Abstract
Identifying novel NF-κB-regulated immune genes in the human genome is important to our understanding of immune mechanisms and immune diseases. We fit logistic regression models to the promoters of 62 known NF-κB-regulated immune genes, to find patterns of transcription factor binding in the promoters of genes with known immune function. Using these patterns, we scanned the promoters of additional genes to find matches to the patterns, selected those with NF-κB binding sites conserved in the mouse or fly, and then confirmed them as NF-κB-regulated immune genes based on expression data. Among 6440 previously identified promoters in the human genome, we found 28 predicted immune gene promoters, 19 of which regulate genes with known function, allowing us to calculate specificity of 93%–100% for the method. We calculated sensitivity of 42% when searching the 62 known immune gene promoters. We found nine novel NF-κB-regulated immune genes which are consistent with available SAGE data. Our method of predicting gene function, based on characteristic patterns of transcription factor binding, evolutionary conservation, and expression studies, would be applicable to finding genes with other functions.
The human immune system is our host defense system against microbes and environmental hazards, and thus understanding immune mechanisms is of great interest in medical and biological research. NF-κB is a transcription factor (TF) that is known to be an important mediator of immune responses (Baeuerle and Baltimore 1996; Ghosh et al. 1998), and the 62 known NF-κB-regulated immune genes play fundamental roles in both innate immunity and adaptive immunity. A draft of the human genome sequence was published recently (International Human Genome Sequencing Consortium 2001; Venter et al. 2001), and it is estimated that there are about 35,000 genes in the human genome. More than half of these genes are novel, and it is reasonable to hypothesize that some of these novel genes have immune functions. Because of the importance of NF-κB signaling in the immune system, identifying these novel NF-κB-regulated immune genes will advance our understanding of human immunity and immune diseases.
Gene promoters are regulatory regions that are integral components of genes. For the purposes of this study, a promoter is the regulatory region of the gene that is proximal to the transcription start site (TSS). Eric Davidson used the patterns of TF binding sites in the promoters of sea urchin developmental genes to build a computational model to accurately predict their expression (Yuh et al. 1998). In Drosophila, TF binding-site patterns in regulatory regions have been used to search the fly genome to find developmental genes (Berman et al. 2002; Markstein et al. 2002). In mammals, efforts using logistic regression analysis (LRA) models of regulatory regions to find muscle and liver genes have also seen some success (Wasserman and Fickett 1998; Krivan and Wasserman 2001).
For most genes in the human genome, the precise locations of TSSs and their proximal promoters are still unknown. As such, finding the promoters is often a necessary first step in studying gene regulation, and previous work in our lab has involved finding these sites (Liu et al. 2001; Liu and States 2002). For genes with known mRNAs, our prediction method, CONPRO, has been shown to identify promoters with 70% sensitivity and over 90% specificity. Applying CONPRO to the human genome, we found 6440 promoters for genes with known mRNAs.
To identify novel NF-κB-regulated immune gene promoters among these 6440 promoters, we used the patterns of TF binding sites in the promoters of known NF-κB-regulated immune genes and evolutionary conservation of the NF-κB binding sites, then confirmed the predictions based on available expression data. We retrieved 62 known NF-κB-regulated immune response genes and their promoters (Baeuerle and Baichwal 1997), and we found that five TF families (including NF-κB) have binding sites overrepresented by a factor of at least two in these immune gene promoters. This overrepresentation suggests that these TFs are important in coregulating immune genes. We fit two LRA models, based on the patterns of binding sites for these five TFs and the positions of the NF-κB binding site within these promoters, then searched the 6440 promoters to find preliminary candidates for immune gene promoters. To improve the specificity of our predictions, these preliminary candidates were checked for NF-κB binding-site conservation between the human and mouse genome, or the human and Drosophila genome when mouse genomic data were not available. Serial analysis of gene expression (SAGE) data on myeloid, lymphoid, and microvascular endothelial cells are consistent with nine of these genes being activated by NF-κB in vivo, and we identified them as putative novel NF-κB-regulated immune response genes. Our method has sensitivity of 42%, and our two LRA models show specificity of 93% and 100%, respectively.
RESULTS
Positions of NF-κB Binding Sites in Immune Gene Promoters
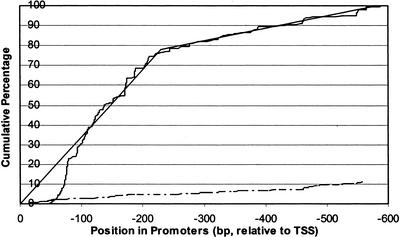
We first looked for positional preferences in the NF-κB binding sites of 62 known NF-κB-regulated mammalian immune gene promoters (Baeuerle and Baichwal 1997). In Figure 1, the cumulative percentage of NF-κB binding sites in these 62 promoters is plotted against the position relative to the TSS (solid curve, Fig. 1). For reference, among a set of mammalian nonimmune promoters, NF-κB binding sites appear as random noise without positional preference (dotted curve, Fig. 1). Among immune genes, the region immediately upstream of the TSS contains the highest density of NF-κB sites, whereas the region further upstream also contains an enrichment of NF-κB sites relative to nonimmune promoters, but not as high as in the proximal region. To find the breakpoint between these two regions, we used piecewise linear regression (S-Plus 6.0) to find the turning point in the curve at −230bp (solid lines, Fig. 1). Over 75% of NF-κB binding sites in the immune gene promoters are within 230 bp upstream of the TSS, and NF-κB binding is more significant in these genes than in immune genes with the NF-κB binding site farther upstream. We built separate LRA models for these two groups of genes.
Figure 1.
Position distribution of NF-κB binding sites. The x-axis is the position of NF-κB binding sites relative to the transcription start site. The y-axis is the cumulative percentage of NF-κB binding sites. The upper (solid) curve is for NF-κB sites in immune promoters, and the lower (dotted) curve is for NF-κB sites in nonimmune promoters (percentage relative to total NF-κB binding sites in the 62 immune promoters). The upper curve has roughly two phases; one is between −1 and −230 bp, and the other is from −231 to −600 bp. The turning point is revealed by piecewise linear regression (solid lines). Over 75% of NF-κB sites are within the 230-bp region immediately upstream of TSS. From the dotted curve, it appears that NF-κB sites in nonimmune promoters do not have positional preference.
