cu-EDTA in 100 mM Tris, 10 mM EDTA at pH 8, which was transferred into a calibrated EPR tube.
Thermal-stability assays.
The thermal denaturation of the different protein batches was performed at 1 mg/ml in the buffers described above and mixed with Sypro orange (Life Technologies) solution at the concentration recommended by the manufacturer in a final volume of 25 μl.
The thermal stability was assayed in 96-well, thin-wall PCR plates (Bio-Rad) sealed with optical-quality sealing tape (Bio-Rad) and heated with an iCycler iQ real-time detection system (Bio-Rad) from 20 to 92°C in increments of 0.2°C (according to the work of Coutard et al. [47]). Thermal denaturation was monitored using Sypro orange as the external fluorescent probe, and fluorescence intensity was measured at 490/530-nm excitation/emission wavelengths, respectively. Protein denaturation was monitored by following the increase of the fluorescence emitted by the probe progressively binding to exposed hydrophobic regions. The melting temperature (Tm) was calculated as the midpoint of the phase transition from the native to the denatured protein using a Boltzmann model (Origin software).
Enzymatic activity measurements.
SOD activities of the various forms of the Mg277 protein were measured in an indirect assay using the SOD assay kit WST (Sigma catalog number 19160), made up of xanthine, xanthine oxidase, and WST [2-(4-iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt], which produces formazan dye upon reduction with a superoxide anion. Because the rate of WST reduction is inhibited by SOD activity, the 50% inhibitory concentration (IC50) of WST reduction was determined by a colorimetric method by measuring the absorbance changes at 560 nm, as previously described (48).
The SOD activity of the nonsupplemented Mg277b1 enzyme in 20 mM Tris (pH 8) was determined using serial dilutions of the enzyme and was reproduced three times in independent experiments. Another batch of nonsupplemented selenomethionylated Mg277 was produced and was desalted in 20 mM Tris (pH 8) (Mg277b2). Mg277b2 activity was measured with a serial dilution of the enzyme in the two buffers to confirm that the enzyme was equally active with the nonselenomethionylated Mg277 protein. The Cu/Zn-supplemented Mg277 enzyme activity in 10 mM CAPS (pH 10.5) (Mg277b3) was measured on serial dilutions of the enzyme, and results were reproduced three times in independent experiments. The resulting activities were compared to that of the commercially available bovine superoxide dismutase (Sigma-Aldrich catalog number S9697). The IC50s were calculated from three independent experiments for Mg277b1, Mg277b3, and the commercial bovine superoxide dismutase.
Protein structure accession number.
The megavirus chilensis structure has been deposited in the PDB under the accession number 4U4I.
RESULTS
Sequence and structural analysis of the viral SOD protein.
