Abstract
In this communication, we describe the use of specialized transposons (Tn5 derivatives) to create deletions in the Escherichia coli K-12 chromosome. These transposons are essentially rearranged composite transposons that have been assembled to promote the use of the internal transposon ends, resulting in intramolecular transposition events. Two similar transposons were developed. The first deletion transposon was utilized to create a consecutive set of deletions in the E. coli chromosome. The deletion procedure has been repeated 20 serial times to reduce the genome an average of 200 kb (averaging 10 kb per deletion). The second deletion transposon contains a conditional origin of replication that allows deleted chromosomal DNA to be captured as a complementary plasmid. By plating cells on media that do not support plasmid replication, the deleted chromosomal material is lost and if it is essential, the cells do not survive. This methodology was used to analyze 15 chromosomal regions and more than 100 open reading frames (ORFs). This provides a robust technology for identifying essential and dispensable genes.
[Supplemental material is available online at www.genome.org and is supplied as an extended table enumerating genes lost in two multiple round deletion strains (Δ20-1 and Δ20-4). These data are summarized in Table 1.]
Transposons are powerful tools for performing genomic structure/function studies. They have long been used in the generation of knockout mutations. With the advent of in vitro transposition systems (Devine and Boeke 1994; Gwinn et al. 1997; Akerley et al. 1998; Goryshin and Reznikoff 1998; Griffin IV et al. 1999; Haapa et al. 1999), the use of transposons in genome analysis has been greatly expanded to include, for instance, their being applied as mobile primer binding sites in high-throughput sequencing efforts (Butterfield et al. 2002; Shevchenko et al. 2002). In this communication, we will describe another powerful application of DNA transposition that combines in vitro and in vivo Tn5-based technologies to generate random deletions in the Escherichia coli K12 genome. As will be described, the Tn5 deletion formation system can be used in essentially any bacterial species to define essential genes, to generate minimal essential genomes, and to clone genes.
The entire DNA sequences of many bacterial genomes have been determined. However, the experimentalist is still faced with the challenge of determining the role of various putative genes. Transposition-based strategies have been developed recently for identifying essential genes (for review, see Judson and Mekalanos 2000a; Hamer et al. 2001; Gerdes et al. 2002). Conceptually, these methods are based on the fact that transposon insertion into a gene causes loss of gene function (gene knockout), and insertion into an essential gene is lethal to the organism and cannot be observed (Akerley et al. 1998; Hare et al. 2001). By generating large libraries with chromosomal transposon insertions, followed by sequencing or PCR analysis of inserts in surviving cells, it can be assumed that any gene that is not found to contain a transposon insertion is essential. In essence, these methods catalog observed nonessential genes and assume that other genes must therefore be essential. An important exception is the development of a method using a transposon with regulated outward-facing promoters. Insertion of this transposon into the promoter region of an essential gene can be recovered under conditions in which the transposon promoters are turned on to support expression of the essential gene. Insertions can then be screened on medium that does not turn on the transposon promoter, resulting in loss of viability of those cells that depend on the conditional transposon promoter for growth. This results in the ability to positively identify essential genes and has been used for this purpose (Judson and Mekalanos 2000b). However, the technique is hindered by the fact that few of the transposon inserts within a population will be inserted into gene promoter regions.
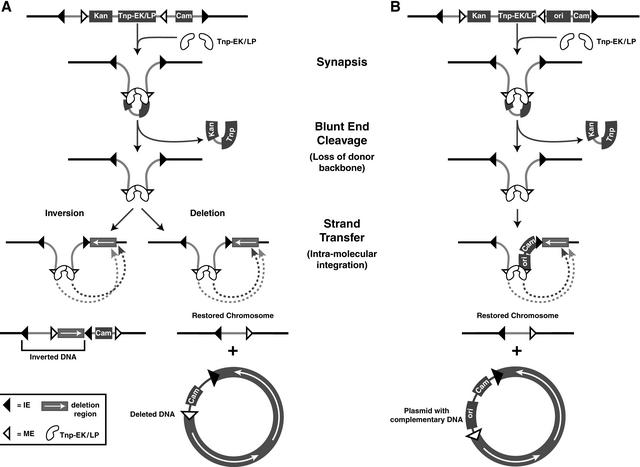
In addition to insertions, transposons are also capable of promoting other types of DNA rearrangements. Deletion (and inversion) formation is a natural feature of composite transposons. Composite transposons can create deletions or inversions by an intramolecular transposition mechanism (Fig. 1). Internal transposon ends are oriented in such a way that DNA between the ends can be considered as the donor DNA, which is released by transposase-catalyzed cleavage. The rest of the DNA (a plasmid or a chromosome) is recognized as a transposon that can undergo self-integration, leading to a deletion or inversion event. During this process, two deletions are formed, removal of the internal part of the transposon by double-ended cleavage at the transposon ends and deletion of a portion of the chromosome by the integration event.
Figure 1.
Strategy for recursive deletion and coupled deletion/plasmid formation systems. The strategy for deletion formation can be used after integration of the transposon into the host's genome. The internal transposon ends (MEs) are used in the second transposition event. Two deletions result from this transposition event in vivo, the first leading to the removal of the internal part of the transposon, and the second resulting in the deletion of a portion of the chromosome. TnpEK/LP binds to the MEs, resulting in blunt-end cleavage and loss of the donor DNA. Tnp-EK/LP then facilitates intra molecular strand transfer into the chromosome. (A) Not all events during strand transfer will result in deletion. Inversions may also result in this mechanism. Deletions may happen to the left or to the right, defining the loss of the corresponding part of the transposon. Deletions to the right will result in loss of all transposon DNA, with the exception of a neutral linker. This event can be detected by replica plating (under conditions of high transposition frequency). Traditional transposition by use of the external pair of transposon ends (IEs) does not occur because the expressed Tnp does not interact with the external ends. (B) The addition of a conditional origin of replication allows for the capture of the deleted chromosomal material into a complementary self-replicating plasmid. The presence of IPTG in the medium results in the capture of these circular DNAs as plasmids that are complementary to the chromosome.
