Abstract
Plants respond to herbivory by mounting a defense. Some plant-eating spider mites (Tetranychus spp.) have adapted to plant defenses to maintain a high reproductive performance. From natural populations we selected three spider mite strains from two species, Tetranychus urticae and Tetranychus evansi, that can suppress plant defenses, using a fourth defense-inducing strain as a benchmark, to assess to which extent these strains suppress defenses differently.
We characterized timing and magnitude of phytohormone accumulation and defense-gene expression, and determined if mites that cannot suppress defenses benefit from sharing a leaf with suppressors.
The nonsuppressor strain induced a mixture of jasmonate- (JA) and salicylate (SA)-dependent defenses. Induced defense genes separated into three groups: ‘early’ (expression peak at 1 d postinfestation (dpi)); ‘intermediate’ (4 dpi); and ‘late’, whose expression increased until the leaf died. The T. evansi strains suppressed genes from all three groups, but the T. urticae strain only suppressed the late ones. Suppression occurred downstream of JA and SA accumulation, independently of the JA–SA antagonism, and was powerful enough to boost the reproductive performance of nonsuppressors up to 45%.
Our results show that suppressing defenses not only brings benefits but, within herbivore communities, can also generate a considerable ecological cost when promoting the population growth of a competitor.
Keywords: defense suppression, herbivore communities, hormonal crosstalk, jasmonic acid (JA), salicylic acid (SA), Solanum lycopersicum (tomato), Tetranychus spp. (spider mite)
Introduction
Higher plants possess sophisticated means to prevent or hamper herbivore feeding (Walling, 2000; Wu & Baldwin, 2010). Such defenses can be constitutive and/or induced upon attack by herbivores. In general, induced defenses may include morphological reinforcements as well as the accumulation of toxins and inhibitors of herbivore digestion (Kessler & Baldwin, 2002), but may also involve hypersensitive responses (Klingler et al., 2009) and resource allocation (Gomez et al., 2012). The first critical step to mount antiherbivore defenses is the perception of herbivory, but how this takes place and whether receptors are involved is not well understood (Bonaventure et al., 2011). It is clear that some characteristics of the response can be attributed to mechanical feeding damage (Mithöfer et al., 2005) but others can only be attributed to herbivory-derived signals referred to as elicitors (Howe & Jander, 2008). Most of these emanate from herbivore saliva or regurgitant and, when applied as pure compounds, elicit defined herbivory-induced changes, such as phytohormone accumulation, transcription of defense genes, and emission of volatiles (Wu & Baldwin, 2010).
The central regulators of plant defense responses are a set of phytohormones that mediate between signal recognition and activation of defenses. Although most of the known plant hormones have been found to influence the establishment of defenses in one way or another (Pieterse et al., 2012), there are three, jasmonic acid (JA), salicylic acid (SA) and ethylene (Et), which play primary roles, as interference with their biosynthesis or perception results in strong defense deficiencies (Wu & Baldwin, 2010). While JA, SA and Et have distinct effects on the type of defenses a plant displays, they also modulate each other's individual actions, that is, ‘crosstalk’ (Pieterse et al., 2009), in a nonlinear way (Mur et al., 2006). While SA is essential for defense against biotrophic pathogens (Vlot et al., 2009), JA and, in particular, its amino acid conjugate JA-isoleucine (JA-Ile) are essential for defenses against herbivores (Howe & Jander, 2008) and necrotrophic pathogens (Glazebrook, 2005), whereas Et most probably modulates these two (Diezel et al., 2009). Defense responses induced by stylet-feeding herbivores appear to involve a cocktail of JA and SA responses (Kaloshian & Walling, 2005). In tomato (Solanum lycopersicum) JA accumulation is upstream of the expression of several defense genes, commonly used as markers for JA defenses, such as Wound-Induced Proteinase Inhibitor I (WIPI-I) and II (WIPI-II) (Farmer et al., 1992), Threonine Deaminase 2 (TD-2) (Gonzales-Vigil et al., 2011) and the activities of defensive enzymes such as polyphenol oxidases (PPOs) and peroxidases (Felton et al., 1989). SA defenses, in turn, are marked by the expression of pathogenesis-related (PR) proteins (Van Loon & Van Strien, 1999) and, in many different plant species, by the accumulation of reactive oxygen species (ROS), sometimes followed by apoptosis (Walling, 2000). Collectively, these are referred to as direct defenses. In addition, JA regulates the biosynthesis and release of an induced blend of volatiles, in part depending on SA (Ament et al., 2004, 2010), which can attract foraging natural enemies of herbivores and is therefore referred to as indirect plant defense (Kant et al., 2009).
The guild of stylet-feeding arthropods can be divided into two subguilds, those that feed predominantly on vascular sap, usually phloem sap, and those that feed from cytoplasm only (Miles, 1972). The latter applies to spider mites (Tetranychus ssp.): the adults use stylets of c. 150 μm long for lacerate-and-flush feeding on mesophyll cells, predominantly parenchyma, of which they can empty up to 18–22 min−1 (Jeppson et al., 1975), leading to c. 1 mm2 of visible chlorotic leaf surface area per adult mite d–1 on tomato (Kant et al., 2004). The two-spotted spider mite T. urticae is highly polyphagous and has been recorded to feed from over 1100 plant species, among them tomato (Dermauw et al., 2012). This mite species is endemic to Europe. The red spider mite, Tetranychus evansi, is a specialist on Solanaceae in Brazil and Africa and a recent invasive pest in Europe (Boubou et al., 2012), where it has extended its host range and has displaced T. urticae on several host plant species in southern Europe (Ferragut et al., 2013). Adult females of both species produce, on tomato, between five and 15 eggs d−1 which will develop into fertile adults within c. 2 wk, resulting in exponential population growth and, subsequently, host–plant overexploitation (Sarmento et al., 2011a). Spider mites produce silk webbing across the host-plant surface which shields them and their eggs from natural enemies. However, while biological control of T. urticae is well feasible, that of T. evansi is troublesome, as the webbing it produces is extraordinarily dense while, in addition, many biological control agents have a poor reproductive performance when preying on it (Sarmento et al., 2011b; Navajas et al., 2013).
When feeding on tomato leaves, most genotypes of T. urticae simultaneously induce expression of the JA- and SA-dependent marker genes WIPI-II and PR-P6, respectively (Li et al., 2002; Ament et al., 2004; Kant et al., 2004). However, some genotypes of T. urticae and T. evansi were found to suppress expression of these marker genes (Kant et al., 2008; Sarmento et al., 2011a). The use of the JA-perception mutant jasmonic acid-insensitive-1 (jai-1; Li et al., 2004) and of the biosynthesis mutant defenseless-1 (def-1; Li et al., 2002; Ament et al., 2004; Kant et al., 2008) has demonstrated that spider mites reach their maximal reproductive performance in the absence of JA signaling, while on 35S::Prosystemin tomato, which is primed to display exceptionally strong JA defenses (Chen et al., 2006; Kandoth et al., 2007), reproductive performance is minimal. Although this strongly suggests that JA defenses are key anti-mite defenses for tomato, it appears that some spider mites have acquired resistance to them (Kant et al., 2008). However, such direct resistance against JA defenses was absent in the defense-suppressing T. urticae genotype (Kant et al., 2008). Taken together, these data suggest that the traits that enable some mite genotypes to suppress plant defenses are not likely to be allelic variants of the same traits that enable other mite genotypes to resist the same defenses.
Suppression of plant defenses is a phenomenon that is especially well known from plant pathogens (Abramovitch et al., 2006; Kamoun, 2006), but also herbivores, such as nematodes (Haegeman et al., 2012) and insects (Musser et al., 2002, 2005; Will et al., 2007; Zarate et al., 2007; Weech et al., 2008; Zhang et al., 2009, 2011; Bos et al., 2010; Consales et al., 2012; Stuart et al., 2012; Wu et al., 2012), were found to manipulate plant defenses. Spider mites and insects do not share a recent history: the Chelicerates (among which the mites evolved) and the Uniramians (among which the insects evolved) diverged early in the arthropod lineage, probably well over 400 million yr ago, from an aquatic ancestor (Weygoldt, 1998), suggesting that traits that allow some of the current plant-eating insect and mite species to suppress host defenses may have evolved independently. Hence we reasoned that the distinct intraspecific and heterospecific variation among Tetranychid mites (Matsushima et al., 2006; Kant et al., 2008; Sarmento et al., 2011a) forms an ideal basis for assessing some of the ecological costs and benefits of defense suppression within herbivore communities and for determining which processes are targeted by suppression. Therefore, we selected several putative defense-suppressing spider mites from natural populations, determined how tomato plants responded to them, and to what extent these responses modulate the mite's interactions with its natural defense-inducing competitors within two species communities.
Materials and Methods
Plants
Tomato (Solanum lycopersicum L. cv Castlemart, 35S::prosystemin and def-1) and bean (Phaseolus vulgaris L. cv Speedy) were germinated and grown in a glasshouse (16 : 8 h, 25 : 18°C, day : night, 50–60% relative humidity (RH)). Experiments involving plants were carried out in a climate room (25°C, 16 : 8 h, light : dark, 60% RH, 300 μmol m−2 s−1), to which plants were transferred 3 d in advance.
Spider mites
Tetranychus evansi Baker & Pritchard Viçosa-1 (Supporting Information, Notes S1; Fig. S1a; Sarmento et al., 2011a), T. evansi Algarrobo-1 (Fig. S1b; this paper), T. urticae Koch DeLier-1 (Fig. S1c; this paper, see the section ‘Selection of T. urticae DeLier-1’) and T. urticae Santpoort-2 (Fig. S1d; ‘KMB’ in Kant et al., 2008) were reared on detached leaves of S. lycopersicum cv Castlemart (for T. evansi) or P. vulgaris cv Speedy (for T. urticae) in a climate room (25°C 16 h : 8 h, light : dark, 60% RH, 300 μmol m−2 s−1). The species identity of all four strains was confirmed on the basis of a phylogenetic reconstruction using their mitochondrial cytochrome oxidase subunit 1 (COI) sequences (Fig. S2). For all infestation experiments and performance assays, we used adult female mites (3 ± 1 d old).
