Abstract
Localization of mRNAs to subcellular domains can enrich proteins at sites where they function. Coordination with translational control can ensure that the encoded proteins will not appear elsewhere, an important property for factors that control cell fate or body patterning. Here I focus on two aspects of mRNA localization. One is the question of how mRNAs that undergo directed transport by a shared mechanism are bound to the transport machinery, and why localization signals from these mRNAs have very diverse sequences. The second topic concerns the role of particles, in which localized mRNAs often appear. Recent evidence highlights the importance of such assemblies, and the possibility that close association of mRNAs confers community effects and a novel form of regulation.
Introduction
Many mRNAs are localized to specific regions within the cell. The widespread nature of this phenomenon was most dramatically revealed by examination of thousands of Drosophila embryonic mRNAs, with over 70% non-randomly distributed in dozens of different patterns [1]. A complementary approach is to identify mRNAs associated with a specific subcellular structure, again providing many localized mRNAs [2].
One rationale for mRNA localization is to efficiently target the encoded protein to a subcelluar site or region. Many mRNAs will spend most of their lives at the site of localization, and efficient protein localization can be obtained without any need for coordinated translational control. However, translational regulation significantly expands the utility of mRNA localization, and is particularly important in two types of situations.
The first is where inclusion and exclusion are equally important: the protein must be provided in a particular location, but it must also be excluded from other locations. This feature is crucial for patterning or cell fate determinants. Such proteins direct a particular path of development, and their presence at inappropriate locations can be lethal [3–6]. Repression of translation during the localization process, as well as repression of any mRNA that escapes localization, prevents protein from accumulating at inappropriate sites.
A second general use of combining translational regulation with mRNA localization is to allow regulation at the site of localization: translation is only activated upon proper stimulation, and localization alone is not sufficient. This form of regulation is common in neurons, extremely large cells in which regional changes in protein expression appear to be important for axon guidance and synaptic plasticity [7–10].
There have been numerous recent advances in our understanding of localization mechanisms and their coordination with translation, too many to cover in this short review. These include numerous examples of live imaging and high resolution microscopy to address a range of topics: characterization of an early step in localization, nucleocytoplasmic transport [11]; defining the roles of different motors in driving long range movements in the Drosophila oocyte [12]; and establishing proximity of protein and RNA molecules to obtain mechanistic insights [13, 14]. Here I focus on how mRNAs are associated with the localization machinery, a process that may offer special challenges when the mode of localization is directed transport (Fig. 1), and how association of localized mRNAs with each other can influence their expression.
Figure 1. Challenges in RNA recognition for mRNA localization.
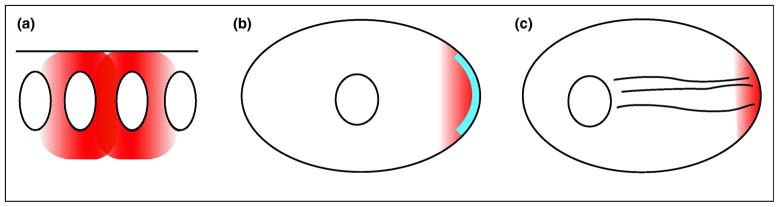
Mechanisms for mRNA localization are shown schematically. A, local transcription in multinucleate cells. A portion of a syncytial blastoderm embryo with four of its nuclei is shown. Transcripts (grey) made only in the central two nuclei will be concentrated around those nuclei. B, diffusion and trapping, and degradation/local stabilization. Although the mechanisms are different, both rely on a localized component: a stabilization protein or an anchoring protein, shown as a thick black line. In either case, an enormous number of possible localization patterns are possible. C, directed transport. A cytoskeletal element (microtubules as wavy lines) serves as the tracks along which motors carry cargo mRNAs. A single mechanism could localize many different mRNAs, but the number of possible destinations would be very limited unless different cargo adaptors recognize different populations of microtubules. The work discussed in this review shows how one class of localization element is recognized for directed transport.
