Abstract
Recent discoveries have revealed that the genes for the biosynthesis of a variety of plant specialized metabolites are organized in operon-like clusters within plant genomes. Here we identify a regulatory process that is required for normal expression of metabolic gene clusters in Arabidopsis thaliana.
Comparative gene expression analysis of a representative clustered gene was performed in a set of chromatin mutant lines. Subsequently, metabolite levels were analysed by GC-MS and the local chromatin structure was investigated by chromatin immunoprecipitation and nucleosome positioning.
We show that the transcript levels of genes within two metabolic clusters are coordinately reduced in an arp6 and h2a.z background. We demonstrate that H2A.Z enrichment in the clusters is positively correlated with active cluster expression. We further show that nucleosome stability within the cluster regions is higher in the arp6 background compared with the wild-type.
These results implicate ARP6 and H2A.Z in the regulation of metabolic clusters in Arabidopsis thaliana through localized chromatin modifications that enable the coordinate expression of groups of contiguous genes. These findings shed light on the complex process of cluster regulation, an area that could in the future open up new opportunities for the discovery and manipulation of specialized metabolic pathways in plants.
See also the Commentary by Fernie and Tohge
Keywords: Arabidopsis thaliana, ARP6, H2A.Z, nucleosome positioning, plant metabolic gene clusters, specialized metabolism, triterpenes
Introduction
In eukaryotes the arrangement of the genes along the chromosome is not as random as was first thought, and distinctive clusters of functionally related but nonhomologous genes can be found in the genomes of certain animals and fungi (Hurst et al., 2004; Michalak, 2008). These include the major histocompatibility complex in mammals and gene clusters for the synthesis of specialized metabolites (such as penicillin and toxins) in filamentous fungi (Osbourn & Field, 2009). Genes for specialized metabolic pathways in plants have until recently been assumed to be dispersed throughout plant genomes, and certainly there are well-known examples of pathways for which this is true (e.g. the phenylpropanoid and glucosinolate pathways). However, over 20 examples of biosynthetic gene clusters for the synthesis of specialized metabolites have now been reported from diverse plant species (Boycheva et al., 2014; Nützmann & Osbourn, 2014). These clusters span regions of c. 35–270 kb and consist of 3 to 10 genes. They include clusters for the synthesis of hydroxamic acids in maize; terpenes in Arabidopsis thaliana, castor, rice and oats; cyanogenic glycosides in Lotus japonicus, sorghum and cassava; and alkaloids in poppy, tomato and potato (Frey et al., 1997; Qi et al., 2004; Wilderman et al., 2004; Shimura et al., 2007; Field & Osbourn, 2008; Field et al., 2011; Nützmann & Osbourn, 2014; Takos et al., 2011; Winzer et al., 2012; Itkin et al., 2013; Krokida et al., 2013; Matsuba et al., 2013; King et al., 2014). The natural functions of these specialized metabolites are likely to be in defence against herbivores and pathogens. Some of these compounds also have major pharmaceutical and agricultural importance, for example, noscapine (an antitumor alkaloid from poppy), and the anti-nutritional steroidal glycoalkaloids found in tomato and potato. Although plant specialized metabolic gene clusters share some features in common with bacterial operons (physical clustering of genes and coordinate regulation of expression) there is a compelling body of evidence to indicate that they have arisen de novo by genome reorganization, and that they are not a consequence of horizontal gene transfer from microbes (Field & Osbourn, 2008; Field et al., 2011). The regulatory mechanisms that govern coordinate expression of metabolic gene clusters in plants are, for the most part, unknown. So far only one transcriptional regulator has been identified for clustered pathway genes (for diterpene biosynthesis in rice) and it is not clear whether this is pathway specific or whether it has a broader role in regulation of metabolism in rice (Miyamoto et al., 2014).
Physical clustering has the potential to enable coordinate regulation of the genes within plant metabolic gene clusters at the level of chromatin. In filamentous fungi there is evidence that metabolic gene clusters are controlled by chromatin-based mechanisms involving histone acetyltransferases, deacetylases, and methyl transferases (Bok et al., 2009; Reyes-Dominguez et al., 2010; Nützmann et al., 2011; Brakhage, 2013). In yeast (Saccharomyces cerevisiae) the histone 2 variant H2A.Z is required for the coordinate regulation of the DAL cluster, a catabolic gene cluster for allantoin utilization (Wong & Wolfe, 2005). H2A.Z is also involved in the regulation of adaptive gene clusters in other organisms, including the virulence (vir) cluster in the malaria parasite (Plasmodium falciparum) and the Hox gene cluster in animals (Meneghini et al., 2003; Creyghton et al., 2008; Petter et al., 2013).
Here we demonstrate that the histone variant H2A.Z is essential for normal expression of metabolic gene clusters and regulation of metabolite synthesis in Arabidopsis thaliana. We show by mutation that inability to incorporate H2A.Z into nucleosomes leads to reduced cluster expression. We show by chromatin immunoprecipitation (ChIP) analysis that activation of clusters is accompanied by increased H2A.Z deposition into nucleosomes in the genic and intergenic regions within the clusters and, by measurement of nucleosome occupancy, with loosening of the local nucleosome structure. Thus we propose that H2A.Z-mediated chromatin modification leads to localized opening of the nucleosome structure and facilitates expression of gene clusters for the synthesis of specialized metabolites in plants.