The Mg277 gene encodes a 158-amino-acid-long protein predicted to be a Cu,Zn-SOD. Homologues of this are found in several megavirus chilensis relatives (megaviruses and moumouviruses), with 70 to 99% of residues being identical at the protein sequence level, but are absent from other Mimiviridae, including mimivirus. Homologues of Mg277 are also present in some baculoviruses, chloroviruses, and poxviruses.
We chose the spinach SOD structure (PDB accession number 1SRD [45]) (47% of residues are identical over 135 amino acids) as a reference for structural comparison of Mg277 with its viral and eukaryotic homologues (Fig. 1 and 2).
FIG 1.
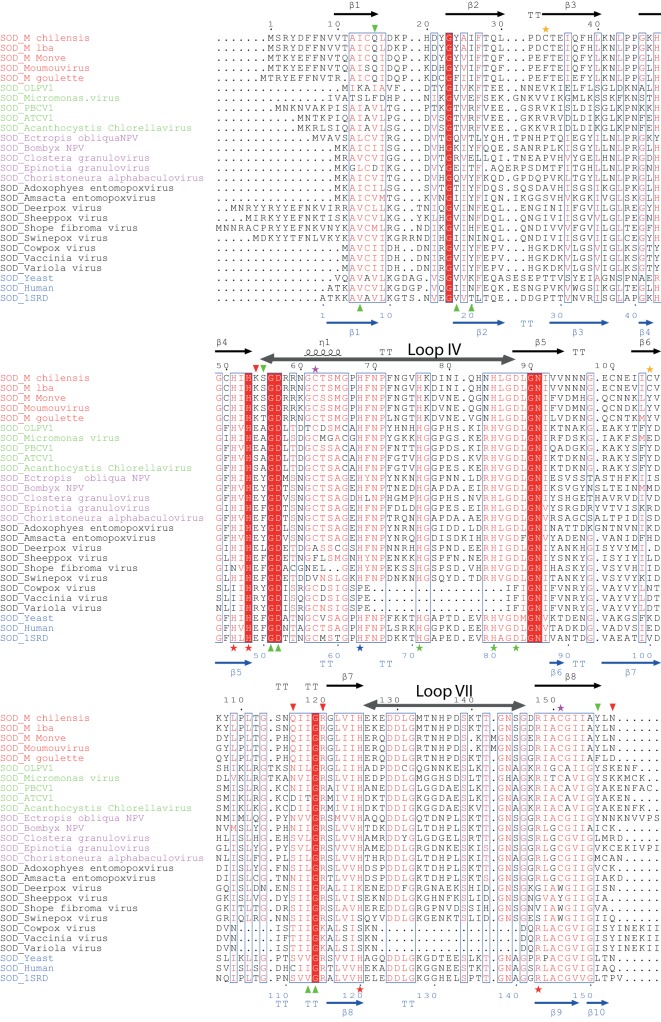
Structural alignment of the viral Mg277 structure with the 1SRD spinach Cu,Zn-SOD eukaryotic structure. A selection of Mimiviridae (red), poxvirus (black), baculovirus (pink), and chlorovirus (green) SOD sequences has been included, as has the eukaryotic (blue) human and Saccharomyces cerevisiae sequences. Copper binding residues are indicated by a red asterisk, Zn binding residues are indicated by a green asterisk, and residues coordinating the two metal ions are marked in blue. The green triangles correspond to the residues involved in dimer formation. Special residues involved in the dimer stabilization of Mg277 are marked by red triangles. The crucial disulfide bond conserved in all SOD sequences is marked by a cyan asterisk, and the disulfide bridge unique to megavirus is marked by an orange asterisk. The structural alignment was produced using the 3DCoffee server (45), and the figure was produced using the ESPript 3.0 server (58).
FIG 2.
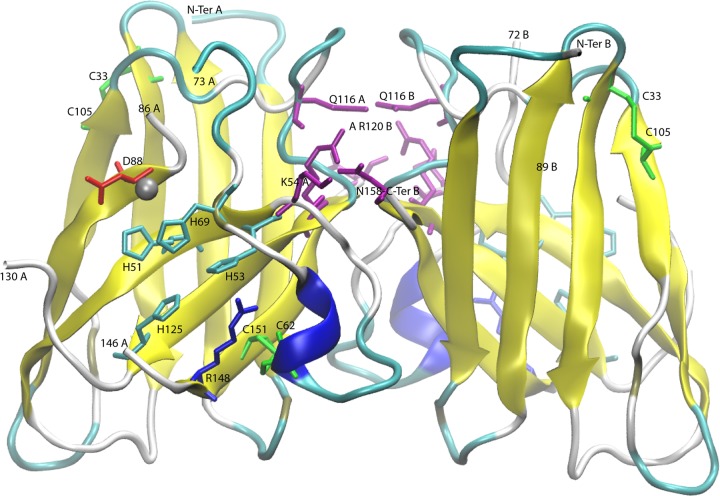
Ribbon representation of the Mg277 dimer. The residues involved in metal coordination and enzymatic activity are indicated on the figure (cyan-red) as well as the disulfide bridges (green). The partially occupied Zn site is marked as a gray sphere. Specific residues involved in dimer stabilization are indicated in purple. This includes the 2 salt bridges and the Q116 residues connecting the two monomers through H bonds at the exact location of the heptasulfane in the human structure. The rendering was made using VMD (59).
As with other Cu,Zn-SODs, Mg277's structure is dimeric and the crystal contains one dimer per asymmetric unit. Each Cu,Zn-SOD monomer folds as an eight-stranded, Greek key β-barrel with seven connecting loops, of which loops IV and VII, termed the “zinc” and “electrostatic” loops, respectively, are functionally important. The C-α root mean square deviation (C-αRMSD) is 0.54 Å between the two Mg277 monomers. Two salt bridges and 19 hydrogen bonds stabilize the dimer and result in a 1,851.6-Å2 buried surface area and a 926-Å2 interface area (Pisa server) (49). The two salt bridges involve the N158-terminal residue carboxylate of one monomer and the Nz-K54 and NH1-R120 residues of the second monomer, providing a very stable dimer. Interestingly, the dimer interface area is smaller in 1SRD (interface, ∼700 Å2; 8 H bonds) and involves different amino acids. The structural superimposition of Mg277 on the 1SRD dimer produced an overall C-αRMSD of 1.6 Å over 240 amino acids and a C-αRMSD of ∼1.4 Å for the monomers over 122 amino acids.
