Abstract
Three active-site components in rhodopsin play a key role in the stability and function of the protein: 1) the counter-ion residues which stabilize the protonated Schiff base, 2) water molecules, and 3) the hydrogen-bonding network. The ionizable residue Glu-181, which is involved in an extended hydrogen-bonding network with Ser-186, Tyr-268, Tyr-192, and key water molecules within the active site of rhodopsin, has been shown to be involved in a complex counter-ion switch mechanism with Glu-113 during the photobleaching sequence of the protein. Herein, we examine the photobleaching sequence of the E181Q rhodopsin mutant by using cryogenic UV-visible spectroscopy to further elucidate the role of Glu-181 during photoactivation of the protein. We find that lower temperatures are required to trap the early photostationary states of the E181Q mutant compared to native rhodopsin. Additionally, a Blue Shifted Intermediate (BSI, λmax = 498 nm, 100 K) is observed after the formation of E181Q Bathorhodopsin (Batho, λmax = 556 nm, 10 K) but prior to formation of E181Q Lumirhodopsin (Lumi, λmax = 506 nm, 220 K). A potential energy diagram of the observed photointermediates suggests the E181Q Batho intermediate has an enthalpy value 7.99 KJ/mol higher than E181Q BSI, whereas in rhodopsin, the BSI is 10.02 KJ/mol higher in enthalpy than Batho. Thus, the Batho to BSI transition is enthalpically driven in E181Q and entropically driven in native rhodopsin. We conclude that the substitution of Glu-181 with Gln-181 results in a significant perturbation of the hydrogen-bonding network within the active site of rhodopsin. In addition, the removal of a key electrostatic interaction between the chromophore and the protein destabilizes the protein in both the dark state and Batho intermediate conformations while having a stabilizing effect on the BSI conformation. The observed destabilization upon this substitution further supports that Glu-181 is negatively charged in the early intermediates of the photobleaching sequence of rhodopsin.
Keywords: Rhodopsin, Photobleaching sequence, Photointermediates, Glu-181, E181Q, Blue-Shifted Intermediate (BSI), Absorption spectroscopy, Low-Temperature trapping
Introduction
Rhodopsin, a member of the G Protein-Coupled Receptor (GPCR) visual opsins, is located in the rod photoreceptor cells, which are responsible for scotopic (low-light) vision [1]. The visual pigment consists of a seven transmembrane helical apoprotein and an organic chromophore covalently bound to a conserved Lysine residue (Lys-296) in helix VII via a Protonated Schiff Base (PSB) linkage. Upon absorption of light, the chromophore, 11-cis retinal, isomerizes to an all-trans conformation, initiating a series of conformational changes within the protein, which are associated with the formation of a series of spectrally discrete photointermediates with known lifetimes [order of magnitude shown for 298 K]: Bathorhodopsin (Batho) [ns], Blue-Shifted Intermediate (BSI) [ns], Lumihodopsin (Lumi) [μs], Metahodopsin I (Meta I) [ms], and Metahodopsin II (Meta II). The Meta II intermediate is stable on the timescale of minutes and activates the heterotrimeric G protein, transducin [2–4].
The photointermediate Batho is the first intermediate stable at low temperatures and stores the energy needed (~32 Kcal/mol photon energy) to propagate the structural and conformational changes necessary to form the active state of the protein (Meta II), which in turn catalyzes the visual transduction process [5–7]. Although the exact mechanism of energy storage is unknown, it has been proposed that the energy storage in Batho involves conformational strain within the chromophore and electrostatic interactions between the chromophore and the protein [8]. At low temperatures, Batho is converted directly to Lumi by gradually warming rhodopsin in the dark. However, at room temperature, the formation of a BSI is observed after formation of Batho and prior to the formation of Lumi [9]. The first observation of a BSI during the Batho to Lumi transition occurred during nanosecond photolysis experiments on various artificial visual pigments, including 13-desmethyl rhodopsin and 5,6-dihydrorhodopsin [10,11]. These rhodopsin analogs displayed a destabilization in the Batho intermediate as well as the accumulation of the BSI to sufficient concentrations as to be observed at low temperatures [8,10–13]. Time-resolved resonance Raman analysis suggests the Batho → BSI transition involves the relaxation of the chromophore in the C10-C11 = C12-C13 region [14], and it is important to note that Glu-181 is located directly above this region of the chromophore. The formation of the BSI in native rhodopsin has only been observed at room temperature, which is due to the equilibrium constant shifting towards the Batho intermediate resulting in too little formation of the BSI to be experimentally observed at low temperatures [8,9]. While changes in the steric and/or electrostatic interactions between the chromophore and the protein have been associated with the formation of BSI, the decay rate appears to be largely dependent on conformational changes within the protein, or protein relaxation [8].
Models of rhodopsin developed from recent crystal structure data provide evidence of an extended hydrogen-bonding network around the PSB involving residues Glu-113, Glu-181, Ser-186, Tyr-192, Tyr-268, and select water molecules [15,16], which has been shown to stabilize the dark state conformation of rhodopsin [17]. The spectral shifts observed during the early photointermediates (dark state, Batho, BSI, and Lumi) are due to modulations of the retinal PSB structure in and around this hydrogen-bonding network, in addition to the eletrostatic influence of the charged residues (Glu-113 and Glu-181), π-stacking, and van der Waals interactions with surrounding residues within the protein-binding pocket [18]. Titration experiments on native rhodopsin and site-directed mutants reveal that the primary counter-ion to the PSB in Meta I is Glu-181, implying there is a switch in the primary counter-ion from Glu-113 in the dark state to Glu-181 in the Meta I state via a rearrangement of the active site hydrogen-bonding network [19–21] and a rotation of helix VI to accommodate this shift [22]. During the transition from Meta I to Meta II, the PSB is deprotonated and the ionic interaction between the PSB and Glu-113, which serves to lock the protein in an inactive conformation, is broken [23]. This deprotonation of the PSB has been shown to be largely influenced by an enhanced torsional flexibility of the retinal polyene chain and a shift in the relative orientation of the β-ionone ring [24–27].
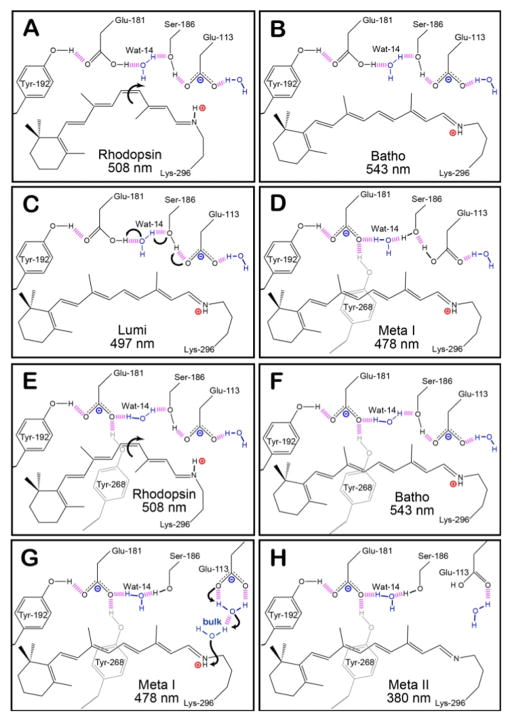
The ionizable residue Glu-181, which is located within the binding pocket along extracellular loop II connecting transmembrane helices IV and V, is directly involved in a hydrogen-bonding network with Tyr-268, Tyr-192, Ser-186, and Water molecule 14 (Wat-14) [28]. The protonation state of the carboxylic acid side chain of this residue during the early intermediates has been debated through numerous experimental and theoretical investigations [20,29–45]. Original models of the counter-ion switch predict that a proton is transferred from Glu-181 to Glu-113 during the Lumi to Meta I transition (Figures 1A–1D), however, our recent report on the excited state manifold of the rhodopsin mutant with a glutamine substitution at position 181 (E181Q) strongly supports a negatively charged Glu-181 during Batho [42]. Figures 1E–1H demonstrate that the counter-ion switch model is still applicable with a negatively charged Glu-181 during the dark state and Batho. Previous time-resolved studies on several Glu-181 mutants also suggest the residue plays a significant role in the early stages of the photobleaching sequence of rhodopsin [39]. At room temperature, the E181Q mutation results in the destabilization of the Batho intermediate and subsequently an accelerated decay from Batho to BSI (< 30 ns), resulting in the formation of a considerable amount of BSI [39].