Materials and Methods
Plant material
Unless otherwise stated all plants used in this study were in the Arabidopsis thaliana Columbia (Col-0) background. The arp6 (Col-0) and vin3-8 lines were provided by Ali Pendle (John Innes Centre, Norwich, UK) and Jean Finnegan (CSIRO Plant Industry, Clayton South, Australia). The arp6 (Ler), hta9/hta11 and hta11-GFP lines were donated by Vinod Kumar (John Innes Centre). Haf1 (SALK_070585C), hda18 (SALK_006938), ref6 (SALK_001018C), vim1 (SALK_050903C), atjmj4 (SALK_135712C), elf6 (SALK_074694C), hda6 (CS66154), otld1 (SALK_037047C), atx1 (SALK_149002C), atxr7 (SALK_149692C), suvh2 (SALK_079574C), prmt10 (SALK_047046C), and prmt5 (SALK_065814C) lines were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000) and homozygous mutants isolated. Further information about these lines is provided in Supporting Information Table S1. Seedlings were grown in Petri dishes on Murashige and Skoog medium containing 1% sucrose (screening of chromatin mutant lines) and Murashige and Skoog plant salt medium supplemented with 0.5% phytagel and 0.75% sucrose (all other experiments) (Murashige & Skoog, 1962). Petri dishes were maintained vertically in growth chambers (22°C, 16 h light) for 6–7 d.
Gene transcript level analysis
Total RNA was isolated from roots of seedlings with the RNeasy Plant Mini Kit (Qiagen, Venlo, The Netherlands) and DNA degradation performed with the TURBO DNA-free kit (Life Technologies, Carlsbad, CA, USA). cDNA was prepared using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA) and quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed with the MyTaq HS Mix 2x (Bioline, London, UK) in combination with Evagreen (Biotium, Hayward, CA, USA) on a CFX96 Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA). The cycling conditions used were: 95°C for 2 min, followed by 45 cycles of DNA denaturation at 95°C for 5 s and primer annealing and extension at 62°C for 35 s. All qRT-PCR experiments were performed with at least three biological replicates and two technical replicates. Data were analysed with the Bio-Rad CFX manager 3.0. Quantification results were normalized against PP2AA3 (At1g13320) (Hong et al., 2010).
Chromatin immunoprecipitation
ChIP assays were carried out as described previously (Song et al., 2014). Sonicated DNA fragments showed average length of 400 bp. Precipitation was performed against anti-GFP antibody (Abcam AB290). All ChIP experiments were quantified by qRT-PCR. H2A.Z abundance at FLOWERING LOCUS C (FLC) was used as an internal control. Data are shown as the ratio of (H2A.Z-GFP locus of interest/input DNA locus of interest) to (H2A.Z-GFP FLC/input DNA FLC). Each ChIP experiment was carried out with at least three biological replicates and qRT-PCR quantification was performed in duplicate.
Nucleosome positioning
Nucleosome positioning was performed as described previously (Kumar & Wigge, 2010). Each positioned nucleosome was determined individually by qRT-PCR using tiled oligos positioned around the putative transcriptional start sites of target genes and in the intergenic region of the gene cluster (Supporting Information Table S2). Stably positioned nucleosomes in intergenic regions were pre-selected based on genome-wide nucleosome positioning data and individually confirmed by qRT-PCR using tiled oligos (Zhang et al., 2007). The −1 nucleosome at At4g07700 was used as an internal control (Kumar & Wigge, 2010). Data are represented as the ΔΔCt value of the region of interest normalized against the −1 nucleosome at At4g07700 and uncut input DNA. The qRT-PCR cycling conditions were adapted to 54°C annealing temperature for 10 s.
Metabolite extraction and quantification
Root tissue (c. 100 mg) from 6 d old seedlings of wild-type and mutant lines was harvested. Extraction was carried out as described (Field & Osbourn, 2008). The internal standard coprostanol (Sigma Aldrich) was incorporated during saponification. GC-MS analysis was performed as previously described (Field & Osbourn, 2008). For quantification, an extracted ion chromatogram was obtained and thalianol (m/z 229), coprostanol (m/z 370) and sitosterol (m/z 357) peaks were integrated with the RTE integrator (Agilent Technologies, Santa Clara, CA, USA).
Results and Discussion
Previously we have characterized two gene clusters for the synthesis of specialized metabolites in Arabidopsis thaliana, the thalianol cluster and the marneral cluster (Field & Osbourn, 2008; Field et al., 2011). The thalianol and marneral clusters have evolved independently and encode enzymes for the synthesis and elaboration of the triterpenes thalianol and marneral, respectively (Fig.1a,b). These pathways have both been implicated in development and in ecological interactions (Field & Osbourn, 2008; Field et al., 2011; Go et al., 2012). The genes within each of these two clusters are co-regulated and are expressed exclusively in roots (Supporting Information Fig. S1) (Field & Osbourn, 2008; Field et al., 2011).
Fig 1.