Linking localized mRNAs to transport machinery
Cis-acting signals within mRNAs direct their localization. For the most part, the localization signals appear to serve as sites for recognition by RNA binding proteins, although RNA/RNA interactions can also make contributions. The roles played by RNA binding proteins differ according to the mechanism of mRNA localization: options include transcript stabilization, decay, anchoring, or association with transport machinery (Fig. 1). Although movement of mRNAs along cytoskeletal elements has been studied extensively, our understanding of the interactions that link an mRNA to a molecular motor – a key component of specificity – is limited. Previously, the only well defined example was for ASH1 mRNA, with an RNA binding protein (She2p) and a myosin cargo adaptor protein (She3p) linking the mRNA to a myosin (Myo4p) [15–18]. Now, a new paradigm for linkage of an mRNA to a motor has emerged. The work is notable in part because it explains how a large class of structurally related localization signals function, and also for new insights into RNA structures and how they are likely to be recognized by proteins.
During oogenesis in Drosophila many mRNAs are synthesized in nurse cells, and then transported via ring canals into the oocyte. One such mRNA is fs(1)K10. Characterization of the Transport and Localization Signal (TLS) of this mRNA provided the first example of a family of localization signals with very similar activities but very diverse sequences [19–23]. Some of the other family members come from mRNAs expressed not the ovary, but in the embryo where they become localized apically. Independent of their origins, signals of this family prove to work well in either setting, directing transport to the oocyte in the ovary and to apical regions of the embryo [24]. The TLS-like signals mediate movement towards the minus ends of microtubules, a process that requires the Bicaudal-D (Bic-D) and Egalitarian (Egl) proteins, which associate with the dynein motor complex [24–27]. Neither Bic-D nor Egl has any of the known RNA binding motifs, implying that recognition of the localization signals relied on other, unidentified, proteins. Furthermore, the lack of any highly conserved sequence elements among the TLS-like signals suggested that multiple recognition factors might be needed. Two recent papers now reveal the identity of the RNA binding protein, define the interactions that link the localization signals to the dynein motor complex, and provide an appealing explanation for how recognition elements are imbedded in the signals with such diverse primary sequences.
Using an affinity tag for capture of embryonic factors bound to the TLS, Dienstbier et al.[28] found that only two proteins – Bic-D and Egl – were consistently recovered and had the appropriate specificity. While purification of these proteins might be expected, since they should be part of the localization complex, the absence of other proteins that would provide the direct link to the TLS was surprising. However, Egl was shown to bind with specificity to functional forms of the TLS and several other TLS-like signals, while Bic-D lacks this activity. Thus, the case is very strong that Egl is the missing recognition factor.
A large part of Egl is required for RNA binding (~80% of the 1004 aa protein), including an RNaseD exonuclease homology domain. Mutation of putative catalytic amino acids within this domain has no effect on Egl function in vivo or RNA binding activity in vitro [29] [28]. Specificity of Egl binding is not high (affinity for the TLS is about three times higher than for nonlocalizing mutant RNAs), a feature that might facilitate binding to diverse sites, but possibly at the expense of relatively promiscuous localization activity. This may well be the case, as many mRNAs are transported to the oocyte or localized apically in the embryo [1, 30]. Moreover, increasing the concentration of Egl in the embryo promotes apical localization of mRNAs that would not normally localize [31].
The work of Dienstbier et al. not only identifies the missing RNA recognition factor, it also builds on previous observations about Bic-D and Egl to provide a model for assembly and activation of a transport particle. In addition to its role in mRNA localization, Bic-D acts in dynein-dependent transport of Golgi vesicles and lipid droplets [32, 33]. For this type of transport, Bic-D is linked via its C terminal domain (CTD) to the cargo by Rab6, a membrane associated GTPase [32]. Egl and Rab6 appear to act as interchangeable adaptors, with each serving three functions: activation of Bic-D by disrupting an autoinhibitory effect; recruiting the cargo; and binding to dynein light chain to augment the Bic-D-mediated interaction with the dynein motor complex [26–28, 34–36](Fig. 2).
