Abstract
Cell division cycle 42 (CDC42), a small GTPase of the Rho-subfamily, regulates diverse cellular functions including proliferation, cytoskeletal rearrangement and even promotes malignant transformation. Here, we found that increased expression of CDC42 correlated with undifferentiated neuroblastoma as compared to a more benign phenotype. CDC42 inhibition decreased cell growth and soft agar colony formation, and increased cell death in BE(2)-C and BE(2)-M17 cell lines, but not in SK-N-AS. In addition, silencing of CDC42 decreased expression of N-myc in BE(2)-C and BE(2)-M17 cells. Our findings suggest that CDC42 may play a role in the regulation of aggressive neuroblastoma behavior.
Keywords: CDC42, AKT2, Twist, N-myc, Transformation, Neuroblastoma
Introduction
Neuroblastoma is the most common solid tumor malignancy in infancy and children with two-thirds of patients presenting with distant organ metastasis at the time of diagnosis [1]. More importantly, metastatic disease is associated with ‘high-risk’ group category and an overall dismal survival of 40% [2,3]. Our ongoing research continues to focus on identifying pathways that promote malignant tumor transformation and progression in neuroblastoma, but much remains to be elucidated. The Rho family of GTPases represents a subgroup of the Ras superfamily of GTPases whose aberrant regulation has been associated with key features of aggressive tumor behavior including increased cell migration, invasion and metastasis [4]. Specifically, Rho GTPases are integral in the regulation of cellular functions such as cell proliferation, survival and migration and cytoskeleton development, which all have important implications in aggressive tumor development [5,6].
CDC42 (cell division cycle 42), a member of the Rho family, is known to contribute to tumorigenesis and cancer progression. Although there are no known activating mutations of CDC42, which result in its proto-oncogenic behavior, it is overexpressed in several different cancers [7]. Specifically, CDC42 has been shown to induce cellular transformation, invasion and metastasis in several tissues types including melanoma, breast and colon cancers [6,8,9]. Furthermore, genetic knockdown of CDC42 results in cell cycle arrest and apoptosis in colorectal cancer [10]. However, the exact role of CDC42 in neuroblastoma tumorigenicity has not been fully elucidated.
Downstream signaling for CDC42 is mediated via many different pathways. One of the effectors of CDC42-induced cell transformation is the PI3K/AKT pathway [11]. It has been well established that AKT2, an isoform of the AKT family, promotes cell survival, invasiveness and metastasis in cancer cells [12]. Similarly, work done in our lab has shown AKT2 is critically important in regulating the metastatic potential of neuroblastoma cells [13]. Another upstream regulator of AKT2 cited in the literature is the transcription factor Twist [14]. Twist is a key regulator on neuron crest cell migration during development [15], and its activation has been linked to aggressive adult cancer by promoting AKT2, SNAIL and SLUG to promote cancer initiation and progression [16,17]. Twist is constantly overexpressed in neuroblastoma and inhibits cell apoptosis by cooperating with oncoprotein N-myc [18]. To date there is no known established link providing evidence that the CDC42 acts in concert with AKT2 or Twist to regulate neuroblastoma tumor progression or transformation. Based on the cited interactions between CDC42/AKT2 and Twist/AKT2, we hypothesized that silencing CDC42 would inhibit neuroblastoma tumorigenesis via the regulation of Twist and AKT2.
Materials and methods
Materials
Antibody against CDC42 was purchased from Cytoskeleton, Inc. (Denver, CO) and Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibody against AKT2, AKT, survivin, N-myc, cleaved caspase-3, cleaved PARP and cell lysis buffer were obtained from Cell Signaling Technology (Beverly, MA). Twist antibody and horseradish peroxidase (HRP)-conjugated secondary antibodies against mouse and rabbit IgG were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Anti β-actin monoclonal antibody and fetal bovine serum (FBS) were from Sigma-Aldrich (St. Louis, MO). NuPAGE Novex 4–12% Bis–Tris Gel and Lipofectamine 2000 were purchased from Invitrogen (Carlsbad, CA). Cell Counting Kit-8 (CCK-8) was purchased from Dojindo Molecular Technologies, Inc. (Santa Clara, CA). Agarose (SeaPlaque®) was from Lonza Inc. (Allendale, NJ). Cell Death Detection ELISAPlus was purchased from Roche Applied Science (Indianapolis, IN).
