Abstract
A characteristic of imprinted genes is that the maternal and paternal alleles show differences in methylation. To perform a genome-wide screen for novel imprinted loci, we applied methylation-sensitive representational difference analysis (Me-RDA) to parthenogenetic mouse embryos, to identify differentially methylated regions (DMRs) methylated specifically on the maternal allele. We isolated a total of 26 distinct clones from known and novel DMRs and identified three novel imprinted genes. Nap1l5 is located on proximal chromosome 6 and encodes a protein with homology with nucleosome assembly proteins (NAPs); it has tissue-specific imprinting with expression from the paternal allele. We identified two DMRs on chromosome 15, a chromosome that was not thought to contain imprinted loci, and demonstrated that each is associated with a paternally expressed transcript. Peg13 gives rise to a noncoding RNA that is highly expressed in the brain and imprinted in all tissues examined. A DMR was also identified at the chromosome 15 Slc38a4 gene, which encodes a system A amino acid transporter; we show that Slc38a4 is imprinted in a tissue-specific manner. Interestingly, two of the three novel genes identified in this screen are located within the introns of other genes; their identification indicates that such “microimprinted” domains may be more common than previously thought.
[The sequence data from this study have been submitted to GenBank under accession nos. AY151252 and AY151253. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: C. Beechey, J. Peters and D. Bodle.]
Genomic imprinting is a parent-of-origin-dependent epigenetic mechanism by which a subset of autosomal genes are expressed from only one allele. Correct regulation of imprinted genes is essential for normal mammalian development, and a number of human disorders are associated with increased or insufficient dosage of imprinted gene products (Morison et al. 2001). An imprinting map of the mouse has been produced (http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html), which describes chromosome (Chr) regions that cause developmental abnormalities when inherited as a uniparental duplication (UPD) and are likely to contain imprinted genes. To date, >50 imprinted genes have been described in the mouse, comprising protein-coding genes, antisense transcripts, small nucleolar RNAs, and other noncoding RNAs. However, the full extent of imprinting in the mouse genome is currently unclear, and in order to determine the full role of imprinted genes in mammalian development, it is essential that additional imprinted genes are identified.
The imprinted genes described to date have been identified by a variety of means. The association of an imprinted phenotype with a genomic region has led to the discovery of a number of imprinted loci; for example, Igf2r was identified owing to its association with the Tme maternal effect on mouse Chr 17 (Barlow et al. 1991). Analysis of regions with a UPD phenotype led to the realization that many imprinted genes occur in clusters, such as that on distal Chr 7, which contains at least 14 imprinted genes (Onyango et al. 2000; Paulsen et al. 2000). Many loci that were initially described as having only one or few imprinted genes are now being reexamined, and neighboring genes are found to be imprinted. For example, the imprinted locus on proximal Chr 17 was initially thought to comprise just the Igf2r transcript (Barlow et al. 1991) and the Air antisense (Wutz et al. 1997). Recent investigations have indicated that this cluster contains at least two further imprinted genes (Zwart et al. 2001). However, examining only genes within known imprinted regions is clearly limiting, so a number of screens have been developed to identify imprinted genes independent of their genomic location.
Many screens for imprinted genes are based on the differences in expression of the maternal and paternal alleles. Subtractive hybridization has been performed on cDNAs from parthenogenetic (Pg) and androgenetic (Ag) embryos, and several imprinted genes, designated Pegs (paternally expressed genes) and Megs (maternally expressed genes), have been identified by this technique (Kaneko-Ishino et al. 1995; Miyoshi et al. 1998). Pg and Ag embryonic fibroblast cell lines have been created and further imprinted genes identified by subtractive hybridization of their cDNA (Piras et al. 2000). Allele-specific expression has also been recognized by differential display (Kikyo et al. 1997) or its derivative allelic message display (Hagiwara et al. 1997). More recently, cDNA microarray screening has been performed by using RNAs from UPD mice (Choi et al. 2001) or from uniparental embryos (Mizuno et al. 2002). Microarray screening has led to the recovery of both known and novel imprinted loci, and because of the ability to examine so many genes at once, holds great promise. Although screens based on imprinted expression have clearly been successful, they are unlikely to be exhaustive because of the limitations imposed by tissue and stage-specificity of expression and low abundance of transcripts. Such screens may also detect downstream targets, as well as the imprinted genes themselves.
Imprinted genes may be identified independently of their expression status by the use of an epigenetic feature of the locus. Of the characteristics described to date, the most amenable for analysis is the region of differential methylation (DMR) that is found at most imprinted loci (Constância et al. 1998). It is thought that all DMRs will indicate the positions of imprinted genes, as for all DMRs currently known, the closest neighboring gene is imprinted. These regions of parental allele-specific methylation are generally maintained in all somatic tissues, irrespective of expression, and the methylation state of an allele may be determined by the use of methylation-sensitive restriction enzymes (Constância et al. 1998). Restriction landmark genome scanning (RLGS) was devised to compare the methylation of parental alleles at sites of rare-cutter restriction enzymes by using two-dimensional gel electrophoresis. RLGS has been applied to human uniparental samples and mouse F1 hybrids, and a number of novel genes have been described (Hayashizaki et al. 1994; Plass et al. 1996; Hayward et al. 1998; Kamiya et al. 2000). One limitation of RLGS is that in order that the two-dimensional gel is not overly complex, frequent cutting restriction enzymes cannot be used, and therefore, each screen analyzes only a small number of CpGs. We have developed a technique for the identification of novel imprinted genes based on their methylation. We use methylation-sensitive restriction enzymes that have a high frequency of cutting in CpG islands, in combination with a subtractive hybridization approach to recover fragments from the unmethylated allele; we have designated this technique methylation-sensitive representational difference analysis (Me-RDA). We have previously applied Me-RDA to the isolation of DMRs from the distal Chr 2 imprinted region and found Gnasxl and Nesp to be novel imprinted transcripts in the mouse (Kelsey et al. 1999; Peters et al. 1999). To extend this research, we have now applied Me-RDA to Pg and Ag embryos in order to perform genome-wide screens for DMRs.
Here we describe the application of Me-RDA to Pg embryos, which contain Chrs of maternal origin only, to isolate DMRs that are methylated specifically on the maternal allele. This screen successfully identified multiple DMRs from known imprinted genes. We also identified a novel potentially expressed imprinted gene, Nap1l5, with homology with nucleosome assembly proteins (NAPs), on proximal Chr 6. Furthermore we have identified two DMRs on Chr 15 and demonstrate that each is associated with an imprinted, paternally expressed transcript; one of these genes, Peg13, gives rise to a noncoding RNA and the other Slc38a4 encodes a system A amino acid transporter.
RESULTS
Isolation of Maternally Methylated DMRs in a Genome-Wide Screen
To isolate DMRs methylated specifically on the maternal allele, we performed Me-RDA essentially as described previously (Kelsey et al. 1999; Peters et al. 1999) by using amplicons from Pg embryos as the driver and amplicons from normally fertilized (N) embryos as the tester. We used two different methylation-sensitive restriction enzymes, Hin6I and HpaII, to increase the number of CpGs examined. After two rounds of subtractive hybridization to enrich the tester-specific fragments from the unmethylated paternal allele, we analyzed and cloned the resulting difference products. To assess the proportion of difference product clones (DPCs) that were tester-specific, we examined their representation in the Pg and N amplicons by Southern blot hybridization. For the Me-RDA using the Hin6I enzyme, three of 12 (25%) DPCs were represented only in the N amplicons, and for the HpaII Me-RDA, one of 12 (8.3%) was differentially represented. To increase the proportion of HpaII DPCs that were differentially represented, we carried out a third round of subtractive hybridization. After this, all HpaII DPCs tested (38 of 38, 100%) were present exclusively in the N amplicons. To identify additional novel DPCs, we pre-screened further DPCs to exclude those that had been previously isolated.
We anticipated that a mouse CpG island library (Cross et al. 1997) would contain most of the DMRs within the genome, and would be expected to have a larger average insert size than the DPCs and therefore contain more useful sequence information. We chose to hybridize this library with total (uncloned) difference products from the second round of subtractive hybridization for both Hin6I and HpaII Me-RDAs, as these would have a good enrichment for maternally methylated DMRs while maintaining fragments that may have been excluded during the HpaII third-round subtractive hybridization. We identified CpG island clones (designated CPGCs) that hybridized with the difference products but not with Pg amplicons, and analyzed their representation in Pg and N amplicons in the same way as for DPCs. Of the 41 CPGCs analyzed from the hybridization with Hin6I difference products, seven (17%) were represented only in the N amplicons, and for the HpaII hybridization, all clones analyzed (six of six, 100%) were differentially represented.