Selection of T. urticae DeLier-1
Adult T. urticae females were collected from three natural populations in the Netherlands in 2009: 125 individuals from spindle tree (Euonymus europaeus L.), 64 from deadnettle (Lamium album L.) and 50 from castor oil (Ricinus communis L.) plants. Mites were individually transferred to def-1 leaves. Their virgin female offspring (F1) were separated again and allowed to produce eggs on def-1. Mothers with a high reproductive performance (≥ 20 eggs per 4 d) were backcrossed with their sons for two more generations to F3 (hereafter referred to as ‘strains’). The fecundity of adult females of all strains was subsequently assessed on def-1, wild-type (WT) and 35S::Prosystemin tomato plants to identify JA defense-suppressing mites (Kant et al., 2008). This yielded one putative suppressor strain from the spindle tree population; three from the deadnettle population and one from the castor oil population (Fig. S3a). After comparing the expression levels of Proteinase Inhibitor IIf (PI-IIf) induced by these strains on tomato with those induced by the benchmark inducer strain T. urticae Santpoort-2 and in uninfested controls, we selected the strain that gave the smallest increase in PI-IIf transcript abundances for further experiments; this was T. urticae DeLier-1 (Fig. S3b).
Performance assays for individual spider mite strains
To establish whether our spider mite strains are affected by JA-mediated defense responses, we assessed their performance on WT and def-1 tomato plants. Adult females were transferred to 21-d-old tomato plants (Methods S1): five mites per leaflet; three leaflets per plant; six plants per treatment. After 4 d, the number of eggs was recorded using a stereo microscope. This experiment was repeated three times. The total number of eggs per female were analyzed for each tomato genotype, and statistically analyzed using the Student's t-test (PASW Statistics 17.0; SPSS Inc., Chicago, IL, USA).
Performance assay for two spider mite strains sharing a leaflet (coinfestation)
To assess the extent to which one strain can influence the reproductive performance of another strain, we followed the setup used in Kant et al. (2008). Leaflets of 21-d-old intact tomato plants were divided into two using a lanolin barrier. Five T. urticae Santpoort-2 females were transferred to the tip-half of the leaflet, whereas the petiolule-half was infested with 15 mites from one of the suppressor strains (five + 15 mites per leaflet; three leaflets per plant; six plants per treatment). After 4 d, the number of eggs laid by the five T. urticae Santpoort-2 females at the tip was recorded. This experiment was repeated three times. The average number of eggs per female 4 d−1 was analyzed using ANOVA and the means of each group were compared by least significant difference (LSD) post hoc test using PASW Statistics 17.0.
Phytohormone and gene expression assay on leaflets infested with 15 mites (time course)
Leaflets of 21-d-old tomato plants were infested with adult female spider mites: 15 mites per leaflet; three leaflets per plant; 12 plants per treatment. At 1, 4 and 7 d postinfestation (dpi); four infested plants from each treatment and four control plants were sampled: infested leaflets and corresponding noninfested leaflets of control plants were excised (without petiolule), flash-frozen in liquid nitrogen and stored at −80°C until we extracted phytohormones and mRNA. The three leaflets obtained from the same plant were pooled to form one biological replicate. Under these standard experimental conditions (Kant et al., 2004), leaflets with T. urticae Santpoort-2 enter senescence 8–9 dpi and die c. 11–12 dpi (J. M. Alba & B. C. J. Schimmel pers. obs.).
Phytohormone and gene expression assay on leaflets simultaneously infested with mites from two different strains (coinfestation)
Leaflets of 21-d-old tomato plants were infested with adult female spider mites: five to 30 mites per leaflet; three leaflets per plant; six to 10 plants per treatment. At 7 dpi, leaflets were harvested and stored as described earlier. The three leaflets obtained from the same plant were pooled. Two types of coinfestation experiments were carried out with different infestation regimes, using T. urticae Santpoort-2 (TuSP-2), T. evansi Viçosa-1 (TeV-1), and/or T. urticae DeLier-1 (TuDL-1). The first experiment consisted of six treatments, in which leaflets were infested with: no mites (control); 15 TuSP-2; 15 TeV-1; 30 TuSP-2; 30 TeV-1; or 15 TuSP-2 + 15 TeV-1 (coinfestation). Ten plants were used per treatment. The second experiment consisted of eight treatments: no mites; five TuSP-2; 15 TuSP-2; 15 TuDL-1; 25 TuDL-1; 15 TuSP-2 + 15 TuDL-1; five TuSP-2 + 15 TuDL-1; and five TuSP-2 + 25 TuDL-1. Six plants were used per treatment. This experiment was repeated twice.
Isolation of phytohormones and analysis by means of LC-MS/MS
Phytohormone analysis was performed using the procedure of Wu et al. (2007) with some minor modifications (Methods S2; Table S1). Amounts were compared across treatments per time point independently using ANOVA with ‘spider mite strain’ as factor. Means of each group were compared by LSD post hoc test using PASW Statistics 17.0.
Gene expression analysis
Total RNA was isolated as described in Verdonk et al. (2003). Two micrograms of DNAse-treated RNA was used for cDNA synthesis and 1 μl of 10-times-diluted cDNA served as a template for a 20 μl quantitative reverse-transcriptase polymerase chain reaction (qRT-PCR) using the Platinum SYBR Green qPCR-SuperMix-UDG kit (Invitrogen) and the ABI 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA). To survey tomato defenses, we analyzed expression of the following genes: PPO-D,PPO-F,JIP-21,GAME-1,TD-2,THM27,LX,PR-1a,PR-P6,PI-IIc and PI-IIf. Actin was used as a reference gene. PCR-generated amplicons were sequenced to verify primer specificity. Gene identifiers, primer sequences and references are listed in Table S2. The normalized expression (NE) data were calculated by the ΔCt method 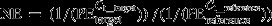 ; in which PE is the primer efficiency and Ct is the cycle threshold. The NE of each target gene was compared per time point independently using a nested ANOVA with ‘spider mite strain’ as factor and ‘technical replicate’ (i.e. two for each reaction) nested into the corresponding biological replicate (cDNA sample). Means of each group were compared by Fisher's LSD post hoc test using PASW Statistics 17.0. To plot the relative expression, NE values were scaled to the treatment with the lowest average NE.
; in which PE is the primer efficiency and Ct is the cycle threshold. The NE of each target gene was compared per time point independently using a nested ANOVA with ‘spider mite strain’ as factor and ‘technical replicate’ (i.e. two for each reaction) nested into the corresponding biological replicate (cDNA sample). Means of each group were compared by Fisher's LSD post hoc test using PASW Statistics 17.0. To plot the relative expression, NE values were scaled to the treatment with the lowest average NE.
Results
Selection of putative suppressor genotypes from natural populations
To identify and isolate putative JA defense-suppressing T. urticae, adult female spider mites were collected from natural populations found on three different host plants. We reasoned that the fecundity of JA-suppressor strains should be equally high on tomato (S. lycopersicum) WT and on the JA-biosynthesis mutant tomato def-1, as suppression will only be favored by natural selection when improving the reproductive performance of mites. Hence we tested the reproductive performance of each strain on these plants. Tetranychus urticae Santpoort-2 mites produced 34 ± 3 eggs on def-1, but only 22 ± 1 on WT (Fig. S4; Student's t-test: P = 0.003), confirming its susceptibility to JA-mediated defenses (Kant et al., 2008). By contrast, mites from the putative suppressor strains T. urticae DeLier-1, T. evansi Viçosa-1, and Algarrobo-1 produced a similar number of eggs on both genotypes of plant (Student's t-test, P > 0.05).
The reproductive performance of defense-susceptible T. urticae Santpoort-2 mites increases when sharing a leaflet with the putative suppressor strains
Using the performance test on def-1 and WT plants, we could not exclude the possibility that a high reproductive performance on WT plants is the result of direct resistance to induced tomato JA defenses. Hence we reasoned that only a genuine defense suppressor should be able to boost the reproductive performance of a defense-susceptible mite when both reside on the same leaflet. Indeed, all three strains clearly and significantly boosted the performance of T. urticae Santpoort-2 (Fig.1). Compared with the control, where T. urticae Santpoort-2 ‘shared a leaflet with itself’, T. urticae DeLier-1 and T. evansi Algarrobo-1 improved the susceptible strains’ fecundity with > 25%, while T. evansi Viçosa-1 did so with over 45%.
Fig 1.
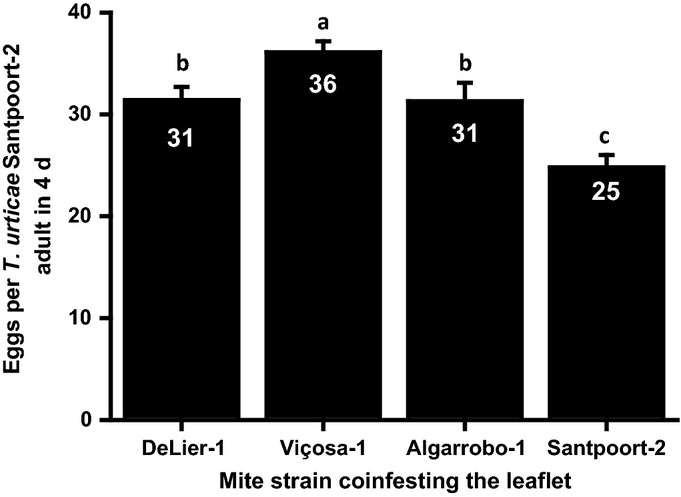
The reproductive performance of the jasmonic acid (JA)-defense-inducing and -susceptible Tetranychus urticae Santpoort-2 increases on tomato (Solanum lycopersicum) leaflets shared with spider mites from suppressor strains. The figure shows the average (+ SEM) number of eggs produced by adult female mites of strain T. urticae Santpoort-2 per 4 d on leaflets simultaneously coinfested with 15 adult females of T. urticae DeLier-1, T. evansi Viçosa-1, or T. evansi Algarrobo-1, or with T. urticae Santpoort-2 as a control. Numbers within the bars indicate the average egg production. Bars annotated with different letters were significantly different according to Fisher's least significant difference (LSD) test (P ≤ 0.05) after ANOVA.
