Abstract
Gene Ontology Annotation (GOA) is a project run by the European Bioinformatics Institute (EBI) that aims to provide assignments of terms from the Gene Ontology (GO) resource to gene products in a number of its databases (http://www.ebi.ac.uk/GOA). In the first stage of this project, GO assignments have been applied to a data set representing the complete human proteome by a combination of electronic mappings and manual curation. This vocabulary has also been applied to the nonredundant proteome sets for all other completely sequenced organisms as well as to proteins from a wide range of organisms where the proteome is not yet complete.
Continual advancement in proteome research has led to an increase in sequences from a wide range of species requiring addition to the SWISS-PROT Protein Knowledgebase and its supplement, TrEMBL (Bairoch and Apweiler 2000), the majority of these lacking functional characterization. To fully exploit the potential of this vast quantity of data, the SWISS-PROT group has intensified its efforts to capture all available biological information related to these sequences and, in particular, to the human proteome.
Crucial to this work is the integration of in-house resources with those of external database groups. Integration and data exchange involve resolving the complexities that exist between databases. For example, the use of different vocabularies to describe gene function can hinder searching across multiple proteins and species for common characteristics. The use of a common vocabulary facilitates the identification of relationships and common properties between gene products from different species.
This problem has been addressed by the creation of the Gene Ontology (GO) resource (The Gene Ontology Consortium 2001), a dynamic, controlled vocabulary that can be applied to all organisms even as protein knowledge is accumulating and changing. The GO Consortium has developed three separate ontologies: molecular function, biological process, and cellular component. These help to describe gene products in a standardized way and allow the annotation of molecular characteristics across species. Each vocabulary is structured as a directed acyclic graph (DAG), wherein any term may have more than one parent as well as zero, one, or more children. This makes attempts to describe biology much richer than would be possible with a hierarchical graph. Currently, the GO vocabulary consists of >13,000 terms, which will, in time, all have strict definitions of their intended usage.
SWISS-PROT has joined the GO Consortium and has adopted its structured vocabulary to characterize the activities of proteins in the SWISS-PROT, TrEMBL, and InterPro (Apweiler et al. 2001a) databases. It has initiated the Gene Ontology Annotation (GOA) project to provide assignments of GO terms to gene products for all organisms with completely sequenced genomes by a combination of electronic assignment and manual annotation. By annotating all characterized proteins with GO terms and facilitating the transfer of this knowledge to similar uncharacterized proteins, the SWISS-PROT group will make a valuable contribution to biological and biotechnological research through a better understanding of all proteomes.
METHODS AND RESULTS
Automatic GO Annotation of SWISS-PROT, TrEMBL, and InterPro
The first phase of the GOA project involved the large-scale assignment of GO terms to SWISS-PROT and TrEMBL entries using electronic methods. This strategy was based on the use of existing properties of the entries including the presence of keywords and Enzyme Commission (EC) numbers. Mapping of InterPro entries to GO also allowed further associations of GO terms to entries to be made, based on the presence of InterPro cross-references in SWISS-PROT and TrEMBL. “Mapping” is used here to refer to the linking of various classification systems to GO terms, while the word “association” refers to a connection between a database object (which may represent a gene, transcript, or protein) and a GO term that describes the gene product. The electronic mappings described in detail below were used to generate associations in SWISS-PROT and TrEMBL.
Mapping SWISS-PROT Keywords to GO
SWISS-PROT and TrEMBL entries contain keywords that serve as a subject reference for each sequence and assist in the retrieval of specific categories of data from the database. Currently, SWISS-PROT maintains a controlled list of ∼840 keywords, each with a definition to clarify its biological meaning and intended usage. This list is available at http://www.expasy.org/cgi-bin/keywlist.pl and is updated on a regular basis. Seventy-four percent of SWISS-PROT keywords have been manually mapped to a high-level GO term. Keywords that were not mapped include those that have multiple usages, have no equivalent GO term, or are beyond the scope of the GO project, such as keywords describing domains.
To enable data transfer, an index file containing the SWISS-PROT keyword to GO mappings (spkw2go) is shared on the GO home page (http://www.geneontology.org/external2go/spkw2go). This is frequently updated with the latest version, helping users to keep track of changes for local use. During these updates, more specific GO mappings may be added and obsolete GO terms and SWISS-PROT keywords removed.
During manual annotation of a SWISS-PROT entry, curators assign keywords based on literature and sequence analysis checks. Keywords are also added to TrEMBL entries during automatic annotation of the TrEMBL database (Apweiler 2001). This procedure utilizes a novel system of standardized transfer of annotation from well-characterized proteins in SWISS-PROT to unannotated TrEMBL entries (Fleischmann et al. 1999). Consequently, the accuracy of the association of GO terms to SWISS-PROT and TrEMBL entries based on the keywords in the entries is assured by the annotation quality standards already existing in SWISS-PROT. To associate GO terms to SWISS-PROT and TrEMBL entries, the spkw2go mapping is combined with a mapping of protein accession numbers to SWISS-PROT keywords. In-house annotation tools and browsers are updated automatically as the new data is loaded.