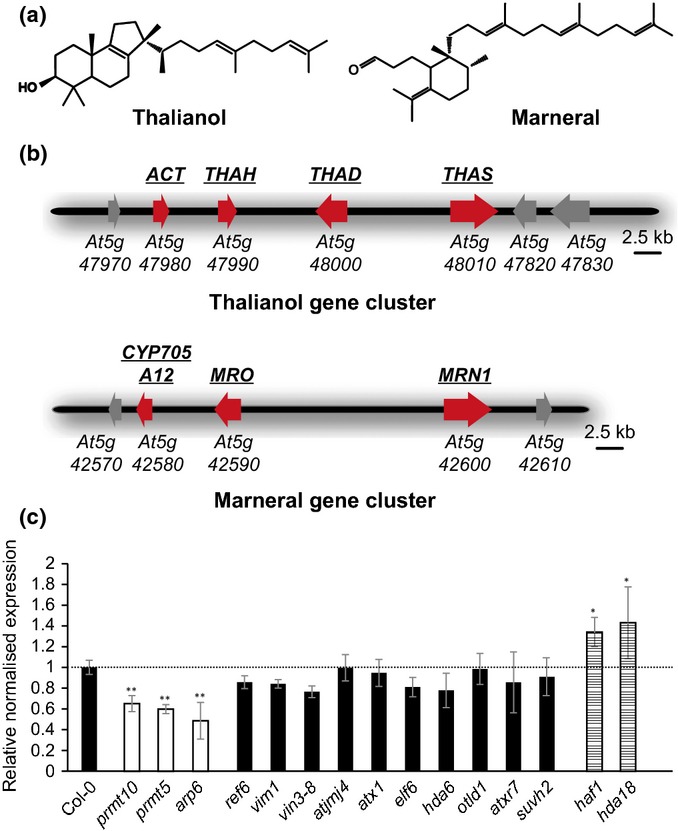
Steady state transcript levels of THAH, a representative thalianol cluster gene, in different Arabidopsis thaliana chromatin mutant lines. (a) The structures of thalianol and marneral. (b) The thalianol and marneral gene clusters (red, cluster genes; grey, flanking genes). The thalianol cluster consists of an oxidosqualene cyclase gene (thalianol synthase, THAS), two nonhomologous cytochrome P450 genes (thalianol hydroxylase, THAD; thalian-diol desaturase, THAH) and an acyltransferase gene (ACT); the marneral cluster consists of one oxidosqualene cyclase gene (marneral synthase, MRN1) and two nonhomologous cytochrome P450 genes (CYP705A12; marneral oxidase, MRO). (c) The relative transcript levels of the thalianol cluster gene THAH was measured in roots of seedlings of the Col-0 wild-type and chromatin mutant lines (see Supporting Information Table S1). Gene expression was measured by qRT-PCR and the wild-type transcription level set as 1. PP2AA3 (At1g13320) was used as an internal control (Hong et al., 2010). Error bars indicate standard deviation for three biological replicates. Statistical significance (t-test): *P < 0.05; **P < 0.01.
To assess the potential role of chromatin in regulation of plant metabolic gene clusters we first evaluated the transcript levels of a representative gene from one of these clusters, the thalianol cluster (the gene encoding thalianol hydroxylase (THAH)) in a set of Arabidopsis thaliana mutants affected in histone modifications and chromatin remodelling (Supporting Information Table S1). THAH transcript levels were clearly reduced in three of these lines relative to the Col-0 wild-type (the histone methyltransferase mutants prmt5 and prmt10; and arp6, which is mutated in a gene encoding a component of a chromatin remodelling complex) and elevated in two (the histone acetyltransferase mutant haf1 and the histone deacetylase mutant hda18) (Fig.1c). The arp6 mutant line had the most pronounced negative effect on THAH gene transcript levels of all the mutant lines investigated. ARP6 is part of the SWR1 chromatin remodelling complex and is required for the deposition of the histone variant H2A.Z into nucleosomes (Deal et al., 2007; Kumar & Wigge, 2010). The potential involvement of H2A.Z in regulation of plant metabolic gene clusters raises intriguing parallels with the DAL gene cluster in yeast, where H2A.Z deposition is associated with activation of cluster gene expression (Meneghini et al., 2003; Wong & Wolfe, 2005). We therefore focused our subsequent investigations on ARP6.
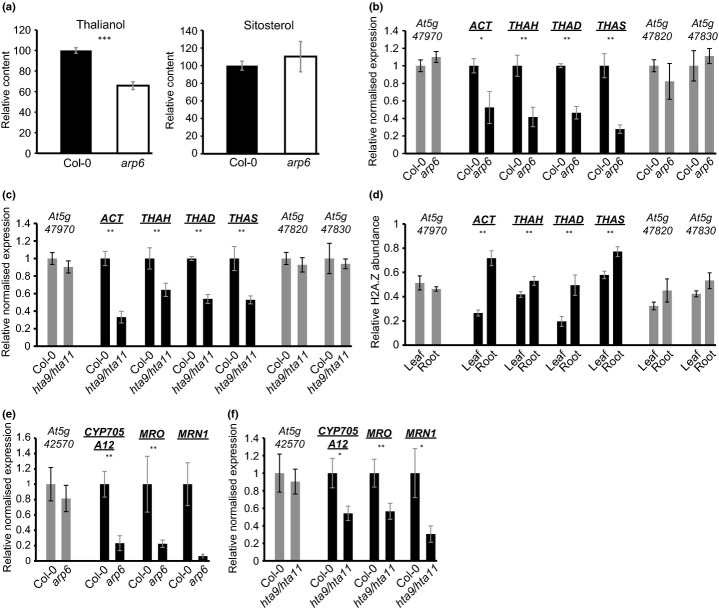
We next investigated the importance of ARP6 for the expression of metabolic gene clusters in Arabidopsis thaliana in detail. Thalianol levels were significantly reduced in root extracts of arp6 mutant seedlings when compared with the wild-type, while levels of sitosterol (a representative sterol that is a product of primary metabolism and that has a common precursor with thalianol) were unaltered (Fig.2a, Supporting Information Fig. S2). Investigation of the transcript levels for all four thalianol cluster genes revealed that these were reduced in the arp6 mutant background while those of the genes immediately flanking the cluster were not, indicating that ARP6 is required for normal cluster regulation (Fig.2b). These experiments were carried out in the Colombia (Col-0) accession of A. thaliana. Similar results were obtained with the A. thaliana Landsberg (Ler) accession and its respective arp6 mutant line (Martin-Trillo et al., 2006) (Supporting Information Fig. S3).
Fig 2.