The system that we will describe makes use of both types of deletions associated with intramolecular transposition (Fig. 1A). Double-ended cleavage eliminates the transposase gene and the selectable marker used for the prior transposon insertion selection. The self-integration reaction results in formation of the second deletion that starts at the point of transposon location and extends to the point on the chromosome or plasmid defined by the second transposition event. This deletion is the deletion of interest. An important feature of this scenario is that the protocol eliminates all selectable markers and the transposase gene and, thus, can be performed repetitively for an accumulation of deletions.
In this communication, we will describe the use of the transposon-based deletion strategy as a tool for both defining essential genes and for removing nonessential genes from the chromosome.
RESULTS
Deletion Modules: Composition and Strategy
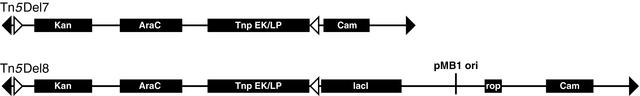
The transposons we used exploit the observation that composite transposons make deletions by use of internal transposon ends. The structures of transposons used in this study are shown in Figure 2. The transposon Tn5Del7 is designed for immediate deletion of chromosomal DNA and does not contain an origin of replication. The Tn5Del8 transposon contains a conditional origin of replication that allows the capture of deleted chromosomal DNA in the cell as a complementary plasmid. Elimination of this plasmid is triggered by the removal of IPTG from the medium.
Figure 2.
Structure and features of transposons used in this work. The structure of Tn5Del7 and Tn5Del8 is conceptually the same. Both transposons are defined by IE sequences CTGTCTCTTGATCAGATCT (open triangle indicates IE sequence). Two ME sequences CTGTCTCTTATACACATCT (filled triangle indicates ME sequence) are faced toward each other, defining donor DNA for transposition using these ends. Distance between the tips of the IE and ME ends on the left is 64 bp. This is the size of the linker that remains in the chromosome after deletion. Donor DNA encodes the Tnp gene for ME end-mediated transposition under the control of an arabinose inducible promoter and also a KmR gene (Kan). Between the right pair of transposon ends, both transposons have a selectable marker (Cam). The only difference is that Tn5Del8 has a conditional origin of replication. Lac repressor encoded by the lacI gene controls the origin. Moderate plasmid (after its formation) copy number is ensured by Rop function.
The particular features of the transposons used in these experiments are as follows. Both transposons are flanked by 19-bp inside ends (IEs) and can be separated from the donor DNA component of the vector by digestion with PshAI. The Tn5 IE ends are capable of participating in synaptic complex formation in the presence of mutant Tnp protein Tnp sC7v2.0 (Naumann and Reznikoff 2002). The synaptic complexes are delivered into cells by electroporation (Goryshin et. al. 2000). The second set of 19-bp end sequences in the deletion modules are artificially constructed mosaic ends (MEs) (Zhou et. al. 1998). We use MEs in combination with Tnp EK/LP for the second transposition step, as this was the most efficient Tnp:DNA end combination available when these experiments were performed. A high efficiency of in vivo transposition events is absolutely critical for their identification by screening.
The deletion formation protocol is as follows (also see Fig. 1). Cells are electroporated with preformed transposome complexes, and transposon inserts are selected for by plating on LB medium containing both kanamycin and chloramphenicol. Then, either a single colony or a collection of colonies is used to start a liquid culture. At early exponential phase, arabinose is added to induce Tnp EK/LP synthesis. In the case of Tn5Del8, IPTG and chloramphenicol are added to activate the transposon origin of replication and to select for the presence of the excised plasmid, respectively. After a few hours, cells are subcultured in the same medium with a 500-fold dilution and grown overnight with shaking. Cells are then diluted and plated on agar medium containing the same components to obtain single colonies for replica plating. In the case of Tn5Del7, colonies that are sensitive to both kanamycin and chloramphenicol are picked as deletions. In the case of Tn5Del8, cells are checked for sensitivity to kanamycin on plates that contain IPTG. Plasmid DNA is then isolated for further analysis.
Deletions in the Lactose Operon Region
The maximum size of chromosomal DNA that can be deleted in a single transposition-mediated event can be limited by the relative location of essential DNA. To analyze the distribution of deletion sizes created by Tnp without limitations imposed by the presence of essential genes, we chose to isolate a Tn5Del7 insert in the lactose operon, as it is known that deletions exceeding 100 kb can be isolated in this region with no negative impact on cell growth in rich medium (Bachmann 1996). After electroporation of the deletion module into MG1655 and plating on Lactose-MacConkey agar with kanamycin and chloramphenicol, we selected a few white colonies among thousands of red colonies. One colony was isolated, and the transposon insert site was sequenced. The insert was located at 362,522 bp on the E. coli chromosome within the lacZ gene.
Transposition events were stimulated by inducing Tnp EK/LP synthesis. The resulting colonies were tested for transposition-associated events by replica plating as described above and in the Methods section. As was typical of experiments using Tn5Del7, transposition events had occurred in ∼50% of the surviving cells with 1/4 of these events being deletions in the desired direction, as indicated by loss of resistance to chloramphenicol. The large fraction of cells that had undergone transposition-associated events is presumably due to the hyperactive nature of the transposase end-sequence combination that was used. In addition, Tnp expression is known to be relatively toxic (Weinreich et al. 1994), and, therefore, cells that undergo transposition and loose the Tnp gene would have a selective advantage.
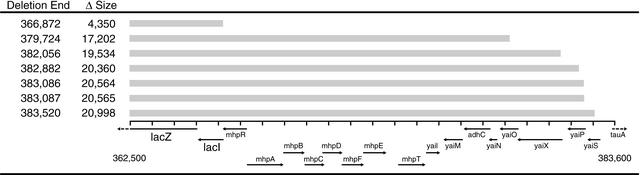
A total of nine independent deletions were selected and analyzed by DNA sequencing either directly from chromosomal DNA or by the use of inverse PCR. Two deletions ended within the Cam gene. The other seven deletions are described in Figure 3. The deletion sizes varied from 4–23 kb, with most deletion sizes being around 20 kb. This large deletion size makes it feasible to create a minimal genome in a reasonable amount of time by using our method recursively.
Figure 3.