Cell culture and transfection
Human neuroblastoma cell line, BE(2)-C was purchased from American Type Culture Collection (Manassas, VA). Cells were maintained in RPMI 1640 medium with L-glutamine (Cellgro Mediatech, Inc. Herndon, VA) supplemented with 10% FBS. The cells were maintained at 37 °C in a humidified atmosphere of 95% air and 5% CO2. For transfection, cells were plated in 6-well plates and transfected with plasmids (total of 4 μg) using Lipofectamine 2000 according to the manufacturer’s protocol. Plasmid pCDNA3.1 was obtained from Invitrogen (Carlsbad, CA), and pcDNA3-Myr HA-AKT2 (Addgene plasmid 9016, Cambridge, MA), pCMV6-XL5-Twist, and pCMV6-XL4-N-myc were purchased from OriGene (Rockville, MD). Plasmid pLKO.1-shRNA against CDC42 and its non-targeting control vector SHC002 were purchased from Sigma-Aldrich (St. Louis, MO). For stable transfection, cells were selected with puromycin (2.5 μg/mL). Cells were seeded on 6-well plates for protein preparation and 96-well plates for DNA fragmentation or cell growth assays. The experiments were repeated on at least three separate occasions.
Western blot analysis
Whole-cell lysates were prepared using cell lysis buffer with 1 mM PMSF. Total protein (30 μg/lane) was resolved on NuPAGE Novex 4–12% Bis–Tris gels and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes (Bio-Rad Laboratories, Hercules, CA). Nonspecific binding sites were blocked with 5% milk in TBS-T (120 mM Tris–HCl, pH 7.4, 150 mM NaCl, and 0.05% Tween 20) for 2 h at room temperature or overnight at 4 °C. Target proteins were detected by using rabbit or mouse anti-human antibodies (1:500–1000 dilution) for 2 h at room temperature or overnight at 4 °C. The membranes were washed three times and incubated with secondary antibodies (1:5000 dilution) conjugated with HRP. Immune complexes were visualized using the ECL system. Equal loading and transfer were confirmed with β-actin. Data are representative of three independent experiments.
Reverse transcription, semi-quantitative and quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using Trizol and reverse-transcribed to cDNA using High Capacity cDNA Reverse Transcription Kit according to manufacturer’s protocol (Applied Biosystems, Foster City, CA). Semi-quantitative PCR was performed using a Peltier Thermal Cycler (PTC-200) using specific 3′ and 5′ primers for CDC42, MYCN, TWIST and AKT2 and final product was visualized on 1% agarose gel using a Gel Doc (Bio-Rad). qRT-PCR was performed using the Bio-Rad Thermocycler CFX96. SsoFAST EvaGreen Supermix, cDNA and specific 3′ and 5′ primers were incubated together using the manufacturer’s protocol (Bio-Rad). GAPDH and β-actin were used as internal controls.
DNA fragmentation assay
Apoptosis was measured using a DNA fragmentation assay as previously described [19]. Briefly, 5 × 103 cells per well were plated in triplicate. Cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) were detected using a Cell Death Detection ELISAplus kit according to manufacturer’s recommended protocol. The experiments were repeated on at least three separate occasions.
Cell proliferation and cell death assay
Cells were seeded in 96-well plates at a density of 3 × 103 cells per well in RPMI 1640 culture medium with 10% FBS and grown for up to 4 days. Cell proliferation was assessed using CCK-8 daily. Each assay point was performed in triplicate, and the experiment was repeated three times. The values, corresponding to the number of viable cells, were read at OD450 with the FlexStation 3 Microplate Reader (Molecular Devices, Sunnyvale, CA). Cell death induced by CDC42 shRNA transfection was evaluated by trypan blue exclusion assay. Cells were stained with 0.25% trypan blue solution and the percentages of dead cells were determined by TC10™ Automated Cell Counter (Bio-Rad).
Soft agar colony formation assay
Using a 12-well plate, cells were trypsinized and resuspended in RPMI 1640 containing 0.4% agarose and 7.5% FBS and then overlaid onto a bottom layer of solidified 0.8% agarose in RPMI 1640 containing 5% FBS at a concentration of 3 × 103 cells per well and incubated for 3 weeks. Colonies were stained with 0.05% crystal violet dissolved in 70% methanol before being photographed and quantified.
Cell migration assay
For Transwell migration assay, polycarbonate Transwell filters (8 μm; Corning Inc., Corning, NY) were coated on the lower side with 5 μg/mL collagen type I (BD Biosciences) overnight and then blocked with 2.5% BSA in PBS for 1 h. 50,000 cells in 100 μL of serum-free media were added to the Transwell and allowed to migrate for 24 h at 37 °C under tissue culture conditions. Media with 10% FBS was added to the lower chamber. Cells that failed to migrate through the filter after incubation were scraped out using a sterile cotton swab. Cells that migrated to the bottom surface of the filter were fixed with 4% paraformaldehyde, stained with DAPI, and counted. Each substrate was repeated in duplicate wells, and within each well counting was done in five randomly selected microscopic fields (200× magnification).