Mutation of the ARP6 and histone H2A.Z genes leads to reduced transcript levels of the genes within the thalianol and marneral clusters. (a) Analysis of thalianol and sitosterol levels in root extracts of Arabidopsis thaliana Col-0 wild-type and arp6 mutant lines. Measurements were made relative to an internal standard, coprostanol, added before extraction. Extracts were analysed by GC-MS and peak areas of individual compounds were quantified (Supporting Information Fig. S2). Error bars indicate standard deviation of three biological replicates. Significant differences between wild-type and mutant lines (t-test): ***P < 0.001. (b, c) Relative transcript levels of the thalianol cluster genes in the Col-0 wild-type and arp6 mutant line (b) and hta9/hta11 double mutant line (c). Black bars, thalianol pathway genes; grey bars, flanking genes. Transcript levels were measured by qRT-PCR as for Fig.1. Error bars indicate standard deviation of three biological replicates. Statistical significance (t-test): *P < 0.05; **P < 0.01. (d) H2A.Z abundance at thalianol cluster genes in chromatin preparations from roots and leaves of seedlings of A. thaliana accession Col-0. H2A.Z abundance was measured at the transcriptional start sites of the cluster genes and was quantified relative to FLOWERING LOCUS C (FLC). Error bars indicate standard deviation of three biological replicates. Significant differences in H2A.Z abundance between leaves and roots (t-test): *P < 0.05; **P < 0.01. (e, f) Relative transcript levels of the marneral cluster genes in the Col-0 wild-type and arp6 mutant line (e) and hta9/hta11 double mutant line (f). Black bars, marneral pathway genes; grey bars, flanking genes. Transcript levels were measured by qRT-PCR as for Fig.1. Error bars indicate standard deviation of three biological replicates. Statistical significance (t-test): *P < 0.05; **P < 0.01.
Since ARP6 is a SWR1 remodeller required for deposition of H2A.Z into nucleosomes and this histone variant has previously been shown to be important for regulation of the DAL gene cluster in yeast, we next investigated the role of H2A.Z in thalianol cluster regulation. To do this we used the Arabidopsis thaliana double mutant hta9/hta11, which is mutated in two of the three A. thaliana H2A.Z genes and has previously been shown by others to phenocopy the arp6 mutant line (March-Diaz et al., 2008; Kumar & Wigge, 2010). The transcript levels of all four thalianol cluster genes were significantly reduced in the hta9/hta11 mutant line compared with the wild-type, while those of the genes immediately flanking the clusters were unaffected (Fig.2c). The effects of the arp6 and hta9/hta11 mutations are therefore localized and specific to the cluster genes when compared with the flanking genes. These results implicate H2A.Z in ARP6-mediated cluster expression.
The role of H2A.Z in positive regulation of the thalianol cluster may be direct, or alternatively could be an indirect result of modifications elsewhere in the genome. We therefore analysed H2A.Z abundance in the thalianol gene cluster region to investigate whether H2A.Z is likely to directly regulate cluster expression. To do this we carried out ChIP experiments using an HTA11:GFP transgenic Arabidopsis thaliana line (Kumar & Wigge, 2010). H2A.Z deposition in A. thaliana is generally thought to be restricted to genic sequences, with peaks at the 5′ ends of target genes (Zilberman et al., 2008; Coleman-Derr & Zilberman, 2012). We initially chose the transcriptional start sites of the thalianol cluster genes as targets for our ChIP analysis. Comparison of H2A.Z abundance at these positions in chromatin isolated from roots and leaves revealed H2A.Z deposition in nucleosomes from both tissues, but with significantly higher levels in chromatin isolated from roots (the organ in which the thalianol cluster is expressed), suggesting a positive correlation between H2A.Z nucleosome incorporation and gene cluster transcription (Fig.2d). There were no significant organ-specific differences in H2A.Z levels in the genes immediately flanking the thalianol cluster. We next investigated whether the modifiable H2A.Z deposition was restricted to the genic regions within the cluster by measuring H2A.Z abundance using probes localized in different parts of the thalianol gene cluster, including the intergenic regions. We detected significantly higher H2A.Z levels throughout the whole cluster in roots when compared with leaves (Fig.3a). Thus H2A.Z deposition spans the entire cluster region and is not restricted to the individual cluster genes. By contrast, in nonclustered eukaryotic genes H2A.Z deposition is usually limited to gene bodies and the immediate vicinity of the transcriptional start site (Guillemette et al., 2005; Creyghton et al., 2008; Whittle et al., 2008; Coleman-Derr & Zilberman, 2012). As controls, we analysed H2A.Z levels at three loci previously reported to show either presence (At2g27880 and At2g39210) or absence (At4g07700) of H2A.Z (March-Diaz et al., 2008; Kumar & Wigge, 2010). We detected constant H2A.Z deposition at At2g27880 and At2g39210 and precipitated only minor amounts of DNA at the transcriptionally silent locus At4g07700, as expected (Fig.3b).
Fig 3.