The application of SWISS-PROT keywords in the electronic assignment of GO terms to gene products continues to be a large-scale success. As of November 2002, spkw2go has been used to generate over 1,023,969 GO associations with 376,845 SWISS-PROT and TrEMBL entries (see http://www.ebi.ac.uk/GOA/SPTR_release.html). It has also been used successfully by a number of external databases such as the Mouse Genome Database (MGD) (Hill et al. 2001).
Mapping of EC Numbers to GO
A second electronic strategy takes advantage of an existing mapping (ec2go) of GO terms from the molecular function ontology to the nomenclature of the EC as contained in the ENZYME database (Bairoch 2000). EC numbers are consistently annotated in SWISS-PROT and TrEMBL enzyme entries as part of the description line. To associate GO terms to the SWISS-PROT and TrEMBL data, the ec2go mapping available from the GO home page was updated and combined with a mapping of protein accession numbers to EC numbers. This strategy was very successful, generating 164,205 GO associations in 72,496 SWISS-PROT and TrEMBL proteins.
Mapping of the InterPro Resource to GO
InterPro is an integrated documentation resource for protein families, domains, and sites that combines the complementary efforts of the PROSITE (Falquet et al. 2002), PRINTS (Attwood et al. 2002), Pfam (Bateman et al. 2002), ProDom (Corpet et al. 2000), SMART (Letunic et al. 2002), and TIGRFAMs (Haft et al. 2001) databases. Individual signatures from the member databases, which describe the same protein family or domain, are grouped together into a single InterPro entry. Each entry provides comprehensive annotation describing a set of related proteins, some of which may have identical molecular functions, be involved in the same processes, and perform their function in the same cellular locations. Furthermore, each entry also contains a match list of the SWISS-PROT and TrEMBL proteins that hit the signatures in that entry. Mapping InterPro entries to GO terms thus provides an automatic means of assigning GO terms to the protein sequences that form the match table of a particular InterPro entry. An additional advantage is that multifunctional proteins can be mapped to multiple GO terms through associations with more than one InterPro entry.
The assignment of GO terms to InterPro entries was performed manually by inspecting all available information. In each case, the abstracts and the annotation of proteins within the match lists were read and an appropriate GO term was mapped if it applied to the whole protein. Some entries could be mapped to very deep level (specific) GO terms, while entries describing wider families or common domains could only be mapped to higher level terms or could not be mapped at all. The associated GO term therefore applies to all proteins with true hits to all signatures in the InterPro entry. As of November 2002, the electronic application of these InterPro mappings has led to 1,333,215 GO associations with 442,293 SWISS-PROT and TrEMBL proteins. The integrity of the InterPro to GO mappings is maintained by running regular sanity checks on the data. These checks include searching for mappings from secondary or deleted InterPro accession numbers and mappings to obsolete or nonexistent GO terms. The reports are manually verified and corrected.
For each associated term, the name of the term and GO accession number is given, and these are available in InterPro entries directly from the database at http://www.ebi.ac.uk/interpro/ (Fig. 1). A file listing InterPro entries and their corresponding GO terms is also available from the EBI FTP site at ftp://ftp.ebi.ac.uk/pub/databases/interpro/interpro2go and, on the GO home page at http://www.geneontology.org/interpro2go. InterPro includes a sequence search facility that allows users to search a sequence against the database and to retrieve all InterPro matches for that sequence. As well as an SRS-based version, there is also a stand-alone Perl-based package available for local installation that returns GO terms as part of the results.
Figure 1.
InterPro entry IPR000040 (acute myeloid leukemia 1 protein [AML 1]/Runt) that shows the GO terms that have been manually mapped to the entry.
GO Annotation of the Human Proteome
As part of a consortium agreement to fast-track GO annotation of human data, SWISS-PROT curators have manually assigned GO terms to a SWISS-PROT/TrEMBL/Ensembl nonredundant human proteome set. The dataset was created by combining all human protein sequences in the SWISS-PROT database and supplementing these with a nonredundant selection of human proteins from the TrEMBL and Ensembl (Hubbard et al. 2002) databases. Full details of the preparation of the set are available at http://www.ebi.ac.uk/proteome/SPTREnsembl.html. This set contained 28,736 sequences, of which 7146 came from SWISS-PROT, 14,659 from TrEMBL and 6931 from Ensembl.
Before manual annotation, the dataset was annotated with GO terms using the various electronic methods described above. Next, assignments of GO terms to LocusLink (Pruitt and Maglott 2001) were added. LocusLink is a database of genetic loci, many of which had GO terms assigned to them in a one-off annotation marathon by Proteome Inc. LocusLink loci were therefore mapped to SWISS-PROT and TrEMBL entries by identifier tracking, and relevant assignments were transferred.
Entries for which no assignments were available or for which only electronically applied GO terms existed were identified as potential candidates for manual annotation. This set was then filtered to remove those entries with references, which were unlikely to contain useful content. For example, entries whose only references describe a DNA sequencing project were not considered for manual annotation at this stage. A reduced set of 3042 entries was thus identified for priority manual annotation by the SWISS-PROT curation team.