Informative Transcription Factors in NF-κB-Regulated Immune Promoters
Regulation of eukaryotic gene expression is normally the consequence of interactions among a network of TFs (Kadonaga 1998; Lemon and Tjian 2000), and thus, in addition to NF-κB, binding sites for other TFs may be informative in identifying immune genes. We searched for these other informative TFs in NF-κB-regulated immune gene promoters by looking for TFs with binding sites that are overrepresented in NF-κB-regulated immune gene promoters. To find overrepresented TF binding sites, we compared the number of promoters with at least one binding site for a TF between two groups of promoters: the group of 62 known immune gene promoters and a null group that includes four sets of 62 mammalian nonimmune promoters taken from the Eukaryotic Promoter Database (EPD; Perier et al. 2000). Evaluating all TFs in the TRANSFAC database (Wingender et al. 2000), we selected those with binding sites that are at least twice as frequent in the NF-κB-regulated group as in the null group. The TFs that meet this criterion are AP1, IK1, IK3, IRF1, IRF2, ISRE, NF-κB, and STAT (Fig. 2, left). For comparison, the number of promoters containing binding sites for eight other TFs (Fig. 2, right) is not significantly different between immune and nonimmune promoters. This result is consistent with biological observations, because NF-κB has been shown to interact with AP1 and IRF1 to regulate genes (Thanos and Maniatis 1995), and the other five overrepresented TFs also have documented roles in the immune system. We grouped these TFs into five families by binding site sequence similarity: IK1 and IK3 are combined into IK, whereas IRF1, IRF2, and ISRE are combined into IRF, leaving informative TFs: NF-κB, AP1, IK, IRF, and STAT.
Figure 2.
Abundance of transcription factor binding sites. Comparisons are made between the 62 NF-κB-regulated promoters and four sets of 62 mammalian nonimmune promoters from the EPD. The y-axis is the number of promoters with at least one binding site for the transcription factor. The white bar is the number of NF-κB-regulated immune gene promoters with the TF, and the gray bar is the mean count of promoters with the TF in the four sets of nonimmune promoters. The error bars represent one standard deviation in the nonimmune promoter data. Note that among the 62 NF-κB-regulated promoters, the number of promoters with NF-κB sites is 53, instead of 62. In this analysis, we set a high PWM threshold for detecting NF-κB binding sites (0.9), and thus we did not detect variant sites.
Logistic Regression Models
We used logistic regression to model the probability that a given promoter fits into the group of immune promoters, versus the group of nonimmune promoters, given the pattern of informative TF binding sites in the promoter. Based on our earlier observation that there are proximal and distal regions in the 62 known immune gene promoters, representing different levels of NF-κB regulation, we built one LRA model for immune promoters with NF-κB binding sites within 230 bp upstream of the TSS (Table 1, left) and one model for promoters with more distal NF-κB binding sites (Table 1, right). For both models, the most informative TF is NF-κB, which is expected for NF-κB-regulated immune genes, but the presence or absence of the other informative TF binding sites also helps categorize promoters.
Table 1.
The LRA Models for NF-κB-Regulated Immune Gene Promoters
| LRA model | Model variables | Promoters with NF-κB sites from −1 to −230bp | Promoters with NF-κB sites only from −231 to −600bp | ||||
| Coeff. | S.E. | P-value | Coeff. | S.E. | P-value | ||
| Model w/o number of | Intercept | −3.45 | 0.38 | 6.1E-17 | −5.80 | 0.98 | 8.8E-09 |
| TF binding sites | NF-κB | 3.49 | 0.44 | 4.3E-14 | 4.19 | 0.95 | 1.5E-05 |
| AP1 | 0.70 | 0.47 | 0.135 | 1.35 | 0.96 | 0.160 | |
| IK | 0.87 | 0.49 | 0.074 | 1.73 | 0.81 | 0.034 | |
| IRF | 0.93 | 0.64 | 0.146 | 2.67 | 0.91 | 0.003 | |
| STAT | 1.00 | 0.44 | 0.022 | 0.70 | 0.75 | 0.355 | |
The columns labeled Coeff. are the coefficients in the logistic regression model. S.E. is the standard error in the fit for the coefficient, and P-value is the probability of observing a coefficient this far from 0 in a fit to random data. The weight matrix score cutoff for NF-κB and AP1 sites is 0.95; that for the IK1, IK3 binding sites is 0.94; for STAT, IRF1, IRF2, and ISRE, the score cutoff is 0.91.
Searching for NF-κB-Regulated Immune Genes
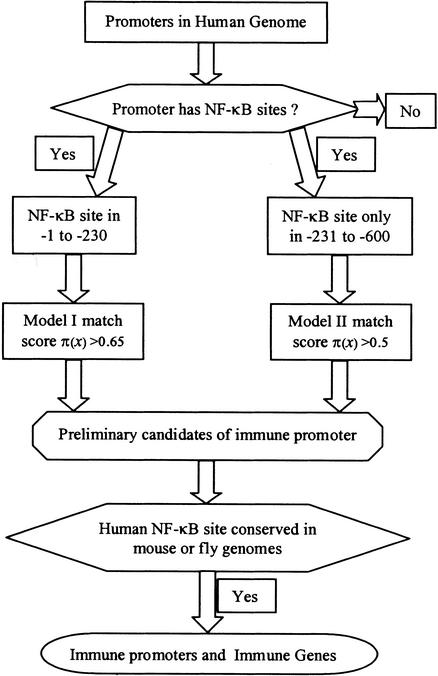
Our algorithm for finding NF-κB-regulated immune gene promoters is shown in Figure 3. For each promoter, we first check for an NF-κB binding site in the first 600 bp upstream of the TSS. If none is found, we exit the algorithm. If an NF-κB binding site is found and it is in the −1 to −230 bp window we use model I; otherwise we use model II. If a promoter has a pattern of TF binding sites yielding a probability, π(x) (probability of being from the immune gene group), which exceeds the threshold established for the LRA model used, we consider the promoter a preliminary candidate.
Figure 3.
Algorithm for searching for immune genes. The promoters with NF-κB binding sites are checked for positions of the binding sites. Model I or model II is used to estimate the probability that a promoter fits in the group of immune promoters. The last step of the procedure is verification of NF-κB binding site conservation in the mouse or fly genome. The promoters found by this method are predictions of immune gene promoters.
