SUMMARY
The Beijing/W lineage strains are the major prevalent strains in China. The prevalence, mortality and drug-resistant rates of tuberculosis in Xinjiang, Northwestern China are higher than in other parts of the country. Our previous study results showed that the dominant strains of Mycobacterium tuberculosis (MTB) were ‘Beijing/W lineage’ MTB in Xinjiang; those strains had no significant correlation with drug resistance. We investigated whether the prevalence of ‘Beijing/W lineage’ sublineage strains was associated with drug resistance. We collected 478 sputum specimens from patients with pulmonary tuberculosis. Beijing/W strains and their sublineages were identified by distinguishing five specific large sequence polymorphisms, using polymerase chain reaction. All strains were subjected to a drug susceptibility test using the proportion method on Löwenstein–Jensen culture medium. In total, 379 clinical isolates of MTB were isolated and identified, 57·26% of these isolates were identified as Beijing/W strains, of which 11·06% isolates were in sublineage 105, 14·74% isolates in sublineage 207, 69·59% isolates in sublineage 181, and 4·61% isolates in sublineage 150. None of the isolates was in sublineage 142. Our data showed there were four sublineages of Beijing/W isolates in Xinjiang province, China. However, there were no correlations between drug resistance and the sublineages of Beijing/W strains.
Key words: Beijing/W strains sublineages, drug resistance, Mycobacterium tuberculosis, Xinjiang province
INTRODUCTION
The World Health Organization (WHO) estimates that currently more than one-third of the world's population is asymptomatically infected with tuberculosis (TB). China occupies second place behind India among the 22 countries considered to be high-burden countries that account for about 80% of new TB cases worldwide each year. Large sequence polymorphisms (LSPs) represent unique event polymorphisms that can be used to construct robust phylogenies for Mycobacterium tuberculosis (MTB) [1]. LSPs (robust phylogenetic markers) classify MTB into six major lineages, each of which is strongly associated with specific geographically distinct human populations [2].The East Asian lineage is one of these six lineages, and it includes the Beijing/W strains [3]. Beijing/W lineage strains were first identified in MTB isolates from the Beijing area of China after which it was named. These strains are defined by a spoligotype in which direct variable repeats 1–34 are missing [4]. Beijing/W lineage strains can also be recognized by several LSPs unique to Beijing/W lineage strains using genomic microarray approaches, which are further subdivided into five sublineages using specific regions of difference (RDs) [5]. One LSP (RD105 region) is 3467 bp in size and completely deletes Rv0072 and Rv0073 and truncates Rv0071 and Rv0074. The RD105 region serves as a useful marker for distinguishing Beijing/W lineage strains from non-Beijing/W lineage strains, because the RD105 region deletion was found in all Beijing/W lineage strains [6], but has not been found in any non-Beijing/W lineage strains [2]. RD142 (2851 bp), RD150 (2487 bp), and RD181 (711 bp) regions are variably deleted in Beijing/W lineage strains but are monophyletic for subdivisions of the Beijing/W lineage group [6].
Beijing/W lineage strains of MTB are endemically prevalent in eastern Asia [7], South Africa [8], and Russia [9]. Previous studies have revealed that Beijing/W lineage strains are associated with outbreaks [10] and treatment failures [11]. Studies of genetic relatedness suggest that Beijing/W strains also show evidence of clonal expansion [12], and the selective advantage of the Beijing clonal family may be an elevated mutation rate that facilitates the rapid accumulation of antibiotic resistance [13]. Beijing/W lineage strains are the major prevalent strains in China [14]. Xinjiang is a province in the northwestern area of China, and the prevalence, death rates and drug-resistant rates of TB are higher there than in other parts of the country. The WHO surveillance report on MTB drug resistance in Xinjiang showed the proportions of overall drug resistance, drug resistance in new cases, and re-treated smear-positive cases, were 26%, 26% and 31%, respectively, while the multidrug resistance (MDR) rate was 5% [15]. Therefore, the quality of TB control in this region clearly needs to be strengthened [15]. The main aim of this study was to classify the Beijing/W lineage strains of MTB in Xinjiang by identifying specific LSPs. We also sought to determine the drug susceptibility patterns of the sublineages of Beijing/W lineage strains and to determine whether the drug resistance of epidemic MTB is associated with the individual sublineages of Beijing/W lineage strains.