The assignments to this set of 3042 entries were based initially on abstract information. GO terms from each of the three ontologies were assigned to each entry in the set, using the GO evidence codes described at http://www.geneontology.org/GO.evidence.html. During this initial manual annotation phase, a large number of new GO terms were requested by the SWISS-PROT and InterPro curators, thus extending the coverage of GO and increasing its utility in the analysis of human proteins. At the end of this phase of the project, 9927 manual associations of GO terms to the human proteome set had been made. Together, these assignments represent the first stage of the GOA project at EBI, released in November 2001. Complete references for each entry are now being read so that additional or deeper-level GO terms can be assigned. Current numbers of electronic and manual assignments to the human data set are shown at http://www.ebi.ac.uk/GOA/release.html. The SWISS-PROT group at EBI will continue to prioritize the fast-tracking of human GO annotation.
Manual Annotation of GO to Proteins From All Organisms
One of the distinguishing features of the SWISS-PROT database is the high level of annotation it provides in each entry. This is achieved by a team of biologists who extract up-to-date information from a variety of sources, including published literature and compile this information into a concise but comprehensive report. SWISS-PROT curators are therefore well placed to contribute to the work of the GO consortium by assigning GO terms during the annotation process and now assign GO terms to every entry that they annotate. As these entries come from a wide range of organisms (50,000 different species), they also continue to contribute to the expansion of the ontologies by requesting new terms when necessary, thus extending the scope of the GO ontologies beyond those terms required to describe the proteins of the model organism databases, SGD (Dwight et al. 2002), FlyBase (The FlyBase Consortium 2002), and MGD (Blake et al. 2002), the founding members of the GO consortium.
Data Searching and Retrieval
The EBI contributes to functional studies by distributing and updating GO mappings and associations generated in-house. This data is displayed via the QuickGO browser, Gene Association file, EBI and GO FTP servers, SRS and Proteome Analysis pages as detailed below.
QuickGO
QuickGO (http://www.ebi.ac.uk/ego/) is a fast, Web-based browser that was developed at the EBI to allow users to search and browse GO data and associated links to other data sets. It has access to the core GO data comprising the terms of the three GO ontologies, the relationships between these terms, their synonyms, and definitions where such exist. In addition, QuickGO accesses the manually curated annotations and mappings of SWISS-PROT keywords, InterPro entries, and the EC and Transport Commission (http://tcdb.ucsd.edu/tcdb/) classification schemes to GO terms, as well as electronically and manually curated associations of GO terms to SWISS-PROT and TrEMBL entries (GOA). There are also links to the Expression Profiler GO browser (EP:GO) (http://ep.ebi.ac.uk/EP/GO/), which allows the extraction of genes associated with each GO category and the analysis of gene expression, regulatory sequence, and protein–protein interaction data for these genes. QuickGO is updated on a weekly basis so that all electronic and manual mappings and associations displayed reflect the current status.
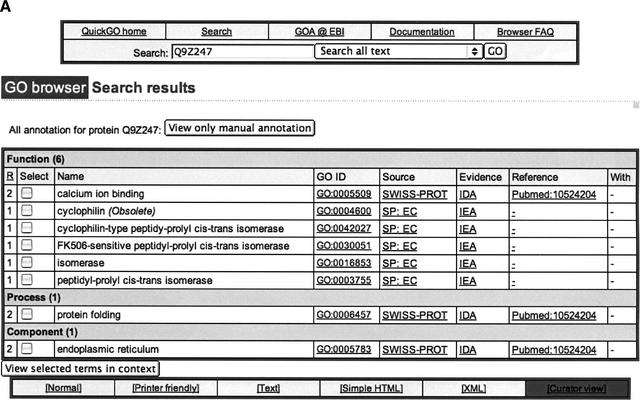
The QuickGO search interface has recently been updated. For example, querying by protein accession number will show all terms mapped to that SWISS-PROT entry and the source of each term association (Fig. 2A). The default setting will retrieve all associations but it is also possible to display only manually assigned GO terms. Searches may return multiple results, in which case an exploded view of the subset of GO that contains all or selected results can be seen according to their position within the DAG structure (context view).
Figure 2.
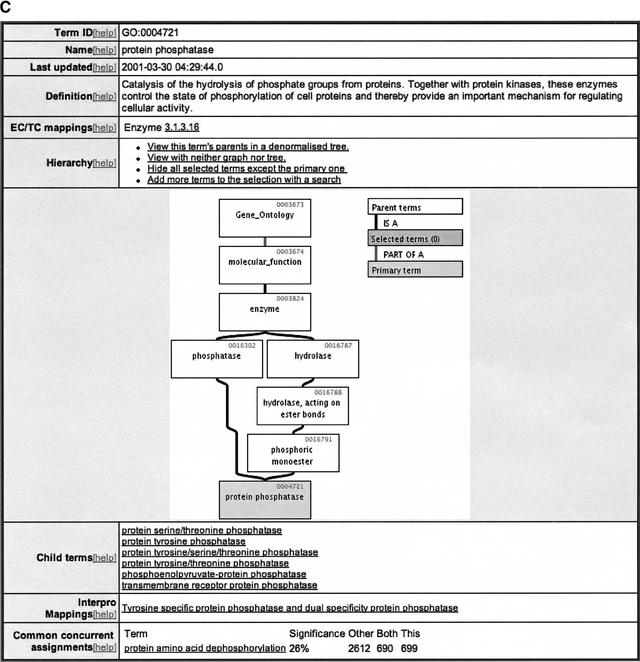
(A) Sample of QuickGO display page showing all GO terms that have been mapped electronically and manually to SWISS-PROT entry Q9Z247. (B) Sample of QuickGO display screen showing all information currently available through EBI for the GO term “protein phosphatase,” using the denormalized tree view. (C) Sample of QuickGO display screen showing all information currently available through EBI for the GO term “protein phosphatase,” using the graphical tree view.