Deletions characterized for the lactose operon region. Genes deleted in E. coli MG1655 with the deletion transposon Tn5Del7 are shown by a gray line for each of the seven deletions in the lac operon. The insertion site was located 362,522 bp in the E. coli chromosome within the lacZ gene.
Recursive Deletion Formation
An important feature of this transposition/deletion system is that after the deletion is generated, all components of the transposon are lost except for a short linker (64 bp). The loss of the transposase gene ensures the stability of the chromosome (in terms of transposition). The loss of all selectable markers provides an opportunity for recursive deletion formation using transposon insertion and transposase-mediated deletion formation, repetitively.
We performed 20 rounds of recursive deletion formation in strain MG1655. Rather than focusing on one deletion at a time, we isolated and mixed at least 10 colonies following each round. This was then used as the starting material for the subsequent rounds. It was assumed that strains with debilitating deletions would be lost because other deletion strains with unimpaired growth would out compete them during growth of the mixed culture. We obtained 10 final strains after 20 rounds that had a growth rate equal to that of the parental strain (data not shown).
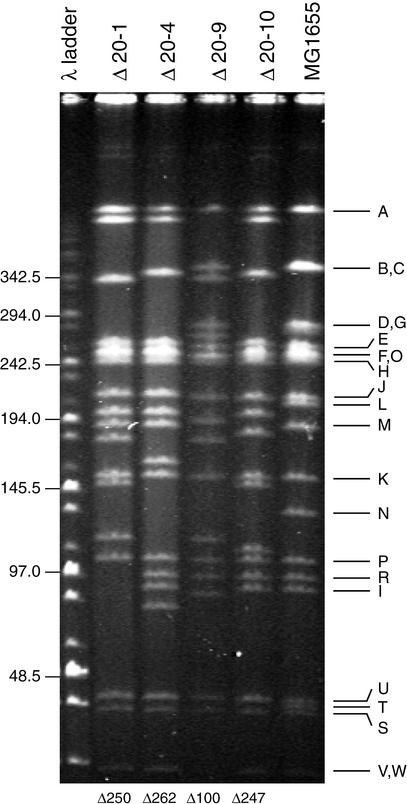
Pulsed-field gel electrophoresis analysis of NotI-digested chromosomal DNA was used to analyze the diversity among the 10 deletion strains and to estimate the average deletion size that occurred. An example of such a gel is shown in Figure 4. The differences and similarities between samples are evident by inspection of the gel. In some cases, bands disappear from their original position (in the MG1655 DNA digestion) and are substituted by shorter fragments. In other cases, longer fragments appear, presumably due to the loss of NotI site(s), which results in the formation of a large band coupled with the loss of two or more smaller bands. We calculated the total amount of DNA deleted for 4 strains that had undergone 20 rounds of deletion by estimating the size of each chromosomal band compared with marker DNAs. From several gels that were run under various conditions to resolve different regions in the DNA pattern, we calculated that the four strains whose DNAs were analyzed in Figure 4 contain deletions of 250, 262, 100, and 247 kb. This indicates that the average deletion size per round is 11 kb, which is in reasonable agreement with the results obtained for the lactose operon region in the previous experiment.
Figure 4.
Pulsed-field gel electrophoretic analysis of MG1655 deletion derivatives after 20 rounds of transposition and deletion. Transpositional deletion of chromosomal DNA was performed recursively for a total of 20 cycles. After these 20 cycles, chromosomal DNA was prepared from 4 deletion strains as well as the initial strain MG1655. The chromosomal DNA was digested to completion with NotI restriction endonuclease and analyzed by pulsed-field gel electrophoresis. Visualization by ethidium bromide staining and UV irradiation revealed the pattern of DNA bands. The pattern for MG1655 matches that reported previously (Heath et al. 1992), and the identification letter for each fragment is shown at right. For each of the deletion strains, the pattern is altered and indicates a loss of DNA. The size of each new band in the four deletion strains was estimated by comparison with DNA markers. The estimation for total amount of deleted DNA in each of the strains is shown at bottom.
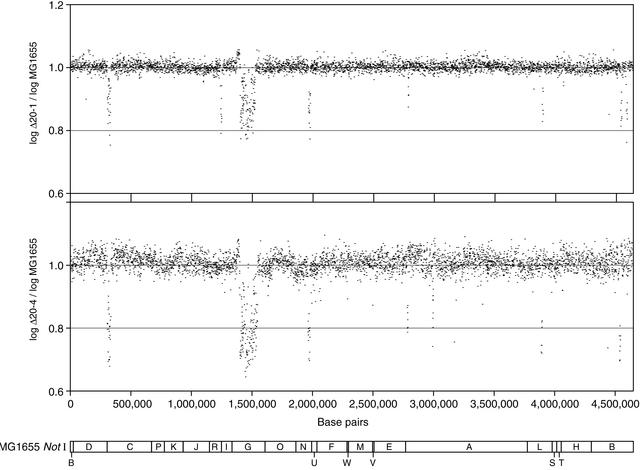
Microarray Mapping of Deletions
The locations of deletions found in strains Δ20-1 (as judged by the pulsed-field gel electrophoresis to be missing ∼250 kb) and Δ20-4 (missing ∼262 kb) were mapped by microarray hybridization experiments versus the progenitor strain. This technique provided an excellent means to determine which genes were removed by the transposition/deletion procedure. Figure 5 presents the results schematically and Table 1 describes the results in more detail. The two strains are apparently each lacking four common sets of genes. Δ20-1 has seven unique deletions and Δ20-4 has five unique deletions. The data suggests that the two strains diverged after the fourth common deletion was generated. The two strains contain a wide range of deletion sizes. The deletion sizes presumably reflect the locations of the essential genes nearest the different transposon inserts and/or differences in localized DNA condensation near various inserts (see below). The mapped deletions are consistent with both the pulsed-field gel electrophoresis pattern and the total amount of DNA missing, as determined by the pulsed-field gel electrophoresis patterns.
Figure 5.
Microarray deletion map of deletions found in strains Δ20-1 and Δ20-4. Each dot represents the log ratio of normalized signal intensities for a particular ORF from the deletion strain to its control strain counterpart. Deletion regions were determined by consecutive signal ratios of E. coli ORFs below 1 and P-values <0.05. These hybridization results were plotted against base pair location for comparison with a NotI restriction map (Heath et al. 1992).