The T. evansi strains suppress expression of tomato genes that mark JA, SA and senescence, but the suppressor T. urticae strain only suppresses that of genes induced late in the interaction
In order to narrow down the mechanisms that underlie the positive effect of putative suppressor strains on the fecundity of the susceptible strain, we assessed the magnitude and timing of defense-related phytohormone and transcript accumulation in response to each of the strains.
In general, T. urticae Santpoort-2 induced a significant increase of the oxylipins 12-oxo-phytodienoic acid (OPDA, the JA-precursor), JA, and JA-Ile at 7 dpi, with JA and JA-Ile already being significantly higher than uninfested controls at 4 dpi (Fig.2a–c). The accumulation of free SA upon infestation with T. urticae Santpoort-2 mites was even more rapid, that is, significantly higher than controls after 1 d, and appeared continuous (Fig.2d). Notably, the basal SA concentration in control plants increased as they grew older (Fig.2d: F2,8 = 6.010, P = 0.025). Phytohormone accumulation induced by suppressor T. urticae DeLier-1 followed the same temporal pattern, albeit consistently at c. two to five times lower levels (Fig.2). Whereas concentrations of the defense-related phytohormones clearly peaked at 7 dpi with either of the two T. urticae strains, only minor, nonsignificant, increases were observed for any of these hormones after prolonged infestation with the T. evansi strains (Fig.2), even though they induced SA to concentrations similar to those induced by T. urticae DeLier-1 at 1 dpi.
Fig 2.
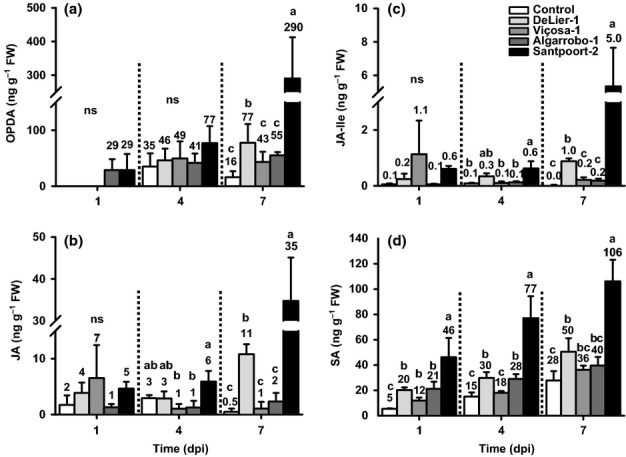
The amounts of 12-oxo-phytodienoic acid (OPDA), jasmonic acid (JA), jasmonic acid-isoleucine (JA-Ile), and free salicylic acid (SA) accumulated in spider mite-infested tomato (Solanum lycopersicum) leaflets during the course of 7 d. The figure shows the average (+ SEM) amounts of OPDA (a), JA (b), JA-Ile (c), and free SA (d) at 1, 4, and 7 d postinfestation (dpi) of tomato leaflets with 15 adult females from strain Tetranychus urticae DeLier-1, T. evansi Viçosa-1, T. evansi Algarrobo-1, or T. urticae Santpoort-2. Uninfested leaflets were used as controls. OPDA was not detected at 1 dpi in control, T. urticae DeLier-1, and T. evansi Viçosa-1 samples. Bars annotated with different letters were significantly different according to Fisher's least significant difference (LSD) test (P ≤ 0.05) after ANOVA. Bars marked with ‘ns’ did not test differently in the ANOVA. Data were log-transformed before the statistical analysis.
We then selected 10 genes related to plant defenses for a detailed time-course expression analysis via qRT-PCR using the same samples. We selected Polyphenol-oxidase-D (PPO-D) and PPO-F (Newman et al., 1993; Thipyapong et al., 2004), Glycoalkaloid metabolism-1 (GAME-1) (Itkin et al., 2011), the AtMYB4/PhMYB4 homolog THM27 (Mintz-Oron et al., 2008), the Cathepsin-D-inhibitor/chymotrypsin inhibitor encoding gene Jasmonate-inducible Protein-21 (JIP-21) (Lisón et al., 2006), Threonine Deaminase-2 (TD-2) (Gonzales-Vigil et al., 2011), the senescence-associated T2-like RNAse ribonuclease LX (LX) (Lers et al., 2006), Pathogenesis-related protein 1a (PR-1a) (Tornero et al., 1997), PR-P6 (Van Kan et al., 1992), and Proteinase Inhibitor IIc (PI-IIc) (Gadea et al., 1996)
Using T. urticae Santpoort-2 as a benchmark, the expression pattern of the selected genes clustered into three groups (Fig.3, black bars): those with the highest transcript abundance at 1 dpi (referred to as ‘early’; PPO-D and PPO-F), 4 dpi (‘intermediate’; GAME-1;THM27;JIP-21 and TD-2), or 7 dpi (‘late’; LX;PI-IIc;PR-1a and PR-P6). Except for GAME-1,T. urticae Santpoort-2 mites induced expression of all nine other genes in the tomato leaflets.
Fig 3.
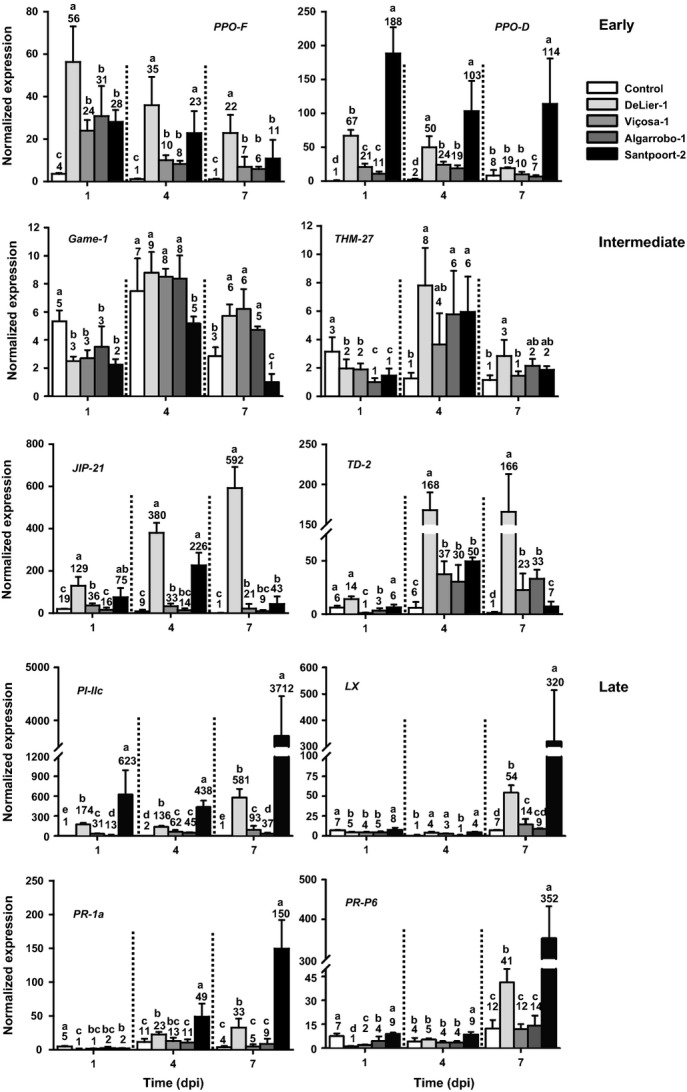
Relative transcript abundances of various defense-related genes in spider mite-infested tomato (Solanum lycopersicum) leaflets during the course of 7 d. Based on the Tetranychus urticae Santpoort-2 samples, the genes separated into three groups: those whose transcript abundances were highest at 1 d postinfestation (dpi) were annotated as ‘early’; those with a peak at 4 dpi as ‘intermediate’; and those with a peak at 7 dpi as ‘late’. Compared with T. urticae Santpoort-2, both Tetranychus evansi Viçosa-1 and T. evansi Algarrobo-1 mites suppressed genes from all three groups, while T. urticae DeLier-1 mites only moderately suppressed the ‘late’ defense genes. Uninfested leaflets were used as controls. The bars represent the means (+ SEM) of the normalized transcript abundances scaled to the lowest mean value per 7 d gene panel. Transcript abundances were normalized to actin. Numbers above the bars indicate the mean value represented by the bar. Expression data were statistically evaluated per day and bars annotated with different letters were significantly different according to Fisher's LSD test (P ≤ 0.05) after ANOVA. Gene identifiers and corresponding references can be found in Table S2. GAME-1,Glycoalkaloid Metabolism-1;JIP-21,Jasmonate-inducible protein-21;LX,RNase Lycopersicon extravacuolar;PI-IIc,Proteinase Inhibitor IIc;PPO-D,Polyphenol-oxidase-D;PPO-F,Polyphenol-oxidase-F;PR-1a,Pathogenesis-related protein 1a;PR-P6,Pathogenesis-related protein 6;TD-2,Threonine Deaminase-2;THM27,Tomato Hypocotyl Myb 17.
Rapid increased expression of the ‘early’ genes was evident after infestation with suppressor T. urticae DeLier-1 (Fig.3). Expression levels of PPO-F were even higher in the DeLier-1 samples than in the Santpoort-2 ones. This was also observed for the ‘intermediate’ genes JIP-21 and TD-2. Moreover, contrary to Santpoort-2-infested leaflets, transcript abundances of all ‘intermediate’ genes in DeLier-1-infested leaflets remained above control values at 7 dpi. The expression patterns of the ‘late’ genes resulting from DeLier-1 and Santpoort-2 feeding, respectively, were similar, but in general DeLier-1 mites induced lower transcript abundances.