To increase the specificity of our predictions, we compared the preliminary candidates with their mouse or fly homologs. If possible, they were checked for conserved NF-κB binding sites in the mouse genome, but we do not have genomic sequences for all mouse genes. In such cases, we compared the preliminary candidates with the Drosophila genome. Although flies have only a simple form of immune response (innate immunity), conservation of regulatory regions across such evolutionary distance is likely to be functionally important. On searching the 62 known NF-κB-regulated immune genes, our method selected 18 promoters by model I and eight promoters by model II, yielding sensitivity of 26/62, or 42%.
Next, we searched for NF-κB-regulated immune gene promoters among the 6440 human promoters previously identified by CONPRO (Liu and States 2002). CONPRO finds promoters that are associated with mRNA transcripts, and thus finding an immune promoter is equivalent to finding an immune gene. Among the 6440 promoters, we found 28 immune genes, 22 genes by model I and six genes by model II.
Specificity: Predicted Immune Genes With Known Function
Among the 22 predicted immune gene promoters found by model I, 15 regulate well characterized human genes (Table 2). Eleven of these promoters have been cloned, and mutagenesis studies confirmed that the genes are NF-κB-regulated (Promoter Study, Table 2). Two other genes, MAP3K8 and MIP-3α, have gene expression data consistent with NF-κB regulation (Gene; Array; Table 2): MAP3K8 is regulated by activation of NF-κB under five different experimental conditions, and MIP-3α shows increased expression after NF-κB is activated by seven different stimulators in different cell lines.
Table 2.
Model 1 Predictions of Immune Genes Among Well Characterized Genes
| Gene name | Acc. no. | Evidence of NF-κB regulation | Immune gene | Model score | Functions in immune system |
| RANTES | AF088219 | Promoter study(1) | +(2) | 0.935 | Activator of variety of leukocytes |
| ELAM-1 | AL021940 | Promoter study(3) | +(4) | 0.833 | Migration of leukocytes |
| HLA-B | AJ250917 | Promoter study(5) | + | 0.935 | MHC class I molecule |
| IL8 | M26383 | Promoter study(3) | +(6) | 0.850 | Acute inflammation |
| GROγ | M36821 | Promoter study(3) | +(7) | 0.850 | Macrophage inflammatory protein |
| CD83 | NM_004233 | Promoter study(8) | +(9) | 0.833 | Dendritic cells maturation |
| NF-κB2 | S76638 | Promoter study(3) | +(3) | 0.739 | Regulator of immune response |
| MAJCAM1 | U43628 | Promoter study(3) | +(10) | 0.714 | Migration of leukocytes |
| IL6 | X04430 | Promoter study(3) | +(11) | 0.677 | Acute immune response |
| TNF-β | D12614 | Promoter study(3) | +(12) | 0.739 | Lymphoid organ development and chronic inflammation |
| CD69 | Z22576 | Promoter study(3) | +(13) | 0.871 | T-cell activation and function in other hematopoietic lineage cells |
| MAP3K8 | D14497 | Gene(14) | +(15) | 0.739 | Regulator of NF-κB in T cells; Involved in T-cell activation |
| MIP-3α | U64197 | Gene(16); array(17) | + | 0.714 | Macrophage inflammatory protein |
| IP3KC | D38169a | N/A | +(18) | 0.714 | T/B cell activation/differentiation; Signaling in monocyte/ macrophage |
| Nuclear matrix p84 | L36529 | N/A | − | 0.850 | N/A |
NF-κB binding site conserved in Drosophila, otherwise conserved in the mouse genome.
In the third column, experimental evidence on whether the promoters are regulated by NF-κB is found in the literature. “Promoter study” means the promoters have been cloned upstream of a reporter gene and expression assays demonstrated that the promoters are regulated by NF-κB. “Gene” means that a single gene expression assay of the candidate, usually by Northern blot, shows increased expression in cells after activating NF-κB with the stimulating factors. “Array” refers to microarray experiments showing that expression of the gene increases after NF-κB is activated in the cells. In the fourth column, a plus sign means the genes play a role in the immune system. The fifth column gives the score (π[x]) for the promoter in the LRA model. Ref. (1) Lee et al. 2000; Moriuchi et al. 1997. (2) Appay and Rowland-Jones 2001. (3) Baeuerle and Baichwal 1997. (4) Elangbam et al. 1997. (5) Girdlestone 2000; Gobin et al. 1998. (6) Harada et al. 1994. (7) Driscoll 1994. (8) McKinsey et al. 2000. (9) Robinson et al. 1998. (10) Elangbam et al. 1997. (11) Tilg et al. 1997; Akira and Kishimoto 1992. (12) Ruddle 1999. (13) Marzio et al. 1999; Ziegler et al. 1994. (14) Expression data in fibroblasts: OA, IL-1 (Chan et al. 1993); Spleen cells: Con A (Patriotis et al. 1993); T:αCD3, phorbol ester (Sanchez-Gongora et al. 2000). (15) Belich et al. 1999; Patriotis et al. 1993. (16) Expression data in THP-1:PMA; PBMC:LPS; I-HUVEC:TNFα (Hromas et al. 1997); Macrophage: Influenza A, Sendai virus (Matikainen et al. 2000). (17) Expression data in THP1:L. monocytogenes (Cohen et al. 2000); Macrophages:LDL (Shiffman et al. 2000). (18) Marshall et al. 2000; Chow et al. 1995; Ward and Cantrell 2001.
Inositol 1,4,5-triphosphate kinase C (IP3KC) was cloned (Dewaste et al. 2000), but data from expression studies are not yet available. However, the IP3 second messenger signaling pathway is involved in T-cell and B-cell activation and differentiation, as well as in monocyte and macrophage signaling. There are three IP3 kinase isoforms, IP3KA, IP3KB, and IP3KC. It was initially observed that the IP3 kinase in thymus and lymphocytes has a higher molecular weight than IP3KA and IP3KB (D'Santos et al. 1994). It was later shown that the IP3 kinase in thymus is isoform C (Dewaste et al. 2000). Furthermore, the IP3KC promoter has binding sites for IK1, AP1, STAT, and NF-κB;, whereas the IP3KB and IP3KA promoters do not have any of the above sites. Based on these studies, we believe that IP3KC is the isoform expressed in the immune system, regulating T-cell and B-cell activation. Model I also predicts p84 as an immune gene, but we found no experimental evidence to confirm this, yielding specificity of 14/15, or 93% for model I.