Figure 1.
The hydrogen-bonding network of the protein-binding site of native rhodopsin at select intermediates during the photobleaching sequence. Panels A–D depict the original model of the counter-ion switch, in which Glu-181 is initially neutral. The primary photochemical event involves the isomerization of 11-cis retinal in rhodopsin (A) to all-trans retinal in Batho (B). During these early intermediates, Glu-113 serves as the primary counter-ion for the PSB. The transition from Lumi (C) to Meta I (D) is characterized by the transfer of a proton from Glu-181 to the hydrogen bonding network, which subsequently leads to the protonation of Glu-113. Panels E–H demonstrates our model, [42] which predicts that Glu-181 is also negatively charged during the early intermediates (E and F), and the hydrogen-bonding network rearranges to allow Glu-181 to serve as the primary counter-ion after the transition to Meta I (G). During the Meta II state (H), the PSB is deprotonated within the protein-binding site and the protein is activated in order to catalyze the visual transduction cascade. The purple dashed lines represent hydrogen bonding, and the positively and negatively charged species are indicated using red and blue labels, respectively.
Herein, the consequence of the mutational exchange of residue Glu-181 with Gln-181 on the structural stability and photoactivation mechanism of rhodopsin is explored using low-temperature trapping methods [46–48]. The mutant pigment E181Q was genetically engineered and spectroscopically characterized at cryogenic temperatures following purification. The formation and decay of each photointermediate was analyzed using UV-visible spectroscopy and notable differences were observed between the photobleaching sequences of E181Q and native rhodopsin. Plotting the potential energy surface of each intermediate of the photobleaching sequences provides insight into the observation of BSI in the rhodopsin mutant E181Q at low temperatures and possible mechanisms of BSI stability are discussed. The combination of experimental and theoretical data shown below lead us to conclude that a negatively charged carboxylic acid side chain at position 181 is crucial for the stability of the Batho and Lumi states in rhodopsin.
Materials and Methods
Visual pigment expression and purification
The pigment was constructed and isolated as previously reported [49]. The E181Q mutant was expressed in mammalian COS1 cells and purified by immunoaffinity chromatography techniques. The pigment was eluted in buffer Y1 [50 mM HEPES, 140 mM NaCl, 3 mM MgCl2, pH 6.6] with 20% glycerol and 0.1% N-dodecyl-β-D-Maltoside (DM) and stored at 193 K until used.
Cryogenic experiments
The spectra of each photointermediate in the photobleaching sequence of rhodopsin and E181Q were measured using standard methods [42,47,48,50,51]. The samples were prepared in 67% glycerol, buffer Y1 [50 mM HEPES, 140 mM NaCl, 3 mM MgCl2, pH 6.8], and 0.05% DM. Low-temperature experiments were conducted from 10 K to 220 K in a closed-cycle helium-refrigerated cryostat (APD Cryogenics) coupled to a Cary 50 UV-visible spectrophotometer (Varian, Inc.). For the mutant E181Q, a temperature below 20 K was required due to the instability of the E181Q Batho photoproduct. The 10 K temperature was chosen to prevent formation of any intermediates other than Batho. To generate each Photostationary State (PSS), samples were equilibrated to 10 K prior to illumination with a Photomax system equipped with a 200 W arc lamp and a monochromator (Oriel Instruments) tuned 20 nm to the blue of the absorption maximum (λmax) of the dark state. Once photoconversion to the Batho photoproduct was complete, the temperature was then raised to 220 K in increments of 10 K. To avoid artifacts arising from temperature-dependent baseline shifts, the temperature of the sample was cooled down to the starting temperature (10 K) before each spectrum was recorded. Semi-low temperature experiments were carried out from 233 K to 293 K in a Cary 5000 UV-visible spectrophotometer (Varian, Inc.) equipped with a temperature controlled sample holder (Quantum Northwest, Inc.). Samples were illuminated with 495 nm light until no further spectral shift was observed. The temperature of the sample was then raised to 293 K in increments of 5 K once photoconversion was completed.
Two methods were used in combination to determine the composition of each PSS. The first method involved warming the sample to ambient temperature to allow for the formation of Meta II, which has a λmax for both rhodopsin and E181Q rhodopsin of approximately 380 nm. The spectrum of the resulting sample contains a mixture of the dark state and Meta II, separated in wavelength to a sufficient extent to allow reliable spectral deconvolution. The integral of the λmax band, when compared to that observed for the pure dark state at the same temperature, permits accurate assignment of the amount of rhodopsin converted. Additionally, retinal oximes were extracted and analyzed using High Performance Liquid Chromatography (HPLC) following the methods and procedures reported previously [50] and described below. The isomeric compositions of each PSS are then used to deconvolute the measured spectra and determine the λmax of the photointermediates of E181Q. The pure dark state is assumed to be comprised of 100% 11-cis retinal for spectral deconvolution, as previously described [52–54]. All absorption spectra presented are the average of three spectra normalized with respect to the protein aromatic residue band at 280 nm. Each difference spectrum was calculated by subtracting the selected PSS from the corresponding dark state or relevant PSS.
Chromophore extraction
The isomeric ratio of the chromophore was determined as follows. A mixture of 150 μL of ice-cold 1.0 M hydroxylamine (pH 7) solution, 1 mL of methanol, and 1 mL of dichloromethane was added to each glycerol/protein sample. The mixture was shaken vigorously for 1 min and put on ice. Hexane (1 mL) was then added and the sample was shaken and spun in a clinical centrifuge for 45 s. The hexane layer was removed, and another 5 mL hexane extraction was performed. The combined hexane layers were then dried (Na2SO4), filtered through a 0.2 μm filter, and evaporated in a clean tube under vacuum. The total volume of the hexane layer was brought to 100 μL.
A portion of the hexane layer (50 μL) was injected into an HPLC instrument (Waters Corporation), which was equipped with two HPLC columns (Waters Prep Nova-Pak HR silica columns, 3.9×300 mm, Waters catalog no. WAT038501) and a Waters 2487 dual wavelength absorbance detector monitoring at 360 nm. The mobile phase used to separate the retinal isomers was composed of 96% hexane, 3% tert-butyl methyl ether, 0.5% 1-octanol, and 0.5% 1,4-dioxane. All solvents used were HPLC grade (Fisher Scientific). The flow rate was fixed at 2.5 mL/min. Retinal oxime standards (all-trans, 11-cis and 9-cis retinal oxime) were used to assign the retention times of the peaks observed following the chromophore extractions. The syn oxime enantiomers were favored due to the low temperature and concentrations used for the extraction, and these peaks were used exclusively to determine the isomeric ratios of retinal within the chromatograms. The anti oxime enantiomers were observed, however, these species were found at longer retention times and were ignored for this analysis.