Table 1.
Genes Deleted in Strains Δ20-1 and Δ20-4
| Deletion | B-Numbera | Map Positionb | Estimated Sizec (bp) | |
| Δ20-1 | Δ20-4 | |||
| 1.1 | b0116 | 2.8 | 1,424 | |
| 1.2 | 4.1 | b0288–b0315 | 6.5–7.2 | 29,606 |
| 1.3 | b1188–b1197 | 26.6–26.9 | 11,667 | |
| 1.4 | b1335–b1452 | 30.1–32.8 | 124,647 | |
| 4.2 | b1336–b1471 | 30.1–33.3 | 145,467 | |
| 1.5 | 4.3 | b1878–b1892 | 42.3–42.6 | 15,625 |
| 4.4 | b2379 | 53.8 | 1,238 | |
| 1.6 | b2649–b2655 | 60.0 | 3,604 | |
| 4.5 | b2648–b2655 | 60.0 | 4,117 | |
| 4.6 | b2853–b2861 | 64.5–64.6 | 4,293 | |
| 4.7 | b3027 | 68.3 | 332 | |
| 1.7 | b3647 | 82.3 | 1,688 | |
| 1.8 | 4.8 | b3708–b3715 | 83.8–84.0 | 8,724 |
| 1.9 | b4216 | 95.6 | 554 | |
| 1.10 | 4.9 | b4308–b4315 | 97.7–97.9 | 9,252 |
| 1.11 | b4349–b4354 | 98.7–98.9 | 2,165 | |
Deletion span based on b-number (Blattner et al. 1997).
Map position of genes lost within deletion span.
Size is estimated from left end of first deleted ORF to right end of last detected ORF.
There are a few surprises. First, we found positive hybridization results interrupting what would otherwise appear to be a contiguous set of deleted genes. This observation is likely due to a phenomenon observed by Richmond et al. (1999) that cross hybridization occurs between IS or paralog-containing sequences. The majority of these positive hybridization signals do represent IS sequences found elsewhere in the genome or genes that have known paralogs in the genome. Another surprising result is that there are fewer apparent deletions in each of the strains than might be expected from 20 cycles (9 and 11 deletions were found in the two strains). It is possible that some deletions may have ended within the transposon as found in our lac insert-deletion analysis and, thus, they would not be detected. Alternatively, some deletions may be too small to have been picked up by the microarray hybridization technique. In some cases, what looks like one deletion by the array data may really represent two or more adjacent deletions. Finally, both of the strains each have one extremely large deletion (∼125,000 and 146,000-bp long) in NotI fragment G, near the chromosome replication terminus. These two deletions have one end in common (near the NotI I/G fragment boundary), which suggests that they arose from the same insertion event, but had separate deletion events. Thus, it is likely that the two strains diverged with the formation of these deletions. The observation that these deletions were both relatively large indicates that the transposition/deletion system is capable of generating extremely large deletions, that there are no essential genes in this region, and that the chromosome condensation of this region of the chromosome may favor large deletion formation.
Coupled Chromosomal Deletion/Plasmid Formation System
The obvious limitation of the deletion formation system described above is the inability to introduce deletions involving essential genes. To address this, we developed a technique to conditionally save deleted DNA in the same cell and to attempt its elimination later. In this manner, the essentiality of various sections of the chromosome can be directly tested by removal of the deleted DNA from the cell and testing for viability. Tn5Del7 was modified by inserting a conditional origin of replication to generate Tn5Del8 (Fig. 2). When a deletion is formed, the deleted material is excised as a circle (Fig. 1B). Because the circle contains an origin of replication, the deletion has formed a replicating plasmid.
The protocol used for the Tn5Del8 experiments is described above, in the Methods section, and in Figure 1B. The critical feature is that IPTG is present during all steps, commencing with the induction of transposase in order to support replication of plasmids formed during deletion formation. In addition to supporting replication of the newly formed plasmids, addition of IPTG activates origins of replication that remain on the chromosome in cells that have not undergone deletion formation. This activation presumably depresses cell growth and creates selection against cells that had not undergone excision of the transposon-encoded origin of replication, as in these experiments, >95% of the resulting colonies contained the desired class of deletions.
We used a coupled deletion/plasmid system for the search of essential genes. A total of 15 insertion events were treated independently as above to isolate deletion events. Final induced cultures for each were diluted and plated on agar medium containing chloramphenicol and IPTG. For each insert, a total of 20 colonies were analyzed by isolation of plasmid DNA, followed by an estimation of plasmid size by agarose gel electrophoresis. In each case, the largest plasmid isolated was analyzed by DNA sequencing of the two transposon/chromosomal DNA junctions to determine the portion of the chromosome that was deleted. Cells from the colony were then streaked onto plates containing only LB (no IPTG) to determine whether cells could survive without the deleted chromosomal material. The colonies were also streaked on LB plates with chloramphenicol and no IPTG to confirm loss of plasmid without integration into the chromosome. Growth of cells on this medium was not detected in any case, indicating that reintegration of the plasmid did not occur.
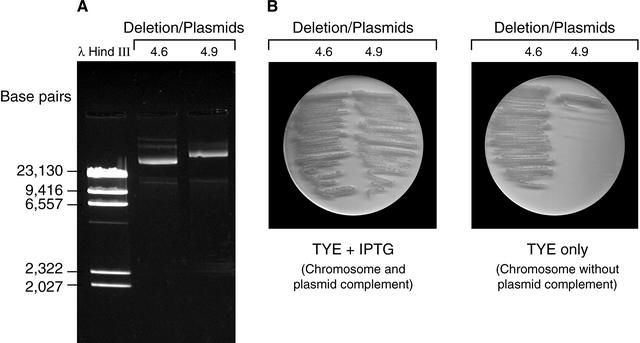
Results of this analysis are shown in Table 2. Of 15 insertion strains, 11 yielded viable cells following plasmid loss from the largest isolated deletion. This indicated that these regions of DNA are dispensable. In the other four cases, cells were unable to grow without the deleted plasmid DNA, indicating the presence of at least one essential gene in the deleted material. An example of this is deletion 4.9. In this case, a smaller deletion, 4.6, was analyzed and found to survive in the absence of the plasmid (Fig. 6). In this case, the result indicates that genes glyS and/or glyQ are essential.