Model II predicts six immune gene promoters, with four of them regulating well characterized genes (Table 3). The first three of these genes have been shown in promoter studies to have immune functions and NF-κB regulation. Additionally, MyD118 (gadd45beta) is a response gene in myeloid differentiation and is upregulated when NF-κB is activated in both T and B cells. Ecto-ATPase provides signals for activating cytokine secretion in T cells and antibody secretion in B cells, as well as signals for B-cell proliferation. As such, all four of the model II predictions are supported by experimental data, yielding specificity of 4/4, or 100%.
Table 3.
Model 2 Predictions of Immune Genes Among Well Characterized Genes
| Gene name | Acc. no. | Evidence of NF-κB regulation | Immune gene | Model score | Functions in immune system |
| M-CSF-1 | M37435 | Promoter Study(1) | +(2) | 0.586 | Macrophage differentiation |
| B factor | S67310 | Promoter Study(1) | +(3) | 0.975 | MHC class III gene |
| MyD118 | NM_015675 | Promoter Study(4) Gene(5) Array(6) | +(7) | 0.500 | Myeloid cell differentiation |
| Ecto-ATPase | U91510 | Gene(8) | +(9) | 0.586 | T: cytokine secretion, cytolytic activity; B: Ab secretion, proliferation |
For table legend, please see Table 2. Ref. (1) Baeuerle and Baichwal 1997. (2) Hamilton 1997. (3) Campbell et al. 1986. (4) De Smaele et al. 2001. (5) Expression data in T:TNF, PMA (De Smaele et al. 2001). (6) Expression data in 3T3:CSF1 (Fambrough et al. 1999); B:LPS, IL1 (Li et al. 2001). (7) Vairapandi et al. 1996.(8) Expression data in NK:IL2 (Dombrowski et al. 1998); HUVEC:CMV (Kas-Deelen et al. 2001); hepatoma cells: Dioxin (Gao et al. 1998). (9) Dombrowski et al. 1998.
Novel Predictions of NF-κB-Regulated Immune Genes
Our method predicts nine novel immune genes (Table 4). Although these genes are not well characterized, there is some evidence to support immune function in these genes. Two of these nine genes have well studied homologs in model organisms. The first one is , and the Drosophila homolog is four-jointed (fj), which is a putative intercellular signaling protein (Brodsky and Steller 1996). It has also been shown that fj may be a downstream gene in the JAK/STAT pathway in Drosophila (Zeidler et al. 2000). The role of the JAK/STAT pathway in the immune system is well established (Leonard and O'Shea 1998), so we have reason to believe that encodes an immune-related gene in humans. Another gene having characterized homologs is (sir2 homolog 2), which is one of seven human sir2 homologs. The yeast sir2 homolog is a histone deacetylase, which causes gene silencing and is related to aging (Shore 2000). However, in humans the protein product of primarily locates in the cytoplasm, rather than the nucleus, and overexpression of the gene in human cells has no effects on cell growth or chromosome stability (Afshar and Murnane 1999). These experiments suggest that the human homolog has functions that are different from yeast sir2 unrelated to gene silencing and aging, and we propose that the human gene functions in the immune system.
Table 4.
Predicted Immune Related Genes Among Not Well Characterized Genes
| Acc. no. | Homologs | TF sites in promoter | Model score | SAGE data after activation of NF-κB |
| AB007869 | Homologs in mice | AP1, STAT, NF-κB | 0.850 | Th1:PMA(1:9, 1.1e-2)(1); Th2:PMA(1:3, 3.1e-1)(1); HMVEC:VEGF (19:34, 2.8e-2)(2) |
| AF055016a | Homologs in mice, and fly | AP1, STAT, NF-κB | 0.850 | Th1:PMA(0:6, 1.6e-2)(1); Th2:PMA(0:6, 1.6e-2)(1); HMVEC:VEGF (0:51, < e-10)(2) |
| AJ245599 | Homologs mice, and fly (four-jointed) | AP1, IK1, NF-κB | 0.833 | HMVEC:VEGF(0:34, < e-10)(2) |
| AK022822a | Homologs in fly, and yeast | AP1, IK1, NF-κB | 0.833 | Th1:PMA(0:54, < e-10)(1) Th2:PMA(0:120, < e-10)(1); HMVEC:VEGF (4930:6471)(2) |
| AK023143 | Homolog fly | AP1, IK1, NF-κB | 0.833 | Th1:PMA(0:3, 1.3e-1)(1); Th2:PMA(0:6, 1.6e-2)(1); HMVEC:VEGF(19:34, 2.8e-2)(2) |
| AK025876 | Homologs in mice, fly, worm, yeast (sir2) | AP1, IK1, NF-κB | 0.833 | Mo:LPS(0:5, 3.1e-2)(4); Mo:M-CSF(0:7, 7.8e-3)(3); Mo:GM-CSF(0:7, 7.8e-3)(3); Th1:PMA(0:3, 1.3e-1)(1); Th2:PMA(0:9, 1.9e-3)(1); HMVEC:VEGF(19:103, < e-10)(2) |
| NM_007229 | Homologs in mice, fly, and yeast | AP1, STAT, NF-κB | 0.850 | Mo:M-CSF(1:4, 1.9e-1)(3) |
| AK000731b | Homolog in mice, and fly | AP1, STAT, NF-κB | 0.586 | Mo:LPS(0:3, 1.3e-1)(4); HMVEC:VEGF(19:34, 2.8e-2)(2) |
| NM-016185b | Homolog in mice | AP1, IK1, NF-κB | 0.796 | Th1:PMA(0:6, 1.6e-2)(1); Th2:PMA(0:6, 1.6e-2)(1); Mo:GM-CSF+TNFα+IL4(0:16, 1.5e-5)(5) |
NF-κB binding site conserved in Drosophila, otherwise conserved in mouse.
Predictions by Model 2, the rest are predictions by model 1.
The second column indicates whether homologs of the genes are found in other model organisms. The third column shows the transcription factors that may regulate the gene. The fourth column gives score (π[x]) for the promoter in the LRA model. The last column indicates the SAGE data. Among the data on monocytes (Mo), results are number of tags per 57,000 tags. With T cells (Th1 and Th2), they are tags per 93,000 tags. For HMVEC, data are presented as tags per million tags. The SAGE data are presented as cell-line: NF-κB-activator. In parentheses, the first number is expression level before stimulation, the second number is expression level after the stimulation, and the last number is the p-value of the change. The comparisons of gene expression in Th1 and Th2 are made with IEL T cells. Ref. (1) Nagai et al. 2001, (2) Lash et al. 2000 (http://www.ncbi.nlm.nih.gov/SAGE), (3) Hashimoto et al. 1994, (4) Suzuki et al. 2000, (5) www.prevent.m.u-tokyo.ac.jp/SAGE.html.