The structural alignment highlights some common features, as well as differences, between the viral SODs and the eukaryotic enzyme. First, the residues involved in metal coordination and enzymatic activity of all Cu,Zn-SODs are conserved in the mimiviridae, the baculoviruses, the chloroviruses, and the entomopoxviruses but are absent or mutated in the other poxvirus SOD sequences. In eukaryotic cells, the activation of the SOD enzyme involves several posttranslational modifications, including the formation of an intramolecular disulfide bond that contributes to its dimerization. The cysteine residues forming this disulfide bridge, critical for eukaryotic SOD activation (C57-C146 in 1SRD), are conserved in all the viral enzymes (Fig. 1), and the bridge is clearly visible in our Mg277 crystal structure (C62-C151). The two entomopoxvirus enzymes and the ones encoded by the other viruses, excluding the other poxviruses, might thus correspond to active Cu,Zn-SOD enzymes.
The second step of SOD activation corresponds to the insertion of the zinc and copper ions. In the metal-bound form of eukaryotic Cu,Zn-SOD, the zinc atom is known to stabilize the zinc binding loop (loop IV, residues 50 to 82 in 1SRD, 55 to 87 in Mg277) and the electrostatic loop (loop VII, residues 121 to 142 in 1SRD, 126 to 145 in Mg277). These loops form the active-site lid, limiting the solvent's access to the metal binding sites. They are also involved in the first step of Cu,Zn-SOD activation through the crucial intrachain disulfide bridge formation, which stabilizes the dimer. In the Mg277 structure, the histidine residues involved in zinc atom coordination (H76, H85, and D88 in Mg277) are disordered. However, the fluorescence spectrum acquired on the crystal that provided the three-dimensional (3D) structure showed peaks at wavelengths specific for Zn and Cu. We thus suspected that the residual density visible in monomer A, at the exact location of the zinc atom in the reference 1SRD structure, might correspond to a partially occupied zinc site. This was confirmed by the metal dosage on the Mg277 proteins, which returned one zinc and two copper atoms for 10 molecules of Mg277b2 (Table 2).
TABLE 2.
Metal content of the M. chilensis Mg277 protein determined by ICP-MS
| SOD, desalting buffer, pH | Protein concn | Growth culture | Concn (μM) of: |
[Cu]/[Mg277] (μM) | [Zn]/[Mg277] (μM) | EPRa | |
|---|---|---|---|---|---|---|---|
| Cu | Zn | ||||||
| Mg277b1, 20 mM Tris, pH 8 | 1.1 mM | 2YT | 20 | 60 | 1/65 | 1/19 | No signalCu2+ |
| Mg277b2, 20 mM Tris, pH 8 | 400 μM | M9-SeMet (20) | 80 | 40 | 1/5 | 1/10 | No signalCu2+ |
| Mg277b3, 10 mM CAPS, pH 10.5 | 350 μM | 2YT with 1.2 mM CuSO4, 0.4 mM ZnSO4 | 360 | 395 | 1/1 | 1/1 | Type II Cu2+ ion, 165 μM |
EPR, electron paramagnetic resonance.
On the other hand, the residues involved in copper coordination are conserved in Mg277 (H51, H53, H68, H125, and R148) (Fig. 1 and 2) and are superimposable on the equivalent residues in the 1SRD structure, but the copper ion is conspicuously absent from the Mg277 structure, and the type II Cu2+ signal was not identified by EPR on the selenomethionyl substitution protein (Fig. 3a). Our structure thus corresponds to a metal-free form of a Cu,Zn-SOD enzyme. In eukaryotic SODs, the enzyme activation can be performed through CCS-dependent (50, 51) or CCS-independent (Caenorhabditis elegans) (52) pathways. The fact that we obtained the apo form of the enzyme with the proper disulfide bridge in the Mg277 structure suggests that the viral enzyme, as with the C. elegans SOD and to a lesser extent the human enzyme, may not need a specific chaperone for its activation.
FIG 3.
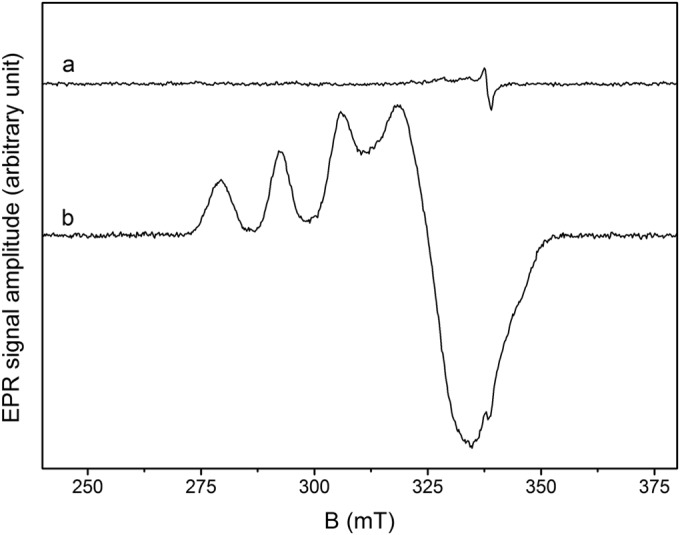
EPR spectra of megavirus chilensis SOD purified from cultures supplemented (trace b) or not (trace a) with Cu and Zn. Recording conditions were as follows: a T of 50,000, a microwave frequency of 9.476 GHz, microwave power of 1 mW, a modulation amplitude of 1 mT, and numbers of accumulations of 36 (a) and 10 (b), where T is temperature in Kelvin. The protein concentrations were 350 μM in 20 mM Tris buffer, pH 8 (a), and 400 μM Tris buffer in 10 mM CAPS buffer, pH 10.5 (b). B, magnetic field in millitesla (mT).