Table 2.
The Summary of Analyzed Deletion/Plasmid Combinations for Different Insertions
| Insert No. | Δ Νο. | Insert End | Δ End | Size | Partial ORFs | Complete ORFs | Grow w/o IPTG |
| 1 | 1.3 | 1073109 | 1094823 | 21714 | b1028; b1012 | b1013; putA; putP; b1016–1018; ycdB; phoH; b1021–1025; tra5Δ3; b1027 | Y |
| 2 | 2.8 | 2776930 | 2770104 | 6826 | b2638; b2647 | b2639–b2646 | Y |
| 3 | 3.1 | 4407731 | 4403605 | 4126 | yjfl | yjeB; vacB; yjfH | Y |
| 4 | 4.2 | 3735047 | 3725227 | 9821 | yiaB | xylB; xylA; xylF; xylG; xylH; xylR; b3570 | Y |
| 4 | 4.6 | 3735047 | 3723521 | 11526 | none | yiaH; yiaA; yiaB; xylB; xylA; xylF; xylG; xylH; xylR;b3570 | Y |
| 4 | 4.9 | 3735047 | 3719490 | 15557 | yi5b | glyS; glyQ; yiaH; yiaA; yiaB; xylB; xylA; xylF; xylG;xylH; xylR;b3570 | N |
| 5 | 5.1 | 1224154 | 1210042 | 14112 | mcrA; minD | b1160-b1173; minE | Y |
| 6 | 6.2 | 3045069 | 3042244 | 2825 | bglA; gcvP | ygfF | Y |
| 7 | 7.1 | 2779269 | 2767042 | 12227 | b2633; b2647 | b2634–B2646 | Y |
| 8 | 8.4 | 3882592 | 3882500 | 92 | none | none | Y |
| 9 | 9.7 | 4589240 | 4595093 | 5853 | mdoB; tsr | yjjN; yjjM; b4356 | Y |
| 10 | 10.3 | 4141018 | 4117696 | 23322 | menA; frwC | hsIU; hsIV; ftsN; cytR; priA; rpmE; yiiX; metJ; metB; metL; metF; katG; yijE; yijF; gldA; talC; ptsA; yijl | N |
| 11 | 11.3 | 2793099 | 2795902 | 2803 | gabP; b2668 | ygaE; b2665–b2667 | Y |
| 12 | 12.17 | 14166 | 12088 | 2078 | none | dnaK | N |
| 13 | 13.19 | 4542104 | 4546611 | 4507 | fimC; fimH | fimD; fimF; fimG | Y |
| 14 | 14.17 | 3821524 | 3818998 | 2526 | spoT | rpoZ; gmk | N |
| 15 | 15.17 | 1570686 | 1544492 | 26194 | yddC; yddG | fdnG; fdnH; fdnl; b1477; b1478; sfcA; rpsV;b1481; osmC; b1483–1491; xasA; gadB | Y |
Figure 6.
An example of plasmid dependent strains. (A) The gel showing two plasmids generated from the same insertion of deletion module Tn5Del8. (B) Strains 4.6 and 4.9 are streaked on TYE medium with and without IPTG. The difference between plasmids 4.6 and 4.9 lies in the absence of glyS and glyQ in plasmid 4.6. The elimination of both plasmids may indicate that either glyS or glysQ, or both, are essential.
In the case of insert 8, only a short chromosomal DNA deletion was isolated. This transposon insert resides between open reading frames (ORFs), and the deletion does not contain any predicted ORFs. The inability to isolate larger deletions from this insert could be due to the fact that the area contains a large region of essential DNA that cannot be bypassed by typical deletion lengths. Alternatively, the localized condensation of the DNA may not be amenable to the formation of large deletions. A final possibility is that the capture of DNA immediately next to the insert, when transferred to the moderate copy number plasmid, may be lethal to the cell. We cannot distinguish between these options, but this area contains a stretch of essential genes, including gyrB and dnaA.
The data shown in Table 2 were compared with known information about the essentiality of E. coli ORFs by using the PEC database (www.shigen.nig.ac.jp/ecoli/pec/). Of the four deletions that we determined to contain essential DNA, all but one contained an ORF(s) that had been reported previously to be essential. In the remaining case, the deletion occurs in a single ORF, dnaK. Mutations in dnaK give rise to a conditionally essential phenotype at 37°C (Bukau and Walker 1989; Wild et al. 1992). Of the nonessential cases, all ORFs have been reported as either nonessential or unknown. The deletions that fall under this category indicate that 40 ORFs of unknown essentiality can now be considered nonessential.
DISCUSSION
In this communication, we describe a simple and reliable system for making chromosomal deletions in E. coli K-12. This system should also be of use in any other bacterial species for which the Tn5 transposome/electroporation strategy can be utilized. We used transposition driven by Tn5 transposase derivatives in such a way that we can separate two transposition events, each using different Tnp:transposon end combinations (Naumann and Reznikoff 2002). The first transposition event delivers components for the second transposition event into the chromosome via transposase–DNA complex codelivery (Goryshin et al. 2000). The second transposition event generates a deletion or inversion event. By simple screening for loss of antibiotic resistance, deletions with only a small portion of the transposon left in the chromosome can be chosen. As a variation of this technique, deleted material can be saved in the same cell as a conditional plasmid.
Determining the minimal genome content is a topic of interest for many laboratories. One approach used for determining the minimal genome is to assemble a theoretical minimal genome in silico by the comparison of a variety of different microbial genomes. This method assumes that all essential DNA exists in homologous form in all genomes, and that all nonessential DNA will be absent in one or more cases. Alternatively, the smallest genome among existing genomes (mycoplasma) has been analyzed by transposon knockout mutagenesis and large-scale sequencing (Hutchison III et al. 1999). Any genes that are found as knockout mutants are considered nonessential, and genes that do not contain knockouts are labeled as essential. It is then assumed that a chromosome containing only the essential genes would be sufficient for survival. However, this analysis assumes that genes that are not found to contain transposon knockouts are essential when they may, instead, be poor targets for the transposon and, more importantly, that removal of one gene has no effect on the essentiality of remaining genes. This second assumption likely results in an artificially low estimate of the number of genes that are needed to form a viable organism. In any event, the construction of a living cell with a minimal genome is desirable.