SAGE data on monocytes and T cells recently became available (Hashimoto et al. 1999; Suzuki et al. 2000; Nagai et al. 2001; Shires et al. 2001), and a SAGE study was performed on human microvascular endothelial cells (HMVEC; G.J. Reggins). Changes in the expression levels of these nine genes after the activation of NF-κB in these cell lines, along with estimates of the statistical significance (P-values) of these changes are summarized in the last column of Table 4 (Audic and Claverie 1997). The activators of NF-κB include LPS, M-CSF, or GM-CSF on monocytes; phorbol myristate acetate (PMA) on T cells; and vascular endothelial growth factor (VEGF) on HMVEC (Kim et al. 2001). In these cell lines, after activation of NF-κB we observe that the expression level of these nine genes increases significantly, and thus SAGE data are consistent with the prediction that these novel genes are regulated by NF-κB. Given the evidence that our method is very specific, we are convinced that these nine genes are novel NF-κB-regulated immune response genes.
DISCUSSION
Given the fundamental roles of the NF-κB signaling pathway in the immune system, identifying novel NF-κB-regulated immune genes in the human genome will undoubtedly advance the investigation of immune mechanisms. Although experimental methods such as DNA microarrays are available to characterize patterns of gene expression, these methods are expensive and require biological materials which may be difficult to obtain for some tissues and conditions. Thus, computational methods can complement experimental methods by identifying candidate genes, maximizing the effectiveness of expression studies. We build LRA models for immune promoters with NF-κB binding sites within the −1 to −230-bp window (model I), and for promoters with NF-κB binding sites between −231 and −600 bp (model II). This method shows sensitivity of 42%, with specificity of 93%–100%.
We found 28 immune gene promoters with an LRA model score above the threshold and with NF-κB binding sites conserved in the mouse or Drosophila genome. These 28 genes are predicted to be immune response genes, with nine genes among them being novel NF-κB-regulated immune genes. For all of the nine predicted novel immune genes, SAGE data on microvascular endothelial, myeloid, or lymphoid cells suggest that they are NF-κB-regulated genes. Because our search procedure is very specific, we propose that these nine genes are NF-κB-regulated immune response genes.
One of these novel NF-κB-regulated immune genes (sir2 homolog 2) is notable. The yeast sir2 protein is believed to be functional in the nucleus and is involved in gene silencing and aging (Shore 2000). However, the human homolog 2 primarily locates in cytoplasm, and overexpression of the gene has no affect on cell growth or chromosome stability (Afshar and Murnane 1999). Our LRA analysis suggests that it is an immune gene in humans, and SAGE data further demonstrate that the gene is expressed in both lymphoid and myeloid lineages after the cells are stimulated by NF-κB activators. We anticipate that the gene plays a fundamental role which is common to both lymphoid and myeloid cells.
The score thresholds (π[x]) for the two models are set so that for model I (π[x] > 0.65), the promoter must have binding sites for NF-κB, and at least one other informative TF (AP1, IK, IRF, or STAT) to be considered a preliminary candidate. To meet the threshold for model II (π[x] > 0.50), the promoter must have binding sites for NF-κB and either IK or IRF, else NF-κB and both AP1 and STAT.
Because our goal is to develop a specific method for immune gene prediction, we set the position weight matrix (PWM) thresholds high to reduce false predictions. The high specificities of model I (93%) and model II (100%) tend to justify expensive and time-consuming experimental characterization of the nine predicted immune genes. Because the sensitivity of our method is 42% and we have 28 predictions from 6440 genes, we would expect that there are about 70 NF-κB-regulated immune genes among the 6440 genes and about 400 NF-κB-regulated immune genes among the 35,000 genes in the entire human genome.
This is the first successful attempt at a genome-scale computational search for immune genes by promoter analysis. As the sequencing of the human genome is finished, assigning functions to novel genes in a high-throughput manner becomes increasingly important. We demonstrate here that regulatory genomics can be applied to genome-scale prediction of the functions for novel genes in specific physiological pathways or biological systems.
METHODS
Promoters and TF Binding Sites
Transcription factor binding sites are analyzed by MatInspector (Quandt et al. 1995). The weight matrices for transcription factor binding sites are from TRANSFAC (Wingender et al. 2000), and the weight matrix score cutoffs are 0.95 for AP1 and 0.9 for other transcription factors, unless otherwise specified in the text.
Piecewise Linear Regression
Piecewise linear regression analysis (S-Plus6.0) was used to reveal the turning point in the NF-κB binding-site positional distribution curve (Fig. 1). In a two-segment curve, piecewise linear regression analysis fits both sections with lines after a knot is specified. We choose the knot yielding the minimal sum of squared errors (−270 bp).
Logistic Regression Analysis
We also used S-Plus6.0 to perform a multivariate logistic regression analysis (LRA). In the LRA, maximum likelihood was used to estimate the relative effect of each TF binding site on the probability that a promoter is associated with an immune gene. We first tested for possible synergistic effects of multiple-copy TF binding by fitting two linear models: One model uses the count of binding sites for each TF in each promoter, and the other model uses an indicator variable for the existence of at least one binding site for each TF in a promoter, regardless of the TF copy number. The count of TFs was not found to be significant, so we used indicator variables for the presence of binding sites for AP1, IK (IK1, IK3), IRF (IRF1, IRF2, ISRE), NF-κB, or STAT. Modeling was performed on the 62 NF-κB-regulated immune promoters and the 248 mammalian nonimmune promoters extracted from the EPD. The probability that a given promoter is from the group of immune genes, π(x), is estimated by:
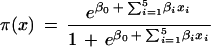 |
1 |
where β0 is the intercept, coefficient βi is the effect of TF xi, and i indexes the five TFs.
Assessing the Significance of SAGE Data
Expression levels of the predicted novel NF-κB-regulated genes, with and without NF-κB activation, is obtained by data mining from SAGE databases. We measured the significance of expression level changes after NF-κB is activated, by the method of Audic and Claverie (1997). In this method, the probability of a change of expression level from x to y is:
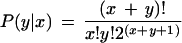 |
2 |
and P-values are calculated as the probability of a change at least as extreme as the observed.
Genomes
Human genomic sequence data are downloaded from the UCSC Genome Server (http://genome.ucsc.edu, Golden Path assembly April 2001 release). Mouse genomic sequence data were retrieved from the NCBI mouse trace database. The Drosophila genome was retrieved from the NCBI genome data set.