The GO term page displays all information currently held at the EBI for that term including the term name, term ID, and definition. Two different views for each term are available: a denormalized tree view of the GO structure ascending from the term (Fig. 2B) or a graphical tree view (Fig. 2C), which makes it easier to visualize the position of a GO term within the hierarchy. The concise, denormalized view can be selected as the default view. New GO users may prefer the graphical output for tracing more complex paths.
Another useful and unique feature of the QuickGO browser is its display of common concurrent assignments, that is, GO terms that are frequently assigned in tandem. Although, as an explicit part of the GO design there are no relations that span the three ontologies, there are clearly links between terms in different ontologies. By applying data-mining techniques, a large number of pairs of GO terms that are commonly associated with one another were found (S. Clelland, unpubl. 2001). For example, “heavy-metal ion transporter”, which is part of the process ontology and “heavy-metal ion transport”, which is part of the function ontology, are often found assigned together. Therefore, the QuickGO entry for the GO term “heavy-metal ion transporter” lists “heavy-metal ion transport” as a common concurrent assignment. These data act not only as a curation guide but also point to potential problems with the GOA data in its current state.
Gene Association File
The most common form of data transfer within the GO Consortium is a tab-delimited file of the associations between gene products and GO terms referred to as a gene association file. Because SWISS-PROT annotates proteins rather than genes, the semantics of some fields are slightly different to gene association files produced by other consortium members, and these details are fully documented at http://www.ebi.ac.uk/GOA. Currently, the SWISS-PROT group at EBI produces two GOA files: GOA-Human contains the GO assignments for the proteins in the SWISS-PROT, TrEMBL, and Ensembl nonredundant human proteome set, and GOA-SPTR contains all GO annotations to SWISS-PROT and TrEMBL. For each association, cross-references are supplied to SWISS-PROT, TrEMBL, Ensembl, the International Protein Index (IPI) (http://www.ebi.ac.uk/IPI/), and GO, along with evidence for its annotation according to GO evidence codes. Data provided by the GOA project is available on the EBI FTP site at ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/ as well as the GO FTP site at ftp://ftp.geneontology.org/pub/go/gene-associations/. In addition to the human gene association file, the EBI releases a file of cross references that displays the relationship between the entries in the GOA data set with other databases, such as the EMBL-Bank/GenBank/DDBJ nucleotide sequence databases (Stoesser et al. 2003), HUGO, and LocusLink and RefSeq (Pruitt and Maglott 2001) at the NCBI. This tab-delineated file is available at ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/HUMAN/xrefs.goa and is updated with each GOA-Human release. Monthly releases of GOA will include the regular replacement of electronic associations with experimentally verified evidence codes. In between GOA releases, we recommend use of the QuickGO browser for the latest curated associations.
Sequence Retrieval System (SRS)
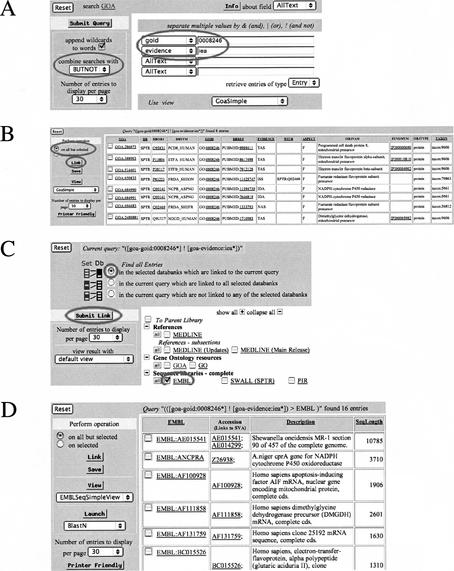
The EBI's SRS server (Zdobnov et al. 2002) at http://srs.ebi.ac.uk/ acts as a central resource for biological databases and now includes the EBI's gene association file of the GOA project as well as a mirror of the GO Consortium repository. This is a welcomed development to the query form, as it allows more functional questions to be asked across a range of databases, quickly and efficiently. Under SRS, the gene association file can be searched using a range of fields including GO ID, SWISS-PROT, or TrEMBL accession number, and GO evidence type. The sample query shown in Figure 3A illustrates how a user asks the question, “How many protein sequences in GOA have been manually assigned the GO function ‘electron transfer flavoprotein’?” The user can simply create a query that searches for all proteins linked to the GO term “electron transfer flavoprotein” (GO:0008246) and filters out any associations that have an IEA evidence code. A further facility of SRS is its ability to link to databases that do not contain direct references to each other. As such, the user can perform a second search to gather all the sequences in his previous search with accession numbers for the EMBL/GenBank/DDBJ nucleotide databases (Fig. 3B,C,D).
Figure 3.