In total, we tested a total of 269 DPCs and CPGCs, of which 160 (59.5%) were represented exclusively in N amplicons and comprised 26 distinct clones. We sequenced these clones and performed BLASTN searches against available sequence databases to determine whether the fragments had arisen from known DMRs. A total of 18 fragments arose from regions known to be maternally methylated DMRs, at the imprinted genes Peg5/Nnat, Gnas (Gnasxl and exon1A), Peg1/Mest, Peg3/Pw1, Snrpn, Zac1, and U2af1-rs1 (Table 1). The identification of Zac1 is published elsewhere (Smith et al. 2002). In addition, we identified a novel DMR at the imprinted gene Sgce and isolated a clone mapping close to the imprinted region containing Peg1 on proximal Chr 6, but not within Peg1 (Table 1). We found that the remaining DPCs and CPGCs identified three novel DMRs, each of which is associated with an imprinted, paternally expressed transcript.
Table 1.
Difference Product Clones (DPCs) and CpG Island Clones Isolated in a Methylation-Sensitive Representational Difference Analysis (Me-RDA) Screen for Maternal Methylation
| Me-RDA enzyme | Me-RDA clone ID | Gene or transcript name | Chromosomal location | GenBank accession | |
| (Ensembl) | (Celera) | ||||
| Known DMR at known imprinted gene | |||||
| HpaII | |||||
| DPC#2-11 | Peg5/Nnat | 2H2 | 2H2 | AB041829, AF303656, AJ245740 | |
| DPC#26-1 | Peg1/Mest | 6A3 | 6A3.1 | AF017994 | |
| DPC#26-2 | Peg1/Mest | 6A3 | 6A3.1 | AB007765, AF017994 | |
| DPC#26-5 | Peg3 | 7A1 | 7A1 | AF105262 | |
| DPC#28-5 | Gnasxl | 2H3 | 2H4 | AL593857, AJ251761, AJ245739 | |
| CPGC#13 | Zac1 | 10A2 | 10A1 | AF314094, AJ308559, AJ308560 | |
| CPGC#43 | U2af1-rs1 | 11A3.3 | 11A3.1 | AF309654, D17407, D26474, NM_011663, S69507 | |
| CPGC#49 | Peg1/Mest | 6A3 | 6A3.1 | AF017994 | |
| Hin6I | |||||
| DPC#4-7 | Peg3 | 7A1 | 7A1 | AF105262 | |
| DPC#4-8 | Peg1/Mest | 6A3 | 6A3.1 | AB007765, AF017994 | |
| DPC#4-10 | Peg3 | 7A1 | 7A1 | AF105262 | |
| DPC#12-13 | Peg1/Mest | Unmapped | 6A3.1 | AF017994 | |
| DPC#12-14 | Peg3 | 7A1 | 7A1 | AF015262 | |
| DPC#17-6 | Peg1/Mest | 6A3 | 6A3.1 | AB007765, AF017994 | |
| DPC#19-15 | Gnas exon 1A | 2H3 | 2H4 | AL593857 | |
| DPC#19-23 | Snrpn | 7B5 | Unmapped | AF081460, AF130843, AF332579 | |
| CPGC#1 | Peg3 | 7A1 | 7A1 | AF105262 | |
| CPGC#2 | Peg1/Mest | 6A3 | 6A3.1 | AB007765, AF017994 | |
| Novel DMR at known imprinted gene/novel DMR in imprinted region | |||||
| Hin6I | |||||
| DPC#17-60 | Sgce | 6A2 | 6A1 | — | |
| DPC#17-65 | Novel | 6A3 | 6A3.1 | — | |
| Novel DMR and imprinted gene | |||||
| HpaII | |||||
| DPC#26-7 | Slc38a4 | 15F2 | 15F1 | — | |
| CPGC#29 | Slc38a4 | 15F2 | 15F1 | — | |
| Hin6I | |||||
| DPC#17-27 | Peg13 | 15E1 | 15D3 | AY151252, AY151253 | |
| DPC#19-2 | Peg13 | 15E1 | 15D3 | AY151252, AY151253 | |
| DPC#24-1 | Nap1l5 | 6C1 | 6B3 | AB041556, AK003875, NM_021432 | |
| CPGC#17 | Peg13 | 15E1 | 15D3 | AY151252, AY151253 | |
DMR indicates differentially methylation region.
Each Me-RDA clone represents a distinct Hin6I or HpaII fragment or CpG island library clone. In many cases, the clones were isolated many times.
GenBank accession nos. indicate the best BLASTN matches with database sequences; more than one entry is given for similar BLASTN scores. No GenBank accession is given for Slc38a4 as the DPC and CPGC were not within GenBank database sequence.
Nap1l5, a Novel Imprinted Gene on Chr 6 Encoding a Protein With Homology With NAPs
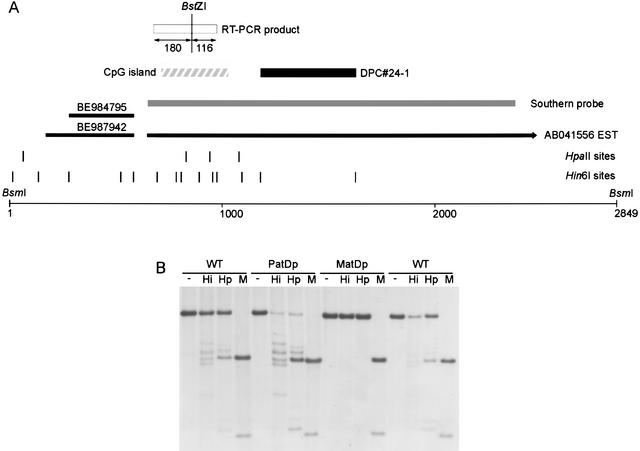
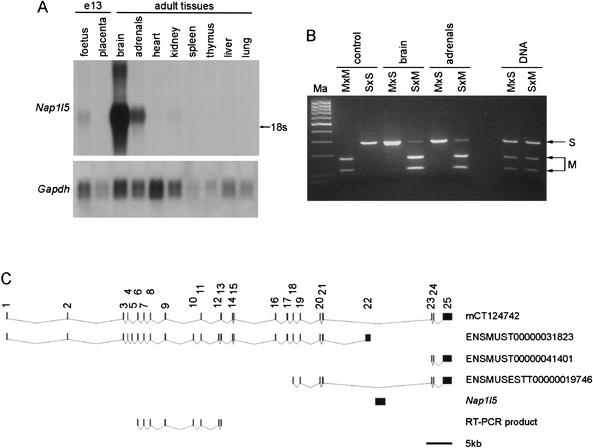
We isolated DPC#24–1, a 449-bp fragment from the Hin6I Me-RDA that maps to Chr 6, to chromosomal band C1 (Ensembl, http://www.ensembl.org) or B3 (Celera, http://www.celera.com). Its location is close to the T6Ad translocation breakpoint in 6B3 that defines the distal end of the imprinted region containing Peg1 (http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html), and it is currently unclear whether DPC#24–1 is located within this defined imprinted region. DPC#24–1 is located near a CpG island (Fig. 1A), and the region has many EST matches. Most of the EST matches are contained within AB041556, the full insert sequence of brain cDNA clone MNCb-0385 (Fig. 1A). The transcript at this locus is intron-less and contains an ORF that is predicted to encode a protein (BAA95041) containing a NAP domain (Pfam accession no. PF00956). We call this locus Nap1l5 (NAP 1 like 5). To investigate whether there is a DMR at Nap1l5, we analyzed the methylation profile in T77H translocation UPD animals in comparison to their wild-type littermates (Fig. 1B). This indicated that DPC#24–1 was isolated from a maternally methylated DMR, as expected from the Me-RDA strategy. We investigated the expression profile of the transcript at this locus by Northern blot hybridization and found that it is highly expressed in adult brain and adrenal glands and is also detected in day-13 fetuses (Fig. 2A). To assess whether this gene is imprinted in the tissues in which it is highly expressed, we performed an RT-PCR assay by using a BstZI RFLP between Mus musculus (C57BL/6) and M. spretus to identify the parental alleles (Fig. 1A). Nap1l5 is imprinted and expressed exclusively or predominantly from the paternal allele in brain and adrenal glands (Fig. 2B). In other tissues, we found no evidence of paternal allele-specific expression (data not shown).