Effects of Stringency in Accepting TF Binding Sites
The search described above was designed to emphasize specificity, so we set the thresholds for accepting TF binding sites accordingly. To test the effects on specificity and sensitivity when we lower the stringency in classifying sequences as TF binding sites, both models I and II were recalculated with lower PWM score cutoffs (Table 5). As shown in Table 5, the specificity is very high (18/19 overall) using our high threshold for accepting TF binding sites, but sensitivity is only 42% (26/62). With medium stringency (described in Table 5), the sensitivity increases to 61%, but the specificity among these additional predictions drops to 40% (4/10 additional predictions with known function being NF-κB-regulated immune genes). Lowering the stringency further brings the specificity down dramatically, because only one of the 17 additional predicted genes with known functions is likely to be an immune gene, and the sensitivity is only slightly improved (71%). We conclude that the high stringency search has detected most of the immune genes which could be detected by this method and should be applied when we take a genomic approach to search for genes with specific functions.
Table 5.
Effects of Stringency of TF Sites on Specificity
| Stringency | Sensitivity | Model 1 | Model 2 |
| High | 42% | 14/15 | 4/4 |
| Medium | 61% | 2/5 | 2/5 |
| Low | 71% | 0/7 | 1/10 |
High stringency is the original model I and II, as in the above analyses. In medium stringency, we lower the matrix cutoff scores to NF-κB, 0.95; AP1, 0.95; IK1, 0.94; STAT, 0.91; IK3, IRF1, IRF2, ISRE, 0.86. Low stringency sets the matrix cutoff scores to NF-κB, 0.92; AP1, 0.95; IK1, 0.94; STAT, 0.91; IK3, IRF1, IRF2, ISRE, 0.86. As we lower the matrix cutoffs, only the additional predictions are shown in the table.
WEB SITE REFERENCES
http://genome.ucsc.edu; Golden path genome sequences.
www.prevent.m.u-tokyo.ac.jp/SAGE.html; SAGE data.
http://www.ncbi.nlm.nih.gov/SAGE; SAGE data.
Acknowledgments
We thank Tom Blackwell and the rest of the group for insightful discussions, and the reviewers for their constructive comments. This work was supported in part through grants from the NIH (HG-R01-01391), the Department of Energy (DE-FG02-94ER61910), and the Merck Foundation for Genome Research (grant #225)
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL dstates@umich.edu; FAX (734) 615-6553.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.911803. Article published online before print in March 2003.
REFERENCES
- 1.Afshar G. and Murnane, J.P. 1999. Characterization of a human gene with sequence homology to Saccharomyces cerevisiae SIR2. Gene 234: 161-168. [DOI] [PubMed] [Google Scholar]
- 2.Akira S. and Kishimoto, T. 1992. IL-6 and NF-IL6 in acute-phase response and viral infection. Immunol. Rev. 127: 25-50. [DOI] [PubMed] [Google Scholar]
- 3.Appay V. and Rowland-Jones, S.L. 2001. RANTES: A versatile and controversial chemokine. Trends Immunol. 22: 83-87. [DOI] [PubMed] [Google Scholar]
- 4.Audic S. and Claverie, J.M. 1997. The significance of digital gene expression profiles. Genome Res. 7: 986-995. [DOI] [PubMed] [Google Scholar]
- 5.Baeuerle P.A. and Baichwal, V.R. 1997. NF-κ B as a frequent target for immunosuppressive and anti-inflammatory molecules. Adv. Immunol. 65: 111-137. [PubMed] [Google Scholar]
- 6.Baeuerle P.A. and Baltimore, D. 1996. NF-κ B: Ten years after. Cell 87: 13-20. [DOI] [PubMed] [Google Scholar]
- 7.Belich M.P., Salmeron, A., Johnston, L.H., and Ley, S.C. 1999. Tpl-2 kinase regulates the proteolysis of the NF-κB-inhibitory protein NF-κB1 p105. Nature 397: 363-368. [DOI] [PubMed] [Google Scholar]
- 8.Berman B.P., Nibu, Y., Pfeiffer, B.D., Tomancak, P., Celniker, S.E., Levine, M., Rubin, G.M., and Eisen, M.B. 2002. Exploiting transcription factor binding site clustering to identify cis-regulatory modules involved in pattern formation in the Drosophila genome. Proc. Natl. Acad. Sci. 99: 757-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brodsky M.H. and Steller, H. 1996. Positional information along the dorsal-ventral axis of the Drosophila eye: Graded expression of the four-jointed gene. Dev. Biol. 173: 428-446. [DOI] [PubMed] [Google Scholar]
- 10.Campbell R.D., Carroll, M.C., and Porter, R.R. 1986. The molecular genetics of components of complement. Adv. Immunol. 38: 203-244. [DOI] [PubMed] [Google Scholar]
- 11.Chan A.M., Chedid, M., McGovern, E.S., Popescu, N.C., Miki, T., and Aaronson, S.A. 1993. Expression cDNA cloning of a serine kinase transforming gene. Oncogene 8: 1329-1333. [PubMed] [Google Scholar]
- 12.Chow C.W., Grinstein, S., and Rotstein, O.D. 1995. Signaling events in monocytes and macrophages. New Horiz. 3: 342-351. [PubMed] [Google Scholar]
- 13.Cohen P., Bouaboula, M., Bellis, M., Baron, V., Jbilo, O., Poinot-Chazel, C., Galiegue, S., Hadibi, E.H., and Casellas, P. 2000. Monitoring cellular responses to Listeria monocytogenes with oligonucleotide arrays. J. Mol. Biol. 275: 11181-11190. [DOI] [PubMed] [Google Scholar]
- 14.De Smaele E., Zazzeroni, F., Papa, S., Nguyen, D.U., Jin, R., Jones, J., Cong, R., and Franzoso, G. 2001. Induction of gadd45β by NF-κB downregulates proapoptotic JNK signalling. Nature 414: 308-313. [DOI] [PubMed] [Google Scholar]
- 15.Dewaste V., Pouillon, V., Moreau, C., Shears, S., Takazawa, K., and Erneux, C. 2000. Cloning and expression of a cDNA encoding human inositol 1,4,5-trisphosphate 3-kinase C. Biochem. J. 352: 343-351. [PMC free article] [PubMed] [Google Scholar]