MATERIALS AND METHODS
Isolates selection
Sputum specimens (n = 478) were collected from pulmonary TB patients between June 2007 and June 2012 from Xinjiang province in China. The patients were all diagnosed as having pulmonary TB according to the national guidelines of China, and were treated in local lung disease hospitals or TB hospitals. Of these patients, 45 were from Akesu, 32 from Chang ji, 44 from Dushanzi, 32 from Hotan, 146 from Kashigar, nine from Kizilsu kirghiz, 14 from Manasi, 89 from Shihezi, 28 from Tumushuk, 25 from Tacheng, and 14 from Wusu. Following verbal and written consent, three sputum samples from each patient were collected at the same time. Information was obtained on sex, age, place of birth, recent smear-positive sputum tests, previous history of TB, and current address by a structured questionnaire. The study was performed with the approval of the Ethics Committee of the First Hospital of Shihezi University and performed in compliance with the Helsinki Declaration. Informed consent was obtained from all subjects before commencement of the study.
MTB isolation and drug susceptibility test
Sputum samples were cultured on Löwenstein–Jensen (LJ) culture medium. The culture, identification and drug susceptibility tests of all strains were performed according to the TB diagnosis bacteriology test criteria of the China Antituberculosis Association [16] at the Ministry of Education Key Laboratory of Xinjiang Endemic and Ethnic Disease. Drug susceptibility testing was performed using the proportion method. Four first-line anti-TB drugs [isoniazid (INH), rifampicin (RFP), streptomycin (SM), ethambutol (EMB)] plus seven second-line anti-TB drugs [Ofloxacin (Ofx), Capreomycin (Cm), Kanamycin (Km), Amikacin (Am), P-aminosalicylic acid (PAS), Ethionamide (Eto) and Cycloserine (Cs)] were incorporated into LJ medium, at the following concentrations: 0·2 μg/ml INH, 40·0 μg/ml RFP, 4·0 μg/ml SM, 2·0 μg/ml EMB, 2·0 μg/ml Ofx, 40·0 μg/ml Cm, 30·0 μg/ml Km, 40·0 μg/ml Am, 1·0 μg/ml PAS, 40·0 μg/ml Eto, 40·0 μg/ml Cs, respectively, and used to detect drug resistance. Isolates were scored as resistant to a specific drug when the growth rate was >1% compared to the control, or as sensitive when the growth rate was < 1% compared to the control as previously described [17].
Genomic DNA extraction and molecular identification of MTB isolates
Mycobacterial genomic DNA was extracted from colonies growing on LJ medium. Scraped colonies were resuspended in 200–300 μl distilled water and inactivated at 85°C for 30 min, before centrifugation at 5900 g for 5 min. The pellets were re-suspended in 300 μl TE (pH 8·3), boiled for 30 min, and centrifuged at 9300 g for 5 min. Supernatants were collected and stored at –20°C until further use [18]. Molecular identification of the mycobacterial isolates was performed using polymerase chain reaction (PCR) amplification of the 16S rRNA and MTP40 genes [19]. The PCR mixtures consisted of 0·2 μg DNA template, 3 μl buffer, 4 μl 10 mm DNTP, 1 μl of each primer (10 pmol/μl), and 1 μl DNA Taq polymerase (2·5 U/μl). The amplification cycle was 5 min at 95 °C; followed by 30 cycles of 40 s at 95°C, 50 s at 65°C, and 40 s at 72°C; with a final 10 min extension at 72°C. PCR products were analysed on a 2% agarose gel against a 100-bp DNA ladder.
Identification of genomic deletions by PCR and multiplex PCR
PCR and multiplex PCR were performed to identify the sub-population structure of the MTB Beijing/W lineage strains. The RD105 LSP was deleted in all Beijing/W lineage strains and was able to be used as a marker to define the Beijing/W lineage strains [6]. A three-step PCR experiment was conducted to determine the presence or absence of the RD105 region. Primers were designed as described previously [20].