Figure 1.
Identification and methylation analysis of Nap1l5 locus. (A) Map of locus at novel imprinted gene Nap1l5. DPC#24–1 identified the locus and is indicated by a solid box; the CpG island, by a hatched box. The Nap1l5 transcript is contiguous with the genomic DNA; EST AB041556 at Nap1l5 is shown as a black line, and the transcription orientation is indicated by the arrowhead. EST matches (BE984795, BE987942) neighboring AB041556 are indicated as black lines and form part of the same transcription unit as determined by RT-PCR (data not shown). The probe used for methylation analysis by Southern blot hybridization is indicated as a gray bar, and the BsmI fragment analyzed is shown. The RT-PCR product used for imprinted expression analysis by using a BstZI RFLP (Fig. 2B) is shown as a white box, and fragment lengths are given. Hin6I and HpaII sites are given as vertical lines; in some instances, individual sites could not be resolved. (B) Methylation analysis of region containing Nap1l5 in kidney DNA from T77H UPD (PatDp.dist6 and MatDp.dist6) adult mice. The DNA was digested with BsmI alone (−) or in combination with Hin6I (Hi), HpaII (Hp), or MspI (M). Wild-type (WT) controls are shown alongside their PatDp.dist6 and MatDp.dist6 littermates. MatDp.dist6 DNA is highly methylated in comparison to PatD.dist6 DNA, indicating that the region is a maternally methylated DMR.
Figure 2.
Tissue-specific and imprinted expression of Nap1l5. (A) Northern blot hybridization of total RNA from adult tissues and day-e13 foetus and placenta, using an antisense RNA probe from within the sequence of AB041556. The size of the transcript on the Northern blot is, in comparison to 18S/28S rRNA, consistent with the expected full-length transcript sequence. Hybridization with Gapdh gives an indication of the amount of RNA loaded. (B) RT-PCR/RFLP analysis of imprinted expression of Nap1l5 in hybrids and backcross animals between Mus musculus C57BL/6 (M) and M. spretus (S). BstZI specifically digests PCR products from the M. musculus allele and assigns the parental origin of alleles in RT-PCRs from reciprocal hybrid RNA. Samples analyzed are brain and adrenal gland RNAs from a cross between C57BL/6 female and M. spretus male (M × S, maternal inheritance of M allele, paternal inheritance of S allele) and from a cross between an F1 (C57BL/6 × M. spretus) female and C57BL/6 male (S × M, maternal inheritance of S allele, paternal inheritance of M allele at this locus). The DNA amplification indicates the presence of both alleles in the animals examined. No product amplified in control reactions with no reverse transcriptase (not shown). The controls are amplified M. musculus C57BL/6 (M × M) or M. spretus (S × S) DNA to indicate the digestion products. Ma is a 100-bp molecular weight marker. (C) Map indicating the position of Nap1l5 within a large intron of Herc3. mCT124742 is a transcript described in the Celera database; ENSMUST00000031823, ENSMUST00000041401, and ENSMUSEST00000019746 are Ensembl identifiers. mCT124742 and ENSMUSEST00000019746 indicate that an intron of Herc3 transcript spans the region containing Nap1l5; Herc3 and Nap1l5 are transcribed in the opposite orientation. The RT-PCR product used to investigate imprinting of Herc3 in T77H uniparental duplication tissues is shown.
To determine whether Nap1l5 might be part of a cluster of imprinted genes, we initially examined the genomic sequence (Ensembl and Celera databases) to identify neighboring genes. This revealed that Nap1l5 is entirely contained within the intron of another gene, Herc3, which is transcribed in the opposite orientation (Fig. 2C). Herc3 (NM_028705) encodes a protein with a domain homologous to E6-associated protein (E6-AP) C terminus (HECT domain) and a region with homology with the regulator of chromosome condensation (RCC1). We investigated the expression of Herc3 in MatDp.dist6 or PatDp.dist6 tissues by RT-PCR, amplifying the RT-PCR product indicated in Figure 2C. Herc3 was expressed in brain, heart, and kidney from MatDp.dist6 and PatDp.dist6 adults, with no expression detected in liver (data not shown). This result does not preclude imprinted expression in other cell types or developmental stages; however, the lack of imprinting in brain, in which Nap1l5 is expressed from the paternal allele, suggests that Herc3 is not imprinted.
Peg13, a Novel Noncoding Imprinted Transcript on Chr 15
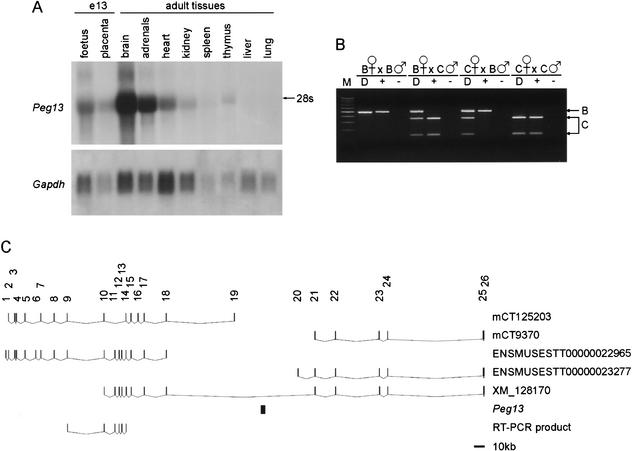
We found that two DPCs and a CPGC (which encompasses one of the DPCs) from the Hin6I Me-RDA screen are located on Chr 15, within band E3 (at 94.13 Mb, Ensembl data). The region to which these clones map contains a CpG island and multiple ESTs, predominantly from brain cDNA libraries (Fig. 3A). We analyzed the methylation at this locus in day-13 fetuses from reciprocal crosses between M. musculus (C57BL/6) and M. musculus castaneus (CAST/Ei), by using a KpnI RFLP to distinguish the alleles, and we found that the region is a maternally methylated DMR (Fig. 3B). The transcript is abundantly expressed in adult brain and adrenal glands and is expressed at lower levels in other adult tissues, day-13 fetus, and placenta (Fig. 4A). We screened a neonatal brain cDNA library and performed 5′ and 3′ RACE on adult brain mRNA to isolate the full-length transcript, which we call Peg13. The transcript is 4397-nt long, excluding polyA tail (accession no. AY151253), with a minor form extended at the 5′ end and length 4723 nt (accession no. AY151252), which was detected by 5′ RACE. The longest ORF in either transcript form is 321 nt; the polypeptide that this would encode does not have homology with any known proteins, and the sequence surrounding the ATG is not similar to the Kozak consensus sequence, indicating that this transcript is not translated. The transcript runs contiguous to the genomic DNA sequence and is therefore intron-less, as well as apparently noncoding. BLASTN analysis of the transcript identifies multiple ESTs, but there is no homology with any known genes. We analyzed the expression of Peg13 in reciprocal crosses between C57BL/6 and CAST/Ei by using a KspI RFLP, and found that it is expressed exclusively from the paternal allele in day-13 fetuses (Fig. 4B) and exclusively or predominantly from the paternal allele in the seven adult tissues tested (data not shown).
Figure 3.
Identification and methylation analysis of the locus containing Peg13. (A) Map of locus containing Hin6I Me-RDA products DPC#19–2 and DPC#17–27 and CPGC#17 (solid boxes). The region contains a CpG island, indicated by a hatched box. Sequence matches to ESTs are indicated by gray boxes. The black arrows indicate the paternally expressed noncoding transcript, Peg13. Peg13 is contiguous with the genomic DNA and was identified in two isoforms, with lengths 4419 nt and 4723 nt, which differ at their 5′ ends. The HindIII sites flank the region analyzed by Southern blot hybridization; the KpnI site is polymorphic between C57BL/6 and CAST/Ei and specifically digests the C57BL/6 allele. The RT-PCR product used for expression analysis by using a KspI RFLP is shown as a white box; the restriction fragment lengths are given. Hin6I and HpaII sites are given as vertical lines; in some instances, individual sites could not be resolved. (B) Methylation analysis in day-13 fetuses from reciprocal crosses (B × C and C × B) between C57BL/6 (B) and CAST/Ei (C). The DNA was digested with HindIII and KpnI (−) or in combination with Hin6I (Hi), HpaII (Hp), or MspI (M). DNA from fetuses of the parental strain are included as controls.