Searching Gene Ontology Annotation (GOA) database with Sequence Retrieval System (SRS). (A) To find all annotated proteins that function as electron transfer flavoproteins and that have an experimental evidence code (Non-IEA), the “goid” field is searched for the GO identifier “0008246”in the GOA database. In the “combined searches with” section of the tool bar, the “BUTNOT” option is selected and ”IEA” (the GO evidence code for “inferred from electronic annotation”) is entered in the “evidence” field. (B) This produces a query result, which displays all proteins manually assigned the function of “electron transport flavoprotein” using published literature. Associations made by electronic inference are filtered out and results displayed in the “gene association file” format. (C, D) A further facility of SRS is its ability to link to databases that may or may not contain direct references to each other. As such, the last search can be extended to display EMBL/GenBank/DDBJ accession numbers by selecting the “link” option and choosing the EMBL database and “submit link.”
Applications of GOA in Proteome Analysis
An application of the GO mappings at the EBI can be seen on the Proteome Analysis pages (http://www.ebi.ac.uk/proteome/) produced by the SWISS-PROT group, where GO is used for classification of proteins belonging to each complete proteome. The aim of the Proteome Analysis project (Apweiler et al. 2001b) is to provide proteome sets for whole genomes with comprehensive statistical and comparative analyses. Nonredundant sets of SWISS-PROT and TrEMBL entries are produced for each complete proteome, based on genome sequence submissions and additional knowledge researched by SWISS-PROT curators. The statistical and comparative analyses are compiled using InterPro, CluSTr (Kriventseva et al. 2001), and GO and contain structural information derived from the HSSP (Dodge et al. 1998) and PDB (Westbrook et al. 2002) databases.
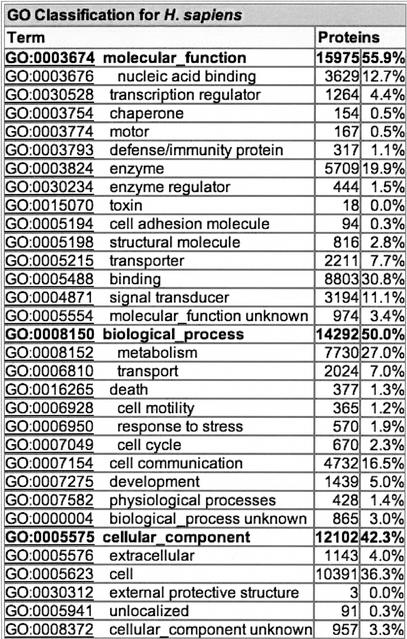
For each proteome, there is an automatically generated table showing the general statistics for the number of proteins that can be assigned to a selection of high-level terms from each of the GO ontologies. These terms have been selected to cover most aspects of the ontologies without overlapping in paths in the GO hierarchy and are described as “GO Slim”. Using the mappings of proteins within a proteome set to GO, which are derived from assignments based on InterPro, SWISS-PROT keywords, and EC numbers as well as from manual assignments, the mappings are collapsed to the selected high-level terms. The number of proteins mapped to each selected term is calculated to provide a table of statistics of the relative percentage of proteins in the proteome mapped to each term (Fig. 4). Because proteins may be assigned to more than one GO term, some proteins will have been counted more than once. The GO Slim used by EBI is archived at ftp://ftp.geneontology.org/pub/go/GO_slims/goslim_goa.2002.
Figure 4.
Example of a table from the proteome analysis database showing the general statistics for the number of proteins in the human proteome that can be assigned to a selection of high-level terms (GO Slim) from each of the three gene ontologies.
The functional classification and mapping of InterPro families and domains, as well as SWISS-PROT keywords and EC numbers, to GO provides a simple method for determining whole proteome composition and provides a basis for comparative analysis. In addition, the CluSTr database has links to InterPro and, from there, to the corresponding functional classification codes and GO terms, making it is possible to identify protein functions within clusters.
Applications of GOA in Genome Analysis
To support the mapping of biological knowledge, and especially to facilitate the interpretation of genomic data, the GOA project annotations have been cross linked to the coding regions directly in EMBL-Bank flatfiles, which contain the nucleotide sequences of the international collaboration EMBL-Bank/GenBank/DDBJ. GOA project annotations have also been integrated into Ensembl, a joint project between EMBL-EBI and the Sanger Institute, which produces automatic annotation on eukaryotic genomes. GOview provides an interface to the Ensembl gene database via the GO hierarchy. GO annotations (GOA-Human) have been mapped to Ensembl human genes and the GO hierarchy can be navigated directly or searched to identify matching loci. The resulting gene matches are displayed in their tribe families and graphically as locations on the human genome. Ensembl to GOA mappings are currently only available for human, although these will be extended to mouse at the next release. As GOA and Ensembl releases are not synchronized, users should check the version of GOA-Human, which has been used within GOview. An example GOview can be seen at http://www.ensembl.org/Homo_sapiens/geneview?gene=ENSG00000139618.