- 16.Dombrowski K.E., Ke, Y., Brewer, K.A., and Kapp, J.A. 1998. Ecto-ATPase: An activation marker necessary for effector cell function. Immunol. Rev. 161: 111-118. [DOI] [PubMed] [Google Scholar]
- 17.Driscoll K.E. 1994. Macrophage inflammatory proteins: Biology and role in pulmonary inflammation. Exp. Lung. Res. 20: 473-490. [DOI] [PubMed] [Google Scholar]
- 18.D'Santos C.S., Communi, D., Ludgate, M., Vanweyenberg, V., Takazawa, K., and Erneux, C. 1994. Identification of high molecular weight forms of inositol 1,4,5-trisphosphate 3-kinase in rat thymus and human lymphocytes. Cell. Signal. 6: 335-344. [DOI] [PubMed] [Google Scholar]
- 19.Elangbam C.S., Qualls, C.W.J., and Dahlgren, R.R. 1997. Cell adhesion molecules—Update. Vet. Pathol. 34: 61-73. [DOI] [PubMed] [Google Scholar]
- 20.Fambrough D., McClure, K., Kazlauskas, A., and Lander, E.S. 1999. Diverse signaling pathways activated by growth factor receptors induce broadly overlapping, rather than independent, sets of genes. Cell 97: 727-741. [DOI] [PubMed] [Google Scholar]
- 21.Gao L., Dong, L., and Whitlock, J.P., Jr. 1998. A novel response to dioxin. Induction of ecto-ATPase gene expression. J. Biol. Chem. 273: 15358-15365. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh S., May, M.J., and Kopp, E.B. 1998. NF-κ B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol. 16: 225-260. [DOI] [PubMed] [Google Scholar]
- 23.Girdlestone J. 2000. Synergistic induction of HLA class I expression by RelA and CIITA. Blood 95: 3804-3808. [PubMed] [Google Scholar]
- 24.Gobin S.J., Keijsers, V., van Zutphen, M., and van den Elsen, P.J. 1998. The role of enhancer A in the locus-specific transactivation of classical and nonclassical HLA class I genes by nuclear factor κB. J. Immunol. 161: 2276-2283. [PubMed] [Google Scholar]
- 25.Hamilton J.A. 1997. CSF-1 signal transduction. J. Leukoc. Biol. 62: 145-155. [DOI] [PubMed] [Google Scholar]
- 26.Harada A., Sekido, N., Akahoshi, T., Wada, T., Mukaida, N., and Matsushima, K. 1994. Essential involvement of interleukin-8 (IL-8) in acute inflammation. J. Leukoc. Biol. 56: 559-564. [PubMed] [Google Scholar]
- 27.Hashimoto S., Suzuki, T., Dong, H.Y., Yamazaki, N., and Matsushima, K. 1999. Serial analysis of gene expression in human monocytes and macrophages. Blood 94: 837-844. [PubMed] [Google Scholar]
- 28.Hromas R., Gray, P.W., Chantry, D., Godiska, R., Krathwohl, M., Fife, K., Bell, G.I., Takeda, J., Aronica, S., Gordon, M., et al. 1997. Cloning and characterization of exodus, a novel β-chemokine. Blood 89: 3315-3322. [PubMed] [Google Scholar]
- 29.International Human Genome Sequencing Consortium 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- 30.Kadonaga J.T. 1998. Eukaryotic transcription: An interlaced network of transcription factors and chromatin-modifying machines. Cell 92: 307-313. [DOI] [PubMed] [Google Scholar]
- 31.Kas-Deelen A.M., Bakker, W.W., Olinga, P., Visser, J., de Maar, E.F., van Son, W.J., The, T.H., and Harmsen, M.C. 2001. Cytomegalovirus infection increases the expression and activity of ecto-ATPase (CD39) and ecto-5′nucleotidase (CD73) on endothelial cells. FEBS Lett. 491: 21-25. [DOI] [PubMed] [Google Scholar]
- 32.Kim I., Moon, S.O., Kim, S.H., Kim, H.J., Koh, Y.S., and Koh, G.Y. 2001. Vascular endothelial growth factor expression of intercellular adhesion molecule 1 (ICAM-1), vascular cell adhesion molecule 1 (VCAM-1), and E-selectin through nuclear factor-κ B activation in endothelial cells. J. Biol. Chem. 276: 7614-7620. [DOI] [PubMed] [Google Scholar]
- 33.Krivan W. and Wasserman, W.W. 2001. A predictive model for regulatory sequences directing liver-specific transcription. Genome Res. 278: 167-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lash A.E., Tolstoshev, C.M., Wagner, L., Schuler, G.D., Strausberg, R.L., Riggins, G.J., and Altschul, S.F. 2000. SAGEmap: A public gene expression resource. Genome Res. 10: 1051-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee A.H., Hong, J.H., and Seo, Y.S. 2000. Tumor necrosis factor-α and interferon-γ synergistically activate the RANTES promoter through nuclear factor κB and interferon regulatory factor 1 (IRF-1) transcription factors. Biochem. J. 350: 131-138. [PMC free article] [PubMed] [Google Scholar]
- 36.Lemon B. and Tjian, R. 2000. Orchestrated response: A symphony of transcription factors for gene control. Genes & Dev. 14: 2551-2569. [DOI] [PubMed] [Google Scholar]
- 37.Leonard W.J. and O'Shea, J.J. 1998. Jaks and STATs: Biological implications. Annu. Rev. Immunol. 16: 293-322. [DOI] [PubMed] [Google Scholar]
- 38.Li J., Peet, G.W., Balzarano, D., Li, X., Massa, P., Barton, R.W., and Marcu, K.B. 2001. Novel NEMO/IκB kinase and NF-κ B target genes at the pre-B to immature B cell transition. J. Biol. Chem. 276: 18579-18590. [DOI] [PubMed] [Google Scholar]
- 39.Liu R. and States, D.J. 2002. Consensus promoter identification in the human genome utilizing expressed gene markers and gene modeling. Genome Res. 12: 462-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu R., Blackwell, T.W., and States, D.J. 2001. Conformational models for binding site recognition by the E. coli MetJ transcription factor. Bioinformatics 17: 622-633. [DOI] [PubMed] [Google Scholar]