Further identification of the sublineages of MTB Beijing/W lineage strains were made on the basis of the variable appearance of the RD207, RD181, RD150, and RD142 deletions. In this study, we refer to sublineage 105 strains as those containing only deletions of RD105 region, while sublineage 207 strains also contained concurrent deletions of RD105 and RD207 regions, sublineage 181 strains had concurrent deletions of RD105, RD207 and RD181 regions, sublineage 150 strains had concurrent deletions of RD105, RD207, RD181, and RD150 regions, while sublineage 142 strains had concurrent deletions of RD105, RD207, RD181, and RD142 regions. PCR was used to identify sublineage RD181 and sublineage RD150, while multiplex PCR was used to identify sublineage RD207and sublineage RD142. Primers were designed as described previously [21] (Table 1). The amplification cycle was 4 min at 94°C; followed by 30 cycles of 45 s at 94°C, 45 s at 58°C, and 50 s at 72°C; with a final 10 min extension at 72°C. PCR products were analysed on a 1·5% agarose gel against a 100-bp DNA ladder. We randomly selected the PCR products of ten specimens from each sublineage for DNA sequencing.
Table 1.
Primers used in large sequence polymorphisms (LSPs) and DNA sequence analysis for sublineages of Mycobacterium tuberculosis Beijing strains
| LSP | Primer | Sequence (5′–3′) | Product size (bp) | |
|---|---|---|---|---|
| Intact* | Deleted† | |||
| RD105 | RD105-F | CGTGCACAGTTGGGTGTTTA | 547 | |
| RD105-R | CGTCGTTTTCTGCCGATACT | 283 | ||
| RD207 | RD207-F | GTCTGACGACTGACAGGGTG | ||
| RD207-Rint‡ | CACGATGGCCACCTCCATG | 516 | ||
| RD207-Rdel¶ | CGTTGCGTGCTCGACGCTG | 682 | ||
| RD181 | RD181-F | CAACAGCACCAGCATCGGAC | 326 | |
| RD181-R | CTTGCTATCGGCGTCGTTGC | 537 | ||
| RD150 | RD150-F | CCATCCTGGCGTTGGTTGG | 505 | |
| RD150-R | CCGAGGACCTTACTGCGTG | 300 | ||
| RD142 | RD142-F | GGAGGACACATGTCGCAACAC | ||
| RD142-Rint | CGTGCAGCACGAACACCAC | 400 | ||
| RD142-Rdel | GTCGCAGCGCGAGTAGATC | 577 | ||
Intact = possessing an intact specific LSP region, e.g. RD105 region, RD207 region, etc.
Deleted = specific LSP region was deleted.
RD207-Rint, Reverse primer of intact RD207 region by multiplex PCR.
RD207-Rdel, Reverse primer of the deleted RD207 region by multiplex PCR.
Statistical analysis
The statistical analyses were performed using the χ2 test and the Fisher's exact test.
RESULTS
Characteristics of patients and samples
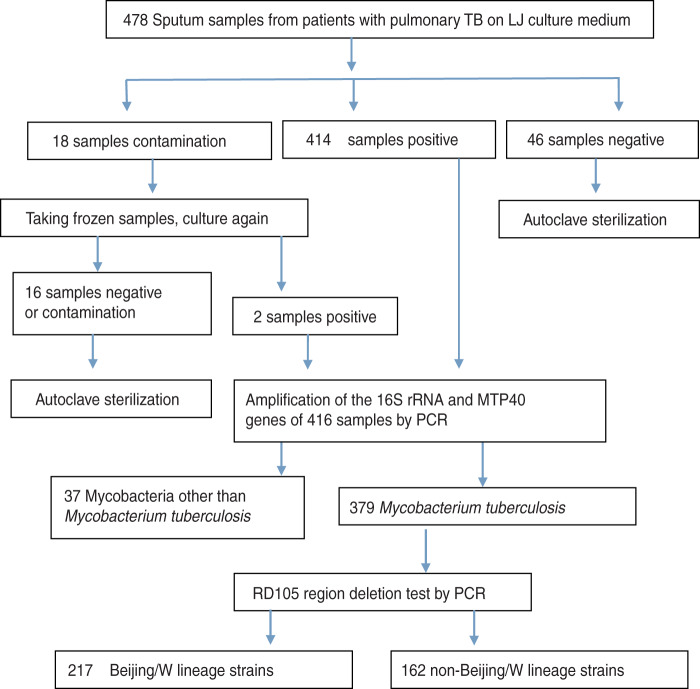
Among the 478 samples, 12·97% (62/478) of the specimens were either culture-negative or culture-contaminated and were therefore excluded, the remaining 87·03% (416/478) of the samples tested positive for mycobacterium. Of these, 8·89% (37/416) specimens tested positive for mycobacteria other than MTB and were excluded. Therefore, a total of 379 MTB culture-positive specimens were used for this study. A flow diagram shows the numbers of eligible patients, negative cultures, contamination, non-TB mycobacteria, and molecular tests (see Fig. 1).