Computational methods
All molecular orbital calculations were carried out using Gaussian 09 [55]. The heavy atom coordinates for dark state rhodopsin, Batho, Lumi, and Meta II were taken from the 1U19 [16], 2G87 [56], 2HPY [57], and 2I37 [58] crystal structures of rhodopsin, respectively. Hydrogen atoms were added to all relevant atoms of residues within 5.6Å of the chromophore by using Anamol 5.6.4. The hydrogen atoms and these local residues, in addition to the chromophore, were optimized by using the Parameterized Model 3 (PM3) methods [59,60] in Gaussian 09, while holding all other heavy atoms at the crystal geometry coordinates. During these optimizations, the oxygen atoms of the nearby water molecules were locked while the hydrogen atoms were allowed to fully optimize. Subsequently, seven iterations of B3LYP/6-31G(d) procedures [61,62] in Gaussian 09 were used to generate the ground state structures of the photointermediates. The configuration of the glutamine residue was optimized by minimizing the two possible rotational geometries and selecting the geometry with the lowest energy. The electrostatic charge shifts of the protein-binding sites were generated in MathScriptor 3.5.0 (www.mathscriptor.org).
The rhodopsin photobleaching energy surface was taken from reference [63], and was generated based on photocalorimetry as the primary experimental method. The E181Q energy surface was generated by reference to the rhodopsin surface using the temperature ramping experiments to assign the barriers, and the theoretical calculations to estimate the energy minima. The energy barriers separating the various intermediates in E181Q, , were assigned by reference to the temperature of appearance, , of the next intermediate:
| Equation 1 |
where and are the corresponding barrier and temperatures in the rhodopsin photobleaching sequence. Because the above method and the theoretical models are approximate, the E181Q surface should be viewed as qualitative.
Results
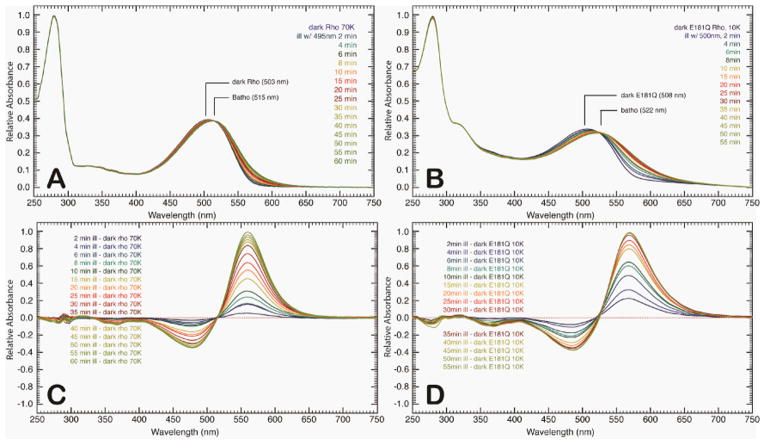
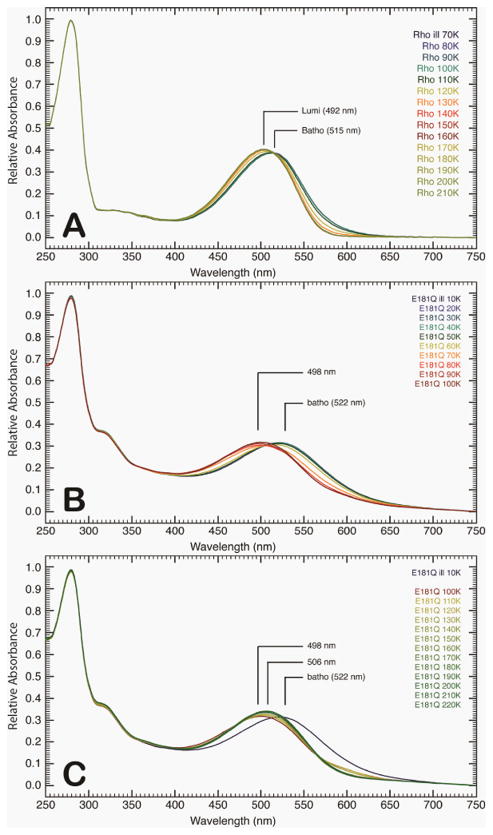
The raw spectra of the low-temperature and semi-low temperature spectroscopy studies of native rhodopsin and E181Q are described in detail below. At room temperature, E181Q displayed a significant red-shift in absorption maximum (λmax = 508 nm) relative to native rhodopsin (λmax = 499 nm) [42]. Lowering the temperature to 10 K resulted in a negligible shift for E181Q and a bathochromic shift of 4 nm for native rhodopsin (λmax = 503 nm) at 70 K (Figure 2). Illumination of E181Q with 500 nm light initiated the formation of the first PSS, a mixture of resting state and Batho, which was red-shifted to a λmax of 522 nm (PSS522). Figures 2A-2D demonstrate that the photoconversion to Batho for native rhodopsin and E181Q was complete in 1 hour at 70 K and 10 K, respectively, and involved the formation of a single species with the disappearance of the dark state. As the temperature of PSS522 was raised in 10 K increments, no spectral shift was observed from 10 K to 50 K (Figure 3). At 60 K, the spectrum begins to shift to form a second PSS at 498 nm (PSS498). The temperature was then gradually raised to 220 K, and the formation of a third intermediate was observed at 506 nm (PSS506). No further spectral change was measured after 200 K, indicating the formation of PSS506 was complete. HPLC analysis of PSS506 (Figure 4) revealed a retinal composition of 32% 11-cis, 61% all-trans, and 7% 9-cis retinal isomers.
Figure 2.
Time-resolved absorption spectra of rhodopsin at 70 K (A) and E181Q at 10 K (B) following illumination by using a 495 nm and 500 nm light source, respectively. The absorption spectra were collected at the given time points following illumination. The difference spectra of the absorption profiles are provided for rhodopsin (C) and E181Q (D) and were obtained by subtracting the respective spectra of the dark state from the corresponding time point throughout the experiment.
Figure 3.
Absorption spectra of rhodopsin (A) and E181Q (B and C) post-illumination as the temperature is ramped to 220 K via 10 K increments. The initial formation of the Batho photointermediate for native rhodopsin was achieved at 70 K (A), and as the temperature was increased, the formation of a single Lumi photointermediate (492 nm) was observed. The absorption spectra for E181Q was first collected at 10 K, in which the PSS522 consisted of a mixture of the dark and Batho states of the mutant protein (B). As the temperature was increased, a second PSS formed (PSS498) at 60 K. The E181Q sample was then allowed to warm to 220 K (C), where a third PSS (PSS506) evolved. The PSS498 was found to be a mixture of Batho and BSI, whereas the PSS506 was a Lumi photointermediate of the mutant protein (see text).
Figure 4.
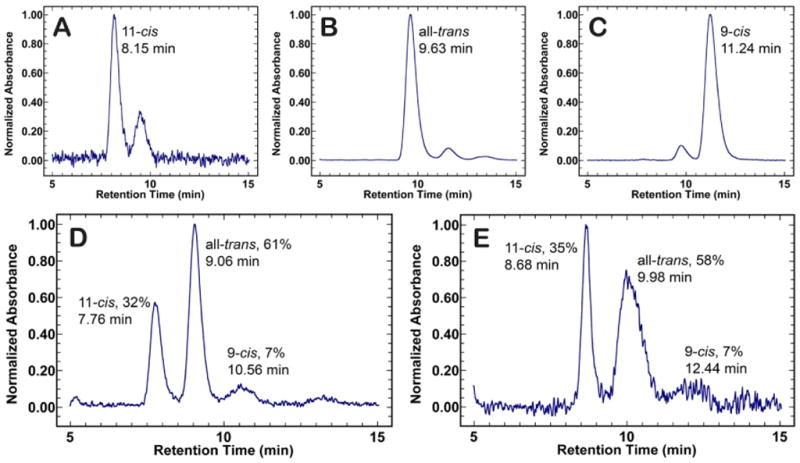
HPLC chromatograms of retinal oxime standards and retinal oxime extractions of PSS506 and PSS383. Panels A, B, and C show the chromatograms for the 11-cis, all-trans, and 9-cis retinal oxime standards, respectively. Each standard peak represents the retention time for the syn oxime enantiomer of the retinal isomers. The percent of retinal isomers formed for PSS506 (D) and PSS383 (E) were determined by integrating under each peak.