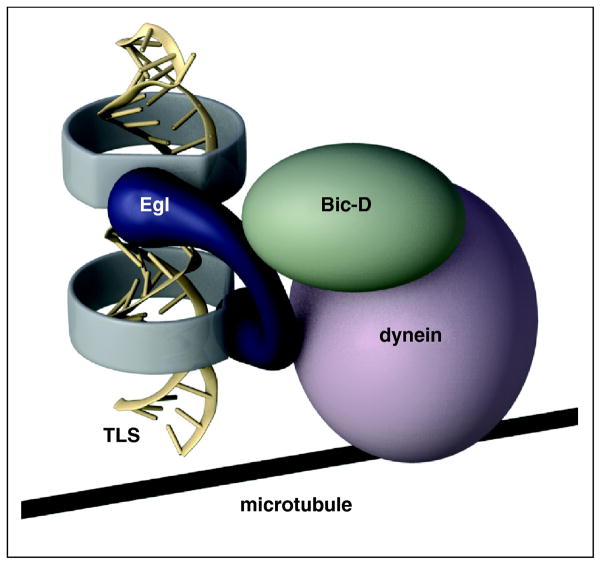
Figure 2. TLS structure and recruitment to the dynein motor.
The TLS consists of an extended helical region in A' form, with two subdomains in which the major groove (darkest shading) are 90° out of register with one another. Egl binds the TLS, presumably recognizing the offset A' helices, as well as Bic-D. Egl activates Bic-D, and Bic-D and Egl contact components of the dynein motor complex, allowing transport along microtubules.
A major remaining puzzle for TLS mediated localization is how Egl can recognize a variety of seemingly very diverse RNA signals. A key part of the answer has come from the structure of the other partner in the reaction, the RNA. The TLS has the potential to adopt a stem-loop structure, with the stem consisting primarily of A:U base pairs. Mutational analysis of the TLS highlighted the importance of basepairing within the stem. By contrast, elimination of unpaired nucleotides within the stem or mutation of the loop did not disrupt activity [19, 37]. Thus, it seemed that features of a double-stranded segment of RNA were being recognized. The problems with this model are twofold. First, although other members of the family of TLS-like signals also have predicted helical regions (and a portion of the orb signal is identical in sequence), most have little sequence homology. Second, double-stranded RNA typically adopts an “A-form” conformation, in which the major groove is not accessible to ligands. In DNA it is the major groove that provides numerous base-specific contacts for sequence-specific binding. In the absence of such interactions it was difficult to see how specific binding would be obtained.
Structural analysis of the TLS by NMR now provides the answers [38]. The key elements of the TLS are indeed helices, but with a twist. Unexpectedly, the RNA adopts the “A'-form” conformation. There are two subdomains within an extended helical region, with the major groove of each positioned on different faces of the helix. A beautiful series of experiments, making use of the traditional combination of directed mutagenesis and functional assays, addresses two questions: what makes the TLS helix adopt the A' form? and which of the structural features of the TLS are important for function? What makes the work so remarkable, and the answers so compelling, is that design of the mutants is guided by the actual structure, and not a predicted secondary structure. Moreover, structures are obtained for a subset of the mutants, confirming the nature of the structural changes.
Adoption of the A' form, rather than the predicted A form, is due to stacking interactions of multiple purines, with 5 stacked purines in the lower helix and 7 in the upper helix. This feature nicely illustrates the difficulties in recognizing shared properties of localization signals with diverse sequences, since stacking does not depend on a specific sequence, and not all stacked purines derive from the same strand of the RNA. Nevertheless, predicted stem loops from other minus end-directed signals contain multiple stretches of three or more contiguous purines, and thus may have structures similar to the TLS.