Fig. 1.
Flow diagram of numbers of eligible patients.
Use of RD deletions to differentiate between Beijing/W and non-Beijing/W lineage strains
A three-step PCR experiment was conducted to determine the presence or absence of RD105 region in the 379 isolates. Two hundred and seventeen isolates contained the RD105 region deletion and were therefore identified as Beijing/W lineage strains. These isolates were from 59·45% (129/217) males and 40·55% (88/217) females. One hundred and sixty-two isolates had no RD105 region deletion and were therefore identified as non-Beijing/W lineage strains (Fig. 1). Between Beijing/W lineage strains and non-Beijing/W lineage strains, there was no significant correlation with regard to gender by χ2 text (χ2 = 0·269, P = 0·604). Neither the Beijing/W lineage strains nor the non-Beijing/W lineage strains showed any significant correlation between new cases and re-treated cases by χ2 text (χ2 = 2·454, P = 0·117) (Table 2). Of 217 Beijing/W lineage strains, 85·71% (186/217) were from farmers, followed by workers (8·76%), students (3·23%), etc. (Table 2).
Table 2.
Characteristics of patients infected by different sublineages of the Beijing strains
| Groups … | 105 (n = 24) | 207 (n = 32) | 181 (n = 151) | 150 (n = 10) | |
|---|---|---|---|---|---|
| Characteristic | |||||
| Sex | |||||
| Female (n = 88) | 9 | 9 | 66 | 4 | |
| Male (n = 129) | 15 | 23 | 85 | 6 | P = 0·429 |
| Treated cases and new cases | |||||
| New cases | 7 | 12 | 52 | 3 | |
| Retreated cases | 17 | 20 | 99 | 7 | P = 0·917 |
| Age (years) | |||||
| 0–24 | 3 | 5 | 16 | 1 | |
| 25–44 | 7 | 7 | 42 | 4 | |
| 45–64 | 10 | 13 | 46 | 3 | |
| ⩾65 | 4 | 7 | 47 | 2 | P = 0·788* |
| Occupation | |||||
| Farmer (n = 186) | 24 | 22 | 134 | 6 | 85·71% |
| Worker (n = 19) | 3 | 4 | 10 | 2 | 8·76% |
| Administrator (n = 4) | 1 | 3 | 1·84% | ||
| Student (n = 7) | 1 | 2 | 4 | 3·23% | |
| Teacher (n = 1) | 1 | 0·5% | |||
| Beijing/W lineage strains (n = 217) | non-Beijing/W lineage strains (n = 162) | ||||
| Males (n = 129) | Females (n = 88) | Males (n = 92) | Females (n = 70) | P = 0·604 | |
| New TB cases (n = 74) | Re-treated cases (n = 143) | New TB cases (n = 68) | Re-treated cases (n = 94) | P = 0·117 | |
Value by Fisher's exact test.