The two T. evansi suppressor strains both significantly induced the ‘early’ defense marker genes (Fig.3). Timing and magnitude of suppression and subsequent induction of GAME-1 and THM-27 by T. evansi were similar to that of the T. urticae DeLier-1 strain, but the levels of induction differed considerably for JIP-21 and TD-2, as the T. evansi strains induced both genes only slightly after 4 and 7 d. When compared with levels induced by T. urticae Santpoort-2 at 7 dpi, T. evansi inhibited JIP-21, but induced TD-2. Of the ‘late’ genes, analogous to T. urticae DeLier-1, only PI-IIc (a JA marker gene; Fig. S5; Notes S2) was induced at 1 dpi, while the three other genes were suppressed. At later time points, transcript abundances of PI-IIc remained elevated, albeit to a far lower extent than with the T. urticae strains, and those of LX (senescence), PR-1a, and PR-P6 (both SA markers) returned to control values, or slightly higher, that is, for LX after infestation with Viçosa-1 (Fig.3).
Based on these phytohormone and gene expression studies, we conclude that each mite strain triggers a unique defense response pattern in tomato leaflets, but that, at a particular time and compared with inducer T. urticae Santpoort-2, the two T. evansi strains suppressed ‘early’, ‘intermediate’, and ‘late’ genes, while the T. urticae suppressor strain DeLier-1 only suppressed the ‘late’ defense genes.
Induction and suppression of defenses do not correlate with feeding intensity
We explored the mites’ feeding intensities as an alternative explanation for differences in the magnitude of defense induction. To do so, we assessed the total area of chlorotic lesions as a result of mite feeding (Kant et al., 2004). Notably, on leaflets infested with either of the suppressor mites, this typical white-on-green feeding damage pattern persisted at least until 7 dpi (Fig. S6a), while on leaves with T. urticae Santpoort-2 the lesions got increasingly surrounded by small areas of white-yellowish senescence and sometimes small oedema and russeting (Fig. S6b). To only assess feeding damage, we visually excluded these senesced areas as much as possible. The two T. evansi strains produced c. 100 mm2 of feeding damage (Fig. S6c), corresponding to c. 9% of the total leaflet area. The two T. urticae mite strains produced a total feeding damage of c. 40 mm2, corresponding to c. 3.5% of the leaflet area. The T. evansi strains thus inflicted at least twice as much feeding damage as the T. urticae strains. When including the senesced areas, damage inflicted by T. urticae Santpoort-2 equaled that of the T. evansi strains (data not shown). Hence, there was no positive relationship between the extent to which defenses were induced and the total leaf area that was damaged.
Tetranychus evansi suppresses the T. urticae-induced expression of JA and SA marker genes but not the upstream accumulation of JA-Ile or SA
As the most clear-cut evidence for defense suppression by spider mites is demonstrated by the increased reproductive performance of the JA defense-susceptible T. urticae Santpoort-2 on coinfested leaflets (Fig.1), and differences in JA and SA defense-related phytohormone (Fig.2) and transcript abundances (Fig.3) between inducer and suppressor strains in ‘single strain-infested’ leaflets were most apparent at 7 dpi, we combined both experiments to determine whether suppressor mites still manage to inhibit these defense signaling pathways when sharing a leaflet with inducer Santpoort-2.
We first selected the most potent suppressor strain, T. evansi Viçosa-1 (Fig.1), and introduced 15 adult females to a leaflet to which, simultaneously, 15 adult T. urticae Santpoort-2 mites were also introduced. Seven days later we compared the concentrations of JA-Ile and SA plus expression levels of PI-IIc and PR-1a from these coinfested leaflets with those of uninfested leaflets (negative control), as well as with leaflets with only 15 adult T. urticae Santpoort-2 mites or only 15 T. evansi Viçosa-1 mites (positive controls). Finally, infestations with only 30 adult T. urticae Santpoort-2 mites or only 30 T. evansi Viçosa-1 were included to control for density-dependent effects.
In line with the previous results, infestation with 15 T. urticae Santpoort-2 mites resulted in strongly induced JA and SA defenses (Fig.4a,b). The plant's defense responses to Santpoort-2 mites increased in a density-dependent manner (Fig. S7). The 15 T. evansi Viçosa-1 mites caused minor increases in JA-Ile and SA concentrations, but higher densities of T. evansi Viçosa-1 did not further elevate hormone concentrations or PI-IIc expression, while even lowering that of PR-1a (Fig. S7).
Fig 4.
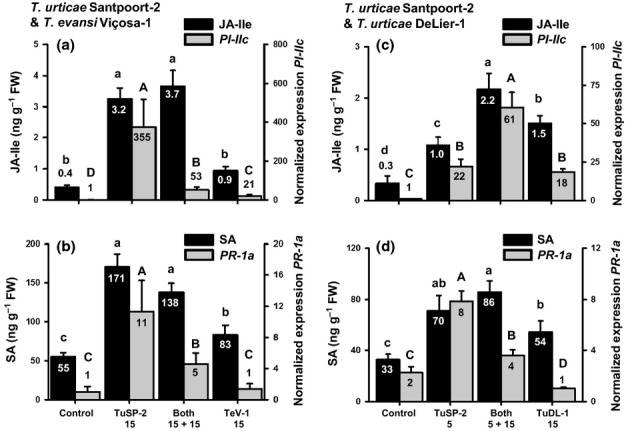
The amounts of jasmonic acid-isoleucine (JA-Ile) and salicylic acid (SA), along with transcript abundances of Proteinase Inhibitor IIc (PI-IIc) and Pathogenesis-related 1a (PR-1a) in tomato (Solanum lycopersicum) leaflets after 7 d of infestation with inducer Tetranychus urticae Santpoort-2, suppressor T. evansi Viçosa-1, and suppressor T. urticae DeLier-1, or a combination of inducer and either of the suppressors. The figure shows the amounts of JA-Ile and PI-IIc transcript (a, c) and the amounts of free SA and PR-1a transcript (b, d). Leaflets were infested with T. urticae Santpoort-2 (TuSP-2), T. evansi Viçosa-1 (TeV-1), or T. urticae DeLier-1 (TuDL-1), or simultaneously with TuSP-2 and either TeV-1 or TuDL-1 (both). Uninfested leaflets were used as controls. The numbers below the x-axis indicate the number of adult female mites used to infest the leaflets. The bars show the means (+ SEM), which are given as numbers within or above the bars. Transcript abundances were normalized to actin and scaled to the lowest mean per panel. Bars annotated with different letters (lowercase for JA-Ile and SA; uppercase for PI-IIc and PR-1a) were significantly different according to Fisher's least significant difference (LSD) test (P ≤ 0.05) after ANOVA.
In leaflets coinfested with 15 T. urticae Santpoort-2 mites and 15 T. evansi Viçosa-1, concentrations of JA-Ile and SA were equal to those only infested with Santpoort-2 (Fig.4a,b). However, expression levels of PI-IIc and PR-1a were intermediate, that is, significantly lower than in leaflets with 15 T. urticae Santpoort-2, but still higher than in the leaflets with 15 T. evansi Viçosa-1 mites. Taken together, in coinfested leaflets, T. evansi Viçosa-1 does not suppress phytohormone accumulation, but only the expression of the downstream marker genes. Hence, suppression by T. evansi Viçosa-1 probably occurs downstream of phytohormone accumulation.
To test whether defense suppression within T. urticae species operates in the same way, we repeated the coinfestation experiment with T. urticae DeLier-1 as the suppressor. Here we used only five T. urticae Santpoort-2 mites, as the magnitude of suppression by DeLier-1 appeared lower than that of the T. evansi strains (Figs3). The tomato JA-defense response induced by the two T. urticae strains together appeared to be additive (Figs4c,d,S8). By contrast, the SA concentrations of coinfested leaflets equaled those infested only with Santpoort-2 and the PR-1a transcript abundances were suppressed down to intermediate values by DeLier-1 (Fig.4c,d). Using 15 instead of five Santpoort-2 mites, or when using 25 DeLier-1 individuals, we did not observe significant cosuppression of defenses (Fig. S8). This indicates that DeLier-1 is a less potent suppressor of SA defenses than T. evansi Viçosa-1 and, although it induced a significantly lower JA response even at higher densities than Santpoort-2, it is unable to significantly suppress the Santpoort-2-induced JA-defense response.
Discussion
Suppression of plant immunity is especially well known from plant pathogens (Abramovitch et al., 2006; Burgyán & Havelda, 2011; De Jonge et al., 2011) and nematodes (Haegeman et al., 2012). In recent years, some herbivorous insects were also found to suppress plant defenses (Hogenhout & Bos, 2011), but defense suppression by Chelicerates is still poorly documented (Kant et al., 2008; Sarmento et al., 2011a). Hence, we have characterized three JA defense-suppressing spider mite strains for the extent to which they are able to lower tomato defenses and to promote the reproductive performance of a JA defense-sensitive competing mite strain. We showed that T. urticae DeLier-1 is a moderate suppressor of induced defenses that improves the reproductive performance of competing Santpoort-2 mites by 25%. Furthermore, we showed that suppression by the strains T. evansi Viçosa-1 and Algarrobo-1 inhibits JA and SA responses simultaneously and, hence, is not depending on the JA–SA antagonism. Moreover, suppression by T. evansi Viçosa-1 most likely occurs downstream of phytohormone accumulation and is powerful enough to cosuppress the expression of defense genes induced by T. urticae Santpoort-2, thereby boosting the reproductive performance of its competitor by 45%.
Induction of JA defenses by T. urticae Santpoort-2 parallels induction of SA defenses, while, the other way around, suppression of JA defenses by the other three mite strains parallels suppression of SA defenses (Figs3). In fact, in tomato leaflets coinfested with T. urticae Santpoort-2 and T. evansi Viçosa-1 mites, PI-IIc (JA-defense marker) and PR-1a (SA-defense marker) were both suppressed, even though JA-Ile and SA were induced to concentrations found in leaflets exclusively infested with Santpoort-2 (Fig.4a,b). We therefore conclude that defense suppression by these spider mites acts downstream of phytohormones and independent of the JA–SA antagonism. By contrast, T. kanzawai (Ozawa et al., 2011) and some other herbivores (Zarate et al., 2007; Weech et al., 2008; Chung et al., 2013) were suggested to manipulate the JA–SA crosstalk mechanism to suppress JA defenses.