Additional mutagenesis and structural determinations show that the key features of the TLS are the two A' form helical regions, and these must be oriented on different faces of the extended helix (Fig. 2). Conversion of either upper or lower helix to A form reduces activity, while the double mutant has no activity. Similarly, changing the orientation of the helices disrupts activity. Notably, it is the structure that is important, not the sequence, as a heterologous A' form helix of unrelated sequence was active in the transport assay. Thus, although the A' form does have an accessible major groove, that feature is not used for discrimination between sequences whose structures are similar. The overall conclusion is that the localization machinery - presumably Egl, at least - primarily recognizes an uncommon structure, rather than searching for a particular set of contacts within a common structure (as for recognition of sequences in double helical DNA).
Linking localized mRNAs to one another
Individual mRNAs exist as members of a community within the cell. Different degrees of proximity are possible, with some mRNAs being concentrated in microscopic ribonucleoprotein particles (RNPs) [7, 39] [40]. That particles might be a vehicle for mRNA transport was initially suggested by experiments involving injection of fluorescently labeled RNAs into cultured oligodendrocytes [41]. Since then many more examples of transport particles have been described. Whether localizing mRNAs congregate by convenience or by necessity is often not known. One example where intimate association of mRNAs is essential is the Drosophila bicoid mRNA: dimerization is required for localization in the embryo [21, 42]. Recent work demonstrates that a protein required for both localization and translational repression of Drosophila oskar (osk) mRNA acts to assemble the mRNA in particles. This assembly appears to underlie an unexpected feature of translational regulation.
Polypyrimidine tract binding protein (PTB) is well known for its role in pre-mRNA splicing [43]. In that context PTB appears to regulate alternate splicing by interacting, via its four RRM RNA binding domains, with different regions of a single pre-mRNA [44]. Recently, Besse et al.[45] characterized Drosophila mutants lacking PTB activity. Localization of osk mRNA was delayed, and translational repression, which normally serves to prevent expression of Osk protein from mRNA not yet localized, was impaired.
Although Drosophila PTB is expected to perform the same splicing function as its mammalian counterpart, that is not its role in osk regulation. Instead, removing PTB activity reduces the size of osk RNP particles in the oocyte. Notably, loss of PTB also dramatically reduces the efficiency of piggybacking, the phenomenon in which osk mRNA can confer localization on another mRNA containing the osk 3' UTR but not itself able to localize [46]. These effects are readily explained by how purified PTB binds to osk mRNA in vitro: there are multiple sites dispersed throughout the osk 3' UTR (as well as a 5' region), and PTB assembles the RNA into large aggregates. Thus, a primary function of PTB is in the assembly of large osk RNPs.
Whether this assembly indirectly influences osk mRNA localization and translational repression, or if PTB acts more directly, remains uncertain. Besse et al suggest that PTB-dependent assembly of osk into densely packed particles renders it inaccessible to the translational machinery, much as previously hypothesized for Bruno-dependent repression [47] [although Besse et al. report that Bruno binding sites are neither required nor sufficient for piggybacking, and thus appear not to oligomerize osk mRNA in vivo].
An unexpected feature of osk translational repression was discovered by Reveal et al.[48]. In the course of characterizing Bruno (Bru) binding sites (BREs), which prove to be required for translational activation, as well as repression, they made use of osk transgenes with mutated BREs. Remarkably, the regulatory defects could be largely suppressed if an endogenous osk mRNA with wild type Bru binding sites (but unable to make Osk protein) was also present. The only simple explanation of this phenomenon is that translational regulation can be conferred in trans, and is facilitated by the PTB-dependent assembly of osk mRNA into particles. To prevent promiscuous trans effects, the particles would need to exclude other mRNAs. Consistent with this notion, osk mRNA is sequestered in subdomains of sponge bodies, a larger class of RNP that harbors many different mRNAs and regulatory factors [49].