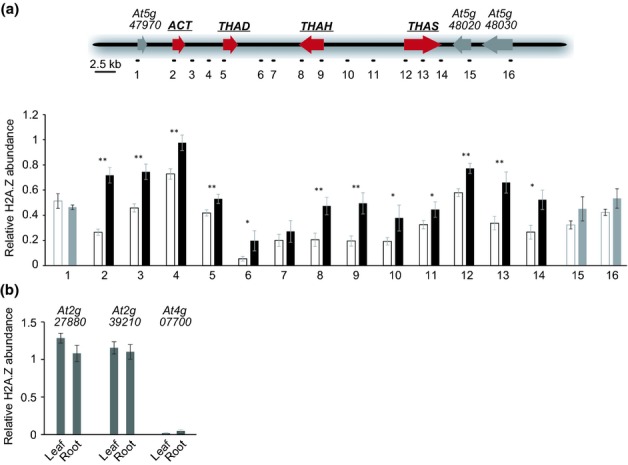
Chromatin immunoprecipitation (ChIP) analysis of H2A.Z abundance in the thalianol gene cluster. (a) H2A.Z abundance within the thalianol gene cluster was measured in chromatin preparations from leaves (white bars) and roots (black bars) of seedlings of Arabidopsis thaliana accession Col-0. H2A.Z abundance in different regions of the cluster was quantified relative to FLOWERING LOCUS C (FLC) (the white bars with grey outlines and the grey bars show data for regions outside the gene cluster). The probes used for this analysis (shown below the histogram) are indicated in the map of the thalianol cluster. Error bars indicate standard deviation of three biological replicates. Significance differences in H2A.Z abundance between leaves and roots (t-test): *P < 0.05; **P < 0.01. (b) H2A.Z abundance at selected control loci. At2g27880 and At2g39210 were used because both have been shown to be marked with the H2A.Z histone variant, have SWR1 dependent expression and do not show differential transcription rate between roots and leaves (based on AtGenExpress) (March-Diaz et al., 2008; Zilberman et al., 2008). The At4g07700 locus was chosen as a negative control for the ChIP experiment, because it has been shown that H2A.Z is not incorporated into nucleosomes of this locus (Kumar & Wigge, 2010). H2A.Z abundance at these loci was quantified relative to FLC. Error bars indicate standard deviation of three biological replicates.
Similar results were obtained for the marneral cluster (Fig.2e,f, Supporting Information Fig. S4). For both the thalianol and marneral clusters, the effects of the arp6 and hta9/hta11 mutations and the incorporation of H2A.Z were localized and specific to the clusters when compared with the genes immediately flanking the clusters. The transcript levels of the thalianol and marneral cluster genes were not completely abolished in arp6 mutants, indicating that additional as yet unidentified regulatory components also contribute to gene cluster expression. However our results provide evidence for a role for ARP6 and H2A.Z in positive regulation of the thalianol and marneral clusters through localized chromatin remodelling.
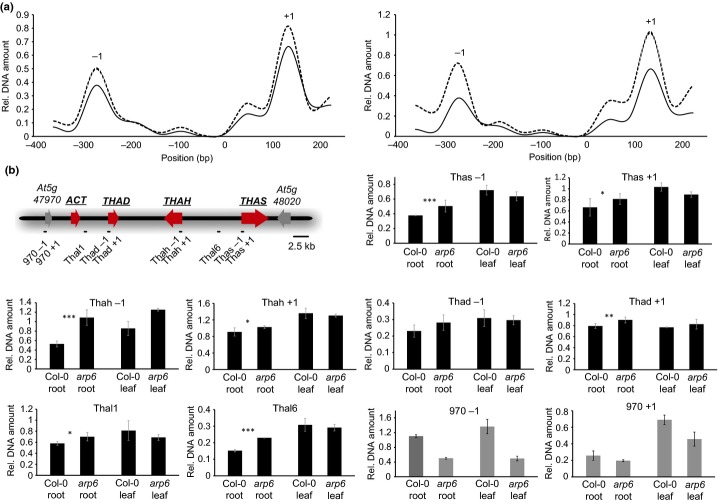
We next analysed the local genome accessibility at the thalianol cluster using micrococcal nuclease accessibility assays. Addition of micrococcal nuclease to nuclear chromatin leads to digestion of nucleosome-free DNA, while nucleosome-bound DNA is protected. Subsequent quantification of undigested DNA enables the identification of nucleosome positions and their stability at these sites. Initially, we compared the nucleosome abundance at the transcriptional start-site of the THAS gene within the thalianol gene cluster. As expected we found two well-positioned nucleosomes, the −1 nucleosome, just upstream of the transcriptional start site and the +1 nucleosome, just downstream (Fig.4a). In preliminary experiments we found that the −1 and +1 nucleosomes were less abundant in chromatin preparations from wild-type roots compared with the transcriptionally less active arp6 roots, or to wild-type leaves (where the cluster is inactive) (Fig.4a). We then broadened our analysis to the whole cluster (Fig.4b). We detected well-positioned nucleosomes around the putative transcriptional start sites of thalianol cluster genes and elsewhere within the gene cluster, and measured the abundance of these nucleosomes in chromatin isolated from the roots and leaves of wild-type and arp6 mutant Arabidopsis thaliana lines. In seven out of the eight positioned nucleosomes analysed we found a significant increase in nucleosome occupancy in the arp6 mutant roots compared with the wild-type (Fig.4b). Surprisingly, we found higher nucleosome occupancy not only at the transcriptional start sites of cluster genes but also at intergenic regions within the cluster, in line with the cluster-wide enrichment of H2A.Z.
Fig 4.
Analysis of nucleosome occupancy within the thalianol cluster. (a) Comparison of nucleosome positioning at the transcriptional start site of the THAS gene in roots of Arabidopsis thaliana Col-0 wild-type (solid line) and arp6 mutant (dashed line) lines (left); Col-0 roots (solid line) and leaves (dashed line) (right).The −1 and +1 indicate the first positioned nucleosome upstream and downstream of the putative transcriptional start site. (b) Relative abundance of stably positioned nucleosomes within the thalianol cluster and at the At5g47970 flanking gene in roots and leaves of seedlings of Col-0 wild-type and arp6 mutant lines. The position of each analysed nucleosome is indicated in the map of the cluster region. The −1 and +1 indicate the first positioned nucleosome upstream and downstream of the putative transcriptional start site of the respective gene. Nucleosome abundance was measured by qRT-PCR relative to the stably positioned −1 nucleosome at the At4g07700 reference locus. Results for representative experiments are shown (error bars indicate standard deviation of two biological replicates). Significant differences between wild-type and mutants lines (five experiments, 10 biological replicates) (t-test): *P < 0.05; **P < 0.01; ***P < 0.001.