The formation and decay of the late photointermediates (Lumi, Meta I, and Meta II) for rhodopsin and E181Q were studied using semi-low temperature spectroscopy, from 233 K to 293 K. Irradiation of E181Q with 495 nm light at 233 K promoted the formation of Lumi (λmax = 495 nm) and no spectral shift was observed after 85 min of continuous illumination (Figure 5A). During the formation of Lumi, the formation and decay of a red-shifted difference spectrum species (λmax = 575 nm) is seen along with the formation of a blue-shifted difference spectrum species (λmax = 385 nm) (Figure 5B). Further temperature ramping experiments illustrated in Figure 6 indicate that the transition to the Meta I intermediate (λmax = 480 nm) began at 238 K and was complete by 243 K. The Meta I spectrum is very broad and may be a mixture of Meta I and Lumi intermediate that has not decayed completely at 233 K. With increasing temperature (in 5 K increments), the spectrum continues to blue shift until the formation of Meta II (λmax = 383 nm; PSS383) is complete. HPLC analysis of the retinal composition of PSS383 (Figure 4) shows the presence of 11-cis (35%) all -trans (58%), and 9-cis (7%) retinal isomers.
Figure 5.
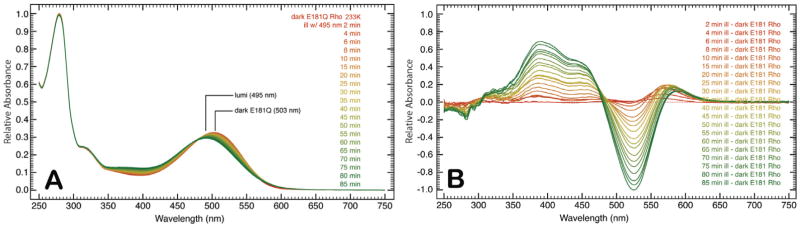
Time-resolved absorption spectra of E181Q at 233 K following illumination with 495 nm light (A). The difference spectra presented in panel (B) was obtained by subtracting the dark state spectrum from the spectra of each time point. While the blue-shifted species is accounted for by the evolution of Lumi (495 nm) over 85 min, the origin of the red-shifted species with a difference spectrum maximum at 575 nm is currently unknown.
Figure 6.
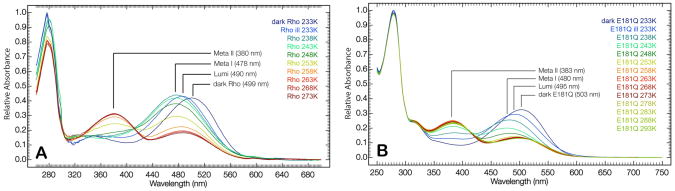
Absorption spectra of rhodopsin (A) and E181Q (B) at semi-low temperatures (233 K to 293 K) following illumination of the dark state to initiate the photobleaching sequence. The higher temperatures allowed for the formation and trapping of the late photointermediates (Meta I and Meta II), in addition to a single Lumi photointermediate.
Discussion
Time-resolved UV-visible spectroscopy has been instrumental in determining the formation and decay of discrete photointermediates of rhodopsin, as well as elucidating the photobleaching pathway of the protein [9]. However, a key disadvantage of room-temperature time-resolved UV-visible spectroscopy for the rhodopsin mutant E181Q is the simultaneous formation and decay of multiple intermediates. Low-temperature trapping experiments, which slow down the photobleaching process and ensure that each photointermediate is observed individually, were performed to avoid this problem [46–48]. In the photobleaching sequence of native rhodopsin, Batho is stable at 70 K and decays directly to Lumi upon gradual warming (Figure 3A). However, in the photobleaching sequence of E181Q, Batho is only stable at temperatures lower than 50 K and thermally equilibrates with BSI when the temperature is raised above 60 K. The final equilibrium mixture (PSS498), which is comprised mainly of BSI, is not established until 90 K. Spectral deconvolution was used to determine that pure BSI for E181Q has a λmax of 479 nm. Raising the temperature to 220 K results in decay of the mixture to Lumi (PSS506), the identity of which is confirmed through the chromophore extraction of the PSS and HPLC analysis. The isomeric composition of PSS506 was found to be 32% 11-cis retinal, 61% all-trans retinal, and 7% 9-cis retinal. Pure Lumi is found to have a λmax of 510 nm (at 220 K) by adding back 32% of the dark state spectrum of E181Q to the difference spectrum of PSS506 minus the dark state spectrum. Contributions from the 9-cis chromophore were ignored for this spectral deconvolution because so little is formed (7%). In comparison to the native rhodopsin spectral data, these observations suggest that the retinylidene binding pocket environment has been significantly perturbed during the early stages of the photoactivation mechanism.
In the semi-low temperature experiments, a single Lumi intermediate is seen for native rhodopsin (λmax = 490 nm) at 233 K. Previous studies have shown that at low temperatures, the equilibrium constant between Lumi I and Lumi II for native rhodopsin is significantly shifted towards Lumi II and thus only one Lumi intermediate is observed [64]. Similarly, we observe the formation of a single E181Q Lumi intermediate (λmax = 495 nm). However, during the formation of E181Q Lumi, the formation and decay of a red-shifted difference spectrum species (λmax = 575 nm) is observed (Figure 5). This species is not seen during Lumi formation in native rhodopsin and warrants further investigation. While the absorption spectra of the later intermediates of E181Q and native rhodopsin are similar (Figure 6), the Meta I and Meta II photointermediates of E181Q form more readily at lower temperatures compared to native rhodopsin. Recall that a negatively charged Glu-181 serves as the primary counter-ion during the late photointermediates [19–21], and thus the substitution with Gln-181 will serve to destabilize the retinal binding site during Meta I and Meta II. The isomeric composition of PSS383 (35% 11-cis retinal, 58% all-trans retinal, and 7% 9-cis retinal) is used in a similar fashion as for PSS506 in order to determine the λmax of pure Meta II (λmax = 384 nm at 273 K).
Lewis et al. [39], investigated the room temperature time-resolved spectra of Glu-181 mutants and observed a destabilization of the Batho intermediate, as well as a significant shift in equilibrium towards BSI in E181Q. During the late stages of the photoactivation mechanism, they also note the absence of a Lumi I to Lumi II transition for the E181Q mutant [39]. Because the formation of these photointermediates are temperature dependent, only the formation of photointermediates that accumulate to appreciable amounts at room temperature are expected to form at low temperatures in observable concentrations. The notion that only a single Lumi intermediate was produced for this mutant at room temperature allows us to predict that we are observing a transition from BSI to Lumi at low temperatures (< 200 K) in E181Q and not a Lumi I to Lumi II transition. The proposed photobleaching sequence pathways of native rhodopsin and E181Q are summarized in Figure 7.
Figure 7.