DISCUSSION
Manual annotation produces reliable GO annotation but is an inefficient approach to tackling the vast amounts of data already accumulated in SWISS-PROT and TrEMBL from the various genome projects. On the other hand, electronic techniques offer a much quicker approach to the assignment of GO terms to new data while enabling a retrofit of GO annotation to previously curated data. To date, 64% of all proteins stored in the SWISS-PROT and TrEMBL databases have been annotated with GO terms using electronic methods. This represents 2.5 million associations covering 544,362 proteins out of a total of 850,795 (http://www.ebi.ac.uk/GOA/SPTR_release.html). In contrast, GO associations generated by biologists cover just 1% of SWISS-PROT and TrEMBL entries. The electronic methods are responsible for assigning GO terms to entries from almost 50,000 different species while manual methods have assigned GO terms to entries from 182 different species. By annotating GO terms to such a wide variety of species, the SWISS-PROT group makes a substantial contribution to the GO Consortium efforts.
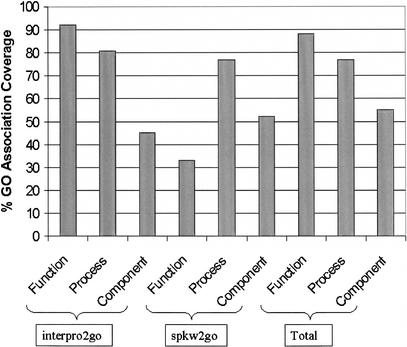
Of the electronic techniques, the InterPro to GO mapping (Interpro2go) has generated the most associations followed closely by the application of SWISS-PROT keywords to GO (spkw2go). GO annotation by electronic techniques assigned GO terms unevenly across the three ontologies (Fig. 5). Interestingly, InterPro associations showed a strong bias toward the assignment of function (92%) and process (81%) terms, whereas the use of SWISS-PROT keyword mappings assigned much fewer function terms (33%) but was a little better than InterPro at assigning component terms (52%). EC numbers have been mapped only to terms from the function ontology so no comment can be made on the success of this method in assigning terms from the process or component ontologies. However, the average depth of terms assigned based on mappings of EC numbers to GO is higher than that of either of the other two electronic methods. The depth of a term is used here to mean the number of terms from the parent term to the assigned term. The average depth of predictions based on EC numbers is 10.54 whereas for InterPro, it is 5.94 and for SWISS-PROT keywords, 4.67. The average overall depth of terms assigned using electronic methods is 5.73. Manual annotation assigned GO terms more evenly across the three ontologies (data not shown) and provided literature references and information about the type of experiments used through GO evidence codes. These results indicate that different methods have their merits and limitations and that combining multiple techniques to assign GO terms increases annotation coverage, an observation also reported by others (Schug et al. 2002).
Figure 5.
Percentage of proteins associated with GO terms from each ontology, using the interpro2go and spkw2go mappings (interpro2go = mapping of InterPro entries to GO terms; spkw2go = mapping of SWISS-PROT keywords to GO).
Although the number of incorrect assignments made electronically was not directly measured in this study, the number of times the different electronic techniques predicted GO assignments in the same lineage has been calculated. In general, electronic techniques assign more general GO terms (higher in the GO hierarchy) than can be applied by manual efforts (data not shown). However, of all SWISS-PROT and TrEMBL entries that had multiple GO associations, 94% had terms assigned by different electronic methods that were from the same lineage, that is, they had the same parent term in GO. As a first-round annotation approach, GO associations by electronic techniques worked extremely well and provided a useful guide to curators who often found evidence to assign more precise terms (deeper in the GO hierarchy) within the same or new lineages. On very few occasions, InterPro GO assignments were inconsistent with manual curation. This discrepancy was related to the fact that not all proteins function according to their membership in a particular family. This membership may only represent their evolutionary origin. An example of such an occurrence is that of the cytokine subunit of the IL-12 heterodimer, p40, which evolved from a primordial IL-6-like receptor (Schoenhaut et al. 1992; Shields et al. 1995) and was assigned the GO term “hematopoeitin/interferon-class (D200-domain) cytokine receptor” (GO:0004896) through an InterPro to GO mapping. In fact, IL-12 p40 does not function as a conventional membrane-bound cytokine receptor. It does, however, bind the second subunit of IL-12, p35 (relative of IL-6), to form a functional heterodimer with very different cytokine functions. This shows that GO terms assigned by electronic means should be treated with a certain amount of caution. It also highlights the need to find real evidence in published literature by either manual or text mining efforts and for users of GO annotation to alert sources of electronic GO associations when inconsistencies occur. It should equally be noted that for proteins of unknown function, GO associations made using InterPro can also offer very precise first-round functional predictions. Such data may help identify new relatives of biologically important proteins and possibly identify candidates for further experimental analysis.
Future Work
To promote database interoperability and provide consistent annotation, the SWISS-PROT group will continue to assign GO terms to the gene products of the SWISS-PROT knowledgebase and its supplementary database, TrEMBL. As the EBI hosts the GO editorial office, the SWISS-PROT curators already work closely with the GO curators in their efforts to expand and improve the GO resource.
Ongoing refinement of automated procedures in the TrEMBL section of the SWISS-PROT knowledgebase is paramount to the success of our large-scale GO annotation. The group will continue to develop these methods and will resolve any in-house complexities that may arise from the integration with other database resources. In collaboration with the Swiss Institute of Bioinformatics (SIB), new GO mappings will be released in 2003 for SWISS-PROT subcellular location as well as from PROSITE (Sigrist et al. 2002) and HAMAP databases.