H2A.Z deposition may be involved in either opening or compaction of chromatin environments in Arabidopsis thaliana (Deal et al., 2007; Kumar & Wigge, 2010). Generally, higher nucleosome density is correlated with a less accessible chromatin structure, which hinders the ability of the transcriptional machinery to associate with its target genes. Our data suggest that ARP6-mediated incorporation of H2A.Z into nucleosomes within the thalianol and marneral gene clusters may lead to localized opening of the chromatin structure within the cluster regions, so facilitating cluster expression. The nucleosomes analysed at the At5g47970 gene, located directly upstream of the thalianol gene cluster, did not show an increase in nucleosome abundance in the arp6 mutant background compared with the wild-type. We also observed more stably positioned nucleosomes in leaf tissues compared with root tissues, consistent with the correlation between transcriptional activity of the gene cluster and more loosely organized nucleosomes. This is in accordance with our previous DNA fluorescence in situ hybridization experiments, which suggested that the avenacin metabolic gene cluster in oat switches between a highly condensed and an open chromatin structure during transition from the repressed to the actively transcribed state (Wegel et al., 2009). We also found a similar decrease in root stable nucleosome positioning at one of the thalianol cluster flanking genes (the At5g47970 gene), which does not show differential regulation in root and leaf tissues. We suggest that the more open nucleosome structure of the highly expressed thalianol gene cluster spreads into this neighbouring region independently of H2A.Z.
The rapidly growing number of reports of metabolic gene clusters for the synthesis of diverse classes of compound from different plant species suggests that this form of genomic organization is common in plants, although a comprehensive overview of the overall relative frequencies of clustered and dispersed pathways for specialized metabolism in plant genomes is not yet known. The physical clustering of genes whose functions are related but that do not share sequence similarity and have not arisen by tandem duplication is intriguing. Clustering should favour co-inheritance of beneficial combinations of alleles that together confer a selective advantage. As we show here, it is also likely to be important for coordinate regulation of expression of pathway enzymes. The synthesis of specialized metabolites in plants is normally tightly regulated. There is evidence for the thalianol and marneral clusters and also for some other plant metabolic clusters that the pathway intermediates are toxic, and so strict regulation may be critical in avoiding build-up of these toxic compounds (Field & Osbourn, 2008; Mylona et al., 2008; Field et al., 2011; Takos et al., 2011). Our data support the hypothesis that clustering facilitates pathway regulation, implicating the SWR1 remodelling complex component ARP6 and the histone 2 variant H2A.Z in the regulation of discrete metabolic gene clusters of Arabidopsis thaliana through highly localized chromatin modifications that enable the coordinate expression of windows of contiguous genes. A possible scenario is that incorporation of H2A.Z into nucleosomes at the thalianol and marneral loci may poise these clusters for rapid expression in an otherwise highly repressive heterochromatin region, so making the genes within them accessible to transcription factors, as has been shown to be the case for the DAL gene cluster for allantoin catabolism in yeast (Meneghini et al., 2003; Guillemette et al., 2005; Wong & Wolfe, 2005). Alternatively ARP6 and H2A.Z may have other indirect roles in cluster regulation through as yet undefined mechanisms. While mutations that affect ARP6-mediated H2A.Z deposition have a clear effect on cluster expression and metabolite production they do not abolish cluster activity completely, indicating that other factors are also likely to be involved. Chromatin regulation of specialized metabolism is not limited to clustered pathways, and several previous studies have shown that mutations in important chromatin regulators can affect the synthesis of compounds such as phenylpropanoids and glucosinolates, pathways for which the genes not regarded as clustered in A. thaliana (Rider et al., 2004; Bennett et al., 2005; Walley et al., 2008). It is not clear, however, whether these effects are a direct consequence of chromatin remodelling at the cognate biosynthetic loci or a secondary effect of modifications elsewhere in the genome. Future work will compare mechanisms of regulation between clustered and nonclustered pathways and investigate the roles of other regulatory processes in cluster regulation, and is expected to open up new opportunities for the discovery and manipulation of specialized metabolic pathways in plants.
Acknowledgments
This work was supported by the UK Biotechnological and Biological Sciences Research Council (BBSRC) Institute Strategic Programme Grant ‘Understanding and Exploiting Plant and Microbial Metabolism’ (BB/J004561/1) and the John Innes Foundation (A.O.), and by Marie Curie Actions and an EMBO Long-Term Fellowship to H-W.N. The authors would like to thank Jean Finnegan for kind provision of chromatin mutant lines, Vinod Kumar for helpful discussions and provision of H2A.Z mutant lines, and Kalliopi Papadopoulou, Paul Ó’Máille, Vinod Kumar and members of the Osbourn laboratory for comments on the manuscript.
Supporting Information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1Relative transcript level of the thalianol and marneral cluster genes in RNA preparations from leaf and root tissue of the Col-0 wild-type.
Fig. S2 GC-MS analysis of Col-0 and arp6 root extracts.
Fig. S3 Relative transcript levels of the thalianol and marneral cluster genes in Ler wild-type and corresponding arp6 mutant line.
Fig. S4 Chromatin immunoprecipitation (ChIP) analysis of H2A.Z abundance at the marneral gene cluster and control loci.