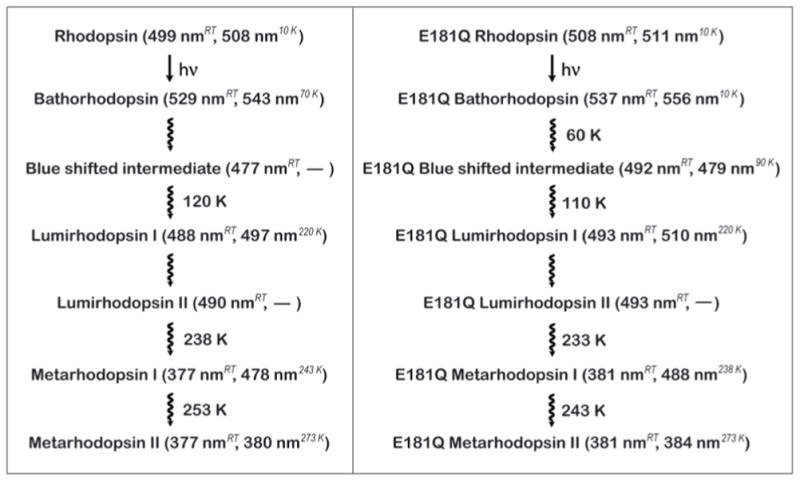
Photobleaching sequences of rhodopsin and E181Q rhodopsin. For each photointermediate, the λmax is shown at both room temperature and low temperatures. The room temperatures are based on the values obtained by Lewis et al. [39] and the low-temperature data is based on the deconvolution of the spectra presented in this study. At low temperatures, no BSI was observed for rhodopsin, and no Lumi II was observed for either rhodopsin or E181Q.
Recall that a destabilized Batho may result from a change in ionization or a disruption in the complex hydrogen-bonding network involving several binding pocket residues. Because Batho and BSI are in equilibrium, changes that destabilize Batho would result in the stabilization of BSI. While Lewis et al. [39] found that E181Q and other Glu-181 mutants lead to an accelerated decay of Batho, replacing Glu-181 with aspartic acid (E181D) resulted in a Batho lifetime similar to that of native rhodopsin. Aspartic acid is one carbon atom shorter than glutamic acid, however, the carboxyl group is maintained. Therefore, it is reasonable to predict that the stability of Batho relies on the presence of the carboxyl atoms. Although there is no clear agreement on the protonation state of Glu-181 [20,29–45], if we assume it is negatively charged in the dark state, then replacing Glu-181 with the neutral residue Gln-181 may provide insight into why the mutant E181Q leads to a faster decay of Batho and stabilizes the BSI. During the primary event, the distance between the C13-methyl group and residue Glu-181 decreases from a distance of 5.7Å to 3.0Å [16,56]. The removal of a charged residue near the highly strained C9-C13 portion of retinal may provide an increased flexibility in the polyene chain of the chromophore, which would allow the chromophore to more readily adopt a planar conformation and shift the equilibrium towards the BSI.
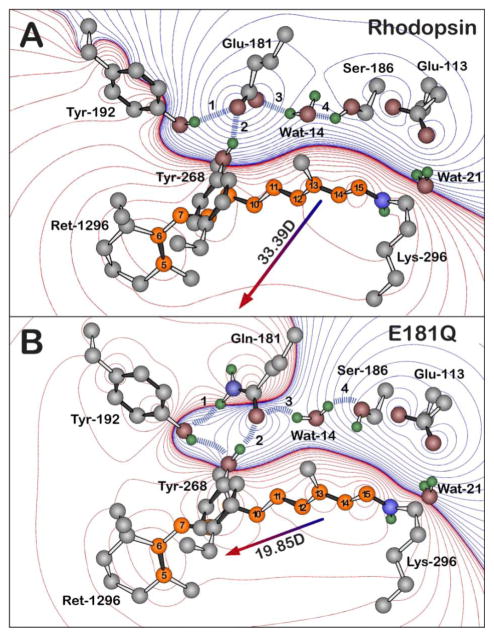
Moreover, substitution of Glu-181 with Gln-181 may also result in a destabilized Batho via perturbation of the active site hydrogen-bonding network. The notion that this mutational change would alter the hydrogen-bonding network is not surprising because the functional groups on these two residues contain different hydrogen-bonding character. Further support for a hydrogen-bonding rearrangement being responsible for the observed destabilized Batho comes from the fact that Wat-14, which is in a direct path of the C13-methyl group of the chromphore during the primary event, is believed to contribute to the stability of Batho [65]. Thus, any perturbation of Wat-14 may also result in a destabilization of Batho. In addition to E181Q, a destabilized Batho intermediate has been observed in several artificial pigments, as well as select rhodopsin mutants and cone-type visual pigments [10,11,66–68]. Previous studies have shown the decay of Batho is dependent on the rotation barrier of the C6-C7 bond, which in turn depends on the steric interaction between the C5-methyl and the C8-hydrogen. In E181Q, a rearrangement in the hydrogen-bonding network during the photobleaching sequence causes a shift in the position of Tyr-268 towards Gln-181 and Tyr-192 (Figure 8). The repositioning of Tyr-268 may lower the barrier to BSI by decreasing the steric interaction between the C5-methyl and C8-H groups. While the E181Q rhodopsin mutant is not the first to display a destabilized Batho [68], it is interesting to note that replacing Ser-186 with an alanine residue results in a normal Batho intermediate, despite the fact that Ser-186 is hydrogen-bonded to Glu-181 via Wat-14 in the dark state and early photointermediates of rhodopsin (Figures 1 and 8) [28]. Thus, in the case of E181Q, the observed destabilization of Batho may be caused in part by a rearrangement of the hydrogen-bonding network involving Tyr-268 and Tyr-192 rather than perturbations in the active site hydrogen-bonding network involving Wat-14 and Ser-186. Although the exact mechanism of Batho destabilization remains unclear, these results demonstrate that the carboxyl group at residue 181 is required for the stabilization of Batho in native rhodopsin. Furthermore, the dramatic influence of the glutamine substituion on the photobleaching kinetics suggests that this carboxyl group is likely negatively charged during the dark and Batho states.
Figure 8.
The hydrogen bonding network of the negatively charged Glu-181 residue of native rhodopsin (A) and of the Gln-181 residue of E181Q (B), both during the Lumi photointermediate. The blue dashed lines and the labels 1–4 highlight the key hydrogen-bonding network between residue 181, Ser-186, Tyr-192, Tyr-268, and Wat-14, which is perturbed upon the glutamine substitution. The Lumi structure (A) is based off of the 2HPY crystal structure [57] and a relaxed conformation of the Lumi photointermediate of E181Q (B) was obtained by minimizing the crystal structure with Gln-181. Polyene atoms of retinal (Ret-1296) are indicated in orange, and the numbering system shown here is used in the text. The water molecules are labeled using the Protein Data Bank (PDB) numbers minus 2000. All hydrogen atoms were included in the calculations and were optimized by using B3LYP/6-31G(d) methods, although only polar hydrogens are shown in the figure. Red and blue contours indicate regions of increased positive and negative charge, respectively. The contours are drawn by using the following first-order electrostatic energies: 0 (black), ± 0.282, ± 2.26, ± 7.63, ± 18, ± 35.3, ± 61, ± 96.9, ± 144, ± 206, ± 282, ± 376, ± 488, ± 621, ± 755 kJ/mol.