How could regulation act in trans? A likely explanation builds on a persuasive model for translational repression of osk [50](Fig. 3). In this model Bru bound to osk mRNA 3' UTR recruits Cup, which in turn binds translation initiation factor eIF4E. Cup binding does not interfere with the ability of eIF4E to bind the cap at the 5' end of the mRNA, but does prevent binding to eIF4G. Because the eIF4E/eIF4G interaction is essential for cap-dependent initiation of translation, translation is repressed. A weak point of this model is that the Bru/Cup/eIF4E complex would need to bind to the 3' UTR and 5' cap of the same RNA molecule for regulation to be specific. However, in the context of an RNP highly and selectively enriched in osk mRNA, the complex could span two RNAs and still provide the observed specificity. By the same reasoning, an osk mRNA lacking BREs could be regulated in trans by wild type osk mRNAs in the RNP (Fig. 3).
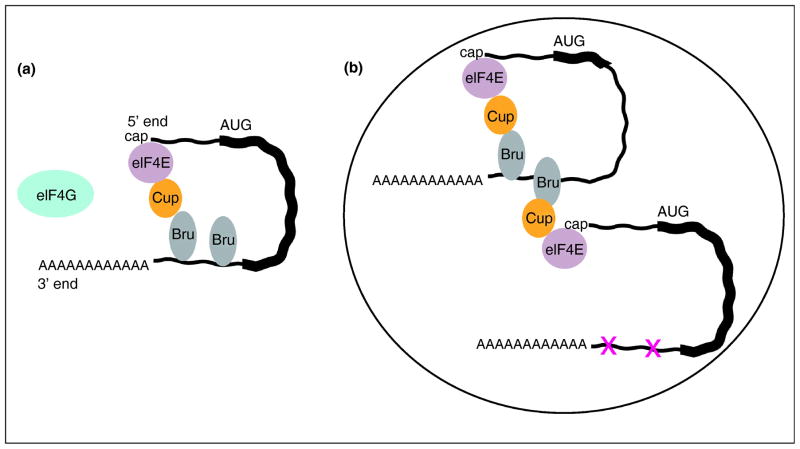
Figure 3. Model for regulation of osk mRNA in trans.
A, model for translational repression by Bru [50]. Bru bound to multiple sites in the 3'UTR recruits Cup, which binds to and inactivates the cap binding protein eIF4E. Specificity requires that the inactivated eIF4E is that bound to the cap.
B, model for how an osk mRNA unable to make Osk protein (the abbreviated coding reigon is shown as a shortened think line) could confer repression on another osk mRNA with mutated Bru binding sites (shown as X's in the thin line of the 3' UTR). In the context of a particle (the encompassing large oval) assembled by PTB, close proximity of the two mRNAs would allow the Cup recruited to the first mRNA to inactive the eIF4E bound to the cap of the second mRNA despite its lack of Bru binding sites.
An attractive feature of trans regulation is that it could function in quality control, increasing the fraction of transcripts that are properly regulated, including any that escape binding by regulatory factors. Such a mechanism would be advantageous for the class of localized mRNAs whose products are damaging if expressed at inappropriate locations. This model is consistent with the possibility that PTB acts only indirectly in regulation of osk mRNA, with its assembly of osk mRNA into particles enhancing community effects and trans regulation. The modest defects in mRNA localization and translational repression (much less severe than when Bru binding sites are mutated) could reflect a less than perfect efficiency of these processes in the absence of trans regulation.
Conclusions
Identification of Egl as a factor that links the TLS localization signal (and probably others) to the dynein motor complex, and determination of the structure of one of these signals, fill a major gap in our understanding of mRNA localization mechanisms. We can eagerly await the next chapter in this story, a description of the structure of Egl and how it binds the TLS. Despite the clear importance of Egl, it seems likely that accessory proteins will also contribute to recognition, perhaps to allow for different final destinations (Fig. 1 legend) or in ensuring that the structure adopted by the isolated element in vitro will be properly folded in its natural setting of the mRNA and in the far more complex environment of the cell. It is in that environment where many RNAs are concentrated in particles. Future studies should reveal if community effects are common when RNAs are in close association, and if this helps ensure that regulation is efficient, especially for mRNAs whose translational repression prior to localization is essential.