Figure 4.
Tissue-specific and imprinted expression of Peg13. (A) Northern blot hybridization of total RNA from adult tissues and midgestation (e13) fetus and placenta, using Peg13 as a probe. The size of the transcript on the Northern blot is, in comparison to 18S/28S rRNA, consistent with the full-length transcript sequence. Hybridization with Gapdh gives an indication of the amount of RNA loaded. (B) RT-PCR/RFLP analysis of imprinted expression of Peg13 in inter-subspecific hybrids of C57BL/6 (B) and CAST/Ei (C) using an RT-PCR product from within the sequence of EST AW125295. KspI digests the PCR product from the CAST/Ei allele and assigns the parental origin of alleles in RT-PCRs (+). Control RT reactions with no reverse transcriptase were included (−). As the transcript runs contiguous to the DNA, PCR amplification from genomic DNA (D) indicates the presence of both alleles in the hybrid embryos. M is a 100-bp molecular weight marker. (C) Map indicating the position of Peg13 within a large intron of the gene encoding the mouse homolog of the predicted protein KIAA1882. mCT125203 and mCT9370 are Celera transcripts; ENSMUSEST00000022965 and ENSMUSEST00000023277 are Ensembl entries. XM_128170 indicates that the KIAA1882 gene spans the region containing the DPC#19–2 transcript; both genes are transcribed in the same transcriptional orientation. The RT-PCR product used to investigate imprinting of mouse KIAA1882 is shown.
Analysis of genomic sequence indicated that Peg13, similar to Nap1l5, is contained entirely within the intron of another gene (Fig. 4C). This gene encodes the mouse homolog of the human gene encoding a novel protein KIAA1882. The mouse KIAA1882 transcript is in the same transcriptional orientation as Peg13. By using reciprocal hybrids between M. musculus and M. spretus, identifying the alleles with a BseLI RFLP, we found that this transcript is not imprinted in the eight adult tissues examined, which included brain and adrenal glands (data not shown).
Slc38a4 Is a Novel Imprinted Gene on Chr 15
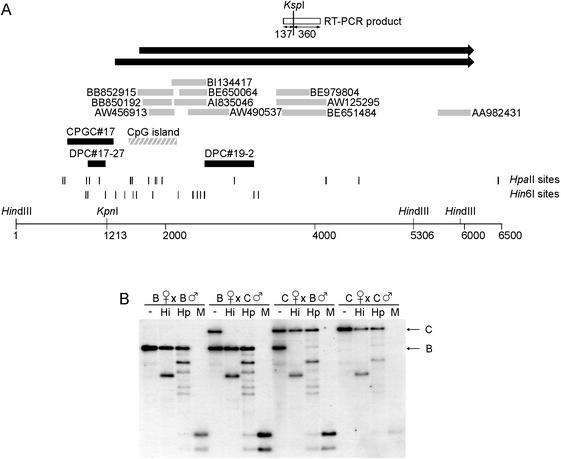
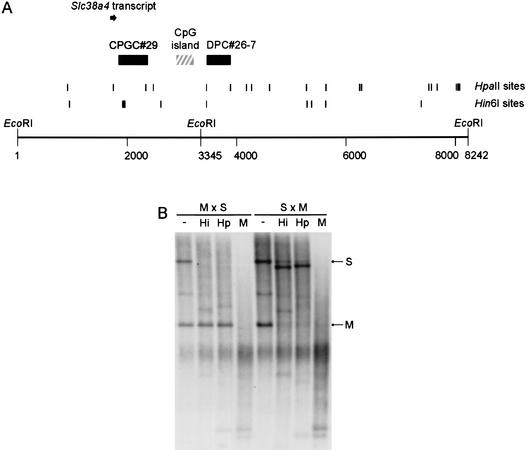
We identified a second imprinted gene on Chr 15, in band F2 (108.95Mb, Ensembl). We isolated a DPC and a CPGC from the HpaII Me-RDA located at a CpG island at the 5′ end of the mouse Slc38a4 gene (Fig. 5A), which encodes a system A amino acid transporter molecule. This gene has been described in human (AF305814) and rat (AF295535) and has also been called ATA3/Ata3 (Sugawara et al. 2000; Hatanaka et al. 2001). We analyzed the methylation of the locus in hybrids between M. musculus and M. spretus by using an EcoRI RFLP to distinguish the parental alleles, and we found that the fragment containing the Me-RDA clones is a maternally methylated DMR (Fig. 5B).
Figure 5.
Identification of Slc38a4 and methylation analysis of the locus. (A) Map of locus indicating relative locations of Me-RDA clones (solid boxes) to a CpG island (hatched box) and the first exon of Slc38a4 (solid arrow). The ATG translation start for Slc38a4 is located in exon 2; downstream exons are not illustrated in this figure. The EcoRI sites indicated are those used in methylation analysis; the site at position 3345 is polymorphic between M. musculus and M. spretus. The RT-PCR assay used to analyze imprinted expression was located in the body of the gene and is not shown here. Hin6I and HpaII sites are given as vertical lines; in some instances, individual sites could not be resolved. (B) Methylation analysis in adult livers from crosses between M. musculus (C57BL/6, M) female and M. spretus (S) male (M × S, maternal inheritance of M alleles, paternal inheritance of S allele), and from backcross animals ([C57BL/6 × M. spretus] × C57BL/6) that inherit a maternal M. spretus allele and a paternal M. musculus allele (S × M) at this locus. The DNA was digested with EcoRI alone (−) or in combination with Hin6I (Hi), HpaII (Hp), or MspI (M).
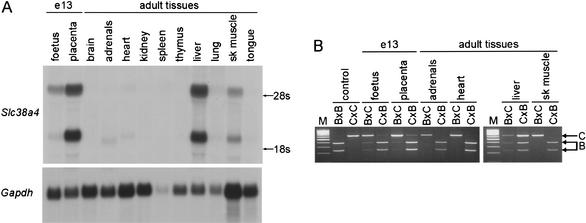
In rat and human, Slc38a4/SLC38A4 is highly expressed in the liver, (Sugawara et al. 2000; Hatanaka et al. 2001) with additional expression reported in kidney (human) and skeletal muscle (rat). We investigated the expression in mouse by Northern blot hybridization and found that Slc38a4 is highly expressed in liver and placenta, with lower expression levels in adrenal glands, heart, lung, and tongue (Fig. 6A). We analyzed the imprinting in these tissues by RT-PCR/RFLP analysis in reciprocal crosses between C57BL/6 and CAST/Ei by using a BseMI RFLP to distinguish the alleles (Fig. 6B). Unexpectedly, we found that the Slc38a4 transcript is biallelically expressed in the liver, the adult tissue in which it is most highly expressed. Slc38a4 is expressed exclusively from the paternal allele in the other adult tissues tested and predominantly from the paternal allele in day-13 fetus and placenta.
Figure 6.
Tissue-specific and imprinted expression of Slc38a4 in the mouse. (A) Northern blot hybridization of adult tissues and day-13 foetus and placenta, using a probe antisense to a portion of the Slc38a4 transcript (146–846 in AX046352). Hybridization with Gapdh gives an indication of the amount of RNA loaded. (B) RT-PCR/RFLP analysis of imprinted expression of Slc38a4 locus in inter-subspecific hybrids of C57BL/6 (B) and CAST/Ei (C). BsmI digests the C57BL/6 allele (B), as indicated in the control reactions performed on cDNA from parental strain midgestation embryos. In each case, the hybrids follow the convention maternal strain × paternal strain. There was no amplification in control reactions performed without reverse transcriptase (data not shown). M is a 100-bp molecular weight marker.