The low concentrations of phytohormones detected in leaflets infested with only suppressor mites thus do not seem to be the cause of suppression of downstream defenses, but rather are a consequence, possibly as a result of altered feedback regulation of hormone biosynthesis (Chini et al., 2007; Paschold et al., 2007 Serrano et al., 2013). The question remains as to why the simultaneous SA and JA responses induced by Santpoort-2 mites do not antagonize each other? One explanation is that these responses might be heterogeneous in space, for example, one may dominate at the feeding site and the other in surrounding tissues. Consequently, by harvesting complete leaflets we mix what in reality is an SA/JA response mosaic. Indeed, in wounded Nicotiana attenuata (Schittko et al., 2000; Wu et al., 2007), Pseudomonas-infected Phaseolus vulgaris (Meier et al., 1993) and elicitor-treated N. tabacum (Dorey et al., 1998) and Zea mays (Engelberth et al., 2012), defense responses were found to be stronger close to the wounded area or infection site. Not all defenses follow this pattern, as PI-I transcript abundances were found to be highest distant to the wound site (Howe et al., 1996). Another explanation might be that simultaneous SA and JA responses actually do antagonize each other and what we observe are intermediate responses, as was also suggested to happen in N. attenuata infested with Manduca sexta (Diezel et al., 2009). Thus, although JA and SA may crosstalk during induction of defenses by mites, their antagonistic interaction is not involved in defense suppression by mites.
Results from coinfestation experiments of Santpoort-2 and DeLier-1 mites suggest that defense suppression by DeLier-1 functionally operates in the same way. The mechanistic evidence, though, is complex, as DeLier-1 triggers a defense response that is clearly distinct from that of the T. evansi strains. The phytohormone accumulation data (Fig.2) and the expression data on the ‘late’ defense genes (Fig.3) suggest that DeLier-1 may delay defenses rather than fully block them. Despite the strong and fast induction of several JA-regulated defense genes, for example, PPO-F,JIP-21, and PI-IIc (Fig.3), suppression of JA-mediated defenses by DeLier-1 was shown to occur within the first 4 d of infestation (Fig. S3a). However, after 7 d of coinfestation with inducer mites, suppression was clear for PR-1a, but not for PI-IIc (Fig.4c,d). Moreover, this suppressive effect on PR-1a was only observed when DeLier-1 outnumbered the inducer mites three to one, which confirms it is a less potent suppressor than Viçosa-1 (Fig.1). Together, this suggests we may be overlooking the (more) relevant defenses and/or that SA defenses play a more important role in the defense response against mites than they do against herbivorous insects.
The defense suppression we observed does not act on the expression of all defense genes in a similar manner and magnitude (Fig.3). Both PPO genes were strongly and rapidly induced by all mite strains, including the suppressors, hence their classification as ‘early’. For PPO-F,T. urticae, DeLier-1 even induced the overall highest transcription. PPOs are believed to act in the guts of herbivores where they may convert plant-derived flavonoids into quinones. These are highly reactive molecules that can make amino acids indigestible, can damage gut enzymes or DNA, and can form reactive oxygen species (Constabel & Barbehenn, 2008).
Two of the ‘intermediate’ response genes, GAME-1 and THM27, are involved in regulation of the secondary metabolism, that is, the alkaloid and flavonoid metabolism, respectively. The same temporal (bell-shaped) expression pattern was observed for both genes in all mite-infested leaflets. GAME-1 is involved in the glycosylation of steroidal alkaloids, in particular aglycon tomatidine, presumably to reduce the autotoxicity of these metabolites (Itkin et al., 2011, 2013). The gene was down-regulated in all leaflets at 1 dpi but remained down-regulated only in T. urticae Santpoort-2-infested leaflets. Tomatidine was found to be toxic to root-knot nematodes and, while most insects can cope with it, the potato aphid suffers from high concentrations (Milner et al., 2011). Hence, whether down-regulation of GAME-1 in T. urticae Santpoort-2 infested leaflets reflects an effective defense response remains to be determined.
THM27 is an R2R3-MYB transcription factor that controls flavonoid metabolism (Adato et al., 2009; Dal Cin et al., 2011) and is homologous to AtMYB4 (Mintz-Oron et al., 2008) and PhMYB4 (Colquhoun et al., 2011). All mite strains down-regulated THM27 at 1 dpi, albeit not all significantly. At 4 dpi, however, it was significantly up-regulated, after which expression levels reduced again.
To put this into perspective, tomato plants might up-regulate the biosynthesis of lignins and flavonoids, including PPO substrates (Constabel & Barbehenn, 2008), especially early in the interaction, but then switch to alternative measures when the infestation progresses. Expression of JIP-21,TD-2, and PI-IIc might be part of such alternative measures. They encode enzymes thought to interfere with the herbivore's digestive processes (Chen et al., 2005; Lisón et al., 2006; Gonzales-Vigil et al., 2011) and were induced at 4 and/or 7 dpi by all mite strains, although in a nonuniform way. For instance, after 7 d of infestation, suppressor mites had induced TD-2 to higher levels than did Santpoort-2, while this pattern was reversed for PI-IIc. Some of the PR genes, which belong to a different class of defense genes (Van Loon & Van Strien, 1999), were sometimes found to be up-regulated upon infestation with DeLier-1, but never by the T. evansi strains.
Using marker genes for drawing accurate conclusions regarding complex processes strongly depends on the correlation between the expression levels of such genes and the associated process. When investigating the correlation between the ‘classical’ tomato JA-marker gene PI-IIf (Notes S2) and JA concentrations, we noticed that, especially at low JA concentrations, the gene was regularly highly expressed (Fig. S5). This suggests that not only JA but also other (hormonal) signals activated by spider mites influence its regulation. However, the correlation between the expression of another family member of this gene (PI-IIc) and JA concentrations was much stronger and hence we used this gene as a marker for JA-related processes induced by spider mites. This underpins the fact that marker genes may require context-specific validation before being used as process indicators.
In summary, each mite strain affects the expression of tomato defense genes differently, but the putative negative effect of each of these genes on the spider mite performance remains largely unknown and is subject to future research. Furthermore, the expression pattern of the senescence-marker LX (Lers et al., 2006) at 7 dpi perfectly reflected the visual development of senescence in the infested leaflets. Possibly as a result of the induced defenses, leaflets infested with Santpoort-2 went into senescence early and in a density-dependent way before they died, while senescence in leaflets infested with DeLier-1 was less severe and came days later and T. evansi-infested leaflets dried out without showing clear signs of senescence before dying.
The mechanism by which spider mites suppress host defenses is still unclear. Some phytopathogens, vectored by arthropods, have been implicated in the suppression of plant defenses, putatively to (indirectly) enhance their own fitness (Belliure et al., 2005; Sugio et al., 2011; Casteel et al., 2012, 2014; Zhang et al., 2012; Chung et al., 2013). Preliminary data, though, indicate that spider mite-associated microbes do not mediate suppression of plant defenses (data not shown). Analogous to phytopathogens (Da Cunha et al., 2007; De Jonge et al., 2011), aphids (Rodriguez & Bos, 2013), and nematodes (Haegeman et al., 2012), spider mites may also secrete effectors via their saliva into plant tissues to interfere with host immune responses (Alba et al., 2011). The spider mite genome (Grbić et al., 2011) encodes at least 293 putative salivary proteins (with an E-value < 1E–20), and thus mites are likely to secrete a rich cocktail of proteins while feeding. Whether (some of) these predicted salivary proteins truly are involved in suppression (and/or induction) of plant defenses – and what their in planta targets are – remains to be demonstrated. Finally, the concurrent suppression of JA and SA defenses hints at manipulation of the redox homeostasis (Koornneef et al., 2008; Gruner et al., 2013), but as inhibition of phytohormone accumulation appears nonessential for suppression of the downstream response, other mechanisms are likely to be at play as well.
Our data suggest that defense-suppression traits are not very rare in natural populations of spider mites, especially not for T. evansi. Judging the effects that suppressors have on tomato, these traits may be diverse across and within species and be intertwined with (unrelated) traits that cause induction of plant defenses (Kant et al., 2008). However, the ecological costs and benefits of defense suppression are still unclear. Rationally, resistance (Kant et al., 2008) seems a ‘safer’ trait than the ability to suppress, as suppression can clearly benefit competing species as well (Fig.1). We found putative suppressor genotypes within all three T. urticae populations we sampled (five putative suppressor strains among the 239 strains tested). This suggests that the trait is either maintained by frequency-dependent selection or results from genetic drift. Given the observation that suppression increases the fitness of these mites in the absence of competitors while – potentially – decreasing it in their presence suggests that competitor-associated fluctuating selection may be a driving force. By contrast, both T. evansi haplotypes suppressed defenses similarly and we did not observe intraspecific variation, suggesting that for this species the suppression trait got to fixation. The natural host range of the T. evansi haplotype from the Brazilian clade (such as T. evansi Viçosa-1; Fig. S2a) appears to be narrower than the ones from the Spanish clade (such as T. evansi Algarrobo-1; Fig. S2a), but both haplotypes are frequently found on several of the same solanaceous species as T. urticae in the same geographical regions (Navajas et al., 2013). Given our observation that T. urticae can increase its reproductive performance up to 45% when sharing a leaflet with T. evansi under laboratory conditions, we would not expect the displacement of natural T. urticae populations by T. evansi as is currently taking place on several host plants in southern Europe (Ferragut et al., 2013). Hence, the key question is how defense-suppressing herbivores manage to prevent or overcome the negative effects such that they themselves receive the largest net benefit from the manipulation? One of the answers may be that T. evansi monopolizes its feeding site by the production of extraordinarily large quantities of silken web, which not only shields the population from acaricides and natural enemies but also makes it hard for competing tetranychid mites to invade (Lemos et al., 2010; Sarmento et al., 2011b). Although speculative, this trait may have been selected under pressure of competitors facilitated by the suppressed defenses. Interestingly, T. urticae DeLier-1 mites do not produce excessive amounts of webbing but do promote the reproductive performance of T. urticae Santpoort-2 and, thus, if and how moderate plant-defense suppressors such as DeLier-1 protect their manipulated resources from competitors warrant more in-depth ecological research.