E. coli K-12 is an ideal subject for the minimal genome construction. It has a short generation time, extensive genetic tools, and a wealth of knowledge of its genome and physiology. It therefore seems attractive to try to generate a minimal or significantly reduced E. coli K-12 genome. In addition to the knowledge gained by the creation of such a strain, the strain itself could be useful. Two examples of the usefulness of such a strain are (1) overproduction of recombinant proteins with fewer E. coli proteins that need to be removed, and (2) as a simplified metabolic model that could be used for mathematical modeling of whole cell metabolism. We are currently going beyond the 20 rounds presented here in an effort to create such a strain. The average size of deletions observed in our experiments for the lac operon area and random deletions gives us hope that we will be able to reduce the size of the E. coli chromosome significantly in a reasonable time frame.
The large average deletion size is striking, because if the genomic DNA were a random coil, the intramolecular transposition site selection would be strongly biased toward distances within a few hundred base pairs of the transposon end sequences. We observed this bias in vitro, in which case, the DNA was free from packing (York at al. 1998). One explanation for the observed large in vivo deletion size is that the chromosomal DNA is not a random coil, but rather is a compact nucleoid body with a supercoiled domain structure that brings distant points into close proximity (Staczek and Higgins 1998). The isolation of two very large deletions (>100 kb) near the replicon terminus suggests that this region of the chromosome was unusually condensed.
Recently, two strategies for repetitive reduction of the E. coli chromosome have been described (Kolisnychenko et al. 2002; Yu et al. 2002). In the first communication, 12 planned deletions were introduced by the use of the efficient λ red recombination system. This approach is different from ours in several fundamental ways. First, the approach described by Kolisnychenko et al. (2002) generates planned deletions and thus uses the prior knowledge of which genes are dispensable, and requires knowledge of the genome sequence. Our system utilizes a random approach and can be used without prior knowledge of which genes are dispensable or prior sequence information. Secondly, the Kolisnychenko et al. (2002) approach requires the use of the λ red recombination system. We do not know whether this system has a limited host range. The strategy described in this communication uses the Tn5 transposase, which has been found to be active in all tested bacterial species (Goryshin at al. 2000). Third, the strategy described in this communication allows one to impose a selection for fast growing cells among several possible deletion strains. Finally, with our system, the deleted material can be saved as a conditional plasmid. Yu and colleges used the Cre/loxP system delivered by Tn5 derivatives and P1 transduction to locate two Tn5 inserts in the same cell (Yu et al. 2002). Importantly, this system depends on E. coli genetic tools (P1 transduction) and involves multiple steps for the formation of each deletion. Furthermore, it fails to save deleted material as a complementary plasmid.
In this communication, we started a list of essential/nonessential genes for E. coli by using the Tn5Del8 system. In the future, it is technically possible to create a representative library of deletions with complementary plasmids that would cover the entire E. coli genome multiple times. Systematic sequencing of deletion borders coupled with survival tests could be used to determine the essentiality of all of the genes in the entire chromosome. This type of analysis could also be adapted, through the use of an appropriate origin of replication, for analyzing the chromosomes of other bacteria.
METHODS
Medium
For all experiments, we used Luria-Bertanii (LB) broth liquid or agar medium or Lactose MacConkey agar medium (Sambrook et. al. 1989) modified to contain the following antibiotics when indicated: chloramphenicol (20 mg/L), kanamycin (40 mg/L), ampicillin (100mg/L).
Bacterial Strains
For making deletions, we used E. coli K12 strain MG1655 (Blattner et al. 1997). For DNA manipulations, we used E. coli K12 strain DH5α (Sambrook et al. 1989).
Plasmids
Plasmid pGT7 was the source of transposon Tn5Del7 (Fig. 2). Plasmid pGT7 was constructed by inserting different components into pGT4 (Nauman and Reznikoff 2002). In many cloning steps, blunt ends were generated using T4 DNA polymerase (Promega). Two ME sequences were taken from pPDM-2 (Epicentre) along with the KmR gene on a HindIII–EaeI fragment that was ligated to the large HindIII–SmaI fragment of pGT4, giving rise to pGT5. The Tnp EK/LP gene and the AraC gene with a portion of the KmR gene were taken from pGRARAK (I. Goryshin, unpubl.) on the BclI–ClaI fragment and ligated to the ClaI–NaeI large fragment of pGT5 to give pGT6. Finally, the CmR gene from pACYC184 was taken by isolating a BsaAI–PshAI fragment and ligating it into the EcoRI site of pGT6. The resulting plasmid contains the Tn5Del7 transposon with the structure shown in Figure 2.
Plasmid pGT8 was the source of transposon Tn5Del8 (Fig. 2). The two KpnI–BssSI fragments of pGT7 were ligated with an AatII–BamHI fragment of pAM34 (ATCC 77185, Hare et al. 2001). Tn5Del8 is similar in structure to Tn5Del7, except that it contains a regulated origin of replication.
Transposase Purification
Transposase protein TnpsC7v2.0 was purified using IMPACT T7 system (NEB) as described previously (Naumann and Reznikoff 2000).
Transposome Complex Preparation
Transposons Tn5Del7 and Tn5Del8 were cut out of the donor plasmid DNA by digestion with PshAI, followed by purification from an agarose gel using the QIAquick Gel Extraction Kit (QIAGEN).
Transposome complexes were assembled by incubation of precut transposon with TnpsC7v2.0 in 20 mM Tris Acetate (pH 7.5), 100 mM K Glutamate for 1 h at 37°C. The DNA:protein molar ratio was 1:5, with a DNA concentration of 0.1μg/μl.
Selection for Initial Transposition Events
Electroporation of complexes (Goryshin et al. 2000) was done using standard recommended conditions (2.5 Kv, 5mS).
After electroporation, cells were recovered by incubation for 1 h in LB broth, and then plated on LB agar containing kanamycin and chloramphenicol.