The comparison of the Mg277 structure with those of its cellular counterparts highlighted specific features of the viral enzyme, such as an additional intrachain disulfide bridge (C33-C105). The corresponding cysteine residues are conserved only in the 99%-identical SOD sequence of megavirus lba, the closest relative of megavirus chilensis. The dimer interfaces are also different between Mg277 and the eukaryotic structures. The human Cu,Zn-SOD dimer was reported as being extraordinarily stable (53), and the megavirus chilensis metal-bound SOD is also hyperstable, with a melting temperature of 76°C measured. The metal-free enzyme has a lower melting temperature of 56°C, comparable to that of the human apo form. In each monomer of the Mg277 crystal structure, a glutamine residue strongly contributes to the dimer interface through 2 H bonds linking the two Q116 residues together. Interestingly, the Q116 residue in Mg277 is replaced by a cysteine residue (C111 in Fig. 1) in the human enzyme, which was reported as being involved in the formation of a covalent polysulfane bridge between the two monomers after the intrachain disulfide bridge was formed (PDB accession number 3K91) (54). Intriguingly, the reactive cysteine residues covalently bound to the heptasulfane of the human enzyme are superimposable on the 2 linked Q116 residues in the megavirus chilensis structure. Altogether, the presence of the additional disulfide bridge in the megavirus chilensis enzyme, the extended interface between the monomers, and the bonds contributing to the dimer interface, especially the two salt bridges (K54 and R120 with the carboxyl-terminal N158) (Fig. 2), may explain the extreme stability of the metal-free form of the megavirus chilensis SOD enzyme. This stability might help the enzyme to remain folded both in the virion and in infected cells. To become active, the viral enzyme requires only zinc and copper ions from the host immediately after protein synthesis, prior to its loading into the viral particles or, conversely, when the virion protein penetrates the cytoplasm at the early stage of the infection.
Metal content and EPR analysis.
The metal content of the Mg277 protein expressed under different conditions was determined by inductively coupled plasma-mass spectroscopy (ICP-MS). As expected from the 3D structure, the Mg277b1 protein produced in 2YT growth medium (Table 2) and the selenomethionylated Mg277b2 protein used to solve the structure (Table 2) contained only substoichiometric quantities of Cu and Zn. The differences observed between the two proteins may originate from the fact that the selenomethionylated Mg277 protein was produced in minimal medium at a much lower bacterial growth rate, yielding less Mg277 protein. Thus, more copper and zinc were available than in the amount of selenomethionylated recombinant protein produced. The metal content analysis of Mg277b3 expressed in bacteria grown with the supplemented medium revealed the presence of Cu and Zn at a ratio of 1:1. Consistently, with the Mg277 protein produced without supplementation of the growth medium with Cu and Zn, no copper signal could be detected by EPR spectroscopy (trace a in Fig. 3). The EPR spectrum of the metal-bound form of the Mg277 protein recorded at 9 GHz displays an axial signal, with g values at a g⊥ of 2.262 and a g// of 2.067 and a hyperfine-coupling (ICu = 3/2) A// of 135 G, characteristic of a Cu2+ (S = 1/2) ion with a type II environment, as found in SODs with a dinuclear Zn,Cu active site (g is a constant of proportionality, whose value is the property of the electron in a certain environment, g// is g parallel, and g⊥ is g perpendicular) (Fig. 3) (55, 56). The shape of the EPR spectrum was found to be insensitive to pH variations in the [8 to 10.5] range, suggesting that there are no measurable differences in the enzyme structures either. Using an external standard, the EPR signal was found to correspond to 0.5 Cu2+ ion per protein.
Despite protein stabilization, we were unfortunately unable to make the metal-bound form crystallize either under the conditions of the metal-free form or under conditions that we routinely use to crystallize new proteins (57). The crystals produced with the nonsupplemented enzyme cracked when soaked with Zn and Cu.
Enzymatic activities of the various forms of Mg277.
As expected from the fluorescence scan, the metal dosage, and the EPR data, the nonsupplemented Mg277 proteins (Mg277b1 and Mg277b2) were not active in the range of concentrations used for the bovine SOD control. However, we were able to observe some activity when increasing concentrations of the Mg277b1 protein were used (Fig. 4), and the analysis of the data provided an IC50 of 180 μg/ml. The supplemented Mg277b2 enzyme produced an IC50 of 5 μg/ml, thus demonstrating that the protein was fully active when the metal-bound form was restored. For comparison, the measured IC50 of the bovine SOD control was 3 μg/ml.