Drug susceptibility patterns of Beijing/W and non-Beijing/W lineage strains
We examined the distribution of drug resistance between Beijing/W lineage strains and non-Beijing/W lineage strains. Of the 217 Beijing/W lineage isolates, 26·73% (58/217) were resistant to at least one drug, and the overall resistance count included 49 isolates for first-line drug resistance and nine isolates for both first- and second-line drug resistance; 6·91% (15/217) of these were MDR-TB strains. Of the 162 non-Beijing/W lineage isolates, 24·69% (40/162) were resistant to at least one drug; 5·56% (9/162) of these were MDR-TB strains. There was no significant difference with regard to drug resistance between Beijing/W lineage strains and non-Beijing/W lineage strains (χ2 = 0·201, P = 0·654) and MDR-TB (χ2 = 0·119, P = 0·731). Two, two, one, and one ‘Beijing/W lineage’ sublineage strains were resistant to Ofx, Cm, Am, and Km respectively; while three, three, and one ‘Beijing/W lineage’ sublineage strains were resistant to PAS, Eto and Cs, respectively. Because the numbers of ‘Beijing/W lineage’ sublineage strains for drug resistance to seven second-line anti-TB drugs is too few, we did not perform statistical analyses for those drug-resistant strains (Table 3).
Table 3.
Distribution of drug resistant strains in different sublineages of the Beijing strains
| Groups … | 105 (n=24) | 207 (n=32) | 181 (n=151) | 150 (n=10) | P |
|---|---|---|---|---|---|
| Resistant to: | |||||
| INH (n = 33) | 2 | 4 | 27 | 0 | 0·402* |
| RFP (n = 16) | 1 | 2 | 13 | 0 | 0·961* |
| SM (n = 32) | 2 | 5 | 25 | 0 | 0·549* |
| EMB (n = 15) | 1 | 1 | 13 | 0 | 0·743* |
| MDR-TB (n = 15) | 1 | 1 | 13 | 0 | 0·743* |
| Ofx (n = 2) | 2 | ||||
| Cm (n = 2) | 1 | 1 | |||
| Am (n = 1) | 1 | ||||
| Km (n = 1) | 1 | ||||
| PAS (n = 3) | 3 | ||||
| Eto (n = 3) | 1 | 1 | 1 | ||
| Cs (n = 1) | 1 | ||||
Value by Fisher's exact test.
Characteristics of MTB isolates in different sublineages
Of the 217 Beijing/W isolates, 11·06% (24/217) had only the RD105 region deletion and were identified as sublineage 105. A group with concurrent deletions of RD105 and RD207 regions was identified as sublineage 207 (n = 32, 14·75%), a group with concurrent deletions of RD105, RD207 and RD181 regions was identified as sublineage 181 (n = 151, 69·59%), and a group with concurrent deletions of RD105, RD207, RD181, and RD150 regions was identified as sublineage 150 (n = 10, 4·60%). PCR verification did not detect the RD142 region deletion in any of the 217 isolates. We therefore concluded that none of the clinical isolates was in sublineage 142 in our specimens. The age, gender, history of treatment and the proportion of resistance to any drug did not show statistically significant correlations to any of the sublineages (Table 2).
DISCUSSION
Large chromosomal deletions, also termed LSPs, are unique event polymorphisms [1]. In recent years, these polymorphisms have been widely used to research the phylogenies of MTB. Whether or not the bacterial lineage influences the development of the disease has been investigated by researchers for many years; however, this still remains unclear. Some studies have shown that the Beijing/W lineage of clinically important MTB strains has led to outbreaks of TB [9, 22, 23] and been associated with drug resistance [24, 25]. However, it is the significance of this association that varies in different countries [9, 26]. This may be due to the different proportions of the sublineages of Beijing/W lineage strains present in different local populations [27–29].
In our study, we examined the frequency of occurrence of each sublineage and drug resistance of Beijing/W lineage strains in Xinjiang, China. MTB isolates were found to be subdivided into four sublineages using specific LSPs as markers. There was no isolate with the RD142 region deletion in our study. Similar reports have shown that 85 isolates from Vietnam [30], 73 isolates from Tuscany, Italy [31], 28 isolates from Myanmar [32], and 14 isolates from Sri Lanka [33] also did not have the RD142 region deletion. However, a study from Beijing in China, found that 26·42% (79/299) of strains of the Beijing/W lineage belonged to sublineage 142 [34], and another study in Arkansas, USA, found that 13/39 (33%) strains of the Beijing/W lineage belonged to sublineage 142 [35].