Table S1Arabidopsis thaliana mutant lines
Table S2 Oligonucleotides used in this study
References
- Bennett RN, Wenke T, Freudenberg B, Mellon FA, Ludwig-Muller J. The tu8 mutation of Arabidopsis thaliana encoding a heterochromatin protein 1 homolog causes defects in the induction of secondary metabolite biosynthesis. Plant Biology. 2005;7:348–357. doi: 10.1055/s-2005-837634. [DOI] [PubMed] [Google Scholar]
- Bok JW, Chiang YM, Szewczyk E, Reyes-Dominguez Y, Davidson AD, Sanchez JF, Lo HC, Watanabe K, Strauss J, Oakley BR, et al. Chromatin-level regulation of biosynthetic gene clusters. Nature Chemical Biology. 2009;5:462–464. doi: 10.1038/nchembio.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boycheva S, Daviet L, Wolfender JL, Fitzpatrick TB. The rise of operon-like gene clusters in plants. Trends Plant Science. 2014;19:447–459. doi: 10.1016/j.tplants.2014.01.013. [DOI] [PubMed] [Google Scholar]
- Brakhage AA. Regulation of fungal secondary metabolism. Nature Reviews Microbiology. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- Coleman-Derr D, Zilberman D. Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genetics. 2012;8:e1002988. doi: 10.1371/journal.pgen.1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Markoulaki S, Levine SS, Hanna J, Lodato MA, Sha K, Young RA, Jaenisch R, Boyer LA. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, Topp CN, McKinney EC, Meagher RB. Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell. 2007;19:74–83. doi: 10.1105/tpc.106.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Fiston-Lavier AS, Kemen A, Geisler K, Quesneville H, Osbourn AE. Formation of plant metabolic gene clusters within dynamic chromosomal regions. Proceedings of the National Academy of Sciences, USA. 2011;108:16116–16121. doi: 10.1073/pnas.1109273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field B, Osbourn AE. Metabolic diversification—independent assembly of operon-like gene clusters in different plants. Science. 2008;320:543–547. doi: 10.1126/science.1154990. [DOI] [PubMed] [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grun S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, et al. Analysis of a chemical plant defense mechanism in grasses. Science. 1997;277:696–699. doi: 10.1126/science.277.5326.696. [DOI] [PubMed] [Google Scholar]
- Go YS, Lee SB, Kim HJ, Kim J, Park HY, Kim JK, Shibata K, Yokota T, Ohyama K, Muranaka T, et al. Identification of marneral synthase, which is critical for growth and development in Arabidopsis. Plant Journal. 2012;72:791–804. doi: 10.1111/j.1365-313X.2012.05120.x. [DOI] [PubMed] [Google Scholar]
- Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biology. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH. Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant and Cell Physiology. 2010;51:1694–1706. doi: 10.1093/pcp/pcq128. [DOI] [PubMed] [Google Scholar]
- Hurst LD, Pal C, Lercher MJ. The evolutionary dynamics of eukaryotic gene order. Nature Reviews Genetics. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science. 2013;341:175–179. doi: 10.1126/science.1240230. [DOI] [PubMed] [Google Scholar]
- King AJ, Brown GD, Gilday AD, Larson TR, Graham IA. Production of bioactive diterpenoids in the Euphorbiaceae depends on evolutionarily conserved gene clusters. Plant Cell. 2014;26:3286–3298. doi: 10.1105/tpc.114.129668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokida A, Delis C, Geisler K, Garagounis C, Tsikou D, Pena-Rodriguez LM, Katsarou D, Field B, Osbourn AE, Papadopoulou KK. A metabolic gene cluster in Lotus japonicus discloses novel enzyme functions and products in triterpene biosynthesis. New Phytologist. 2013;200:675–690. doi: 10.1111/nph.12414. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell. 2010;140:136–147. doi: 10.1016/j.cell.2009.11.006. [DOI] [PubMed] [Google Scholar]
- March-Diaz R, Garcia-Dominguez M, Lozano-Juste J, Leon J, Florencio FJ, Reyes JC. Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant Journal. 2008;53:475–487. doi: 10.1111/j.1365-313X.2007.03361.x. [DOI] [PubMed] [Google Scholar]
- Martin-Trillo M, Larazo A, Poethig RS, Gomez-Mena C, Pineiro MA, Martinez-Zapater JM, Jarillo JA. EARLY IN SHORT DAYS 1ESD1) encodes ACTIN-RELATED PROTEIN 6 (AtARP6), a putative component of chromatin remodelling complexes that positively regulates FLC accumulation in Arabidopsis. Development. 2006;133:1241–1252. doi: 10.1242/dev.02301. [DOI] [PubMed] [Google Scholar]
- Matsuba Y, Nguyen TT, Wiegert K, Falara V, Gonzales-Vigil E, Leong B, Schafer P, Kudrna D, Wing RA, Bolger AM, et al. Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell. 2013;25:2022–2036. doi: 10.1105/tpc.113.111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–736. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- Michalak P. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics. 2008;91:243–248. doi: 10.1016/j.ygeno.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Matsumoto T, Okada A, Komiyama K, Chujo T, Yoshikawa H, Nojiri H, Yamane H, Okada K. Identification of target genes of the bZIP transcription factor OsTGAP1, whose overexpression causes elicitor-induced hyperaccumulation of diterpenoid phytoalexins in rice cells. PLoS ONE. 2014;9:e105823. doi: 10.1371/journal.pone.0105823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Mylona P, Owatworakit A, Papadopoulou K, Jenner H, Qin B, Findlay K, Hill L, Qi X, Bakht S, Melton R, et al. Sad3 and Sad4 are required for saponin biosynthesis and root development in oat. Plant Cell. 2008;20:201–212. doi: 10.1105/tpc.107.056531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nützmann HW, Osbourn A. Gene clustering in plant specialized metabolism. Current Opinion in Biotechnology. 2014;26:91–99. doi: 10.1016/j.copbio.2013.10.009. [DOI] [PubMed] [Google Scholar]