Highlights.
Egl is the elusive factor for recognition of the TLS mRNA localization signal.
The TLS structure consists of two A' form RNA double helices.
PTB regulates oskar mRNA and directs its assembly into large particles.
oskar mRNA is regulated in trans, which may be facilitated by assembly in particles.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1**.Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. The first large scale analysis of mRNA localization. With the discovery that the majority of mRNAs tested display some form of localization, this form of regulation can no longer be considered as a peculiar feature only prominent in germline cells and the nervous system. [DOI] [PubMed] [Google Scholar]
- 2.Lécuyer E, Yoshida H, Krause HM. Global implications of mRNA localization pathways in cellular organization. Curr Opin Cell Biol. 2009;21:409–415. doi: 10.1016/j.ceb.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 3.Paquin N, Chartrand P. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 2008;18:105–111. doi: 10.1016/j.tcb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 4.Johnstone O, Lasko P. Translational regulation and RNA localization in Drosophila oocytes and embryos. Annu Rev Genet. 2001;35:365–406. doi: 10.1146/annurev.genet.35.102401.090756. [DOI] [PubMed] [Google Scholar]
- 5.King ML, Messitt TJ, Mowry KL. Putting RNAs in the right place at the right time: RNA localization in the frog oocyte. Biol Cell. 2005;97:19–33. doi: 10.1042/BC20040067. [DOI] [PubMed] [Google Scholar]
- 6.St Johnston D. Moving messages: the intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 7.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 8.Martin KC, Zukin RS. RNA trafficking and local protein synthesis in dendrites: an overview. J Neurosci. 2006;26:7131–7134. doi: 10.1523/JNEUROSCI.1801-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin AC, Holt CE. Local translation and directional steering in axons. EMBO J. 2007;26:3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11**.Grünwald D, Singer RH. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. This paper highlights the power of high resolution imaging approaches. The authors track nucleocytoplasmic transport of mRNA and propose a model that incorporates docking, transport and release. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell. 2008;134:843–853. doi: 10.1016/j.cell.2008.06.053. Particles containing osk mRNA are tracked in living oocytes. The particles move in both directions along microtubules, but with a posterior bias. The net effect is posterior localization of osk mRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13*.Delanoue R, Herpers B, Soetaert J, Davis I, Rabouille C. Drosophila Squid/hnRNP helps Dynein switch from a gurken mRNA transport motor to an ultrastructural static anchor in sponge bodies. Dev Cell. 2007;13:523–538. doi: 10.1016/j.devcel.2007.08.022. The authors use ultrastructural analysis to distinguish between possible roles for localization factors. The localized transcripts are shown to be in particles. [DOI] [PubMed] [Google Scholar]
- 14*.Weil TT, Xanthakis D, Parton R, Dobbie I, Rabouille C, Gavis ER, Davis I. Distinguishing direct from indirect roles for bicoid mRNA localization factors. Development. 2010;137:169–176. doi: 10.1242/dev.044867. High resolution live imaging is used to distinguish between alternate roles for localization factors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bohl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gonsalvez GB, Urbinati CR, Long RM. RNA localization in yeast: moving towards a mechanism. Biol Cell. 2005;97:75–86. doi: 10.1042/BC20040066. [DOI] [PubMed] [Google Scholar]