FIG 4.
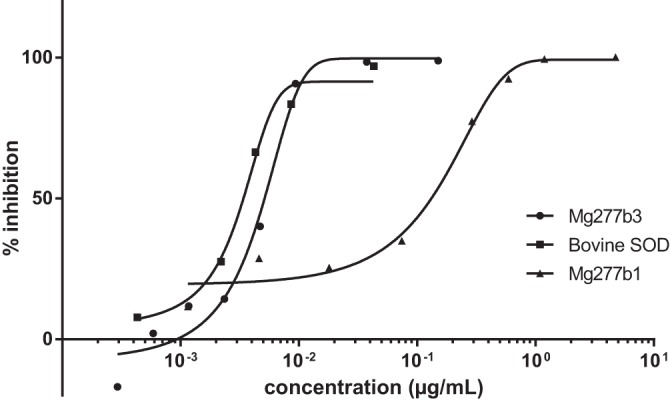
Superoxide dismutase activity measurements at increasing concentrations of Mg277b1 produced in minimum medium (triangles), of Mg277b3 produced in 2YT supplemented with Cu and Zn (circles), and of bovine Cu,Zn-SOD (squares) after inhibition of WST reduction, as indicated in Materials and Methods.
DISCUSSION
Unsurprisingly, given the high level of sequence similarity, this first structure of a viral SOD is strikingly similar to those of its eukaryotic counterparts. However, an in-depth analysis of the structure highlighted some specific characteristics of the viral enzyme. The dimer interfaces are different between the viral and the eukaryotic SODs, and the Mg277 interface area is larger despite the disordered lid in the apo form of the enzyme. Moreover, there is an extra disulfide bridge in the viral enzyme, suggesting that it is more constrained than in the other Cu,Zn-SODs. The reported phylogenetic distance between the mimiviruses and the other viruses (24) might be due to these specific structural features of the mimivirus SOD protein. The viral SODs might thus have different evolutionary histories. Finally, as for other CCS-independent SODs, the supplementation of the bacterial growth medium with copper and zinc was sufficient to recover the Mg277 metal-bound form and to restore its activity, which suggests that megavirus chilensis Mg277 belongs to this group of SODs. As Mg277 protein is part of the virion proteome, megavirus chilensis thus possesses an additional enzyme to resist oxidative stress, either intracytoplasmic or in the host phagocytic vacuole, compared to mimivirus. As it was evidenced for the chloroviruses (24), possessing a viral CCS-independent Cu,Zn-SOD which can be activated by the sole presence of copper and zinc may be an advantage in a host where SOD activation depends on the CCS. Interestingly, A. castellanii seems to possess at least three SOD genes and a CCS protein (XP_004352554, 28% identity over 232 amino acids); unfortunately, no experimental data are available on the dependence of the A. castellanii SOD proteins on CCS activation. On the other hand, other giant viruses infecting the same Acanthamoeba host do not encode a SOD and, yet, replicate as efficiently as megavirus chilensis. One explanation as to this discrepancy may be that mimivirus SOD is dispensable when it infects A. castellanii but is required in another host. Megavirus chilensis was isolated from the environment, and we do not know its natural host in the Pacific Ocean. Interestingly, except for some predicted SOD sequences of poxviruses, viral sequences possess all the key residues involved in metal binding and activity. As for the megavirus chilensis enzyme, they must thus correspond to active Cu,Zn-SODs, suggesting that resistance to oxidative stress is a significant constraint in the evolution of large eukaryotic DNA viruses. As reported for the chloroviruses' SOD, the enzyme packaged inside the virions may lower the concentration of reactive oxygen induced early during virus infection (24).
ACKNOWLEDGMENTS
We thank the BM30, ID14, ID23, ID29 (ESRF), and Proxima (Soleil) beamline teams for expert assistance.
This work was partially funded by the CNRS, Aix-Marseille Université, IBISA, and the Provence-Alpes-Côte d'Azur region.