Acknowledgments
J.M.A. was supported by NWO Earth and Life Sciences (ALW-TOP 854.11.005). B.C.J.S. was supported by NWO Earth and Life Sciences (ALW-TTI Green Genetics 828.08.001). C.A.V. was supported by CONICYT. J.J.G. was supported by NWO Earth and Life Sciences (ALW ‘Meer met Minder’ 847.13.005). M.W.S. was supported by the Royal Academy of Arts and Sciences (KNAW). M.R.K. was supported by NWO (STW-VIDI 13492). The authors wish to thank Ludek Tikovsky, Harold Lemereis and Thijs Hendrix for taking care of the plants; Lin Dong for technical assistance; Arjen van Doorn and Michel de Vries for their help with the LC-MS analyses; and Arne Janssen and Michel Haring for their scientific input.
Supporting Information
Please note: Wiley Blackwell are not responsible for the content or functionality of any supporting information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Fig. S1Photos of adult female spider mites from each of the four strains used for this study.
Fig. S2 Phylogenetic trees based upon the cytochrome oxidase subunit 1 (COI) sequences from spider mites, including sequences from the mite strains used for this study.
Fig. S3 Fecundity of putative JA defense-suppressing T. urticae strains on def-1, wild-type (WT) and 35S::Prosystemin tomato and induction of Proteinase Inhibitor IIf (PI-IIf) by these strains upon infestation of WT plants.
Fig. S4 Reproductive performance of adult female spider mites on wild-type and def-1 tomato.
Fig. S5 Regression analysis of jasmonic acid (JA) content and expression levels of Proteinase Inhibitor IIc (PI-IIc) and PI-IIf upon infestation of tomato leaflets with spider mites.
Fig. S6 Feeding damage inflicted by adult female spider mites on tomato leaflets.
Fig. S7 Concentrations of jasmonic acid-isoleucine (JA-Ile) and salicylic acid (SA), plus transcript abundances of Proteinase Inhibitor IIc (PI-IIc) and Pathogenesis-related protein 1a (PR-1a) in tomato leaflets after 7 d of infestation with spider mites from inducer strain T. urticae Santpoort-2, suppressor T. evansi Viçosa-1 or both strains together.
Fig. S8 Concentrations of jasmonic acid-isoleucine (JA-Ile) and salicylic acid (SA), plus transcript abundances of Proteinase Inhibitor IIc (PI-IIc) and Pathogenesis-related protein 1a (PR-1a) in tomato leaflets after 7 d of infestation with spider mites from inducer strain T. urticae Santpoort-2, suppressor T. urticae DeLier-1, or both strains together.
Table S1 Parameters used for detection of phytohormones by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS)
Table S2 qRT-PCR primer sequences
Methods S1 Protocol for infestation of tomato plants with spider mites.
Methods S2 Protocol for the extraction and quantification of phytohormones from tomato leaves.
Notes S1 Sampling and rearing of spider mites.
Notes S2 The Proteinase Inhibitor II (PI-II) gene family in tomato.
References
- Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nature Reviews. Molecular Cell Biology. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adato A, Mandel T, Mintz-Oron S, Venger I, Levy D, Yativ M, Dominguez E, Wang ZH, De Vos RCH, Jetter R, et al. Fruit-surface flavonoid accumulation in tomato is controlled by a SIMYB12-regulated transcriptional network. PLoS Genetics. 2009;5:e1000777. doi: 10.1371/journal.pgen.1000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba JM, Glas JJ, Schimmel BCJ, Kant MR. Avoidance and suppression of plant defenses by herbivores and pathogens. Journal of Plant Interactions. 2011;6:1–7. [Google Scholar]
- Ament K, Kant MR, Sabelis MW, Haring MA, Schuurink RC. Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiology. 2004;135:2025–2037. doi: 10.1104/pp.104.048694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ament K, Krasikov V, Allmann S, Rep M, Takken FLW, Schuurink RC. Methyl salicylate production in tomato affects biotic interactions. Plant Journal. 2010;62:124–134. doi: 10.1111/j.1365-313X.2010.04132.x. [DOI] [PubMed] [Google Scholar]
- Belliure B, Janssen A, Maris PC, Peters D, Sabelis MW. Herbivore arthropods benefit from vectoring plant viruses. Ecology Letters. 2005;8:70–79. [Google Scholar]
- Bonaventure G, VanDoorn A, Baldwin IT. Herbivore-associated elicitors: FAC signaling and metabolism. Trends in Plant Science. 2011;16:294–299. doi: 10.1016/j.tplants.2011.01.006. [DOI] [PubMed] [Google Scholar]
- Bos JIB, Prince D, Pitino M, Maffei ME, Win J, Hogenhout SA. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (Green Peach Aphid) PLoS Genetics. 2010;6:e1001216. doi: 10.1371/journal.pgen.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boubou A, Migeon A, Roderick GK, Auger P, Cornuet JM, Magalhaes S, Navajas M. Test of colonisation scenarios reveals complex invasion history of the red tomato spider mite Tetranychus evansi. PLoS ONE. 2012;7:e35601. doi: 10.1371/journal.pone.0035601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgyán J, Havelda Z. Viral suppressors of RNA silencing. Trends in Plant Science. 2011;16:265–272. doi: 10.1016/j.tplants.2011.02.010. [DOI] [PubMed] [Google Scholar]
- Casteel CL, Hansen AK, Walling LL, Paine TD. Manipulation of plant defense responses by the tomato psyllid (Bactericerca cockerelli) and its associated endosymbiont Candidatus Liberibacter Psyllaurous. PLoS ONE. 2012;7:e35191. doi: 10.1371/journal.pone.0035191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casteel CL, Yang C, Nanduri AC, De Jong HN, Whitham SA, Jander G. The Nia-Pro protein of Turnip mosaic virus improves growth and reproduction of the aphid vector, Myzus persicae (green peach aphid) Plant Journal. 2014;77:653–663. doi: 10.1111/tpj.12417. [DOI] [PubMed] [Google Scholar]
- Chen H, Jones AD, Howe GA. Constitutive activation of the jasmonate signaling pathway enhances the production of secondary metabolites in tomato. FEBS Letters. 2006;580:2540–2546. doi: 10.1016/j.febslet.2006.03.070. [DOI] [PubMed] [Google Scholar]
- Chen H, Wilkerson CG, Kuchar JA, Phinney BS, Howe GA. Jasmonate-inducible plant enzymes degrade essential amino acids in the herbivore midgut. Proceedings of the National Academy of Sciences, USA. 2005;102:19237–19242. doi: 10.1073/pnas.0509026102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, Adie B, Chico JM, Lorenzo O, Garcia-Casado G, Lopez-Vidriero I, Lozano FM, Ponce MR, et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature. 2007;448:666–671. doi: 10.1038/nature06006. [DOI] [PubMed] [Google Scholar]
- Chung SH, Rosa C, Scully ED, Peiffer M, Tooker JF, Hoover K, Luthe DS, Felton GW. Herbivore exploits orally secreted bacteria to suppress plant defenses. Proceedings of the National Academy of Sciences, USA. 2013;110:15728–15733. doi: 10.1073/pnas.1308867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colquhoun TA, Kim JY, Wedde AE, Levin LA, Schmitt KC, Schuurink RC, Clark DG. PhMYB4 fine-tunes the floral volatile signature of Petunia × hybrida through PhC4H. Journal of Experimental Botany. 2011;62:1133–1143. doi: 10.1093/jxb/erq342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consales F, Schweizer F, Erb M, Gouhier-Darimont C, Bodenhausen N, Bruessow F, Sobhy I, Reymond P. Insect oral secretions suppress wound-induced responses in Arabidopsis. Journal of Experimental Botany. 2012;63:727–737. doi: 10.1093/jxb/err308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constabel CP, Barbehenn RV. Defensive roles of polyphenol oxidase in plants. In: Schaller A, editor. Induced plant resistance to herbivory. New York, NY, USA: Springer Verlag; 2008. pp. 253–269. [Google Scholar]
- Da Cunha L, Sreerekha M-V, Mackey D. Defense suppression by virulence effectors of bacterial phytopathogens. Current Opinion in Plant Biology. 2007;10:349–357. doi: 10.1016/j.pbi.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Dal Cin V, Tieman DM, Tohge T, McQuinn R, de Vos RCH, Osorio S, Schmelz EA, Taylor MG, Smits-Kroon MT, Schuurink RC, et al. Identification of genes in the phenylalanine metabolic pathway by ectopic expression of a MYB transcription factor in tomato fruit. Plant Cell. 2011;23:2738–2753. doi: 10.1105/tpc.111.086975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge R, Bolton M, Thomma BPHJ. How filamentous pathogens co-opt plants: the ins and outs of fungal effectors. Current Opinion in Plant Biology. 2011;14:400–406. doi: 10.1016/j.pbi.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Dermauw W, Wybouw N, Rombauts S, Menten B, Vontas J, Grbic M, Clark RM, Feyereisen R, Van Leeuwen TAF. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proceedings of the National Academy of Sciences, USA. 2012;110:E112–E113. doi: 10.1073/pnas.1213214110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diezel C, von Dahl CC, Gaquerel E, Baldwin IT. Different lepidopteran elicitors account for cross-talk in herbivory-induced phytohormone signaling. Plant Physiology. 2009;150:1576–1586. doi: 10.1104/pp.109.139550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorey S, Baillieul F, Saindrenan P, Fritig B, Kauffmann S. Tobacco class I and II catalases are differentially expressed during elicitor-induced hypersensitive cell death and localized acquired resistance. Molecular Plant-Microbe Interactions: MPMI. 1998;11:1102–1109. [Google Scholar]