- 17.Landers SM, Gallas MR, Little J, Long RM. She3p possesses a novel activity required for ASH1 mRNA localization in Saccharomyces cerevisiae. Eukaryotic Cell. 2009;8:1072–1083. doi: 10.1128/EC.00084-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heuck A, Fetka I, Brewer D, Huls D, Munson M, Jansen R, Niessing D. The structure of the Myo4p globular tail and its function in ASH1 mRNA localization. J Cell Biol. 2010;189:497–510. doi: 10.1083/jcb.201002076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serano TL, Cohen RS. A small predicted stem-loop structure mediates oocyte localization of Drosophila K10 mRNA. Development. 1995;121:3809–3818. doi: 10.1242/dev.121.11.3809. [DOI] [PubMed] [Google Scholar]
- 20.Bullock SL, Zicha D, Ish-Horowicz D. The Drosophila hairy RNA localization signal modulates the kinetics of cytoplasmic mRNA transport. EMBO J. 2003;22:2484–2494. doi: 10.1093/emboj/cdg230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snee MJ, Arn EA, Bullock SL, Macdonald PM. Recognition of the bcd mRNA localization signal in Drosophila embryos and ovaries. Mol Cell Biol. 2005;25:1501–1510. doi: 10.1128/MCB.25.4.1501-1510.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van De Bor V, Hartswood E, Jones C, Finnegan D, Davis I. gurken and the I factor retrotransposon RNAs share common localization signals and machinery. Dev Cell. 2005;9:51–62. doi: 10.1016/j.devcel.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 23.dos Santos G, Simmonds AJ, Krause HM. A stem-loop structure in the wingless transcript defines a consensus motif for apical RNA transport. Development. 2008;135:133–143. doi: 10.1242/dev.014068. [DOI] [PubMed] [Google Scholar]
- 24*.Bullock SL, Ish-Horowicz D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414:611–616. doi: 10.1038/414611a. Evidence that TLS-like localization signals support different programs of mRNA localization in different tissues. [DOI] [PubMed] [Google Scholar]
- 25.Wilkie GS, Davis I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]
- 26.Hoogenraad CC, Akhmanova A, Howell SA, Dortland BR, De Zeeuw CI, Willemsen R, Visser P, Grosveld F, Galjart N. Mammalian Golgi-associated Bicaudal-D2 functions in the dynein-dynactin pathway by interacting with these complexes. EMBO J. 2001;20:4041–4054. doi: 10.1093/emboj/20.15.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol. 2004;6:427–435. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- 28**.Dienstbier M, Boehl F, Li X, Bullock S. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev. 2009;23:1546–1558. doi: 10.1101/gad.531009. This work provides a new paradigm for association of RNA cargoes with cytoskeletal motors. Egl is identified as the RNA binding protein that recognizes the TLS localization signal for dynein-directed transport. Notably, there are striking parallels between transport of RNA cargoes and Golgi vesicles, with Egl and Rab6 competing for binding to Bic-D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mach JM, Lehmann R. An Egalitarian-BicaudalD complex is essential for oocyte specification and axis determination in Drosophila. Genes & Dev. 1997;11:423–435. doi: 10.1101/gad.11.4.423. [DOI] [PubMed] [Google Scholar]
- 30.Dubowy J, Macdonald PM. Localization of mRNAs to the oocyte is common in Drosophila ovaries. Mech Dev. 1998;70:193–195. doi: 10.1016/s0925-4773(97)00185-8. [DOI] [PubMed] [Google Scholar]
- 31.Bullock SL, Nicol A, Gross SP, Zicha D. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr Biol. 2006;16:1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 32.Matanis T, Akhmanova A, Wulf P, Del Nery E, Weide T, Stepanova T, Galjart N, Grosveld F, Goud B, De Zeeuw CI, et al. Bicaudal-D regulates COPI-independent Golgi-ER transport by recruiting the dynein-dynactin motor complex. Nat Cell Biol. 2002;4:986–992. doi: 10.1038/ncb891. [DOI] [PubMed] [Google Scholar]
- 33.Larsen KS, Xu J, Cermelli S, Shu Z, Gross SP. BicaudalD actively regulates microtubule motor activity in lipid droplet transport. PLoS ONE. 2008;3:e3763. doi: 10.1371/journal.pone.0003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoogenraad CC, Wulf P, Schiefermeier N, Stepanova T, Galjart N, Small JV, Grosveld F, De Zeeuw CI, Akhmanova A. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 2003;22:6004–6015. doi: 10.1093/emboj/cdg592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wanschers B, van de Vorstenbosch R, Wijers M, Wieringa B, King SM, Fransen J. Rab6 family proteins interact with the dynein light chain protein DYNLRB1. Cell Motil Cytoskeleton. 2008;65:183–196. doi: 10.1002/cm.20254. [DOI] [PubMed] [Google Scholar]