Immunohistochemistry
Vanderbilt University Institutional Review Board reviewed IRB# 091331 (Surgical Studies on the Role on Gastrin-releasing Peptide in Neuroblastoma) and determined the study does not qualify as “human subject” research per §46.102(f)(2). Samples of discarded tissue without identifiable private information were obtained from Surgical Pathology at Vanderbilt University.
Tissues were fixed in formalin for 3 days and embedded in paraffin wax. Paraffin-embedded sections (5 μm) were deparaffinized in three xylene washes followed by a graded alcohol series, antigen retrieval performed with 10 mM sodium citrate buffer, and then blocked with solution for 1 h at RT. They were incubated with primary antibody against CDC42 overnight at 4 °C, washed with PBS, incubated with secondary antibodies for 30 min at RT, and developed with DAB reagent. All sections were counterstained with hematoxylin, and then dehydrated with ethanol and xylene. Coverslips were mounted and slides observed by light microscopy.
Statistical analysis
Statistical analyses were performed using Student’s paired t-test. A P value of <0.05 was considered significant. Image J was used for densitometric analysis of immunoblot band intensities.
Results
Constitutive CDC42 expression in human neuroblastoma tissues and cell lines
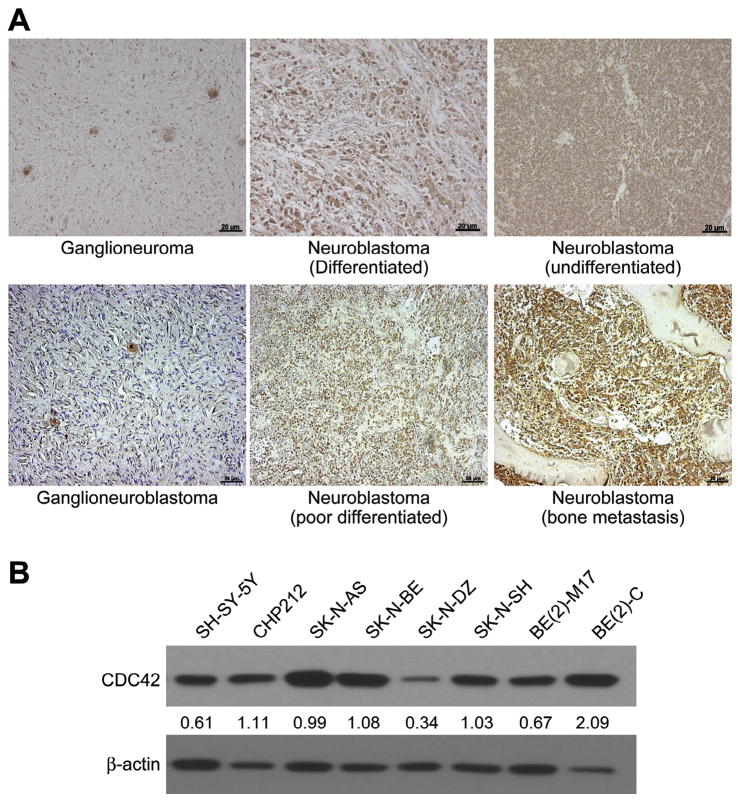
We first wanted to determine whether CDC42 expression correlated with malignant phenotype of neuroblastoma. Using paraffin-embedded tumor sections from five undifferentiated neuroblastomas and two ganglioneuromas, a benign phenotype of neuroblastoma, we performed immunohistochemical analysis to assess for CDC42 expression. We observed an increase in CDC42 expression in undifferentiated neuroblastomas when compared to benign ganglioneuromas (Fig. 1A). Next, we also examined the constitutive protein levels of CDC42 in several human neuroblastoma stable cell lines. Several of the neuroblastoma cell lines demonstrated high CDC42 protein levels (Fig. 1B), with the highest expression found in BE(2)-C. We chose BE(2)-C for further experiments since it overexpresses N-myc and is known to have increased malignant potential and tumorigenic properties [20]. Additionally, we compared BE(2)-M17, another N-myc overexpressing cell line, and SK-N-AS, which does not overexpress N-myc, to better characterize the role of CDC42 in neuroblastoma.
Fig. 1.
CDC42 expression in neuroblastoma. Representative sections show increased CDC42 expression by immunohistochemistry in samples from undifferentiated neuroblastomas as compared to ganglioneuroma (100×, 20 μm bar). (B) Endogenous CDC42 protein expression was examined by immunoblotting in several human neuroblastoma cell lines. β-actin was used as a loading control. The numbers indicate the relative expression of CDC42 to β-actin as measured by densitometry.