REFERENCES
- 1.Iyer LM, Aravind L, Koonin EV. 2001. Common origin of four diverse families of large eukaryotic DNA viruses. J Virol 75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iyer LM, Balaji S, Koonin EV, Aravind L. 2006. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res 117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Abergel C, Rudinger-Thirion J, Giegé R, Claverie JM. 2007. Virus-encoded aminoacyl-tRNA synthetases: structural and functional characterization of mimivirus TyrRS and MetRS. J Virol 81:12406–12417. doi: 10.1128/JVI.01107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parakkottil Chothi M, Duncan GA, Armirotti A, Abergel C, Gurnon JR, Van Etten JL, Bernardi C, Damonte G, Tonetti M. 2010. Identification of an l-rhamnose synthetic pathway in two nucleocytoplasmic large DNA viruses. J Virol 84:8829–8838. doi: 10.1128/JVI.00770-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jeudy S, Abergel C, Claverie JM, Legendre M. 2012. Translation in giant viruses: a unique mixture of bacterial and eukaryotic termination schemes. PLoS Genet 8:e1003122. doi: 10.1371/journal.pgen.1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piacente F, Bernardi C, Marin M, Blanc G, Abergel C, Tonetti MG. 2014. Characterization of a UDP-N-acetylglucosamine biosynthetic pathway encoded by the giant DNA virus Mimivirus. Glycobiology 24:51–61. doi: 10.1093/glycob/cwt089. [DOI] [PubMed] [Google Scholar]
- 7.Piacente F, De Castro C, Jeudy S, Molinaro A, Salis A, Damonte G, Bernardi C, Abergel C, Tonetti M. 2014. Giant virus Megavirus chilensis encodes the biosynthetic pathway for uncommon acetamido sugars. J Biol Chem 289:24428–24439. doi: 10.1074/jbc.M114.588947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arslan D, Legendre M, Seltzer V, Abergel C, Claverie JM. 2011. Distant Mimivirus relative with a larger genome highlights the fundamental features of Megaviridae. Proc Natl Acad Sci U S A 108:17486–17491. doi: 10.1073/pnas.1110889108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raoult D, Audic S, Robert C, Abergel C, Renesto P, Ogata H, La Scola B, Suzan M, Claverie JM. 2004. The 1.2-megabase genome sequence of Mimivirus. Science 306:1344–1350. doi: 10.1126/science.1101485. [DOI] [PubMed] [Google Scholar]
- 10.Legendre M, Arslan D, Abergel C, Claverie JM. 2012. Genomics of Megavirus and the elusive fourth domain of life. Commun Integr Biol 5:102–106. doi: 10.4161/cib.18624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zauberman N, Mutsafi Y, Halevy DB, Shimoni E, Klein E, Xiao C, Sun S, Minsky A. 2008. Distinct DNA exit and packaging portals in the virus Acanthamoeba polyphaga mimivirus. PLoS Biol 6:e114. doi: 10.1371/journal.pbio.0060114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claverie JM, Abergel C, Ogata H. 2009. Mimivirus. Curr Top Microbiol Immunol 328:89–121. [DOI] [PubMed] [Google Scholar]
- 13.Claverie JM, Abergel C. 2010. Mimivirus: the emerging paradox of quasi-autonomous viruses. Trends Genet 26:431–437. doi: 10.1016/j.tig.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 14.Mutsafi Y, Zauberman N, Sabanay I, Minsky A. 2010. Vaccinia-like cytoplasmic replication of the giant Mimivirus. Proc Natl Acad Sci U S A 107:5978–5982. doi: 10.1073/pnas.0912737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renesto P, Abergel C, Decloquement P, Moinier D, Azza S, Ogata H, Fourquet P, Gorvel JP, Claverie JM. 2006. Mimivirus giant particles incorporate a large fraction of anonymous and unique gene products. J Virol 80:11678–11685. doi: 10.1128/JVI.00940-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colson P, De Lamballerie X, Yutin N, Asgari S, Bigot Y, Bideshi DK, Cheng XW, Federici BA, Van Etten JL, Koonin EV, La Scola B, Raoult D. 2013. “Megavirales,” a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol 158:2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desnues C, La Scola B, Yutin N, Fournous G, Robert C, Azza S, Jardot P, Monteil S, Campocasso A, Koonin EV, Raoult D. 2012. Provirophages and transpovirons as the diverse mobilome of giant viruses. Proc Natl Acad Sci U S A 109:18078–18083. doi: 10.1073/pnas.1208835109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoosuf N, Yutin N, Colson P, Shabalina SA, Pagnier I, Robert C, Azza S, Klose T, Wong J, Rossmann MG, La Scola B, Raoult D, Koonin EV. 2012. Related giant viruses in distant locations and different habitats: Acanthamoeba polyphaga moumouvirus represents a third lineage of the Mimiviridae that is close to the megavirus lineage. Genome Biol Evol 4:1324–1330. doi: 10.1093/gbe/evs109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoosuf N, Pagnier I, Fournous G, Robert C, La Scola B, Raoult D, Colson P. 2014. Complete genome sequence of Courdo11 virus, a member of the family Mimiviridae. Virus Genes 48:218–223. doi: 10.1007/s11262-013-1016-x. [DOI] [PubMed] [Google Scholar]