- 36.Nashchekin D, St Johnston D. Egalitarian recruitment of localized mRNAs. Genes Dev. 2009;23:1475–1480. doi: 10.1101/gad.1821509. [DOI] [PubMed] [Google Scholar]
- 37.Cohen RS, Zhang S, Dollar GL. The positional, structural, and sequence requirements of the Drosophila TLS RNA localization element. RNA. 2005;11:1017–1029. doi: 10.1261/rna.7218905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38**.Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ. A'-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Struct Mol Biol. 2010;17:703–709. doi: 10.1038/nsmb.1813. The authors show that the TLS localization signal adopts an unusual structure. Monitoring and modeling the effects of mutations on structure and function, they make a compelling case for recognition of the signal primarily by its structure, rather than a particular sequence. This offers an explanation of the conundrum posed by the existence of multiple localization signals with very similar properties but highly dissimilar sequences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 40.Anderson P, Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol. 2009;10:430–436. doi: 10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 41.Ainger K, Avossa D, Morgan F, Hill SJ, Barry C, Barbarese E, Carson JH. Transport and localization of exogenous myelin basic protein mRNA microinjected into oligodendrocytes. J Cell Biol. 1993;123:431–441. doi: 10.1083/jcb.123.2.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferrandon D, Koch I, Westhof E, Nüsslein-Volhard C. RNA-RNA interaction is required for the formation of specific bicoid mRNA 3' UTR-STAUFEN ribonucleoprotein particles. EMBO J. 1997;16:1751–1758. doi: 10.1093/emboj/16.7.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wagner EJ, Garcia-Blanco MA. Polypyrimidine tract binding protein antagonizes exon definition. Mol Cell Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, Allain FH. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- 45**.Besse F, Lopez de Quinto S, Marchand V, Trucco A, Ephrussi A. Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 2009;23:195–207. doi: 10.1101/gad.505709. PTB is shown to bind to multiple sites in the oskar mRNA, and to act in the cytoplasm to assemble oskar mRNA into large complexes. Loss of PTB activity causes defects in oskar mRNA localization and translational repression. Furthermore, PTB is required for piggybacking, a phenomenon in which an mRNA with the oskar 3' UTR but not competent for localization can be transported together with an intact oskar mRNA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hachet O, Ephrussi A. Splicing of oskar RNA in the nucleus is coupled to its cytoplasmic localization. Nature. 2004;428:959–963. doi: 10.1038/nature02521. [DOI] [PubMed] [Google Scholar]
- 47.Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 48**.Reveal B, Yan N, Snee MJ, Pai C, Gim Y, Macdonald PM. BREs Mediate Both Repression and Activation of oskar mRNA Translation and Act In trans. Dev Cell. 2010;18:496–502. doi: 10.1016/j.devcel.2009.12.021. Bruno is known to repress oskar mRNA translation via binding to sites in the 3' UTR. In this work a subset of these sites are shown to have an additional role in translational activation. Surprisingly, both repression and activation can be conferred in trans, a novel form of regulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snee MJ, Macdonald PM. Dynamic organization and plasticity of sponge bodies. Developmental Dynamics. 2009;238:918–930. doi: 10.1002/dvdy.21914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila Cup Is an eIF4E Binding Protein that Associates with Bruno and Regulates oskar mRNA Translation in Oogenesis. Dev Cell. 2004;6:69–78. doi: 10.1016/s1534-5807(03)00400-3. [DOI] [PubMed] [Google Scholar]