- Engelberth J, Contreras CF, Viswanathan S. Transcriptional analysis of distant signaling induced by insect elicitors and mechanical wounding in Zea mays. PLoS ONE. 2012;7:e34855. doi: 10.1371/journal.pone.0034855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer EE, Johnson RR, Ryan CA. Regulation of expression of proteinase-inhibitor genes by methyl jasmonate and jasmonic acid. Plant Physiology. 1992;98:995–1002. doi: 10.1104/pp.98.3.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton GW, Donato K, Del Vecchio RJ, Duffey SS. Activation of plant foliar oxidases by insect feeding reduces nutritive quality of foliage for noctuid herbivores. Journal of Chemical Ecology. 1989;15:2667–2694. doi: 10.1007/BF01014725. [DOI] [PubMed] [Google Scholar]
- Ferragut F, Garzon-Luque E, Pekas A. The invasive spider mite Tetranychus evansi (Acari: Tetranychidae) alters community composition and host-plant use of native relatives. Experimental and Applied Acarology. 2013;60:321–341. doi: 10.1007/s10493-012-9645-7. [DOI] [PubMed] [Google Scholar]
- Gadea J, Mayda ME, Conejero V, Vera P. Characterization of defense-related genes ectopically expressed in viroid-infected tomato plants. Molecular Plant-Microbe Interactions: MPMI. 1996;9:409–415. doi: 10.1094/mpmi-9-0409. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annual Review of Phytopathology. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- Gomez S, Steinbrenner AD, Osorio S, Schueller M, Ferrieri RA, Fernie AR, Orians CM. From shoots to roots: transport and metabolic changes in tomato after simulated feeding by a specialist lepidopteran. Entomologia Experimentalis et Applicata. 2012;144:101–111. [Google Scholar]
- Gonzales-Vigil E, Bianchetti CM, Philips GN, Jr, Howe GA. Adaptive evolution of threonine deaminase in plant defense against herbivores. Proceedings of the National Academy of Sciences, USA. 2011;108:5897–5902. doi: 10.1073/pnas.1016157108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grbić M, Van Leeuwen T, Clark RM, Rombauts S, Rouzé P, Grbić V, Osborne EJ, Dermauw W, Ngoc PCT, Ortego F, et al. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature. 2011;479:487–492. doi: 10.1038/nature10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruner K, Griebel T, Navarova H, Attaran E, Zeier J. Reprogramming of plants during systemic acquired resistance. Frontiers in Plant Science. 2013;4:252. doi: 10.3389/fpls.2013.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haegeman A, Mantelin S, Jones JT, Gheysen G. Functional roles of effectors of plant-parasitic nematodes. Gene. 2012;492:19–31. doi: 10.1016/j.gene.2011.10.040. [DOI] [PubMed] [Google Scholar]
- Hogenhout SA, Bos JIB. Effector proteins that modulate plant-insect interactions. Current Opinion in Plant Biology. 2011;14:1–7. doi: 10.1016/j.pbi.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Howe GA, Jander G. Plant immunity to insect herbivores. Annual Review of Plant Biology. 2008;59:41–66. doi: 10.1146/annurev.arplant.59.032607.092825. [DOI] [PubMed] [Google Scholar]
- Howe GA, Lightner J, Browse J, Ryan CA. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell. 1996;8:2067–2077. doi: 10.1105/tpc.8.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science. 2013;341:175–179. doi: 10.1126/science.1240230. [DOI] [PubMed] [Google Scholar]
- Itkin M, Rogachev I, Alkan N, Rosenberg T, Malitsky S, Masini L, Meir S, Iijima Y, Aoki K, de Vos R, et al. GLYCOALKALOID METABOLISM1 is required for steroidal alkaloid glycosylation and prevention of phytotoxicity in tomato. Plant Cell. 2011;23:4507–4525. doi: 10.1105/tpc.111.088732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppson LR, Keifer HH, Baker EW. Injurious Tetranychid mites. In: Jeppson LR, Keifer HH, Baker EW, editors. Mites injurious to economic plants. Berkeley, CA, USA: University of California Press; 1975. pp. 127–252. [Google Scholar]
- Kaloshian I, Walling LL. Hemipterans as plant pathogens. Annual Review of Phytopathology. 2005;43:491–521. doi: 10.1146/annurev.phyto.43.040204.135944. [DOI] [PubMed] [Google Scholar]
- Kamoun S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annual Review of Phytopathology. 2006;44:41–60. doi: 10.1146/annurev.phyto.44.070505.143436. [DOI] [PubMed] [Google Scholar]
- Kandoth PK, Ranf S, Pancholi SS, Jayanty S, Walla MD, Miller W, Howe GA, Lincoln DE, Stratmann JW. Tomato MAPKs LeMPK1, LeMPK2, and LeMPK3 function in the systemin-mediated defense response against herbivorous insects. Proceedings of the National Academy of Sciences, USA. 2007;104:12205–12210. doi: 10.1073/pnas.0700344104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant MR, Ament K, Sabelis MW, Haring MA, Schuurink RC. Differential timing of spider mite-induced direct and indirect defenses in tomato plants. Plant Physiology. 2004;135:483–495. doi: 10.1104/pp.103.038315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kant MR, Bleeker PM, Van Wijk M, Schuurink RC, Haring MA. Plant volatiles in defence. In: Van Loon LC, editor. Advances in botanical research (volume 51, plant innate immunity) Burlington, VT, USA: Academic Press; 2009. pp. 613–666. [Google Scholar]
- Kant MR, Sabelis MW, Haring MA, Schuurink RC. Intraspecific variation in a generalist herbivore accounts for differential induction and impact of host plant defences. Proceedings. Biological Sciences/The Royal Society. 2008;275:443–452. doi: 10.1098/rspb.2007.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT. Plant responses to insect herbivory: the emerging molecular analysis. Annual Review of Plant Biology. 2002;53:299–328. doi: 10.1146/annurev.arplant.53.100301.135207. [DOI] [PubMed] [Google Scholar]
- Klingler JP, Nair RM, Edwards OR, Singh KB. A single gene, AIN, in Medicago truncatula mediates a hypersensitive response to both bluegreen aphid and pea aphid, but confers resistance only to bluegreen aphid. Journal of Experimental Botany. 2009;60:4115–4127. doi: 10.1093/jxb/erp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A, Leon-Reyes A, Ritsema T, Verhage A, Den Otter FC, Van Loon LC, Pieterse CMJ. Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiology. 2008;147:1358–1368. doi: 10.1104/pp.108.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos F, Sarmento RA, Pallini A, Dias CR, Sabelis MW, Janssen A. Spider mite web mediates anti-predator behaviour. Experimental and Applied Acarology. 2010;52:1–10. doi: 10.1007/s10493-010-9344-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lers A, Sonego L, Green PJ, Burd S. Suppression of LX Ribonuclease in tomato results in a delay of leaf senescence and abscission. Plant Physiology. 2006;142:710–721. doi: 10.1104/pp.106.080135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Williams MM, Loh YT, Lee GI, Howe GA. Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiology. 2002;130:494–503. doi: 10.1104/pp.005314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhao Y, McCaig BC, Wingerd BA, Wang J, Whalon ME, Pichersky E, Howe GA. The tomato homolog of CORONATINE-INSENSITIVE 1 is required for maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell. 2004;16:126–143. doi: 10.1105/tpc.017954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisón P, Rodrigo I, Conejero V. A novel function for the cathepsin D inhibitor in tomato. Plant Physiology. 2006;142:1329–1339. doi: 10.1104/pp.106.086587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R, Ozawa R, Uefune M, Gotoh T, Takabayashi J. Intraspecies variation in the Kanzawa spider mite differentially affects induced defensive response in lima bean plants. Journal of Chemical Ecology. 2006;32:2501–2512. doi: 10.1007/s10886-006-9159-z. [DOI] [PubMed] [Google Scholar]
- Meier BM, Shaw N, Slusarenko AJ. Spatial and temporal accumulation of defense gene transcripts in bean (Phaseolus vulgaris) leaves in relation to bacteria-induced hypersensitive cell-death. Molecular Plant-Microbe Interactions: MPMI. 1993;6:453–466. doi: 10.1094/mpmi-6-453. [DOI] [PubMed] [Google Scholar]
- Miles PW. The saliva of Hemiptera. Advances in Insect Physiology. 1972;9:183–255. [Google Scholar]
- Milner SE, Brunton NP, Jones PW, O'Brien NM, Collins SG, Maguire AR. Bioactivities of glycoalkaloids and their aglycones from Solanum species. Journal of Agricultural and Food Chemistry. 2011;59:3454–3484. doi: 10.1021/jf200439q. [DOI] [PubMed] [Google Scholar]
- Mintz-Oron S, Mandel T, Rogachev I, Feldberg L, Lotan O, Yativ M, Wang Z, Jetter R, Venger I, Adato A, et al. Gene expression and metabolism in tomato fruit surface tissues. Plant Physiology. 2008;147:823–851. doi: 10.1104/pp.108.116004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mithöfer A, Wanner G, Boland W. Effect of feeding Spodoptera littoralis on lima bean leaves. II. Continuous mechanical wounding resembling insect feeding is sufficient to elicit herbivory-related volatile emission. Plant Physiology. 2005;137:1160–1168. doi: 10.1104/pp.104.054460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mur LAJ, Kenton P, Atzorn R, Miersch O, Wasternack C. The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiology. 2006;140:249–262. doi: 10.1104/pp.105.072348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser RO, Cipollini DF, Hum-Musser SM, Williams SA, Brown JK, Felton GW. Evidence that the caterpillar salivary enzyme glucose oxidase provides herbivore offense in Solanaceous plants. Archives of Insect Biochemistry and Physiology. 2005;58:128–137. doi: 10.1002/arch.20039. [DOI] [PubMed] [Google Scholar]