- 20.Lartigue A, Philippe N, Jeudy S, Abergel C. 2012. Preliminary crystallographic analysis of the Megavirus superoxide dismutase. Acta Crystallogr Sect F Struct Biol Cryst Commun 68:1557–1559. doi: 10.1107/S174430911204657X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marmocchi F, Caulini G, Venardi G, Cocco D, Calabrese L, Rotilio G. 1975. The effect of the presence of the metal prosthetic groups on the subunit structure of bovine superoxide dismutase in sodium dodecyl sulfate. Physiol Chem Phys 7:465–471. doi: 10.1016/0301-0104(75)87030-3. [DOI] [PubMed] [Google Scholar]
- 22.Klug D, Rabani J, Fridovich I. 1972. A direct demonstration of the catalytic action of superoxide dismutase through the use of pulse radiolysis. J Biol Chem 247:4839–4842. http://www.jbc.org/content/247/15/4839. [PubMed] [Google Scholar]
- 23.Lu Z, Li Y, Que Q, Kutish GF, Rock DL, Van Etten JL. 1996. Analysis of 94 kb of the Chlorella virus PBCV-1 330-kb genome: map positions 88 to 182. Virology 216:102–123. doi: 10.1006/viro.1996.0038. [DOI] [PubMed] [Google Scholar]
- 24.Kang M, Duncan GA, Kuszynski C, Oyler G, Zheng J, Becker DF, Van Etten JL. 2014. Chlorovirus PBCV-1 encodes an active copper-zinc superoxide dismutase. J Virol 88:12541–12550. doi: 10.1128/JVI.02031-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomalski MD, Eldridge R, Miller LK. 1991. A baculovirus homolog of a Cu/Zn superoxide dismutase gene. Virology 184:149–161. doi: 10.1016/0042-6822(91)90831-U. [DOI] [PubMed] [Google Scholar]
- 26.Smith GL, Chan YS, Howard ST. 1991. Nucleotide sequence of 42 kbp of vaccinia virus strain WR from near the right inverted terminal repeat. J Gen Virol 72:1349–1376. doi: 10.1099/0022-1317-72-6-1349. [DOI] [PubMed] [Google Scholar]
- 27.Willer DO, McFadden G, Evans DH. 1999. The complete genome sequence of Shope (rabbit) fibroma virus. Virology 264:319–343. doi: 10.1006/viro.1999.0002. [DOI] [PubMed] [Google Scholar]
- 28.Cameron C, Hota-Mitchell S, Chen L, Barrett J, Cao JX, Macaulay C, Willer D, Evans D, McFadden G. 1999. The complete DNA sequence of myxoma virus. Virology 264:298–318. doi: 10.1006/viro.1999.0001. [DOI] [PubMed] [Google Scholar]
- 29.Bawden AL, Glassberg KJ, Diggans J, Shaw R, Farmerie W, Moyer RW. 2000. Complete genomic sequence of the Amsacta moorei entomopoxvirus: analysis and comparison with other poxviruses. Virology 274:120–139. doi: 10.1006/viro.2000.0449. [DOI] [PubMed] [Google Scholar]
- 30.Almazán F, Tscharke DC, Smith GL. 2001. The vaccinia virus superoxide dismutase-like protein (A45R) is a virion component that is nonessential for virus replication. J Virol 75:7018–7029. doi: 10.1128/JVI.75.15.7018-7029.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao JX, Teoh ML, Moon M, McFadden G, Evans DH. 2002. Leporipoxvirus Cu-Zn superoxide dismutase homologs inhibit cellular superoxide dismutase, but are not essential for virus replication or virulence. Virology 296:125–135. doi: 10.1006/viro.2002.1383. [DOI] [PubMed] [Google Scholar]
- 32.Becker MN, Greenleaf WB, Ostrov DA, Moyer RW. 2004. Amsacta moorei entomopoxvirus expresses an active superoxide dismutase. J Virol 78:10265–10275. doi: 10.1128/JVI.78.19.10265-10275.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Teoh ML, Walasek PJ, Evans DH. 2003. Leporipoxvirus Cu,Zn-superoxide dismutase (SOD) homologs are catalytically inert decoy proteins that bind copper chaperone for SOD. J Biol Chem 278:33175–33184. doi: 10.1074/jbc.M300644200. [DOI] [PubMed] [Google Scholar]
- 34.Liu W, Li PW, Li GP, Zhu RH, Wang DC. 2001. Overexpression, purification, crystallization and preliminary X-ray diffraction analysis of Cu,Zn superoxide dismutase from Peking duck. Acta Crystallogr D Biol Crystallogr 57:1646–1649. doi: 10.1107/S0907444901011106. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. 2009. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr 65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grosse-Kunstleve RW, Adams PD. 2003. Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr 59:1966–1973. doi: 10.1107/S0907444903018043. [DOI] [PubMed] [Google Scholar]