- Nützmann HW, Reyes-Dominguez Y, Scherlach K, Schroeckh V, Horn F, Gacek A, Schumann J, Hertweck C, Strauss J, Brakhage AA. Bacteria-induced natural product formation in the fungus Aspergillus nidulans requires Saga/Ada-mediated histone acetylation. Proceedings of the National Academy of Sciences, USA. 2011;108:14282–14287. doi: 10.1073/pnas.1103523108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osbourn AE, Field B. Operons. Cellular and Molecular Life Sciences. 2009;66:3755–3775. doi: 10.1007/s00018-009-0114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petter M, Selvarajah SA, Lee CC, Chin WH, Gupta AP, Bozdech Z, Brown GV, Duffy MF. H2A.Z and H2B.Z double-variant nucleosomes define intergenic regions and dynamically occupy var gene promoters in the malaria parasite Plasmodium falciparum. Molecular Microbiology. 2013;87:1167–1182. doi: 10.1111/mmi.12154. [DOI] [PubMed] [Google Scholar]
- Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A. A gene cluster for secondary metabolism in oat: implications for the evolution of metabolic diversity in plants. Proceedings of the National Academy of Sciences, USA. 2004;101:8233–8238. doi: 10.1073/pnas.0401301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Dominguez Y, Bok JW, Berger H, Shwab EK, Basheer A, Gallmetzer A, Scazzocchio C, Keller N, Strauss J. Heterochromatic marks are associated with the repression of secondary metabolism clusters in Aspergillus nidulans. Molecular Microbiology. 2010;76:1376–1386. doi: 10.1111/j.1365-2958.2010.07051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rider SD, Jr, Hemm MR, Hostetler HA, Li HC, Chapple C, Ogas J. Metabolic profiling of the Arabidopsis pkl mutant reveals selective derepression of embryonic traits. Planta. 2004;219:489–499. doi: 10.1007/s00425-004-1254-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH. Seed and molecular resources for Arabidopsis. Plant Physiology. 2000;124:1477–1480. doi: 10.1104/pp.124.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura K, Okada A, Okada K, Jikumaru Y, Ko KW, Toyomasu T, Sassa T, Hasegawa M, Kodama O, Shibuya N, et al. Identification of a biosynthetic gene cluster in rice for momilactones. Journal of Biological Chemistry. 2007;282:34013–34018. doi: 10.1074/jbc.M703344200. [DOI] [PubMed] [Google Scholar]
- Song J, Rutjens B, Dean C. Detecting histone modifications in plants. In: Spillane C, McKeown PC, editors. Plant epigenetics and epigenomics. New York, NY, USA: Humana Press; 2014. pp. 165–175. [DOI] [PubMed] [Google Scholar]
- Takos AM, Knudsen C, Lai D, Kannangara R, Mikkelsen L, Motawia MS, Olsen CE, Sato S, Tabata S, Jorgensen K, et al. Genomic clustering of cyanogenic glucoside biosynthetic genes aids their identification in Lotus japonicus and suggests the repeated evolution of this chemical defence pathway. Plant Journal. 2011;68:273–286. doi: 10.1111/j.1365-313X.2011.04685.x. [DOI] [PubMed] [Google Scholar]
- Walley JW, Rowe HC, Xiao Y, Chehab EW, Kliebenstein DJ, Wagner D, Dehesh K. The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathogens. 2008;4:e1000237. doi: 10.1371/journal.ppat.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegel E, Koumproglou R, Shaw P, Osbourn A. Cell type-specific chromatin decondensation of a metabolic gene cluster in oats. Plant Cell. 2009;21:3926–3936. doi: 10.1105/tpc.109.072124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle CM, McClinic KN, Ercan S, Zhang X, Green RD, Kelly WG, Lieb JD. The genomic distribution and function of histone variant HTZ-1 during C. elegans embryogenesis. PLoS Genetics. 2008;4:e1000187. doi: 10.1371/journal.pgen.1000187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman PR, Xu M, Jin Y, Coates RM, Peters RJ. Identification of syn-pimara-7,15-diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiology. 2004;135:2098–2105. doi: 10.1104/pp.104.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, et al. A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science. 2012;336:1704–1708. doi: 10.1126/science.1220757. [DOI] [PubMed] [Google Scholar]
- Wong S, Wolfe KH. Birth of a metabolic gene cluster in yeast by adaptive gene relocation. Nature Genetics. 2005;37:777–782. doi: 10.1038/ng1584. [DOI] [PubMed] [Google Scholar]
- Zhang X, Clarenz O, Cokus S, Bernatavichute YV, Pellegrini M, Goodrich J, Jacobsen SE. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biology. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman D, Coleman-Derr D, Ballinger T, Henikoff S. Histone H2A.Z and DNA methylation are mutually antagonistic chromatin marks. Nature. 2008;456:125–129. doi: 10.1038/nature07324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1Relative transcript level of the thalianol and marneral cluster genes in RNA preparations from leaf and root tissue of the Col-0 wild-type.
Fig. S2 GC-MS analysis of Col-0 and arp6 root extracts.
Fig. S3 Relative transcript levels of the thalianol and marneral cluster genes in Ler wild-type and corresponding arp6 mutant line.
Fig. S4 Chromatin immunoprecipitation (ChIP) analysis of H2A.Z abundance at the marneral gene cluster and control loci.
Table S1Arabidopsis thaliana mutant lines
Table S2 Oligonucleotides used in this study