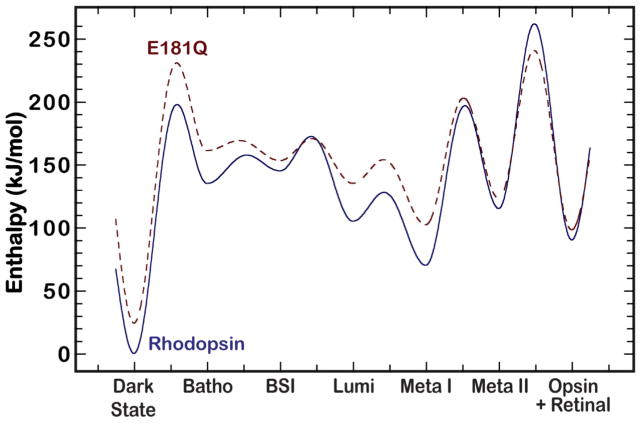
Plotting the potential energy surface of the photobleaching pathway for both rhodopsin and E181Q provides further insight into the stabilization of BSI in E181Q (Figure 9). Our calculations, which utilize the temperature of appearance of the photointermediates and the calculated energies (Equation 1), are in agreement with the literature and predict the BSI in rhodopsin lies higher in energy than Batho [8,63]. When we overlay the energies of the E181Q photointermediates, shown in dashes, it is immediately apparent that the E181Q mutant is less stable than native rhodopsin throughout the photobleaching sequence. All measured photointermediates of E181Q first appeared at lower temperatures than for the corresponding intermediates of native rhodopsin, which correlates with the predicted destabilization. This destabilization is further supported in the model of the Lumi photointermediate for both rhodopsin and E181Q provided in Figure 8, which indicates a significant modulation in the hydrogen-bonding network around the chromophore for the mutant. Most importantly, our model predicts the E181Q Batho intermediate has an enthalpy value 7.99 KJ/mol higher than E181Q BSI, whereas in rhodopsin, the BSI has an enthalpy that is 10.02 KJ/mol higher than Batho. From these results, we conclude the Batho to BSI transition is enthalpically driven in E181Q and entropically driven in native rhodopsin. Note, however, that these enthalpy differences and the potential energy surfaces depicted in Figure 9 are very approximated because they are based on Equation 1. A more rigorous theoretical study using a hybrid Quantum Mechanics/Molecular Mechanics (QM/MM) approach is currently being undertaken for further analysis of these models.
Figure 9.
A potential energy surface of the photobleaching sequence of native rhodopsin (solid blue) and E181Q (dashed red). The rhodopsin surface is adopted from reference [63]. The E181Q surface is approximate and was generated using the methods described in the text.
Conclusions
The photobleaching sequence of the rhodopsin mutant E181Q has been investigated by cryogenic studies in an attempt to further elucidate the role of Glu-181 during the photoactivation process of rhodopsin. We conclude that the photobleaching sequence of E181Q at low temperatures involves a two-step sequential decay from Batho to Lumi that includes an equilibrium between Batho and a subsequent BSI. The present study supports conclusions of other studies that have suggested Batho is destabilized in E181Q and that the Batho to BSI equilibrium lies toward the BSI [39]. The stabilization of the BSI in E181Q can be explained by the thermodynamics of the photobleaching process, which shows the Batho to the BSI transition is enthalpically driven in the rhodopsin mutant. Three key differences are noted for the photobleaching sequence of E181Q compared to rhodopsin collected at cryogenic temperatures: 1) lower temperatures are required to trap the primary photointermediate Batho in E181Q compared to native rhodopsin, 2) the decay of Batho occurs much more rapidly compared to native rhodopsin, and 3) the formation of the BSI is observed during low temperature experiments. Additionally, the formation of the later intermediates (Meta I and Meta II) of E181Q all occur at lower temperatures compared to the formation of the later intermediates of native rhodopsin. We conclude that these differences in the photobleaching sequence of E181Q compared to native rhodopsin provide strong evidence that the negatively charged carboxylic acid side chain of residue Glu-181 plays a critical role in the early intermediates of the photobleaching sequence of native rhodopsin by maintaining the integrity of the active site hydrogen-bonding network which serves to stabilize the protein in the dark state and the primary photointermediate, Batho. Furthermore, by replacing the Glu-181 residue with Gln-181, a key electrostatic interaction is altered concurrent with a rearrangement of the hydrogen-bonding network within the binding pocket, thus allowing for the formation of the BSI to accumulate to concentrations observable at low temperatures.
Acknowledgments
This research was supported in part by grants from the National Institutes of Health to R.R.B. (GM-34548) and B.E.K. (EY-11256 and EY-12975), the National Science Foundation to R.R.B. (EMT-08517), the Harold S. Schwenk Sr. Distinguished Chair funds for support of specialized instrumentation at the University of Connecticut, Research to Prevent Blindness (Unrestricted Grant to SUNY UMU Department of Ophthalmology), and Lions of CNY (B.E.K.).
References
- 1.Baylor D. How photons start vision. Proc Natl Acad Sci USA. 1996;93(2):560–565. doi: 10.1073/pnas.93.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fung BKK, Stryer L. Photolyzed rhodopsin catalyzes the exchange of GTP for bound GDP in retinal rod outer segments. Proc Natl Acad Sci USA. 1980;77(5):2500–2504. doi: 10.1073/pnas.77.5.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann KP, Jager S, Ernst OP. Structure and function of activated rhodopsin. Isr J Chem. 1995;35:339–355. doi: 10.1002/ijch.199500035. [DOI] [Google Scholar]
- 4.Smith SO. Structure and activation of the visual pigment rhodopsin. Annu Rev Biophys. 2010;39:309–28. doi: 10.1146/annurev-biophys-101209-104901. [DOI] [PubMed] [Google Scholar]
- 5.Boucher F, Leblanc RM. Energy storage in the primary photoreaction of bovine rhodopsin. A photoacoustic study. Photochem Photobiol. 1985;41:459–465. doi: 10.1111/j.1751-1097.1985.tb03512.x. [DOI] [PubMed] [Google Scholar]
- 6.Cooper A. Energy uptake in the first step of visual excitation. Nature. 1979;282:531–533. doi: 10.1038/282531a0. [DOI] [PubMed] [Google Scholar]
- 7.Schick GA, Cooper TM, Holloway RA, Murray LP, Birge RR. Energy storage in the primary photochemical events of rhodopsin and isorhodopsin. Biochemistry. 1987;26(9):2556–2562. doi: 10.1021/bi00383a022. [DOI] [PubMed] [Google Scholar]
- 8.Hug SJ, Lewis JW, Einterz CM, Thorgeirsson TE, Kliger DS. Nanosecond photolysis of rhodopsin: Evidence for a new blue-shifted intermediate. Biochemistry. 1990;29(6):1475–1485. doi: 10.1021/bi00458a019. [DOI] [PubMed] [Google Scholar]
- 9.Lewis JW, Kliger DS. Photointermediates of visual pigments. J Bioenerg Biomembr. 1992;24(2):201–210. doi: 10.1007/BF00762678. [DOI] [PubMed] [Google Scholar]
- 10.Einterz CM, Hug SJ, Lewis JW, Kliger DS. Early photolysis intermediates of the artificial visual pigment 13-demethylrhodopsin. Biochemistry. 1990;29(6):1485–1491. doi: 10.1021/bi00458a020. [DOI] [PubMed] [Google Scholar]
- 11.Albeck A, Friedman N, Ottolenghi M, Sheves M, Einterz CM, Hug SJ, et al. Photolysis intermediates of the artificial visual pigment cis-5,6-dihydro-isorhodopsin. Biophys J. 1989;55(2):233–241. doi: 10.1016/S0006-3495(89)82798-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shichida Y, Kropf A, Yoshizawa T. Photochemical reactions of 13-demethyl visual pigment analogs at low temperatures. Biochemistry. 1981;20:1962–1968. doi: 10.1021/bi00510a035. [DOI] [PubMed] [Google Scholar]