The continuous assignment of GO terms to additional SWISS-PROT and TrEMBL entries by manual and electronic strategies will be reflected in subsequent releases of GOA. In each release, electronic associations are replaced by terms that are based on experimental evidence. SWISS-PROT biologists will assign these more detailed terms during literature-based GO curation.
The incorporation of GO data from model organism databases is also planned. Only data with a non-IEA association and nonreview literature reference will be considered for inclusion in the SWISS-PROT GOA releases. In time, we will produce separate GOA files for each proteome. The group also plans to improve displays of GO data on proteome analysis pages and to develop the display pages for the SWISS-PROT/TrEMBL/Ensembl proteome set. The association of GO terms to the clusters in our CluSTr database are also included in the project plans.
Currently, 64% of SWISS-PROT and TrEMBL entries have been mapped to GO terms. We aim to have assigned GO terms to more than 70% of the SWISS-PROT and TrEMBL records by 2004.
How to Submit Updates to Our GO Annotation
Although a careful one-pass annotation is initially useful, it is certain that as our knowledge of biology develops, both the SWISS-PROT annotation and GO vocabulary will grow and change. As such, we envisage that proteins in SWISS-PROT and TrEMBL will need to be updated regularly to keep up with this expanding knowledge. The success and accuracy of our GO annotations rely on frequent electronic and manual checking. As with SWISS-PROT curation, the group actively encourages updates or corrections from the scientific community to improve this shared resource. For all inquiries and corrections to the GOA project, please use the contact e-mail: goa@ebi.ac.uk.
WEB SITE REFERENCES
http://www.geneontology.org/external2go/interpro2go; InterPro to GO mappings.
http://www.geneontology.org/external2go/spkw2go; SWISS-PROT keyword to GO mappings.
http://www.geneontology.org/GO.evidence.html; GO evidence codes.
http://ep.ebi.ac.uk/EP/GO/; Expression Profiler GO browser.
http://www.ensembl.org; Ensembl Home Page.
http://www.ebi.ac.uk/GOA; GO Annotation at EBI home page.
http://www.ebi.ac.uk/IPI/; International Protein Index.
http://www.ebi.ac.uk/interpro/; InterPro home page.
http://www.ebi.ac.uk/interpro/scan.html; InterProScan.
http://www.ebi.ac.uk/proteome/; Proteome Analysis database.
http://www.ebi.ac.uk/proteome/SPTREnsembl.html; SPTr-Ensembl human proteome set.
http://www.ebi.ac.uk/ego/; QuickGO browser.
http://srs.ebi.ac.uk/; SRS server at EBI.
http://www.expasy.org/cgi-bin/keywlist.pl; SWISS-PROT keyword list.
http://tcdb.ucsd.edu/tcdb/; Transport Commission database home page.
http://us.expasy.org/sprot/hamap/; High-Quality Automated and Manual Annotation of microbial Proteomes (HAMAP) database home page.
http://us.expasy.org/prosite; Prosite home page.
Acknowledgments
The GO project at EBI is supported by two European Union contracts BioBabel (QLRT-2000–00981) and TEMBLOR (QLRI-2001–00015, Hermjakob and Apweiler, 2002) and a supplementary NIH grant (1R01HGO2273–01). We are grateful to the SWISS-PROT curation team for the manual assignment of GO terms to SWISS-PROT and TrEMBL entries and to Sarah Clelland for generating the common concurrent assignments in QuickGO as part of an undergraduate research project. We also thank John Maslen for recent improvements to in-house GO annotation tools and GO Consortium members, Michael Ashburner (FlyBase) and David Hill (MGD) for their improvement of the SWISS-PROT keyword to GO mapping (spkw2go). We are also very grateful to Midori Harris, Jane Lomax, Cath Brooksbank, and Amelia Ireland, the GO editorial team at EBI.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL camon@ebi.ac.uk; FAX +44 0 1223 494 468.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.461403. Article published online before print in March 2003.