DISCUSSION
We have developed a screen for imprinted loci based on methylation and describe here its application to Pg embryos to perform a genome-wide screen for maternally methylated DMRs. The screen led to the identification of known imprinted genes, new DMRs in known imprinted regions, and novel DMRs associated with novel paternally expressed transcripts. A particular advantage that Me-RDA has in comparison to other screening strategies is that it samples many restriction fragments at each locus and has the ability to isolate more than one fragment from a particular gene. This increases the likelihood that each gene will be sampled at least once and means that the screen has the potential to be exhaustive. This is demonstrated here by the identification of Peg1/Mest as five DPCs and two CPGCs. With smaller restriction fragments comes the disadvantage that each fragment will yield less sequence information. We decided to use the mouse CpG island library to overcome this; however, with the latest mouse genome sequence, the genomic location of even the smallest Me-RDA clone (DPC#17–27, 236 nt) is readily detected by a BLASTN search. One potential pitfall of the Me-RDA approach is that the different DNA fragments can hybridize with differing efficiency during the subtractive hybridization process, leading to loss of the smaller fragments, as stated before (Kelsey et al. 1999). For this reason, DPCs and CPGCs are not necessarily located within the CpG island defined by sequence composition, in which the Hin6I and HpaII sites are closely clustered. However, all DMRs are likely to have Hin6I or HpaII fragments within the size range suitable for Me-RDA. The large number of restriction fragments analyzed in the Me-RDA screen meant that the pool of difference products was very complex. As such, it is likely that further known and novel imprinted genes are contained within these difference products and would be identified after analysis of additional DPCs. For example, it is known that Igf2r DMR2 was enriched in the difference products (R. Smith, unpubl.); yet, this DMR was not recovered as a DPC in the number examined. In addition, repeating the screen by using different frequently cutting methylation-sensitive restriction enzymes would be expected to recover additional imprinted genes.
Of particular note is the fact that all DPCs and CPGCs recovered in this screen showed the anticipated maternal allele-specific methylation in independent DNA sources (from UPD animals or interspecific/intersubspecific hybrids)—no false positives were encountered. This highlights one of the advantages of a screen based on an epigenetic feature such as DNA methylation, in comparison to an expression-based screen, in which the outcome relies more heavily on how well the RNA sources were matched in terms of developmental stages, tissues examined, and cell growth characteristics. Furthermore, an expression-based screen is likely also to identify genes with expression that may be dependent on imprinted genes, in addition to bona fide imprinted genes. Our Me-RDA screen, because it focuses on an epigenetic property, should provide a more reliable estimate of the number of imprinted genetic elements, namely, DMRs, in the genome. The success of our approach in identifying imprinted genes, therefore, depends on these DMRs being associated with imprinted transcription units. In this screen, there was one fragment (DPC#17–65) that arose from a maternally methylated DMR (R. Smith, unpubl.) that could not be associated with a transcript. Further investigation is required to determine whether there really is no transcription at this locus, or whether there is a transcript that has not been detected previously. We found, however, that all other DPCs did indeed map at imprinted transcripts. Therefore, there is no evidence that there are a large number of DMRs scattered throughout the genome that are not associated with genomic imprinting. As DMRs in many cases coincide with imprinting control elements (Yatsuki et al. 2002), the Me-RDA approach, particularly when applied to early embryonic material, should be directed primarily toward such elements. The identification of a number of new DMRs in our screen, in particular, those at nonclustered imprinted genes or microimprinted domains, should help in understanding the general properties of imprinting control elements.
The most important aspect of the Me-RDA screen was that it led to the identification of a number of novel imprinted genes. One of these genes was Slc38a4 on distal Chr 15, which encodes a system A amino acid transporter and was independently identified as an imprinted gene in a screen using microarray comparison of RNAs from Pg and Ag embryos, while this article was in preparation (Mizuno et al. 2002). It is intriguing that Slc38a4 is imprinted in placenta but not in adult liver, both tissues in which it is highly expressed. Imprinting in the placenta would be in line with the conflict theory of the evolution of genomic imprinting (Moore and Haig 1991). Although Slc38a4 is an imprinted gene on distal Chr 15, UPD of this Chr was not found to have a gross phenotypic effect (Cattanach 1982). It is possible that in this early study, effects of imprinted expression in the placenta, such as fetal growth retardation with catch-up during the early postnatal period, would not have been seen. An alternative explanation is that the presumed lack of expression of Slc38a4 in the placenta of mice with maternal UPD of Chr 15 could have been compensated for by other system A amino acid transporters, such as Slc38a1 or Slc38a2.
A novel imprinted transcript (Nap1l5) was isolated from proximal Chr 6. A human homolog of Nap1l5 has been described and is located at 4q22.1 (Harada et al. 2002); comparable to the mouse locus, it is found within an intron of HERC3 containing a CpG island. The protein encoded by Nap1l5 contains a region shared by members of the nucleosome assembly family of proteins. NAPs chaperone histones to the nucleus and position them onto DNA (Ito et al. 1996; Rodriguez et al. 1997), and are involved in transcriptional activation (Asahara et al. 2002) and mitotic events (Altman and Kellogg 1997). Nap1l5 is intron-less and may have arisen through retrotransposition of another gene from the NAP family, the closest being Nap1l2. At least one other gene encoding a NAP-related protein, Nap1l4 on distal Chr 7, is regulated by genomic imprinting (Engemann et al. 2000). It is possible that Nap1l5 does not have the same function as other NAPs, as it is truncated at the C terminus, a region thought to be important for histone chaperoning (Rodriguez et al. 1997), and lacks a nuclear localization signal (Harada et al. 2002).
A third novel imprinted gene identified in this investigation, Peg13, is located on distal Chr 15. This is the second imprinted gene found on this Chr; however, the large distance (14 Mb) between Peg13 and Slc38a4 indicates that they do not form a cluster of imprinted genes. In addition, the human homologs of Slc38a4 (12q12) and KIAA1882 (8q24.3), which flanks Peg13, are located on separate Chrs. Peg13 is intron-less but does not appear to have arisen by retrotransposition of another gene as there are no related sequences elsewhere in the genome. A genomic region homologous to this transcript is found in the rat genome (AC115133), and two ESTs from rat brain (AT005934, BE106640) arise from this locus. A homologous transcript has not been identified in human; however, it is intriguing that the equivalent intron of KIAA1882 (XM_072493) to that which contains Peg13 in the mouse, contains a CpG island and a number of human EST matches (with no homology with Peg13). Peg13 seem to fall within the class of imprinted genes from which RNAs are transcribed that do not code for a protein. However, it differs from those described to date as it is not associated with an imprinted gene that encodes a protein.
It is intriguing that two of the novel imprinted genes identified in this screen, Nap1l5 and Peg13, are located within the introns of other genes, which are biallelically expressed. Such an arrangement has been described as a “microimprinted domain” (Evans et al. 2001). These imprinted genes may be relatively recent acquisitions and, as such, may help us to understand the evolution of imprinting and the nature of imprinting control elements. Previously described imprinted genes in microimprinted domains are U2af1-rs1, in an intron of Murr1 (Nabetani et al. 1997), and Nnat, which is in an intron of Bc10/Blcap (Evans et al. 2001; John et al. 2001). Nap1l5 is similar to U2af1-rs1, as it is likely that both have arisen through retrotransposition (Nabetani et al. 1997). It is possible that retrotransposons may become imprinted through the retrotransposition process itself, as is seen by the insertion of intracisternal A particles at the agouti locus (Morgan et al. 1999), or may carry with them the elements necessary for imprinting, which has been indicated for the Ins1 locus (Giddings et al. 1994). In each case, it is anticipated that imprinting control elements will be found close to the gene itself. Nnat, in contrast, is proposed to have arisen through a gain of function of intronic sequences (Evans et al. 2001); however, it is thought that in this case also, the imprinting control region is located within close proximity of the gene (John et al. 2001). It may be the case that Peg13 has evolved from intronic sequences in the same way that Nnat is proposed to have done. The identification of two additional genes in microimprinted domains in this investigation indicates that they may be more common than previously thought and that there may be a number of imprinted genes that do not reside in imprinting clusters. The study of microimprinted domains such as these will also aid further investigations into the identification of the minimal elements necessary for imprinting.