Sublineage 142 is one of the more puzzling sublineages of the Beijing/W lineage of MTB. Since there are low isolate numbers in sublineage 142, some scholars have conjectured that it may be the most recently evolved of the sublineages in this monophyletic group and has therefore not yet had time to proliferate and disseminate. It may therefore be less pathogenic [36]. However, another report showed that 80% of the strains from sublineage 142 (8/10 tested) synthesized detectable quantities of phenolic glycolipid (PGL-tb), which is an important virulence factor for a fraction of the Beijing/W lineage isolates that are likely to be associated with pulmonary TB manifestations within the human host [37]. PGL-tb suppresses the release of the pro-inflammatory cytokine, tumour necrosis factor-alpha, as well as interleukins 6 and 12 in vitro [38], and sublineage 142 has been associated with extra-pulmonary TB [37]. These findings imply that sublineage 142 may in fact be more pathogenic than previously thought. Sublineage 150 is another interesting sublineage of the Beijing/W lineage and is worthy of discussion. Two recent reports have shown that the numbers of sublineage 150 have occupied second place, behind sublineage 181, among the top five sublineages of Beijing/W lineage [21, 36]. Sixty percent of the strains from sublineage 150 (6/10 tested) synthesized PGL-tb [37] and sublineage 150 was also associated with extra-pulmonary TB [37]. In 2007, Hanekom et al. used IS6110 DNA fingerprinting, single-nucleotide polymorphisms (SNPs) and LSPs to analyse the genomic structure of the Beijing/W lineage, which suggested that sublineage 150 has the ability to transmit; sublineage 150 has also been associated with MDR-TB in rural settings [27]. The most obvious characteristic of sublineage 181 is the number of strains accounted for absolute advantage in Beijing/W lineage MTB [21, 36]. In our study, 69·59% of isolates belonged to sublineage 181, this result is similar to current reports in San Francisco (sublineage 181: 64·4%) [36]. The 88·74% (134/151) of sublineage 181 in Xinjiang were farmers from 11 different regions. Based on these patients’ current address information, we did not find the means of mutual transmission and direct contact. Therefore, these pulmonary TB patients of sublineage 181 referred to sporadic cases. A study from Reed et al. found that 10% of the sublineage 181 strains tested produced PGL-tb [37]. There are only a few reports on sublineage 207, which is significantly associated with the highest frequency of secondary cases [36]. Reed et al. reported there was only one strain of sublineage 207, and found that it did not produce PGL-tb [37]. Previous studies did not find any associations between drug resistance and pathogenicity for sublineage 105 [27].
Some studies have shown that Beijing/W lineage strains are associated with drug resistance and MDR [9, 39]. MDR is significantly higher in Beijing/W lineage strains compared to non-Beijing/W lineage strains [40]. To further discuss the association between Beijing/W lineage strains and drug resistance, some scholars have subdivided the Beijing/W lineage strains into sublineages using different molecular epidemiological methods and studied the association between these sublineages and drug resistance. Previous studies have shown that several sublineages occur with a significantly higher frequency in the MDR/XDR (extensively drug-resistant) population [9]. These findings suggest that different sublineages of the Beijing family may differ in their mechanisms of adaptation to drug selection pressures. In these studies, the Beijing/W lineage strains were not subdivided into sublineages based only on LSPs, but synonymous SNPs and IS6110 DNA fingerprints were also used at the same time [9, 34]. Other studies have shown that the Beijing/W lineage can be roughly subdivided into ancient sublineages [possessing an intact narcotics task force (NTF) region] and modern sublineages (possessing one or two IS6110 insertions on the right side of the NTF region) by determining whether or not the NTF region is present within the IS6110 sequence [28].
There are only a few reports that analyse the subdivisions of the Beijing/W lineage using LSPs and discuss the association between these Beijing/W lineage and drug resistance [36]. For the low frequency of drug-resistant TB in San Francisco, there are no statistical correlations between drug resistance and the different sublineages. Although our results revealed a high frequency of drug-resistant MTB in Xinjiang, our data still did not demonstrate any association between drug resistance and the different sublineages. The reason for this may because the sample sizes for sublineages 150 and 142 (those most likely to be related to drug resistance and pathogenicity) were too small. The Beijing/W lineage stains have previously been closely associated with nosocomial infections and community outbreaks, global transmission, and drug resistance. Xinjiang is one of the drug-resistant TB hotspots in China, thus the increased drug-resistance rate of MTB remains a serious problem in Xinjiang. Further research using a larger sample of isolates is required to fully explore this association between drug resistance and the different Beijing/W lineage sublineages. Given that no correlation has been found between drug resistance and sublineages, it should be interesting to use molecular epidemiological methods to distinguish individual isolates in order to identify potential outbreaks of drug-resistant isolates.