Inhibition of CDC42 decreased neuroblastoma cell proliferation and increased cell death
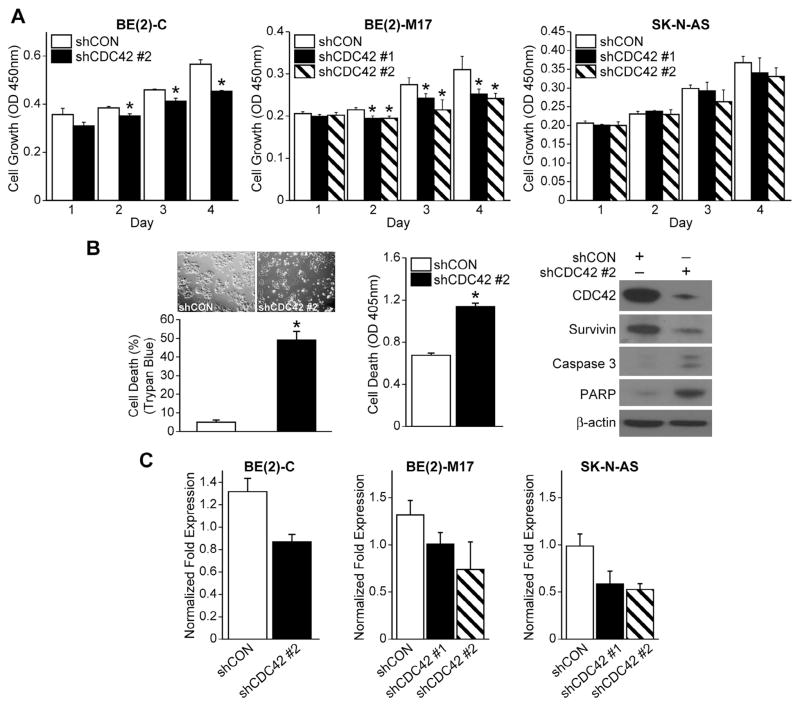
To determine a functional role for CDC42 in neuroblastoma, BE(2)-C cells were transfected with plasmids containing shRNA specific for CDC42 (shCDC42) as well as a separate group of non-targeted shRNA for control, and stable subclonal populations were cultured. After transfection, there was reduced expression of CDC42 utilizing two separate RNA sequences for silencing (shCDC42 #1, shCDC42 #2) as compared to cells transfected with control vector (shCON) (Fig. 2C). We then focused on identifying the phenotypic effects of RNA-mediated silencing of CDC42 in BE(2)-C cells. Cell viability assay using CCK-8 was performed in BE(2)-C, BE(2)-M17 and SK-N-AS cell lines to assess proliferation over a time course (24, 48, 72, and 96 h) after plating the cells. Cell proliferation significantly decreased in CDC42-silenced BE(2)-C and BE(2)-M17 cells when compared to non-targeted controls (shCON), however knockdown of CDC42 did not affect proliferation in the SK-N-AS cell line (Fig. 2A). We then sought to examine the effect of CDC42 silencing on cell death. A significant increase in cell death was detected by trypan blue exclusion assay and measuring levels of DNA fragmentation in BE(2)-C and BE(2)-M17, but not in SK-N-AS (Fig. 2B). Moreover, we found that the expression of survivin, a cell proliferation marker, was decreased, whereas apoptosis markers, cleaved caspase-3 and PARP expression were increased in BE(2)-C/shCDC42 cells when compared to shCON (Fig. 2B). These data suggest that CDC42 plays a critical role in regulating cell growth and death in some, but not all, neuroblastoma cell lines.
Fig. 2.
CDC42 silencing decreases cell growth and increases apoptosis. (A) BE(2)-C and BE(2)-M17 cells treated with shCDC42 exhibited a decrease in cell growth in comparison to control cells, while SK-N-AS cells did not (mean ± SEM; *p < 0.05 vs. shCON). (B) CDC42 inhibition increased apoptosis in BE(2)-C cells as analyzed using Cell Death ELISA (mean ± SEM; *p < 0.05 vs. shCON). Immunoblotting after shCDC42 in BE(2)-C cells. β-actin was used for protein loading control. (C) CDC42 silencing was confirmed in all three cell lines by RT-PCR (mean ± SEM; *p < 0.05 vs. shCON).