METHODS
Pg and N Embryo DNAs
Pg embryos were produced by electroactivation, essentially as described previously (Dean et al. 2002). Mature oocytes collected from superovulated F1 (C57BL/6J × CBA/Ca) females were activated and cultured to the eight-cell stage. Each Pg embryo (diploid) was aggregated with two four-cell tetraploid embryos to improve overall development and reduce variability. The tetraploid embryos (F1 × SD7) were genetically distinct from the Pg embryos. Aggregations of Pg and tetraploid embryos were allowed to develop in vitro to the blastocyst stage, at which point they were transferred to pseudopregnant females. Pg embryos were dissected out at day 10 of Pg development, and all extraembryonic material was removed. N embryos were collected from matings of F1 × F1 at a developmental stage to match that of the Pg embryos.
DNA was extracted from individual embryos by proteinase K digestion and phenol-chloroform extraction. To ascertain that the tetraploid material was restricted to the extraembryonic material and had not contributed to the Pg embryo, the embryo was genotyped by using PCR-RFLP assays for the Nap1l4 (5′-GAAGAATTAATGCCCTGAAGC-3′ and 5′-AACTTGTCCTCCTCCTCATTC-3′ primers, digestion with HphI) and H19 (5′-GTGAAGCTGAAAGAACAGATGGTG-3′ and 5′-GTAGGGCATGTTGAACACTTGATG-3′, digestion with BglI; Sasaki et al. 1995) genes on distal Chr 7. To exclude male embryos from the N embryos, they were genotyped by PCR for the Y Chr Sry (5′-GAGAGCATGGAGGGCCAT-3′ and 5′-CCACTCCTCTGTGACACT-3′), using Hprt (5′-GAA ATGTCAGTTGCTGCGTC-3′ and 5′-GCCAACACTGCTGAAA CATG-3′) as an amplification control (Kaneko-Ishino et al. 1995).
Me-RDA
Me-RDA was carried out essentially as described previously (Kelsey et al. 1999; Peters et al. 1999; Smith and Kelsey 2002). Briefly, DNAs from pooled Pg or N embryos were separately digested with the methylation-sensitive restriction enzymes HpaII (C‘CG_G) or Hin6I (G‘CG_C). Adaptor molecules were ligated to the restriction fragments and amplicons produced in a PCR reaction by using a Tricine buffer (30 mM Tricine at pH 8.4, 2 mM MgCl2) with 200 μM each dNTP, 2 μM (HpaII amplicons) or 4 μM (Hin6I amplicons) primer, 0.05 U/μL Taq DNA polymerase; glycerol (10%) was included in the HpaII PCR. Subtractions were carried out using Pg amplicons (40 μg) as the driver and N amplicons (400 ng) as the tester. For the second and third rounds of subtraction, 100-ng and 2-ng difference products, respectively, were used with 40-μg Pg amplicons. Second- and third-round difference products were cloned into PBS (Stratagene), as previously described. The representation of individual DPCs in N and Pg amplicons was analyzed by hybridizing amplicons blots with inserts amplified by PCR using M13 primers and labeled with [α-32P]dCTP (NEN) by random priming.
Library Screening
Second-round difference products from the HpaII and Hin6I Me-RDA, and corresponding Pg amplicons, were hybridized to gridded filters of 138,240 clones from the mouse CpG island library (library number 123 from RZPD; Cross et al. 1997). Clones that hybridized with difference products, but not with Pg amplicons (CPGCs), were selected for further analysis, and inserts were amplified from colonies by PCR using the recommended primers. The RZPD clone numbers were as follows: CPGC#1 with three equivalent clones EDIUp123E036Q4 (CPGC#1), EDIUp123D22218Q4 (CPGC#26), and EDIUp123J06320Q4 (CPGC#44); CPGC#2 with two equivalent clones EDIUp123J0231Q4 (CPGC#2) and EDIUp123M19334Q4 (CPGC#46); CPGC#13 with three equivalent clones EDIUp123J16109Q4 (CPGC#13), EDIUp123N14229Q4 (CPGC#27), and EDIUp123K10243Q4 (CPGC#32); CPGC#17 with two equivalent clones EDIUp123F12122Q4 (CPGC#17) and EDIUp123D21238Q4 (CPGC#30); CPGC#29 with EDIUp123F08235Q4; CPGC#43 with EDIUp123D10315Q4; and CPGC#49 with EDIUp123B18346Q4. Individual clones were analyzed for their representation in Pg and N amplicons by hybridizing blots of amplicons with labeled inserts. DPC#26–7 (M13 PCR product) was used to isolate a cosmid (RZPD clone MPMGc121F05636Q1) from RZPD library number 121, a gridded 129/Ola mouse cosmid library. A PCR product from within the DPC#19–2 sequence (primers DPC#19–2F, 5′-AGCTGCGTTTGAAAGCCTA-3′; DPC#19–2R, 5′-TTAGTGC ACAGCCACCACAC-3′), which was expressed sequence as determined by RT-PCR, was hybridized to 1.5 × 106 clones from a 129-strain mouse neonatal brain cDNA library in λZapII (Stratagene). A selection of positive plaques were excised to pBluescript SK− by in vivo excision (Stratagene), and individual colonies were picked for minipreparation of DNA (QIAGEN) and sequencing.
Sequencing of Constructs and Computational Sequence Analysis
The sequences of DPCs and CPGCs were obtained by sequencing of purified PCR products (QIAGEN) by using the primers used for PCR amplification. Other clones were sequenced from plasmid DNA by using a primer within the vector sequence. All sequencing was done on an ABI377 (Perkin-Elmer). Sequences were aligned using the Gap4 program in Staden (Staden et al. 2000). Database searching was performed by using the BLASTN (Altschul et al. 1997) and FASTA (Pearson 1994) search tools in the Genetics Computer Group (GCG) suite GCG10 (Womble 2000). BLASTN searches were also performed using the Ensembl genome browser (http://www.ensembl.org) and the Celera discovery system (http://www.celera.com). CpG islands were identified by using the CpGplot program in Emboss (Rice et al. 2000).
Methylation Analysis
Tissues from mice with UPD for the region Chr 6 distal to the T77H breakpoint, (MatDp.dist6 and PatDp.dist6) and normal littermates were a kind gift from C. Beechey, Harwell, Oxfordshire, UK (Beechey 2000). RFLPs among M. musculus (C57BL/6), M. musculus castaneus (CAST/Ei), and M. spretus were used to distinguish the parental alleles. DNAs were extracted from T77H MatDp.dist6 and PatDp.dist6 tissues, and from tissues and fetuses (e13) from interspecific hybrids and backcross animals (which were genotyped to ensure that they carried the appropriate alleles at the locus). DNAs were digested with the RFLP enzyme (when appropriate), in combination with the methylation-sensitive restriction enzymes Hin6I or HpaII, or with MspI, a methylation insensitive isoschizomer of HpaII. An additional restriction enzyme was used to reduce the size of fragment analyzed if required. Southern blots of restriction digests were hybridized with the following probes: DPC#24–1/Napll5 region with PCR product amplified from AB041556 sequence (2–1733); DPC#26–7/Slc38a4 region with 4.8-kb BglII fragment from cosmid MPMGc121F05636Q1 containing DPC#26–7 and CPGC#29, both labeled by random priming; and DPC#19–2/Peg13 region with an RNA probe produced by in vitro transcription from linearized DPC#19–2 incorporating [α-32P]UTP.