- Musser RO, Hum-Musser SM, Eichenseer H, Peiffer M, Ervin G, Murphy JB, Felton GW. Caterpillar saliva beats plant defences. Nature. 2002;416:599–600. doi: 10.1038/416599a. [DOI] [PubMed] [Google Scholar]
- Navajas M, de Moraes GJ, Auger P, Migeon A. Review of the invasion of Tetranychus evansi: biology, colonization pathways, potential expansion and prospects for biological control. Experimental and Applied Acarology. 2013;59:43–65. doi: 10.1007/s10493-012-9590-5. [DOI] [PubMed] [Google Scholar]
- Newman SM, Eannetta NT, Yu HF, Prince JP, Devicente MC, Tanksley SD, Steffens JC. Organization of the tomato polyphenol oxidase gene family. Plant Molecular Biology. 1993;21:1035–1051. doi: 10.1007/BF00023601. [DOI] [PubMed] [Google Scholar]
- Ozawa R, Matsushima R, Maffei M, Takabayashi J. Interaction between Phaseolus plants and two strains of Kanzawa spider mites. Journal of Plant Interactions. 2011;6:125–128. [Google Scholar]
- Paschold A, Halitschke R, Baldwin IT. Co(i)-ordinating defenses: NaCOI1 mediates herbivore-induced resistance in Nicotiana attenuata and reveals the role of herbivore movement in avoiding defenses. Plant Journal. 2007;51:79–91. doi: 10.1111/j.1365-313X.2007.03119.x. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Leon-Reyes A, Van der Ent S, Van Wees SCM. Networking by small-molecule hormones in plant immunity. Nature Chemical Biology. 2009;5:308–316. doi: 10.1038/nchembio.164. [DOI] [PubMed] [Google Scholar]
- Pieterse CMJ, Van der Does D, Zamioudis C, Leon-Reyes A, Van Wees SCM. Hormonal modulation of plant immunity. Annual Review of Cell and Developmental Biology. 2012;28:489–521. doi: 10.1146/annurev-cellbio-092910-154055. [DOI] [PubMed] [Google Scholar]
- Rodriguez PA, Bos JIB. Toward understanding the role of aphid effectors in plant infestation. Molecular Plant-Microbe Interactions: MPMI. 2013;26:25–30. doi: 10.1094/MPMI-05-12-0119-FI. [DOI] [PubMed] [Google Scholar]
- Sarmento RA, Lemos F, Bleeker PM, Schuurink RC, Pallini A, Oliveira MGA, Lima E, Kant M, Sabelis MW, Janssen A. A herbivore that manipulates plant defence. Ecology Letters. 2011a;14:229–236. doi: 10.1111/j.1461-0248.2010.01575.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmento RA, Lemos F, Dias CR, Kikuchi WT, Rodrigues JCP, Pallini A, Sabelis MW, Janssen A. A herbivorous mite down-regulates plant defence and produces web to exclude competitors. PLoS ONE. 2011b;6:e23757. doi: 10.1371/journal.pone.0023757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko U, Preston CA, Baldwin IT. Eating the evidence? Manduca sexta larvae can not disrupt specific jasmonate induction in Nicotiana attenuata by rapid consumption. Planta. 2000;210:343–346. doi: 10.1007/PL00008143. [DOI] [PubMed] [Google Scholar]
- Serrano A, Wang B, Aryal B, Garcion C, Abou-Monsour E, Heck S, Geisler M, Mauch F, Nawrath C, Metraux J-P. Export of salicylic acid from the chloroplast requires the MATE-like transporter EDS5. Plant Physiology. 2013;162:1815–1821. doi: 10.1104/pp.113.218156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JJ, Chen M-S, Shukle R, Harris MO. Gall midges (Hessian flies) as plant pathogens. Annual Review of Phytopathology. 2012;50:339–357. doi: 10.1146/annurev-phyto-072910-095255. [DOI] [PubMed] [Google Scholar]
- Sugio A, Kingdom HN, MacLean AM, Grieve VM, Hogenhout SA. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proceedings of the National Academy of Sciences, USA. 2011;108:E1254–E1263. doi: 10.1073/pnas.1105664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thipyapong P, Melkonian J, Wolfe DW, Steffens JC. Suppression of polyphenol oxidases increases stress tolerance in tomato. Plant Science. 2004;167:693–703. [Google Scholar]
- Tornero P, Gadea J, Conejero J, Vera P. Two PR-1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Molecular Plant-Microbe Interactions: MPMI. 1997;10:624–634. doi: 10.1094/MPMI.1997.10.5.624. [DOI] [PubMed] [Google Scholar]
- Van Kan JAL, Joosten MHAJ, Wagemakers CAM, Van den Berg-Velthuis GCM, de Wit PJGM. Differential accumulation of messenger-RNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium-fulvum. Plant Molecular Biology. 1992;20:513–527. doi: 10.1007/BF00040610. [DOI] [PubMed] [Google Scholar]
- Van Loon LC, Van Strien EA. The families of pathogenesis-related proteins, their activities, and comparative analysis of PR-1 type proteins. Physiological and Molecular Plant Pathology. 1999;55:85–97. [Google Scholar]
- Verdonk JC, de Vos CHR, Verhoeven HA, Haring MA, Van Tunen AJ, Schuurink RC. Regulation of floral scent production in petunia revealed by targeted metabolomics. Phytochemistry. 2003;62:997–1008. doi: 10.1016/s0031-9422(02)00707-0. [DOI] [PubMed] [Google Scholar]
- Vlot AC, Dempsey DA, Klessig DF. Salicylic acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology. 2009;47:177–206. doi: 10.1146/annurev.phyto.050908.135202. [DOI] [PubMed] [Google Scholar]
- Walling LL. The myriad plant responses to herbivores. Journal of Plant Growth Regulation. 2000;19:195–216. doi: 10.1007/s003440000026. [DOI] [PubMed] [Google Scholar]
- Weech MH, Chapleau M, Pan L, Ide C, Bede JC. Caterpillar saliva interferes with induced Arabidopsis thaliana defence responses via the systemic acquired resistance pathway. Journal of Experimental Botany. 2008;59:2437–2448. doi: 10.1093/jxb/ern108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weygoldt P. Evolution and systematics of the Chelicerata. Experimental and Applied Acarology. 1998;22:63–79. [Google Scholar]
- Will T, Tjallingii WF, Thonnessen A, Van Bel AJE. Molecular sabotage of plant defense by aphid saliva. Proceedings of the National Academy of Sciences, USA. 2007;104:10536–10541. doi: 10.1073/pnas.0703535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Baldwin IT. New insights into plant responses to the attack from insect herbivores. Annual Review of Genetics. 2010;44:1–24. doi: 10.1146/annurev-genet-102209-163500. [DOI] [PubMed] [Google Scholar]
- Wu J, Hettenhausen C, Meldau S, Baldwin IT. Herbivory rapidly activates MAPK signalling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell. 2007;19:1096–1122. doi: 10.1105/tpc.106.049353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Peiffer M, Luthe DS, Felton GW. ATP hydrolyzing salivary enzymes of caterpillars suppress plant defenses. PLoS ONE. 2012;7:e41947. doi: 10.1371/journal.pone.0041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate SI, Kempema LA, Walling LL. Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiology. 2007;143:866–875. doi: 10.1104/pp.106.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhu X, Huang F, Liu Y, Zhang J, Lu Y, Ruan Y. Suppression of jasmonic acid-dependent defense in cotton plant by the mealybug Phenacoccus solenopsis. PLoS ONE. 2011;6:e22378. doi: 10.1371/journal.pone.0022378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang PJ, Zheng SJ, Van Loon JJA, Boland W, David A, Mumm R, Dicke M. Whiteflies interfere with indirect plant defense against spider mites in Lima bean. Proceedings of the National Academy of Sciences, USA. 2009;106:21202–21207. doi: 10.1073/pnas.0907890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Luan JB, Qi JF, Huang CJ, Li M, Zhou XP, Liu SS. Begomovirus-whitefly mutualism is achieved through repression of plant defences by a virus pathogenicity factor. Molecular Ecology. 2012;21:1294–1304. doi: 10.1111/j.1365-294X.2012.05457.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1Photos of adult female spider mites from each of the four strains used for this study.
Fig. S2 Phylogenetic trees based upon the cytochrome oxidase subunit 1 (COI) sequences from spider mites, including sequences from the mite strains used for this study.
Fig. S3 Fecundity of putative JA defense-suppressing T. urticae strains on def-1, wild-type (WT) and 35S::Prosystemin tomato and induction of Proteinase Inhibitor IIf (PI-IIf) by these strains upon infestation of WT plants.
Fig. S4 Reproductive performance of adult female spider mites on wild-type and def-1 tomato.
Fig. S5 Regression analysis of jasmonic acid (JA) content and expression levels of Proteinase Inhibitor IIc (PI-IIc) and PI-IIf upon infestation of tomato leaflets with spider mites.
Fig. S6 Feeding damage inflicted by adult female spider mites on tomato leaflets.
Fig. S7 Concentrations of jasmonic acid-isoleucine (JA-Ile) and salicylic acid (SA), plus transcript abundances of Proteinase Inhibitor IIc (PI-IIc) and Pathogenesis-related protein 1a (PR-1a) in tomato leaflets after 7 d of infestation with spider mites from inducer strain T. urticae Santpoort-2, suppressor T. evansi Viçosa-1 or both strains together.
Fig. S8 Concentrations of jasmonic acid-isoleucine (JA-Ile) and salicylic acid (SA), plus transcript abundances of Proteinase Inhibitor IIc (PI-IIc) and Pathogenesis-related protein 1a (PR-1a) in tomato leaflets after 7 d of infestation with spider mites from inducer strain T. urticae Santpoort-2, suppressor T. urticae DeLier-1, or both strains together.
Table S1 Parameters used for detection of phytohormones by liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS)
Table S2 qRT-PCR primer sequences
Methods S1 Protocol for infestation of tomato plants with spider mites.
Methods S2 Protocol for the extraction and quantification of phytohormones from tomato leaves.
Notes S1 Sampling and rearing of spider mites.
Notes S2 The Proteinase Inhibitor II (PI-II) gene family in tomato.