Inhibition of CDC42 suppressed anchorage-independent colony growth and cellular migration
We next wanted to examine the effects of CDC42 inhibition on soft agar colony formation, which is a well-established indicator of tumorigenicity of cancer cells in vitro [21]. Silencing CDC42 resulted in a significant decrease in the number of colonies formed as compared to control (Fig. 3A). To verify the role of CDC42 in aggressive neuroblastoma behavior, we utilized a migration assay as another in vitro assessment of invasive and metastatic potential. Classically, CDC42, as a member of the Rho-GTPase family, plays a critical role in the processes of cellular movement and migration via its regulation of cytoskeletal changes. Consistent with this function, silencing of CDC42 in BE(2)-M17 neuroblastoma cells resulted in decreased migration through an 8 μm Transwell filter compared to controls (Fig. 3B). Interestingly, this same effect was not observed in the SK-N-AS cell line (Fig. 3B), which is consistent with our data evaluating the effect of CDC42 silencing on cell proliferation and death (Fig. 2). The cultured cell lines in which CDC42 appears to regulate aggressive cellular behavior, BE(2)-C and BE(2)-M17, are both N-myc-overexpressing cell lines, while the cell line in which CDC42 does not appear to be a critical regulator of tumorigenicity in vitro, SK-N-AS, does not overexpress MYCN. We therefore hypothesized that CDC42 may play a role in the regulation of MYCN and related signaling mediators.
Fig. 3.
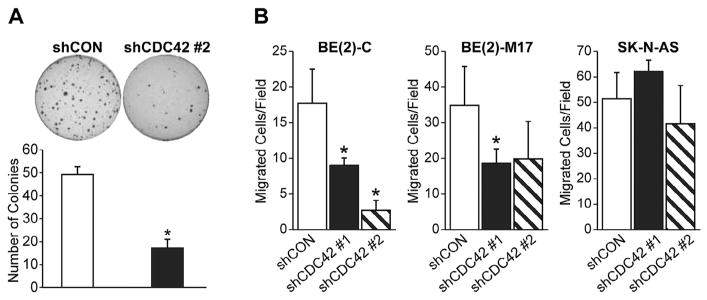
Inhibition of soft agar colony formation and migration after shCDC42. (A) Soft agar colony formation was decreased in BE(2)-C cells as compared to shCON (mean ± SD; *p < 0.05 vs. shCON). (B) Cellular migration through an 8 μm pore Transwell filter system was decreased in BE(2)-C and BE(2)-M17 cells but not SK-N-AS as compared to shCON (mean ± SD; *p < 0.05 vs. shCON).
CDC42-silencing decreases N-myc expression
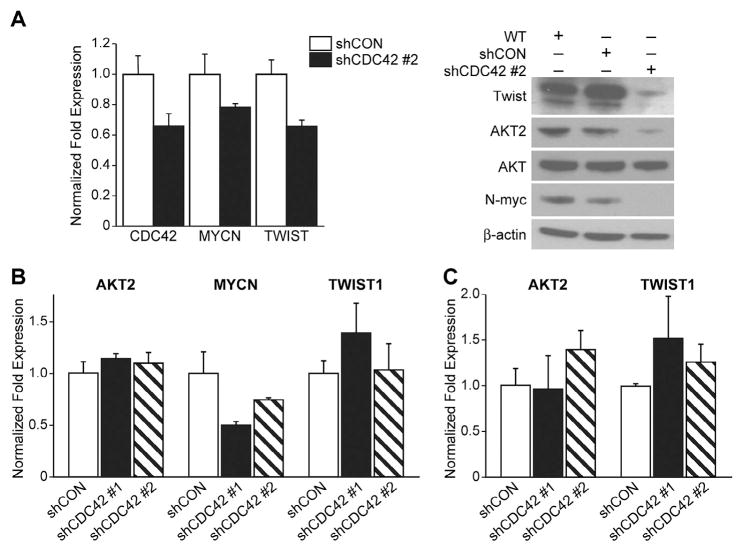
In order to investigate the mechanism by which CDC42 silencing reduces the capacity of cells to proliferate, induce colony formation and migrate, we examined N-myc, AKT2 and Twist after shCDC42 treatment. N-myc is a well-known oncogene in neuroblastoma [22], and, in a previous report, induced N-myc overexpression in SHEP neuroblastoma cells (a cell line without native N-myc overexpression) led to decreased levels of CDC42, implicating a role for N-myc in the regulation of CDC42 expression [23]. Our data show that CDC42-silencing decreases N-myc expression in BE(2)-C and BE(2)-M17 cell lines (Fig. 4A and B), further supporting a mutual interaction for CDC42 and N-myc in neuroblastoma. We further assessed two downstream mediators of N-myc activity AKT2 and Twist. We have previously reported AKT2 to be critical in regulating neuroblastoma metastatic potential [13]. Twist is a transcriptional regulator of AKT2, and is known to be a regulator of metastasis and its increased expression correlates with tumor aggressiveness and poor prognosis in other cancers [17,24,25]. Interestingly, inhibition of CDC42 only decreased AKT2 and Twist expression levels in BE(2)-C cells, and not in BE(2)-M17 and SK-N-AS (Fig. 3B and C). These results suggest that CDC42 may regulate downstream signaling mediators and cellular behavior in only the most aggressive variants of neuroblastoma, those that overexpress the N-myc oncoprotein.