Expression Analysis
RNAs were prepared from total fetuses (e13), placentae (e13), and adult tissues by using RNEasy kits (QIAGEN); mRNA was extracted from adult brain by using Oligotex Direct kit (QIAGEN). Northern blots were hybridized with RNA probes produced by in vitro transcription from linearized plasmids of DPCs or cloned PCR products (Ruppert et al. 1990). Probes specific to each orientation could be made by choice of RNA polymerase promoter used. For RT-PCR analysis, RNAs were reverse transcribed by using SuperscriptII (Invitrogen) and random hexamers. PCR amplification was performed in a 20 μL reaction using the first-strand cDNA template, HotStar Taq DNA polymerase (QIAGEN). The primers used for RT-PCR were as follows: AW125295-F, 5′-TGACCCAGTGGAGCCTT TAC-3′; AW125295-R, 5′-CCGTCGTCACAAAGAACAGA-3′; AB041556–32F, 5′-TCTATTGCAGACTGCGCAAC-3′; AB041556–327R, 5′-GCTTCTGCAGCTTTTTGAGC-3′; AX046352–762F, 5′-TGCATGGTGTTTTTCGTCAG-3′; AX046352–1378R, 5′-GAAGTACGGATCGGGAACAG-3′; mCT124742-e5F, 5′-TTGTGGAGAGGAGCACACAG-3′; mCT124742-e10R, 5′-TAACTGGGGAATGAGGCAAC-3′; KIAA-RFLP-F, 5′-CTCCTTCCTCTTGCAGACCAT-3′; and KIAA-RFLP-R, 5′-GTGCTGATCTGCAGTCTTGG-3′. PCR products were purified when required by using a PCR purification kit (QIAGEN). The AW125295 RT-PCR product (at Peg13) was digested with KspI, the AB041556 product (at Nap1l5) with BstZI; AX046352 RT-PCR product from the Slc38a4 locus was digested with BseMI, and the product from the transcript flanking DPC#19–2 was digested with BseLI. The ends of Peg13 were determined by 5′ and 3′ RACE performed on 250 ng mRNA by using the GeneRacer kit (Invitrogen), according to the manufacturer’s instructions. The gene-specific primers used for 5′ and 3′ RACE were 5′-GTGCGCCACCAAAAC CATTCAAGTG-3′ and 5′-CACGGCTGCATCTGTTTCCCCAG GAT-3′. PCR amplification of cDNA was performed using a touchdown amplification protocol as recommended, and products were cloned into the pCR4-TOPO vector supplied for analysis and sequencing.
WEB SITE REFERENCES
http://www.celera.com; Celera discovery system.
http://www.ensembl.org; Ensembl genome browser.
http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html; Beechey, C.V., Cattanach, B.M., and Blake, A. 2002. Genetic and physical imprinting map of the mouse. MRC Mammalian Genetics Unit, Harwell, Oxfordshire, UK.
Acknowledgments
Data in this publication was generated through use of the Celera Discovery System and Celera Genomics-associated databases. We thank Colin Beechey for providing the T77H UPD mouse tissues, Jo Peters for providing M. spretus material, and Dorothy Bodle for help in screening the Me-RDA clones. This work was supported by a BBSRC studentship to R.J.S. G.K. is a senior fellow of the MRC.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL gavin.kelsey@bbsrc.ac.uk; FAX 44-1223-496022.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.781503.
REFERENCES
- 1.Altman R. and Kellogg, D. 1997. Control of mitotic events by Nap1 and the Gin4 kinase. J. Cell Biol. 138: 119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asahara H., Tartare-Deckert, S., Nakagawa, T., Ikehara, T., Hirose, F., Hunter, T., Ito, T., and Montminy, M. 2002. Dual roles of p300 in chromatin assembly and transcriptional activation in cooperation with nucleosome assembly protein 1 in vitro. Mol. Cell. Biol. 22: 2974-2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barlow D.P., Stöges, R., Herrmann, B.G., Saito, K., and Schweifer, N. 1991. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349: 84-87. [DOI] [PubMed] [Google Scholar]
- 5.Beechey C.V. 2000. Peg1/Mest locates distal to the currently defined imprinting region on mouse proximal chromosome 6 and identifies a new imprinting region affecting growth. Cytogenet. Cell Genet. 90: 309-314. [DOI] [PubMed] [Google Scholar]
- 6.Cattanach B.M. 1982. Non-disjunction tests with Robertsonian translocations. Mouse News Lett. 66: 62-63. [Google Scholar]
- 7.Choi J.D., Underkoffler, L.A., Collins, J.N., Marchegiani, S.M., Terry, N.A., Beechey, C.V., and Oakey, R.J. 2001. Microarray expression profiling of tissues from mice with uniparental duplications of chromosomes 7 and 11 to identify imprinted genes. Mamm. Genome 12: 758-764. [DOI] [PubMed] [Google Scholar]
- 8.Constância M., Pickard, B., Kelsey, G., and Reik, W. 1998. Imprinting mechanisms. Genome Res. 8: 881-900. [DOI] [PubMed] [Google Scholar]
- 9.Cross S.H., Lee, M., Clark, V.H., Craig, J.M., Bird, A.P., and Bickmore, W.A. 1997. The chromosomal distribution of CpG islands in the mouse: Evidence for genome scrambling in the rodent lineage. Genomics 40: 454-461. [DOI] [PubMed] [Google Scholar]
- 10.Dean W.L., Kelsey, G., and Reik, W. 2002. Generation of monoparental enbryos for investigation into genomic imprinting. In Genomic imprinting: Methods and protocols (ed. A. Ward), pp. 1–19. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 11.Engemann S., Strodicke, M., Paulsen, M., Franck, O., Reinhardt, R., Lane, N., Reik, W., and Walter, J. 2000. Sequence and functional comparison in the Beckwith-Wiedemann region: Implications for a novel imprinting centre and extended imprinting. Hum. Mol. Genet. 9: 2691-2706. [DOI] [PubMed] [Google Scholar]
- 12.Evans H.K., Wylie, A.A., Murphy, S.K., and Jirtle, R.L. 2001. The Neuronatin gene resides in a “micro-imprinted” domain on human chromosome 20q11.2. Genomics 77: 99-104. [DOI] [PubMed] [Google Scholar]
- 13.Giddings S.J., King, C.D., Harman, K.W., Flood, J.F., and Carnaghi, L.R. 1994. Allele-specific inactivation of insulin 1 and 2, in the mouse yolk sac, indicates imprinting. Nat. Genet. 6: 310-313. [DOI] [PubMed] [Google Scholar]
- 14.Hagiwara Y., Hirai, M., Nishiyama, K., Kanazawa, I., Ueda, T., Sakaki, Y., and Ito, T. 1997. Screening for imprinted genes by allelic message display: Identification of a paternally expressed gene Impact on mouse chromosome 18. Proc. Natl. Acad. Sci. 94: 9249-9254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada H., Nagai, H., Ezura, Y., Yokota, T., Ohsawa, I., Yamaguchi, K., Ohue, C., Tsuneizumi, M., Mikami, I., Terada, Y., et al. 2002. Down-regulation of a novel gene, DRLM, in human liver malignancy from 4q22 that encodes a NAP-like protein. Gene 296: 171-177. [DOI] [PubMed] [Google Scholar]
- 16.Hatanaka T., Huang, W., Ling, R., Prasad, P.D., Sugawara, M., Leibach, F.H., and Ganapathy, V. 2001. Evidence for the transport of neutral as well as cationic amino acids by ATA3, a novel and liver-specific subtype of amino acid transport system A. Biochim. Biophys. Acta 1510: 10-17. [DOI] [PubMed] [Google Scholar]
- 17.Hayashizaki Y., Shibata, H., Hirotsune, S., Sugino, H., Okazaki, Y., Sasaki, N., Hirose, K., Imoto, H., Okuizumi, H., Muramatsu, M., et al. 1994. Identification of an imprinted U2af binding protein related sequence on mouse chromosome 11 using the RLGS method. Nat. Genet. 6: 33-40. [DOI] [PubMed] [Google Scholar]
- 18.Hayward B.E., Kamiya, M., Strain, L., Moran, V., Campbell, R., Hayashizaki, Y., and Bonthron, D.T. 1998. The human GNAS1 gene is imprinted and encodes distinct paternally and biallelically expressed G proteins. Proc. Natl. Acad. Sci. 95: 10038-10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito T., Bulger, M., Kobayashi, R., and Kadonaga, J.T. 1996. Drosophila NAP-1 is a core histone chaperone that functions in ATP-facilitated assembly of regularly spaced nucleosomal arrays. Mol. Cell. Biol. 16: 3112-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.John R.M., Aparicio, S.A., Ainscough, J.F., Arney, K.L., Khosla, S., Hawker, K., Hilton, K.J., Barton, S.C., and Surani, M.A. 2001. Imprinted expression of Neuronatin from modified BAC transgenes reveals regulation by distinct and distant enhancers. Dev. Biol. 236: 387-399. [DOI] [PubMed] [Google Scholar]