Induction of Deletions (Secondary Transposition Events)
Individual or pooled colonies from this initial selection were inoculated into LB medium with arabinose at a concentration of 0.4%, and grown for 8 h at 37°C. In the case of Tn5Del8, IPTG was also included at a concentration of 1 mM. Cells were subcultured with a 500-fold dilution in LB medium (plus chloramphenicol and IPTG for Tn5Del8), grown overnight, and plated with an appropriate dilution on LB agar (plus chloramphenicol and IPTG for Tn5Del8) to obtain individual colonies for analysis.
Screening for deletions with Tn5Del7 was done by replica plating colonies from LB agar onto agar containing LB, LB-kanamycin, and LB-chloramphenicol. Cells sensitive to kanamycin were considered to have undergone transposition, and cells sensitive to both drugs were considered to have a deletion of adjacent DNA and the correct portion of the transposon with only a small transposon linker being left on the chromosome. In the case of Tn5Del8, the cells of interest were kanamycin sensitive and chloramphenicol resistant, as the replication of excised DNA circles was supported by the presence of 1 mM IPTG in the medium.
Microarray Analysis
We used spotted DNA microarrays containing 95% of the 4290 ORFs from E. coli K-12 genome. These spotted microarrays were supplied by the Gene Expression Center, University of Wisconsin. Genomic DNAs from E. coli strain MG1655, Δ20-1 and Δ20-4 were prepared using a Master Pure DNA Purification Kit (Epicentre) digested with AluI (Promega) and further purified by phenol/chloroform extraction and ethanol precipitation. Genomic DNA labeling with Cy3 and Cy5 dUTP (NEN, Life Science) and hybridizations were performed according to protocols provided by the Gene Expression Center, University of Wisconsin (http://www.gcow.wisc.edu/Gec/index.htm). Microarray images were scanned using Packard BioChip SA5000 and quantitated using Scanalyze 2.1. Normalization of microarray signal intensities involved median background subtraction and calculation of percent signal intensities for each spot (Richmond et al. 1999). The percent signal intensities were used to determine the ratio of deletion strain to the control E. coli MG1655 signals. Values for percent signal intensity were log transformed in order to combine data for paired slide (dye swap) experiments. The ratios of deletion strain to control values were used for subsequent Z-score calculations to determine deletion sites. We established the null hypothesis for the presence of DNA, which was indicated by ratios with values close to 1 and that rejected the null for ratios with P values <0.05, which indicated the deleted material.
DNA Sequence Analysis
DNA sequencing was performed using an ABI PRISM model 377, according to the standard Big Dye protocol. For the lacZinsertion, we performed direct chromosomal DNA sequencing using primer FWD2, 5′-CAGATCTCATGCAAGCTTGA GCTC-3′, which is complementary to the transposon linker. For sequencing of deletions produced from this insertion, we generated DNA using inverse PCR (Ochman et al. 1990) after digestion of chromosomal DNA with FspI followed by ligation and 30 cycles of PCR (30 sec at 94°C, 1 min at 60°C, 1 min at 72°C) with primers 5′-GGTCTGCTTTCTGACAAACTCGGGC-3′ and 5′-ACGCGAAATACGGGCAGACATGGCC-3′ complementary to transposon ends. PCR products were purified from an agarose gel and sequenced with the standard Big Dye protocol.
Pulsed-Field Gel Electrophoresis
Pulsed-field gel electrophoresis was performed using the CHEF-DR II BioRad system according to the protocol found in Heath et al. (1992), with minor technical modifications. Running time was 40 h, voltage = 180 v, ramping – 5 to 80 sec.
WEB SITE REFERENCES
http://www.shigen.nig.ac.jp/ecoli/pec/; Genetic Resource Committee of Japan.
http://www.gcow.wisc.edu/Gec/index.htm; Gene Expression Center, University of Wisconsin-Madison.
Acknowledgments
We thank Barb Schriver for providing all of the medium and reagent preparations for this work. We thank Laura Vanderploeg and the Department of Biochemistry Media Lab for their valuable assistance with the figures and tables. We also thank Kelly Winterberg for her helpful discussion of the manuscript and the other members of our research laboratory for their support. This research was supported in part by NSF grant MCB-0084089, NIH grant GM50692, and a grant from the Robert Draper Technology Fund, Wisconsin Alumni Research Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL Reznikoff@biochem.wisc.edu; FAX (608) 265-2603.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.611403. Article published online before print in March 2003.
REFERENCES
- 1.Akerley B., Rubin, E., Camilli, A., Lampe, D., Robertson, H., and Mekalanos, J. 1998. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. 95: 8927-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachmann B.J., 1996. Derivations and genotypes of some mutant derivatives of the Escherichia coli K-12. In Escherichia coli and Salmonella, pp. 2460–2488. ASM Press, Washington, D.C.