Fig. 4.
CDC42 silencing decreases N-myc expression. (A) Immunoblotting for protein levels of Twist, AKT2 and N-myc in BE(2)-C cells demonstrated a decrease in all three of these signaling mediators with CDC42 inhibition. (B) BE(2)-M17 cells exhibited decreased N-myc expression after shCDC42, and no changes in Twist or AKT2. (C) Neither Twist, AKT2 nor N-myc levels were affected by CDC42 silencing in SK-N-AS cell line. In all cases, β-actin levels were used as a loading control.
Twist, AKT2 or N-myc overexpression did not rescue CDC42 silencing-mediated inhibition of cell growth
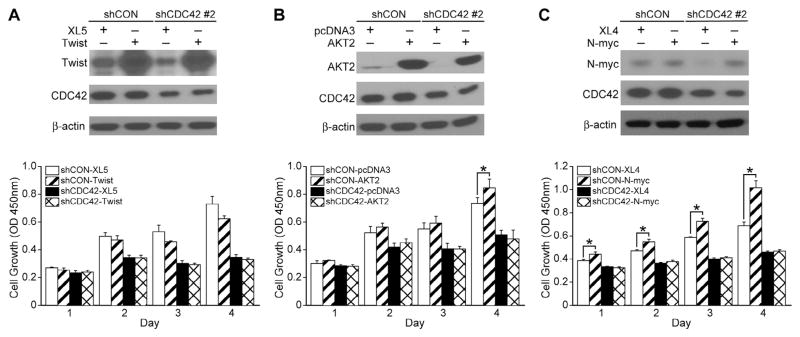
To assess whether Twist, AKT2 or N-myc overexpression can recover the cell growth inhibition induced by CDC42 knockdown in the BE(2)-C cell line, we next performed rescue experiments using either Twist, AKT2 or N-myc plasmid in BE(2)-C/shCDC42 cells. Immunoblotting showed knockdown of CDC42 and overexpression of Twist (Fig. 5A, top panel), AKT2 (Fig. 5B, top panel), or N-myc (Fig. 5C, top panel) after the respective transfections. None of the three mediators were able to reverse the cell growth inhibition seen in CDC42-silenced BE(2)-C cells when overexpressed (Fig. 5A–C). In the control-transfected cells, which express native levels of CDC42, N-myc overexpression increased cell growth (Fig. 5C, bottom panel). Additionally, also in the control-transfected cells, AKT2 overexpression also increased cell growth by the fourth day after transfection (Fig. 5B, bottom panel). These findings suggest that the role of CDC42 in aggressive cellular behavior, which we described above, is not solely dependent on alteration in the levels of N-myc, AKT2 or Twist, and raises the possibility that CDC42 is acting through other, as of yet unidentified, signal transduction pathways.
Fig. 5.
Twist, AKT2 or N-myc overexpression in CDC42-silenced cells does not restore neuroblastoma growth. (A) Immunoblotting confirmed transient Twist overexpression (pCMV6-XL5-Twist) in shCDC42. β-actin indicated relatively equal protein loading. Twist overexpression did not induce increases in shCON cell growth. CDC42 inhibition decreased cell growth; Twist overexpression in the shCDC42 cells did not restore the decreased cell growth. (B) Immunoblotting confirmed transient AKT2 overexpression (pcDNA3- AKT2) in shCDC42. AKT2 overexpression increased cell growth in shCON cells but not in shCDC42 cells. AKT2 overexpression in the shCDC42 cells did not restore cell growth (*p < 0.05 vs. AKT2 control vector). (C) Immunoblotting confirmed transient N-myc overexpression (pCMV6-XL4-N-myc) in shCDC42. N-myc overexpression increased cell growth in shCON cells but not in shCDC42 cells. N-myc overexpression in the shCDC42 cells did not restore cell growth (*p < 0.05 vs. N-myc control vector).
Discussion
CDC42 is overexpressed in multiple types of cancer, in which it promotes cellular transformation, cell cycle dysregulation and tumor progression. In this study, we show that CDC42 expression is higher in patient tumor samples of undifferentiated neuroblastoma compared with more differentiated samples. Targeted silencing of CDC42 using shRNA in some, but not all, cultured neuroblastoma cell lines led to increased cell death and inhibited cell growth. Moreover, CDC42 inhibition resulted in decreased anchorage-independent colony formation in vitro as well as decreased cellular migration. Finally, silencing CDC42 decreased protein expression of N-myc in those cell lines which naturally overexpress this oncogene, and also decreased expression of AKT2 and Twist in a single cell line, indicating that CDC42 plays a role in the regulation of other signaling mediators with proven roles in neuroblastoma progression and metastasis. CDC42-silencing may mediate its effects on cellular behavior via other mechanisms, as overexpression of N-myc, AKT2 or Twist did not rescue the aggressive phenotype of BE(2)-C cells.