- 21.Kamiya M., Judson, H., Okazaki, Y., Kusakabe, M., Muramatsu, M., Takada, S., Takagi, N., Arima, T., Wake, N., Kamimura, K., et al. 2000. The cell cycle control gene ZAC/PLAGL1 is imprinted: A strong candidate gene for transient neonatal diabetes. Hum. Mol. Genet. 9: 453-460. [DOI] [PubMed] [Google Scholar]
- 22.Kaneko-Ishino T., Kuroiwa, Y., Miyoshi, N., Kohda, T., Suzuki, R., Yokoyama, M., Viville, S., Barton, S.C., Ishino, F., and Surani, M.A. 1995. Peg1/Mest imprinted gene on chromosome 6 identified by cDNA subtraction hybridization. Nat. Genet. 11: 52-59. [DOI] [PubMed] [Google Scholar]
- 23.Kelsey G., Bodle, D., Miller, H.J., Beechey, C.V., Coombes, C., Peters, J., and Williamson, C.M. 1999. Identification of imprinted loci by methylation-sensitive representational difference analysis: Application to mouse distal chromosome 2. Genomics 62: 129-138. [DOI] [PubMed] [Google Scholar]
- 24.Kikyo N., Williamson, C.M., John, R.M., Barton, S.C., Beechey, C.V., Ball, S.T., Cattanach, B.M., Surani, M.A., and Peters, J. 1997. Genetic and functional analysis of neuronatin in mice with maternal or paternal duplication of distal Chr 2. Dev. Biol. 190: 66-77. [DOI] [PubMed] [Google Scholar]
- 25.Miyoshi N., Kuroiwa, Y., Kohda, T., Shitara, H., Yonekawa, H., Kawabe, T., Hasegawa, H., Barton, S.C., Surani, M.A., Kaneko-Ishino, T., et al. 1998. Identification of the Meg1/Grb10 imprinted gene on mouse proximal chromosome 11, a candidate for the Silver-Russell syndrome gene. Proc. Natl. Acad. Sci. 95: 1102-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizuno Y., Sotomaru, Y., Katsuzawa, Y., Kono, T., Meguro, M., Oshimura, M., Kawai, J., Tomaru, Y., Kiyosawa, H., Nikaido, I., et al. 2002. Asb4, Ata3, and Dcn are novel imprinted genes identified by high-throughput screening using RIKEN cDNA microarray. Biochem. Biophys. Res. Commun. 290: 1499-1505. [DOI] [PubMed] [Google Scholar]
- 27.Moore T. and Haig, D. 1991. Genomic imprinting in mammalian development: A parental tug-of-war. Trends Genet. 7: 45-49. [DOI] [PubMed] [Google Scholar]
- 28.Morgan H.D., Sutherland, H.G., Martin, D.I., and Whitelaw, E. 1999. Epigenetic inheritance at the agouti locus in the mouse. Nat. Genet. 23: 314-318. [DOI] [PubMed] [Google Scholar]
- 29.Morison I.M., Paton, C.J., and Cleverley, S.D. 2001. The imprinted gene and parent-of-origin effect database. Nucleic Acids Res. 29: 275-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nabetani A., Hatada, I., Morisaki, H., Oshimura, M., and Mukai, T. 1997. Mouse U2af1-rs1 is a neomorphic imprinted gene. Mol. Cell. Biol. 17: 789-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Onyango P., Miller, W., Lehoczky, J., Leung, C.T., Birren, B., Wheelan, S., Dewar, K., and Feinberg, A.P. 2000. Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res. 10: 1697-1710. [DOI] [PubMed] [Google Scholar]
- 32.Paulsen M., El-Maarri, O., Engemann, S., Strodicke, M., Franck, O., Davies, K., Reinhardt, R., Reik, W., and Walter, J. 2000. Sequence conservation and variability of imprinting in the Beckwith-Wiedemann syndrome gene cluster in human and mouse. Hum. Mol. Genet. 9: 1829-1841. [DOI] [PubMed] [Google Scholar]
- 33.Pearson W.R. 1994. Using the FASTA program to search protein and DNA sequence databases. Methods Mol. Biol. 24: 307-331. [DOI] [PubMed] [Google Scholar]
- 34.Peters J., Wroe, S.F., Wells, C.A., Miller, H.J., Bodle, D., Beechey, C.V., Williamson, C.M., and Kelsey, G. 1999. A cluster of oppositely imprinted transcripts at the Gnas locus in the distal imprinting region of mouse chromosome 2. Proc. Natl. Acad. Sci. 96: 3830-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piras G., El Kharroubi, A., Kozlov, S., Escalante-Alcalde, D., Hernandez, L., Copeland, N.G., Gilbert, D.J., Jenkins, N.A., and Stewart, C.L. 2000. Zac1 (Lot1), a potential tumor suppressor gene, and the gene for epsilon-sarcoglycan are maternally imprinted genes: Identification by a subtractive screen of novel uniparental fibroblast lines. Mol. Cell. Biol. 20: 3308-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Plass C., Shibata, H., Kalcheva, I., Mullins, L., Kotelevtseva, N., Mullins, J., Kato, R., Sasaki, H., Hirotsune, S., Okazaki, Y., et al. 1996. Identification of Grf1 on mouse chromosome 9 as an imprinted gene by RLGS-M. Nat. Genet. 14: 106-109. [DOI] [PubMed] [Google Scholar]
- 37.Rice P., Longden, I., and Bleasby, A. 2000. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16: 276-277. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez P., Munroe, D., Prawitt, D., Chu, L.L., Bric, E., Kim, J., Reid, L.H., Davies, C., Nakagama, H., Loebbert, R., et al. 1997. Functional characterization of human nucleosome assembly protein-2 (NAP1L4) suggests a role as a histone chaperone. Genomics 44: 253-265. [DOI] [PubMed] [Google Scholar]
- 39.Ruppert S., Boshart, M., Bosch, F.X., Schmid, W., Fournier, R.E., and Schutz, G. 1990. Two genetically defined trans-acting loci coordinately regulate overlapping sets of liver-specific genes. Cell 61: 895-904. [DOI] [PubMed] [Google Scholar]
- 40.Sasaki H., Ferguson-Smith, A.C., Shum, A.S., Barton, S.C., and Surani, M.A. 1995. Temporal and spatial regulation of H19 imprinting in normal and uniparental mouse embryos. Development 121: 4195-4202. [DOI] [PubMed] [Google Scholar]
- 41.Smith R.J. and Kelsey, G. 2002. Identification of imprinted loci by methylation: Use of methylation-sensitive representational difference analysis (Me-RDA). In Genomic imprinting: Methods and protocols (ed. A. Ward), pp. 113–132. Humana Press, Totowa, NJ. [DOI] [PubMed]
- 42.Smith R.J., Arnaud, P., Konfortova, G., Dean, W.L., Beechey, C.V., and Kelsey, G. 2002. The mouse Zac1 locus: Basis for imprinting and comparison with human ZAC. Gene 292: 101-112. [DOI] [PubMed] [Google Scholar]
- 43.Staden R., Beal, K.F., and Bonfield, J.K. 2000. The Staden package, 1998. Methods Mol. Biol. 132: 115-130. [DOI] [PubMed] [Google Scholar]
- 44.Sugawara M., Nakanishi, T., Fei, Y.J., Martindale, R.G., Ganapathy, M.E., Leibach, F.H., and Ganapathy, V. 2000. Structure and function of ATA3, a new subtype of amino acid transport system A, primarily expressed in the liver and skeletal muscle. Biochim. Biophys. Acta 1509: 7-13. [DOI] [PubMed] [Google Scholar]
- 45.Womble D.D. 2000. GCG: The Wisconsin Package of sequence analysis programs. Methods Mol. Biol. 132: 3-22. [DOI] [PubMed] [Google Scholar]
- 46.Wutz A., Smrzka, O.W., Schweifer, N., Schellander, K., Wagner, E.F., and Barlow, D.P. 1997. Imprinted expression of the Igf2r gene depends on an intronic CpG island. Nature 389: 745-749. [DOI] [PubMed] [Google Scholar]
- 47.Yatsuki H., Joh, K., Higashimoto, K., Soejima, H., Arai, Y., Wang, Y., Hatada, H., Obata, Y., Morisaki, H., Zhang, Z., et al. 2002. Domain regulation of imprinting cluster in Kip2/Lit1 subdomain on mouse chromosome 7F4/F5: Large-scale DNA methylation analysis reveals that DMR-Lit1 is a putative imprinting control region. Genome Res. 12: 1860-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zwart R., Sleutels, F., Wutz, A., Schinkel, A.H., and Barlow, D.P. 2001. Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes & Dev. 15: 2361-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]