- 38.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger TC. 2000. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr 56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:16–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu YK. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, Feolo M, Fingerman IM, Geer LY, Helmberg W, Kapustin Y, Landsman D, Lipman DJ, Lu Z, Madden TL, Madej T, Maglott DR, Marchler-Bauer A, Miller V, Mizrachi I, Ostell J, Panchenko A, Phan L, Pruitt KD, Schuler GD, Sequeira E, Sherry ST, Shumway M, Sirotkin K, Slotta D, Souvorov A, Starchenko G, Tatusova TA, Wagner L, Wang Y, Wilbur WJ, Yaschenko E, Ye J. 2011. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poirot O, Suhre K, Abergel C, O'Toole E, Notredame C. 2004. 3DCoffee@igs: a web server for combining sequences and structures into a multiple sequence alignment. Nucleic Acids Res 32:W37–W40. doi: 10.1093/nar/gkh382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kitagawa Y, Tanaka N, Hata Y, Kusunoki M, Lee GP, Katsube Y, Asada K, Aibara S, Morita Y. 1991. Three-dimensional structure of Cu,Zn-superoxide dismutase from spinach at 2.0 A resolution. J Biochem 109:477–485. [DOI] [PubMed] [Google Scholar]
- 47.Coutard B, Decroly E, Li C, Sharff A, Lescar J, Bricogne G, Barral K. 2014. Assessment of Dengue virus helicase and methyltransferase as targets for fragment-based drug discovery. Antiviral Res 106:61–70. doi: 10.1016/j.antiviral.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 48.Ukeda H, Maeda S, Ishii T, Sawamura M. 1997. Spectrophotometric assay for superoxide dismutase based on tetrazolium salt 3′-{1-[(phenylamino)-carbonyl]-3,4-tetrazolium}-bis(4-methoxy-6-nitro)benzenesulfonic acid hydrate reduction by xanthine–xanthine oxidase. Anal Biochem 251:206–209. doi: 10.1006/abio.1997.2273. [DOI] [PubMed] [Google Scholar]
- 49.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 50.Culotta VC, Klomp LW, Strain J, Casareno RL, Krems B, Gitlin JD. 1997. The copper chaperone or superoxide dismutase. J Biol Chem 272:23469–23472. doi: 10.1074/jbc.272.38.23469. [DOI] [PubMed] [Google Scholar]
- 51.Furukawa Y, O'Halloran TV. 2006. Posttranslational modifications in Cu,Zn-superoxide dismutase and mutations associated with amyotrophic lateral sclerosis. Antioxid Redox Signal 8:847–867. doi: 10.1089/ars.2006.8.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leitch JM, Jensen LT, Bouldin SD, Outten CE, Hart PJ, Culotta VC. 2009. Activation of Cu,Zn-superoxide dismutase in the absence of oxygen and the copper chaperone CCS. J Biol Chem 284:21863–21871. doi: 10.1074/jbc.M109.000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rodriguez JA, Valentine JS, Eggers DK, Roe JA, Tiwari A, Brown RH Jr, Hayward LJ. 2002. Familial amyotrophic lateral sclerosis-associated mutations decrease the thermal stability of distinctly metallated species of human copper/zinc superoxide dismutase. J Biol Chem 277:15932–15937. doi: 10.1074/jbc.M112088200. [DOI] [PubMed] [Google Scholar]
- 54.You Z, Cao X, Taylor AB, Hart PJ, Levine RL. 2010. Characterization of a covalent polysulfane bridge in copper-zinc superoxide dismutase. Biochemistry 49:1191–1198. doi: 10.1021/bi901844d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rotilio G, Finazzi Agrò A, Calabrese L, Bossa F, Guerrieri P, Mondovì B. 1971. Study of the metal sites of copper proteins. Ligands of copper in hemocuprein. Biochemistry 10:616–621. [DOI] [PubMed] [Google Scholar]
- 56.Battistoni A, Folcarelli S, Cervoni L, Polizio F, Desideri A, Giartosio A, Rotilio G. 1998. Role of the dimeric structure in Cu,Zn superoxide dismutase. pH-dependent, reversible denaturation of the monomeric enzyme from Escherichia coli. J Biol Chem 273:5655–5661. doi: 10.1074/jbc.273.10.5655. [DOI] [PubMed] [Google Scholar]
- 57.Abergel C, Coutard B, Byrne D, Chenivesse S, Claude JB, Deregnaucourt C, Fricaux T, Gianesini-Boutreux C, Jeudy S, Lebrun R, Maza C, Notredame C, Poirot O, Suhre K, Varagnol M, Claverie JM. 2003. Structural genomics of highly conserved microbial genes of unknown function in search of new antibacterial targets. J Struct Funct Genomics 4:141–157. doi: 10.1023/A:1026177202925. [DOI] [PubMed] [Google Scholar]
- 58.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Humphrey W, Dalke A, Schulten K. 1996. VMD—Visual Molecular Dynamics. J Mol Graphics 14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]