- 13.Randall CE, Lewis JW, Hug SJ, Bjorling SC, Eisner-Shanas I, Ottolenghi M, et al. A new photolysis intermediate in artificial and native visual pigments. J Am Chem Soc. 1991;113:3473–3485. [Google Scholar]
- 14.Pan D, Ganim Z, Kim JE, Verhoeven MA, Lugtenburg J, Mathies RA. Time-resolved resonance raman analysis of chromophore structural changes in the formation and decay of rhodopsin’s BSI intermediate. J Am Chem Soc. 2002;124(17):4857–4864. doi: 10.1021/ja012666e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 16.Okada T, Sugihara M, Bondar A, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J Mol Biol. 2004;342(2):571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Liu MY, Nguyen JB, Bhagat A, Mooney V, Yan EC. Thermal decay of rhodopsin: Role of hydrogen bonds in thermal isomerization of 11-cis retinal in the binding site and hydrolysis of protonated Schiff base. J Am Chem Soc. 2009;131(25):8750–8751. doi: 10.1021/ja903154u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Campomanes P, Neri M, Horta BA, Röhrig UF, Vanni S, Tavernelli I, et al. Origin of the spectral shifts among the early intermediates of the rhodopsin photocycle. J Am Chem Soc. 2014;136(10):3842–3851. doi: 10.1021/ja411303v. [DOI] [PubMed] [Google Scholar]
- 19.Terakita A, Yamashita T, Shichida Y. Highly conserved glutamic acid in the extracellular IV–V loop in rhodopsins acts as the counterion in retinochrome, a member of the rhodopsin family. Proc Natl Acad Sci USA. 2000;97(26):14263–14267. doi: 10.1073/pnas.260349597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan ECY, Kazmi MA, Ganim Z, Hou JM, Pan D, Chang BS, et al. Retinal counterion switch in the photoactivation of the G protein-coupled receptor rhodopsin. Proc Natl Acad Sci USA. 2003;100(16):9262–9267. doi: 10.1073/pnas.1531970100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan ECY, Kazmi MA, De S, Chang BS, Seibert C, Marin EP, et al. Function of extracellular loop 2 in rhodopsin: Glutamic acid 181 modulates stability and absorption wavelength of metarhodopsin II. Biochemistry. 2002;41(11):3620–3627. doi: 10.1021/bi0160011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eilers M, Goncalves JA, Ahuja S, Kirkup C, Hirshfeld A, Simmerling C, et al. Structural transitions of transmembrane helix 6 in the formation of metarhodopsin I. J Phys Chem B. 2012;116(35):10477–10489. doi: 10.1021/jp3019183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson P, Cohen G, Zhukovsky E, Oprian D. Constitutively active mutants of rhodopsin. Neuron. 1992;9(4):719–725. doi: 10.1016/0896-6273(92)90034-b. [DOI] [PubMed] [Google Scholar]
- 24.Zhu S, Brown MF, Feller SE. Retinal conformation governs pKa of protonated Schiff base in rhodopsin activation. J Am Chem Soc. 2013;135(25):9391–9398. doi: 10.1021/ja4002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leioatts N, Mertz B, Martínez-Mayorga K, Romo TD, Pitman MC, Feller SE, et al. Retinal ligand mobility explains internal hydration and reconciles active rhodopsin structures. Biochemistry. 2014;53(2):376–385. doi: 10.1021/bi4013947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ahuja S, Eilers M, Hirshfeld A, Yan EC, Ziliox M, Sakmar TP, et al. 6-s-cis conformation and polar binding pocket of the retinal chromophore in the photoactivated state of rhodopsin. J Am Chem Soc. 2009;131(42):15160–15169. doi: 10.1021/ja9034768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bartl FJ, Fritze O, Ritter E, Herrmann R, Kuksa V, Palczewski K, et al. Partial Agonism in a G Protein-coupled Receptor: Role of the Retinal Ring Structure in Rhodopsin Activation. J Biol Chem. 2005;280(40):34259–34267. doi: 10.1074/jbc.M505260200. [DOI] [PubMed] [Google Scholar]
- 28.Yan ECY, Epps J, Lewis JW, Szundi I, Bhagat A, Thomas P, Sakmar, et al. Photointermediates of the rhodopsin S186A mutant as a probe of the hydrogen-bond network in the chromophore pocket and the mechanism of counterion switch. J Phys Chem C. 2007;111(25):8843–8848. doi: 10.1021/jp067172o. [DOI] [Google Scholar]
- 29.Birge RR, Murray LP, Pierce BM, Balogh-Nair V, Findsen LA, Nakanishi K. Two-photon spectroscopy of locked-11-cis rhodopsin: Evidence for a protonated Schiff base in a neutral protein binding site. Proc Natl Acad Sci USA. 1985;82(12):4117–4121. doi: 10.1073/pnas.82.12.4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yan EC, Ganim Z, Kazmi MA, Chang BS, Sakmar TP, Mathies RA. Resonance raman analysis of the mechanism of energy storage and chromophore distortion in the primary visual photoproduct. Biochemistry. 2004;43(34):10867–10876. doi: 10.1021/bi0400148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekharan S, Buss V. Glutamic acid 181 is uncharged in dark-adapted visual rhodopsin. J Am Chem Soc. 2008;130:17220–17221. doi: 10.1021/ja805992d. [DOI] [PubMed] [Google Scholar]
- 32.Mollevanger LC, Kentgens AP, Pardoen JA, Courtin JM, Veeman WS, Lugtenburg J, et al. High-resolution solid-state 13C-NMR study of carbons C-5 and C-12 of the chromophore of bovine rhodopsin. Evidence for a 6-s-cis conformation with negative-charge perturbation near C-12. Eur J Biochem. 1987;163(1):9–14. doi: 10.1111/j.1432-1033.1987.tb10729.x. [DOI] [PubMed] [Google Scholar]
- 33.Smith SO, Palings I, Miley ME, Courtin J, de Groot H, Lugtenburg J, et al. Solid state NMR studies of the mechanism of the opsin shift in the visual pigment rhodopsin. Biochemistry. 1990;29(35):8158–8164. doi: 10.1021/bi00487a025. [DOI] [PubMed] [Google Scholar]
- 34.Han M, Smith SO. NMR constraints on the location of the retinal chromophore in rhodopsin and bathorhodopsin. Biochemistry. 1995;34(4):1425–1432. doi: 10.1021/bi00004a037. [DOI] [PubMed] [Google Scholar]
- 35.Honig B, Dinur U, Nakanishi K, Valeria BN, Mary AG, Arnaboldi M, et al. An external point-charge for wavelength regulation in visual pigments. J Am Chem Soc. 1979;101(23):7084–7086. doi: 10.1021/ja00517a060. [DOI] [Google Scholar]
- 36.Nagata T, Terakita A, Kandori H, Shichida Y, Maeda A. The hydrogen-bonding network of water molecules and the peptide backbone in the region connecting Asp83, Gly120, and Glu113 in bovine rhodopsin. Biochemistry. 1998;37(49):17216–17222. doi: 10.1021/bi9810149. [DOI] [PubMed] [Google Scholar]
- 37.Ludeke S, Beck M, Yan EC, Sakmar TP, Siebert F, Vogel R. The role of Glu181 in the photoactivation of rhodopsin. J Mol Biol. 2005;353(2):345–356. doi: 10.1016/j.jmb.2005.08.039. [DOI] [PubMed] [Google Scholar]
- 38.Rohrig UF, Guidoni L, Rothlisberger U. Early steps of the intramolecular signal transduction in rhodopsin explored by molecular dynamics simulations. Biochemistry. 2002;41(35):10799–10809. doi: 10.1021/bi026011h. [DOI] [PubMed] [Google Scholar]