- 41.Markstein M., Markstein, P., Markstein, V., and Levine, M.S. 2002. Genome-wide analysis of clustered Dorsal binding sites identifies putative target genes in the Drosophila embryo. Proc. Natl. Acad. Sci. 99: 763-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marshall A.J., Niiro, H., Yun, T.J., and Clark, E.A. 2000. Regulation of B-cell activation and differentiation by the phosphatidylinositol 3-kinase and phospholipase Cγ pathway. Immunol. Rev. 76: 30-46. [DOI] [PubMed] [Google Scholar]
- 43.Marzio R., Mauel, J., and Betz-Corradin, S. 1999. CD69 and regulation of the immune function. Immunopharmacol. Immunotoxicol. 21: 565-582. [DOI] [PubMed] [Google Scholar]
- 44.Matikainen S., Pirhonen, J., Miettinen, M., Lehtonen, A., Govenius-Vintola, C., Sareneva, T., and Julkunen, I. 2000. Influenza A and sendai viruses induce differential chemokine gene expression and transcription factor activation in human macrophages. Virology 276: 138-147. [DOI] [PubMed] [Google Scholar]
- 45.McKinsey T.A., Chu, Z., Tedder, T.F., and Ballard, D.W. 2000. Transcription factor NF-κB regulates inducible CD83 gene expression in activated T lymphocytes. Mol. Immunol. 37: 783-788. [DOI] [PubMed] [Google Scholar]
- 46.Moriuchi H., Moriuchi, M., and Fauci, A.S. 1997. Nuclear factor-κ B potently upregulates the promoter activity of RANTES, a chemokine that blocks HIV infection. J. Immunol. 158: 3483-3491. [PubMed] [Google Scholar]
- 47.Nagai S., Hashimoto, S., Yamashita, T., Toyoda, N., Satoh, T., Suzuki, T., and Matsushima, K. 2001. Comprehensive gene expression profile of human activated T(h)1- and T(h)2-polarized cells. Int. Immunol. 13: 367-376. [DOI] [PubMed] [Google Scholar]
- 48.Patriotis C., Makris, A., Bear, S.E., and Tsichlis, P.N. 1993. Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc. Natl. Acad. Sci. 90: 2251-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perier R.C., Praz, V., Junier, T., Bonnard, C., and Bucher, P. 2000. The eukaryotic promoter database. Nucleic Acids Res. 28: 302-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quandt K., Frech, K., Karas, H., Wingender, E., and Werner, T. 1995. MatInd and MatInspector: New fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 23: 4878-4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson S.P., Saraya, K., and Reid, C.D. 1998. Developmental aspects of dendritic cells in vitro and in vivo. Leuk. Lymphoma 29: 477-490. [DOI] [PubMed] [Google Scholar]
- 52.Ruddle N.H. 1999. Lymphoid neo-organogenesis: Lymphotoxin's role in inflammation and development. Immunol. Res. 19: 119-125. [DOI] [PubMed] [Google Scholar]
- 53.Sanchez-Gongora E., Lisbona, C., de Gregorio, R., Ballester, A., Calvo, V., Perez-Jurado, L., and Alemany, S. 2000. COT kinase proto-oncogene expression in T cells: Implication of the JNK/SAPK signal transduction pathway in COT promoter activation. J. Biol. Chem. 275: 31379-31386. [DOI] [PubMed] [Google Scholar]
- 54.Shiffman D., Mikita, T., Tai, J.T., Wade, D.P., Porter, J.G., Seilhamer, J.J., Somogyi, R., Liang, S., and Lawn, R.M. 2000. Large scale gene expression analysis of cholesterol-loaded macrophages. J. Biol. Chem. 275: 37324-37332. [DOI] [PubMed] [Google Scholar]
- 55.Shires J., Theodoridis, E., and Hayday, A.C. 2001. Biological insights into TCRγδ+ and TCRαβ+ intraepithelial lymphocytes provided by serial analysis of gene expression (SAGE). Immunity 15: 419-434. [DOI] [PubMed] [Google Scholar]
- 56.Shore D. 2000. The Sir2 protein family: A novel deacetylase for gene silencing and more. Proc. Natl. Acad. Sci. 97: 14030-14032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki T., Hashimoto, S., Toyoda, N., Nagai, S., Yamazaki, N., Dong, H.Y., Sakai, J., Yamashita, T., Nukiwa, T., and Matsushima, K. 2000. Comprehensive gene expression profile of LPS-stimulated human monocytes by SAGE. Blood 96: 2584-2591. [PubMed] [Google Scholar]
- 58.Thanos D. and Maniatis, T. 1995. NF-κ B: A lesson in family values. Cell 80: 529-532. [DOI] [PubMed] [Google Scholar]
- 59.Tilg H., Dinarello, C.A., and Mier, J.W. 1997. IL-6 and APPs: Anti-inflammatory and immunosuppressive mediators. Immunol. Today 18: 428-432. [DOI] [PubMed] [Google Scholar]
- 60.Vairapandi M., Balliet, A.G., Fornace, A.J., Jr., Hoffman, B., and Liebermann, D.A. 1996. The differentiation primary response gene MyD118, related to GADD45, encodes for a nuclear protein which interacts with PCNA and p21WAF1/CIP1. Oncogene 12: 2579-2594. [PubMed] [Google Scholar]
- 61.Venter J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- 62.Ward S.G. and Cantrell, D.A. 2001. Phosphoinositide 3-kinase in T lymphocyte activation. Curr. Opin. Immunol. 13: 332-338. [DOI] [PubMed] [Google Scholar]
- 63.Wasserman W.W. and Fickett, J.W. 1998. Identification of regulatory regions which confer muscle-specific gene expression. J. Mol. Biol. 278: 167-181. [DOI] [PubMed] [Google Scholar]
- 64.Wingender E., Chen, X., Hehl, R., Karas, H., Liebich, I., Matys, V., Meinhardt, T., Pruss, M., Reuter, I., and Schacherer, F. 2000. TRANSFAC: An integrated system for gene expression regulation. Nucleic Acids Res. 28: 316-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yuh C.H., Bolouri, H., and Davidson, E.H. 1998. Genomic cis-regulatory logic: Experimental and computational analysis of a sea urchin gene. Science 279: 1896-1902. [DOI] [PubMed] [Google Scholar]
- 66.Zeidler M.P., Bach, E.A., and Perrimon, N. 2000. The roles of the Drosophila JAK/STAT pathway. Oncogene 19: 2598-2606. [DOI] [PubMed] [Google Scholar]
- 67.Ziegler S.F., Ramsdell, F., and Alderson, M.R. 1994. The activation antigen CD69. Stem Cells 12: 456-465. [DOI] [PubMed] [Google Scholar]