ACKNOWLEDGEMENTS
The authors thank all TB patients participating in this study and the staff of the First Hospital of Shihezi University, Shihezi, China for recruiting the patients. The authors acknowledge the financial support of the National Natural Science Foundation of China (No. 30960356; 81160368).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Hirsh AE, et al. Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proceedings of the National Academy of Sciences USA 2004; 101: 4871–4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gagneux S, Small PM. Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infectious Diseases 2007; 7: 328–337. [DOI] [PubMed] [Google Scholar]
- 3.Gagneux S, et al. Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences USA 2006; 103: 2869–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Soolingen D, et al. Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. Journal of Clinical Microbiology 1995; 33: 3234–3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsolaki AG, et al. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proceedings of the National Academy of Sciences USA 2004; 101: 4865–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsolaki AG, et al. Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. Journal of clinical microbiology 2005; 43: 3185–3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anh DD, et al. Mycobacterium tuberculosis Beijing genotype emerging in Vietnam. Emerging Infectious Diseases 2000; 6: 302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson R, et al. An outbreak of drug-resistant tuberculosis caused by a Beijing strain in the western Cape, South Africa. International Journal of Tuberculosis and Lung Disease 2006; 10: 1412–1414. [PubMed] [Google Scholar]
- 9.Toungoussova OS, et al. Spread of drug-resistant Mycobacterium tuberculosis strains of the Beijing genotype in the Archangel Oblast, Russia. Journal of Clinical Microbiology 2002; 40: 1930–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frieden TR, et al. A multi-institutional outbreak of highly drug-resistant tuberculosis: epidemiology and clinical outcomes. Journal of the American Medical Association 1996; 276: 1229–1235. [PubMed] [Google Scholar]
- 11.Lan NT, et al. Mycobacterium tuberculosis Beijing genotype and risk for treatment failure and relapse, Vietnam. Emerging Infectious Diseases 2003; 9: 1633–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith NH, et al. The population structure of Mycobacterium bovis in Great Britain: clonal expansion. Proceedings of the National Academy of Sciences USA 2003; 100: 15 271–15 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebrahimi-Rad M, et al. Mutations in putative mutator genes of Mycobacterium tuberculosis strains of the W-Beijing family. Emerging Infectious Diseases 2003; 9: 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao XQ, et al. Preliminary analysis of the distribution of Beijing genotype strains of Mycobacterium tuberculosis in parts of China. Chinese Journal of Practical Preventive Medicine 2012; 19: 662–664. [Google Scholar]
- 15.Jia W, et al. The report of WHO drug resistance surveillance on tuberculosis, Xinjiang. Journal of the Chinese Antituberculosis Association 2008; 30: 307–310. [Google Scholar]
- 16.China Antituberculosis Association. The laboratory science procedure of diagnostic bacteriology in tuberculosis. Bulletin of the Chinese Antituberculosis Association 1996; 18: 28–31. [Google Scholar]
- 17.Zhang J, et al. Genotypes and drug susceptibility of Mycobacterium tuberculosis isolates in Shihezi, Xinjiang Province, China. BMC Research Notes 2012; 5: 309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dou HY, et al. Molecular epidemiology and evolutionary genetics of Mycobacterium tuberculosis in Taipei. BMC Infectious Diseases 2008; 8: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huard RC, et al. PCR-based method to differentiate the subspecies of the Mycobacterium tuberculosis complex on the basis of genomic deletions. Journal of Clinical Microbiology 2003; 41: 1637–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jinghua L, et al. A new method for the identification of the ‘Beijing family’ strain of Mycobacterium tuberculosis. Chinese Journal of Microbiology and Immunology 2008; 28: 172–175. [Google Scholar]