- 39.Lewis JW, Szundi I, Kazmi MA, Sakmar TP, Kliger DS. Time-resolved photointermediate changes in rhodopsin glutamic acid 181 mutants. Biochemistry. 2004;43(39):12614–12621. doi: 10.1021/bi049581l. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Mayorga K, Pitman MC, Grossfield A, Feller SE, Brown MF. Retinal counterion switch mechanism in vision evaluated by molecular simulations. J Am Chem Soc. 2006;128(51):16502–16503. doi: 10.1021/ja0671971. [DOI] [PubMed] [Google Scholar]
- 41.Frahmcke JS, Wanko M, Phatak P, Mroginski MA, Elstner M. The protonation state of Glu181 in rhodopsin revisited: Interpretation of experimental data on the basis of QM/MM calculations. J Phys Chem B. 2010;114(34):11338–11352. doi: 10.1021/jp104537w. [DOI] [PubMed] [Google Scholar]
- 42.Sandberg MN, Amora TL, Ramos LS, Chen MH, Knox BE, Birge RR. Glutamic acid 181 is negatively charged in the bathorhodopsin photointermediate of visual rhodopsin. J Am Chem Soc. 2011;133(9):2808–2811. doi: 10.1021/ja1094183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tomasello G, Olaso-Gonzalez G, Altoe P, Stenta M, Serrano-Andres L, Merchán M, et al. Electrostatic control of the photoisomerization efficiency and optical properties in visual pigments: On the role of counterion quenching. J Am Chem Soc. 2009;131(14):5172–5186. doi: 10.1021/ja808424b. [DOI] [PubMed] [Google Scholar]
- 44.Grossfield A, Pitman MC, Feller SE, Soubias O, Gawrisch K. Internal hydration increases during activation of the G-protein-coupled receptor rhodopsin. J Mol Biol. 2008;381(2):478–486. doi: 10.1016/j.jmb.2008.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hall KF, Vreven T, Frisch MJ, Bearpark MJ. Three-layer ONIOM studies of the dark state of rhodopsin: the protonation state of Glu181. J Mol Biol. 2008;383(1):106–121. doi: 10.1016/j.jmb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 46.Yoshizawa T, Shichida Y. Low-temperature spectroscopy of intermediates of rhodopsin. Methods Enzymol. 1982;81:333–54. doi: 10.1016/s0076-6879(82)81051-3. [DOI] [PubMed] [Google Scholar]
- 47.Kusnetzow A, Dukkipati A, Babu KR, Singh D, Vought BW, Knox BE, et al. The photobleaching sequence of a short-wavelength visual pigment. Biochemistry. 2001;40(26):7832–7844. doi: 10.1021/bi010387y. [DOI] [PubMed] [Google Scholar]
- 48.Ramos LS, Chen M-H, Knox BE, Birge RR. Regulation of Photoactivation in Vertebrate Short Wavelength Visual Pigments: Protonation of the retinylidene Schiff base and a counterion switch. Biochemistry. 2007;46(18):5330–5340. doi: 10.1021/bi700138g. [DOI] [PubMed] [Google Scholar]
- 49.Babu KR, Dukkipati A, Birge RR, Knox BE. Regulation of phototransduction in short wavelength cone visual pigments via the retinylidene Schiff base counterion. Biochemistry. 2001;40:13760–6. doi: 10.1021/bi015584b. [DOI] [PubMed] [Google Scholar]
- 50.Vought BW, Dukkipati A, Max M, Knox BE, Birge RR. Photochemistry of the primary event in short-wavelength visual opsins at low temperature. Biochemistry. 1999;38(35):11287–11297. doi: 10.1021/bi990968b. [DOI] [PubMed] [Google Scholar]
- 51.Kusnetzow AK, Dukkipati A, Babu KR, Ramos L, Knox BE, Birge RR. Vertebrate ultraviolet visual pigments: protonation of the retinylidene Schiff base and a counterion switch during photoactivation. Proc Natl Acad Sci USA. 2004;101(4):941–946. doi: 10.1073/pnas.0305206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Birge RR, Einterz CM, Knapp HM, Murray LP. The nature of the primary photochemical events in rhodopsin and isorhodopsin. Biophys J. 1988;53(3):367–385. doi: 10.1016/S0006-3495(88)83114-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suzuki T, Callender RH. Primary photochemistry and photoisomerization of retinal at 77°K in cattle and squid rhodopsins. Biophys J. 1981;34(2):261–270. doi: 10.1016/S0006-3495(81)84848-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vought BW, Dukkipatti A, Max M, Knox BE, Birge RR. Photochemistry of the primary event in short-wavelength visual opsins at low temperature. Biochemistry. 1999;38(35):11287–11297. doi: 10.1021/bi990968b. [DOI] [PubMed] [Google Scholar]
- 55.Frisch MJ, Trucks GW, Schlegel HB, et al. Gaussian 09, Revision A.02. Wallingford, CT: Gaussian, Inc; 2009. [Google Scholar]
- 56.Nakamichi H, Okada T. Crystallographic analysis of primary visual photochemistry. Angew Chem Int Ed Engl. 2006;45(26):4270–4273. doi: 10.1002/anie.200600595. [DOI] [PubMed] [Google Scholar]
- 57.Nakamichi H, Okada T. Local peptide movement in the photoreaction intermediate of rhodopsin. Proc Natl Acad Sci USA. 2006;103(34):12729–12734. doi: 10.1073/pnas.0601765103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Salom D, Lodowski DT, Stenkamp RE, Le Trong I, Golczak M, Jastrzebska B, et al. Crystal structure of a photoactivated deprotonated intermediates of rhodopsin. Proc Natl Acad Sci USA. 2006;103(44):16123–16128. doi: 10.1073/pnas.0608022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stewart JJP. Optimization of parameters for semiempirical methods I. Method. J Comput Chem. 1989;10(2):209–220. [Google Scholar]
- 60.Stewart JJP. Optimization of parameters for semiempirical methods II. Applications J Comput Chem. 1989;10:221–64. [Google Scholar]
- 61.Becke AD. Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys. 1993;98:5648–52. [Google Scholar]
- 62.Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter. 1988;37(2):785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 63.Birge RR, Vought BW. Energetics of rhodopsin photobleaching: photocalorimetric studies of energy storage in the early and later intermediates. Methods Enzymol. 2000;315:143–163. doi: 10.1016/s0076-6879(00)15841-0. [DOI] [PubMed] [Google Scholar]
- 64.Szundi I, Epps J, Lewis JW, Kliger DS. Temperature dependence of the lumirhodopsin I - lumirhodopsin II equilibrium. Biochemistry. 2010;49(28):5852–5858. doi: 10.1021/bi100566r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis JW, Fan GB, Sheves M, Szundi I, Kliger DS. Steric barrier to bathorhodopsin decay in 5-demethyl and mesityl analogues of rhodopsin. J Am Chem Soc. 2001;123(41):10024–10029. doi: 10.1021/ja010724q. [DOI] [PubMed] [Google Scholar]
- 66.Lewis JW, Liang J, Ebrey TG, Sheves M, Kliger DS. Chloride effect on the early photolysis intermediates of a gecko cone-type visual pigment. Biochemistry. 1995;34(17):5817–5823. doi: 10.1021/bi00017a013. [DOI] [PubMed] [Google Scholar]
- 67.Shichida Y, Okada T, Kandori H, Fukada Y, Yoshizawa T. Nanosecond laser photolysis of iodopsin, a chicken red-sensitive cone visual pigment. Biochemistry. 1993;32(40):10832–10838. doi: 10.1021/bi00091a039. [DOI] [PubMed] [Google Scholar]
- 68.Jager S, Han M, Lewis JW, Szundi I, Sakmar TP, Kliger DS. Properties of early photoylsis intermediates of rhodopsin are affected by glycine 121 and phenylalanine 261. Biochemistry. 1997;36(39):11804–11810. doi: 10.1021/bi971122f. [DOI] [PubMed] [Google Scholar]