- 3.Blattner F.R., Plunkett, G., III, Bloch, C.A., Perna, N.T., Burland, V., Rilley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K12. Science 277: 1453-1462. [DOI] [PubMed] [Google Scholar]
- 4.Bukau B. and Walker, G.C. 1989. Cellular defects caused by deletion of the Escherichia coli dnaK gene indicate roles for heat shock protein in normal metabolism. J. Bacteriol. 171: 2337-2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butterfield Y.S.N., Marra, M.A., Chan, S.T., Guin, R., Kryzwinski, M.I., Lee, S.S., MacDonald, K.W.K., Mathewson, D.A., Olson, T.E., Pandoh, P.K., et al. 2002. An efficient strategy for large-scale high-throughput transposon-mediated sequencing of cDNA clones. Nucleic Acids Res. 30: 2460-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devine S.E. and Boeke, J.D. 1994. Efficient integration of artificial transposons into plasmid targets in vitro: A useful tool for DNA mapping, sequencing and genetic analysis. Nucleic Acids Res. 22: 3765-3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerdes S.Y., Scholle, M.D., D'Souza, M., Bernal, A., Baev, M.V., Farrell, M., Kurnasov, O.V., Daugherty, M.D., Mseeh, F., Polanuyer, B.M., et al. 2002. From genetic footprinting to antimicrobial drug targets: Examples in cofactor biosynthetic pathways. J. Bacteriol. 184: 4555-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goryshin I.Y. and Reznikoff, W.S. 1998. Tn5 in vitro transposition. J. Biol. Chem. 273: 7367-7374. [DOI] [PubMed] [Google Scholar]
- 9.Goryshin I.Y., Jendrisak, J., Hoffman, L., Meis, R., and Reznikoff, W. 2000. Insertional transposon mutagenesis by electroporation of released Tn5 transposition complexes. Nat. Biotechnol. 18: 97-100. [DOI] [PubMed] [Google Scholar]
- 10.Griffin T.J., Parsons, L., Leschziner, A.E., DeVost, J., Derbyshire, K.M., and Grindley, N.D.F. 1999. In vitro transposition of Tn552: A tool for DNA sequencing and mutagenesis. Nucleic Acids Res. 27: 3859-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gwinn M.L., Stellwagen, A.E., Craig, N.L., Tomb, J.F., and Smith, H.O. 1997. In vitro Tn7 mutagenesis of Haemophilus influenzae Rd and characterization of the role of atpA in transformation. J. Bacteriol. 179: 7315-7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haapa S., Taira, S., Heikkinen, E., and Salvilahti, H. 1999. An efficient and accurate integration of mini-μ transposons in vitro: A general methodology for functional genetic analysis and molecular biology applications. Nucleic Acids Res. 27: 2777-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamer L., DeZwaan, T.M., Montenegro-Chamorro, M.V., Frank, S.A., and Hamer, J.E. 2001. Recent advances in large-scale transposon mutagenesis. Curr. Opin. Chem. Biol. 5: 67-73. [DOI] [PubMed] [Google Scholar]
- 14.Hare R.S., Walker, S.S., Dorman, T.E., Greene, J.R., Guzman, L., Kenney, T.J., Sulavik, M.C., Baradaran, K., Houseweart, C., Yu, H., et al. 2001. Genetic footprinting in bacteria. J. Bacteriol. 183: 1694-1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heath J.D., Perkins, J.D., Sharma, B., and Weinstock, G.M. 1992. Not I genomic cleavage map of Esherichia coli K-12 strain MG1655. J. Bacteriol. 174: 558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hutchison C.A., Peterson, S.N., Gill, S.R., Cline, R., White, O., Fraser, C.M., Smith, H., and Venter, J. 1999. Global transposon mutagenesis and a minimal Mycoplasma genome. Science 286: 2165-2169. [DOI] [PubMed] [Google Scholar]
- 17.Judson N. and Mekalanos, J.J. 2000a. Transposon-based approaches to identify essential bacterial genes. Trends Microbiol. 8: 521-526. [DOI] [PubMed] [Google Scholar]
- 18.___, 2000b. TnAraOut, a transposon-based approach to identify and characterize essential bacterial genes. Nat. Biotechnol. 18: 740-745. [DOI] [PubMed] [Google Scholar]
- 19.Kolisnychenko V., Plunkett, G., III, Herring, C.D., Feher, T., Posfai, J., Blattner, F.R., and Posfai, G. 2002. Engineering a reduced Escherichia coli genome. Genome Res. 12: 640-647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naumann T.A. and Reznikoff, W.S. 2000. Trans catalysis in Tn5 transposition. Proc. Natl. Acad. Sci. 97: 8944-8949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.___, 2002. Tn5 transposase with an altered specificity for transposon ends. J. Bacteriol. 184: 233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochman H., Mendora, M.M., Garza, D., Hartl, D.L., et al. 1990. Amplification of flanking sequences by inverse PCR. In: In PCR protocols: A guide to methods and applications. (ed. M.A. Innis) Academic Press, San Diego, CA.
- 23.Richmond C.S., Glasner, J.D., Mau, R., Jin, H., and Blattner, F.R. 1999. Genome-wide expression profiling in Escherichia coli K12. Nucleic Acids Res. 27: 3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J., Fritsh, E.F., and Maniatis, T., 1989. Molecular cloning: A laboratory manual Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 25.Shevchenko Y., Bouffard, G.G., Butterfield, Y.S.N., Blakesley, R.W., Hartley, J.L., Young, A.C., Marra, M.A., Jones, S.J.M., Touchman, J.W., and Green, E.D. 2002. Systematic sequencing of cDNA clones using the transposon Tn5. Nucleic Acids Res. 30: 2469-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Staczek P. and Higgins, N.P. 1998. Gyrase and Topo IV modulate chromosome domain size in vivo. Mol. Microbiol. 29: 1435-1448. [DOI] [PubMed] [Google Scholar]
- 27.Weinreich M.D., Mahnke-Braam, L., and Reznikoff, W.S. 1994. A functional analysis of the Tn5 transposase. Identification of domains required for DNA binding and multimerization. J. Mol. Biol. 241: 166-177. [DOI] [PubMed] [Google Scholar]
- 28.Wild J., Kamath-Loeb, A., Ziegelhoffer, E., Lonetto, M., Kawasaki, Y., and Gross, C.A. 1992. Partial loss of function mutations in DnaK, the Escherichia coli homologue of the 70-kDa heat shock proteins, affect highly conserved amino acids implicated in ATP binding and hydrolysis. Proc. Natl. Acad. Sci. 89: 7139-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.York D., Welch, K., Goryshin, I., and Reznikoff, W. 1998. Simple and efficient generation in vitro of nested deletions and inversions: Tn5 intramolecular transposition. Nucleic Acids Res. 26: 1927-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu B.J., Sung, B.H., Koob, M.D., Lee, C.H., Lee, J.H., Lee, W.S., Kim, M.S., and Kim, S.C. 2002. Minimization of the Escherichia coli genome using a Tn5-targeted Cre/loxP excision system. Nat. Biotechnol. 20: 1018-1023. [DOI] [PubMed] [Google Scholar]
- 31.Zhou M., Bhasin, A., and Reznikoff, W.S. 1998. Molecular genetic analysis of transposase-end DNA sequence recognition: Cooperativity of three adjacent base-pairs in specific interaction with a mutant Tn5 transposase. J. Mol. Biol. 276: 913-925. [DOI] [PubMed] [Google Scholar]