CDC42, in its cancer-promoting role, has been shown to act under the control of Ras [26]. Additionally, CDC42 has been shown to bind to the p85 regulatory subunit of PI3K [27]. In fact, PI3K inhibitors reverse changes in cellular morphology induced by a mutant, constitutively active form of CDC42 [28], suggesting that CDC42 may be involved in multiple signaling pathways in a cancer context. Our previous work has shown that PI3K, acting through its classical downstream effector, AKT, critically regulates proliferation, anchorage-independent growth and in vivo metastatic potential via regulation of N-myc expression [13]. Given our laboratory’s previous findings, the reported interactions of CDC42 with the PI3K/AKT signaling pathway and our current findings of CDC42 regulation of aggressive cellular behavior, we hypothesized that CDC42 may play a role in the PI3K–AKT–N-myc signaling pathway in neuroblastoma.
With silencing of CDC42 we were able to show decreased expression of Twist, AKT2 and N-myc, but only in the BE(2)-C cell line, which suggests that it may be an upstream regulator of factors known to be critical for neuroblastoma cell transformation and proliferation [13,18], and may do so in the context of N-myc overexpression. Interestingly, overexpression of Twist, AKT2 or N-myc was not able to rescue the anti-proliferative effect produced by silencing of CDC42. These findings suggest an alternative mechanism by which CDC42 exerts its control on cell behavior in neuroblastoma. Our data reveal a mutual regulation of CDC42 and N-myc, when interpreted in the context of previous reports [23]. It is not clear at present what molecular mechanisms are responsible for these observations. Our findings clearly show that the expression of AKT2 and Twist are only altered by changes in CDC42 in one of two N-myc overexpressing neuroblastoma cell lines, suggesting that regulation of N-myc by CDC42 is likely independent of AKT2 and Twist. This finding represents a potential alternative mechanism of regulation of N-myc expression in neuroblastoma. Further work will need to elucidate whether CDC42 is acting directly to affect N-myc expression levels, or whether this is due to an intermediate signaling pathway.
Acknowledgments
This work was supported by a grant R01 DK61470 from the National Institutes of Health.
Footnotes
Conflict of interest
The authors declare to have no conflict of interest.
References
- 1.Modak S, Cheung NK. Neuroblastoma: therapeutic strategies for a clinical enigma. Cancer Treat Rev. 2010;36:307–317. doi: 10.1016/j.ctrv.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 2.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–2120. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 3.van Noesel MM, Versteeg R. Pediatric neuroblastomas: genetic and epigenetic ‘danse macabre’. Gene. 2004;325:1–15. doi: 10.1016/j.gene.2003.09.042. [DOI] [PubMed] [Google Scholar]
- 4.Rathinam R, Berrier A, Alahari SK. Role of Rho GTPases and their regulators in cancer progression. Front Biosci (Landmark Ed) 2011;16:2561–2571. doi: 10.2741/3872. [DOI] [PubMed] [Google Scholar]
- 5.Tang Y, Olufemi L, Wang MT, Nie D. Role of Rho GTPases in breast cancer. Front Biosci. 2008;13:759–776. doi: 10.2741/2718. [DOI] [PubMed] [Google Scholar]
- 6.Leve F, Morgado-Diaz JA. Rho GTPase signaling in the development of colorectal cancer. J Cell Biochem. 2012;113:2549–2559. doi: 10.1002/jcb.24153. [DOI] [PubMed] [Google Scholar]
- 7.Stengel K, Zheng Y. Cdc42 in oncogenic transformation, invasion, and tumorigenesis. Cell Signal. 2011;23:1415–1423. doi: 10.1016/j.cellsig.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucci MG, Lucarini G, Brancorsini D, Zizzi A, Pugnaloni A, Giacchetti A, et al. Involvement of E-cadherin, beta-catenin, Cdc42 and CXCR4 in the progression and prognosis of cutaneous melanoma. Br J Dermatol. 2007;157:1212–1216. doi: 10.1111/j.1365-2133.2007.08246.x. [DOI] [PubMed] [Google Scholar]