- 21.Reed MB, et al. Major Mycobacterium tuberculosis lineages associate with patient country of origin. Journal of Clinical Microbiology 2009; 47: 1119–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manca C, et al. Virulence of a Mycobacterium tuberculosis clinical isolate in mice is determined by failure to induce Th1 type immunity and is associated with induction of IFN-alpha/beta. Proceedings of the National Academy of Sciences USA 2001; 98: 5752–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toungoussova OS, et al. Molecular epidemiology and drug resistance of Mycobacterium tuberculosis isolates in the Archangel prison in Russia: predominance of the W-Beijing clone family. Clinical Infectious Diseases 2003; 37: 665–672. [DOI] [PubMed] [Google Scholar]
- 24.Cox HS, et al. The Beijing genotype and drug resistant tuberculosis in the Aral Sea region of Central Asia. Respiratory Research 2005; 6: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glynn JR, et al. Worldwide occurrence of Beijing/W strains of Mycobacterium tuberculosis: a systematic review. Emerging Infectious Diseases 2002; 8: 843–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glynn JR, et al. Mycobacterium tuberculosis Beijing genotype, northern Malawi. Emerging Infectious Diseases 2005; 11: 150–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanekom M et al. A recently evolved sublineage of the Mycobacterium tuberculosis Beijing strain family is associated with an increased ability to spread and cause disease. Journal of Clinical Microbiology 2007; 45: 1483–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mokrousov I, et al. Evolution of drug resistance in different sublineages of Mycobacterium tuberculosis Beijing genotype. Antimicrobial Agents and Chemotherapy 2006; 50: 2820–2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strauss OJ, et al. Spread of a low-fitness drug-resistant Mycobacterium tuberculosis strain in a setting of high human immunodeficiency virus prevalence. Journal of Clinical Microbiology 2008; 46: 1514–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thwaites G, et al. Relationship between Mycobacterium tuberculosis genotype and the clinical phenotype of pulmonary and meningeal tuberculosis. Journal of Clinical Microbiology 2008; 46: 1363–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rindi L, et al. Evolutionary pathway of the Beijing lineage of Mycobacterium tuberculosis based on genomic deletions and mutT genes polymorphisms. Infection, genetics and evolution. Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases 2009; 9: 48–53. [DOI] [PubMed] [Google Scholar]
- 32.Stavrum R, et al. Genomic diversity among Beijing and non-Beijing Mycobacterium tuberculosis isolates from Myanmar. PLoS One 2008; 3: e1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rajapaksa US, Perera AJ. Sublineages of Beijing strain of Mycobacterium tuberculosis in Sri Lanka. Indian Journal of Microbiology 2011; 51: 410–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao TJ, et al. Characteristics of epidemiology of Beijing/W lineage Mycobacterium tuberculosis isolates in Changping, Beijing. Chinese Journal of Zhengzhou University: Medical Sciences 2010; 45: 48–50. [Google Scholar]
- 35.Kong Y, et al. Population-based study of deletions in five different genomic regions of Mycobacterium tuberculosis and possible clinical relevance of the deletions. Journal of Clinical Microbiology 2006; 44: 3940–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kato-Maeda M, et al. Differences among sublineages of the East-Asian lineage of Mycobacterium tuberculosis in genotypic clustering. International Journal of Tuberculosis and Lung Disease 2010; 14: 538–544. [PMC free article] [PubMed] [Google Scholar]
- 37.Reed MB, et al. The W-Beijing lineage of Mycobacterium tuberculosis overproduces triglycerides and has the DosR dormancy regulon constitutively upregulated. Journal of Bacteriology 2007; 189: 2583–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reed MB, et al. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 2004; 431: 84–87. [DOI] [PubMed] [Google Scholar]
- 39.Nikolayevskyy VV, et al. Molecular epidemiology and prevalence of mutations conferring rifampicin and isoniazid resistance in Mycobacterium tuberculosis strains from the southern Ukraine. Clinical Microbiology and Infection 2007; 13: 129–138. [DOI] [PubMed] [Google Scholar]
- 40.Kruuner A, et al. Spread of drug-resistant pulmonary tuberculosis in Estonia. Journal of Clinical Microbiology 2001; 39: 3339–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]