REFERENCES
- 1.Apweiler R. 2001. Functional information in SWISS-PROT: The basis for large-scale characterisation of protein sequences. Brief. Bioinform. 2: 9-18. [DOI] [PubMed] [Google Scholar]
- 2.Apweiler R., Attwood, T.K., Bairoch, A., Bateman, A., Birney, E., Biswas, M., Bucher, P., Cerutti, L., Corpet, F., Croning, M.D., et al. 2001a. The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res. 29: 37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apweiler R., Biswas, M., Fleischmann, W., Kanapin, A., Karavidopoulou, Y., Kersey, P., Kriventseva, E.V., Mittard, V., Mulder, N., Phan, I., et al. 2001b. Proteome analysis database: Online application of InterPro and CluSTr for the functional classification of proteins in whole genomes. Nucleic Acids Res. 29: 44-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Attwood T.K., Blythe, M.J., Flower, D.R., Gaulton, A., Mabey, J.E., Maudling, N., McGregor, L., Mitchell, A.L., Moulton, G., Paine, K., et al. 2002. PRINTS and PRINTS-S shed light on protein ancestry. Nucleic Acids Res. 30: 239-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bairoch A. 2000. The ENZYME database in 2000. Nucleic Acids Res. 28: 304-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch A. and Apweiler, R. 2000. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 28: 45-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman A., Birney, E., Cerruti, L., Durbin, R., Etwiller, L., Eddy, S.R., Griffiths-Jones, S., Howe, K.L., Marshall, M., and Sonnhammer, E.L.L. 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blake J.A., Richardson, J.E., Bult, C.J., Kadin, J.A., Eppig, J.T., and The Mouse Genome Database Group 2002. The mouse genome database (MGD): The model organism database for the laboratory mouse. Nucleic Acids Res. 30: 113-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corpet F., Servant, F., Gouzy, J., and Kahn, D. 2000. ProDom and ProDom-CG: Tools for protein domain analysis and whole-genome comparisons. Nucleic Acids Res. 28: 267-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodge C., Schneider, R., and Sander, C. 1998. The HSSP database of protein structure-sequence alignments and family profiles. Nucleic Acids Res. 26: 313-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dwight S.S., Harris, M.A., Dolinski, K., Ball, C.A., Binkley, G., Christie, K.R., Fisk, D.G., Issel-Tarver, L., Schroeder, M., Sherlock, G., et al. 2002. Saccharomyces genome database (SGD) provides secondary gene annotation using the gene ontology (GO). Nucleic Acids Res. 30: 69-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falquet L., Pagni, M., Bucher, P., Hulo, N., Sigrist, C.J.A., Hofmann, K., and Bairoch, A. 2002. The PROSITE database, its status in 2002. Nucleic Acids Res. 30: 235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischmann W., Moeller, S., Gateau, A., and Apweiler, R. 1999. A novel method for automatic and reliable functional annotation. Bioinformatics 15: 228-233. [DOI] [PubMed] [Google Scholar]
- 14.The FlyBase Consortium 2002. The flybase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 30: 106-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Gene Ontology Consortium 2001. Creating the gene ontology resource: Design and implementation. Genome Res. 11: 1425-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haft D.H., Loftus, B.J., Richardson, D.L., Yang, F., Eisen, J.A., Paulsen, I.T., and White, O. 2001. TIGRFAMs: A protein family resource for the functional identification of proteins. Nucleic Acids Res. 29: 41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hermjakob H. and Apweiler, R. 2002. TEMBLOR—Perspectives of EBI database services. A presentation for the ESF workshop “Data integration in functional genomics and proteomics.” Comp. Funct. Genom. 3: 47-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill D.P., Dabis, A.P., Richardson, J.E., Corradi, J.P., Ringwald, M., Eppig, J.T., and Blake, J.A. 2001. Strategies for biological annotation of mammalian systems: Implementing gene ontologies in mouse genome informatics. Genomics 74: 121-128. [DOI] [PubMed] [Google Scholar]
- 19.Hubbard T., Barker, D., Birney, E., Cameron, G., Chen, Y., Clark, L., Cox, T., Cuff, J., Curwen, V., Down, T., et al. 2002. The ensembl genome database project. Nucleic Acids Res. 30: 38-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kriventseva E.V., Fleischmann, W., Zdobnov, E.M., and Apweiler, R. 2001. CluSTr: A database of clusters of SWISS-PROT+TrEMBL proteins. Nucleic Acids Res. 29: 33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letunic I., Goodstadt, L., Dickens, N.J., Doerks, T., Schultz, J., Mott, R., Ciccarelli, F., Copley, R.R., Ponting, C.P., and Bork, P. 2002. Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res. 30: 242-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt K.D. and Maglott, D.R. 2001. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 29: 137-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schoenhaut D.S., Chua, A.O., Wolitzky, A.G., Quinn, P.M., Dwyer, C.M., McComas, W., Familletti, P.C., Gately, M.K., and Gubler, U. 1992. Cloning and expression of murine IL-12. J. Immunol. 148: 3433-3440. [PubMed] [Google Scholar]
- 24.Schug J., Diskin, S., Mazzarelli, J., Brunk, B.P., and Stoeckert, C.J., Jr. 2002. Predicting gene ontology functions from ProDom and CDD protein domains. Genome Res. 12: 648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shields D.C., Harmon, D.L., Nunez, F., and Whitehead, A.S. 1995. The evolution of haematopoietic cytokine/receptor complexes. Cytokine 7: 679-688. [DOI] [PubMed] [Google Scholar]
- 26.Sigrist C.J., Cerutti, L., Hulo, N., Gattiker, A., Falquet, L., Pagni, M., Bairoch, A., and Bucher, P. 2002. PROSITE: A documented database using patterns and profiles as motif descriptors. Brief Bioinform. 3: 265-274. [DOI] [PubMed] [Google Scholar]
- 27.Stoesser G., Baker, W., van den Broek, A., Garcia-Pastor, M., Kanz, C., Kulikova, T., Leinonen, R., Lin, Q., Lombard, V., Lopez, R., et al. 2003. The EMBL nucleotide sequence database: Major new developments. Nucleic Acids Res. 31: 1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westbrook J., Feng, Z., Jain, S., Bhat, T.N., Thanki, N., Ravichandran, V., Gilliland, G.L., Bluhm, W., Weissig, H., Greer, D.S., et al. 2002. The protein data bank: Unifying the archive. Nucleic Acids Res. 30: 245-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zdobnov E.M., Lopez, R., Apweiler, R., and Etzold, T. 2002. The EBI SRS server—recent developments. Bioinformatics 18: 368-373. [DOI] [PubMed] [Google Scholar]