- 9.Ma J, Xue Y, Liu W, Yue C, Bi F, Xu J, et al. Role of activated rac1/cdc42 in mediating endothelial cell proliferation and tumor angiogenesis in breast cancer. PLoS ONE. 2013;8:e66275. doi: 10.1371/journal.pone.0066275. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q, et al. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int J Cancer. 2011;128:1269–1279. doi: 10.1002/ijc.25452. [DOI] [PubMed] [Google Scholar]
- 11.Higuchi M, Masuyama N, Fukui Y, Suzuki A, Gotoh Y. Akt mediates Rac/Cdc42-regulated cell motility in growth factor-stimulated cells and in invasive PTEN knockout cells. Curr Biol. 2001;11:1958–1962. doi: 10.1016/s0960-9822(01)00599-1. [DOI] [PubMed] [Google Scholar]
- 12.Arboleda MJ, Lyons JF, Kabbinavar FF, Bray MR, Snow BE, Ayala R, et al. Overexpression of AKT2/protein kinase Bbeta leads to up-regulation of beta1 integrins, increased invasion, and metastasis of human breast and ovarian cancer cells. Cancer Res. 2003;63:196–206. [PubMed] [Google Scholar]
- 13.Qiao J, Lee S, Paul P, Qiao L, Taylor CJ, Schlegel C, et al. AKT2 regulates metastatic potential in neuroblastoma. PLoS ONE. 2013;8:e56382. doi: 10.1371/journal.pone.0056382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng GZ, Zhang W, Wang LH. Regulation of cancer cell survival, migration, and invasion by Twist: AKT2 comes to interplay. Cancer Res. 2008;68:957–960. doi: 10.1158/0008-5472.CAN-07-5067. [DOI] [PubMed] [Google Scholar]
- 15.Chen ZF, Behringer RR. Twist is required in head mesenchyme for cranial neural tube morphogenesis. Genes Dev. 1995;9:686–699. doi: 10.1101/gad.9.6.686. [DOI] [PubMed] [Google Scholar]
- 16.Eckert MA, Lwin TM, Chang AT, Kim J, Danis E, Ohno-Machado L, et al. Twist1-induced invadopodia formation promotes tumor metastasis. Cancer Cell. 2011;19:372–386. doi: 10.1016/j.ccr.2011.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, et al. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Valsesia-Wittmann S, Magdeleine M, Dupasquier S, Garin E, Jallas AC, Combaret V, et al. Oncogenic cooperation between H-Twist and N-Myc overrides failsafe programs in cancer cells. Cancer Cell. 2004;6:625–630. doi: 10.1016/j.ccr.2004.09.033. [DOI] [PubMed] [Google Scholar]
- 19.Kang JH, Rychahou PG, Ishola TA, Qiao J, Evers BM, Chung DH. MYCN silencing induces differentiation and apoptosis in human neuroblastoma cells. Biochem Biophys Res Commun. 2006;351:192–197. doi: 10.1016/j.bbrc.2006.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walton JD, Kattan DR, Thomas SK, Spengler BA, Guo HF, Biedler JL, et al. Characteristics of stem cells from human neuroblastoma cell lines and in tumors. Neoplasia. 2004;6:838–845. doi: 10.1593/neo.04310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ponten J. Spontaneous and virus induced transformation in cell culture. Virol Monogr. 1971;8:1–253. doi: 10.1007/978-3-7091-8258-1_1. [DOI] [PubMed] [Google Scholar]
- 22.Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- 23.Valentijn LJ, Koppen A, van Asperen R, Root HA, Haneveld F, Versteeg R. Inhibition of a new differentiation pathway in neuroblastoma by copy number defects of N-myc, Cdc42, and nm23 genes. Cancer Res. 2005;65:3136–3145. doi: 10.1158/0008-5472.CAN-04-2469. [DOI] [PubMed] [Google Scholar]
- 24.Hoek K, Rimm DL, Williams KR, Zhao H, Ariyan S, Lin A, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 25.Kwok WK, Ling MT, Lee TW, Lau TC, Zhou C, Zhang X, et al. Up-regulation of TWIST in prostate cancer and its implication as a therapeutic target. Cancer Res. 2005;65:5153–5162. doi: 10.1158/0008-5472.CAN-04-3785. [DOI] [PubMed] [Google Scholar]
- 26.Qiu RG, Abo A, McCormick F, Symons M. Cdc42 regulates anchorage-independent growth and is necessary for Ras transformation. Mol Cell Biol. 1997;17:3449–3458. doi: 10.1128/mcb.17.6.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tolias KF, Cantley LC, Carpenter CL. Rho family GTPases bind to phosphoinositide kinases. J Biol Chem. 1995;270:17656–17659. doi: 10.1074/jbc.270.30.17656. [DOI] [PubMed] [Google Scholar]
- 28.Keely PJ, Westwick JK, Whitehead IP, Der CJ, Parise LV. Cdc42 and Rac1 induce integrin-mediated cell motility and invasiveness through PI(3)K. Nature. 1997;390:632–636. doi: 10.1038/37656. [DOI] [PubMed] [Google Scholar]