Abstract
Using the (near) complete genome sequences of the yeasts Candida albicans, Saccharomyces cerevisiae, and Schizosaccharomyces pombe, we address the evolution of a unique genetic code change, which involves decoding of the standard leucine-CTG codon as serine in Candida spp. By using two complementary comparative genomics approaches, we have been able to shed new light on both the origin of the novel Candida spp. Ser-tRNACAG, which has mediated CTG reassignment, and on the evolution of the CTG codon in the genomes of C. albicans, S. cerevisiae, and S. pombe. Sequence analyses of newly identified tRNAs from the C. albicans genome demonstrate that the Ser-tRNACAG is derived from a serine and not a leucine tRNA in the ancestor yeast species and that this codon reassignment occurred ∼170 million years ago, but the origin of the Ser-tRNACAG is more ancient, implying that the ancestral Leu-tRNA that decoded the CTG codon was lost after the appearance of the Ser-tRNACAG. Ambiguous CTG decoding by the Ser-tRNACAG combined with biased AT pressure forced the evolution of CTG into TTR codons and have been major forces driving evolution of the CTN codon family in C. albicans. Remarkably, most of the CTG codons present in extant C. albicans genes are encoded by serine and not leucine codons in homologous S. cerevisiae and S. pombe genes, indicating that a significant number of serine TCN and AGY codons evolved into CTG codons either directly by simultaneous double mutations or indirectly through an intermediary codon. In either case, CTG reassignment had a major impact on the evolution of the coding component of the Candida spp. genome.
[Supplemental material is available online at http://www.genome.org and at http://www.bio.ua.pt/genomica/Lab/Genomedata.html. The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: N. Federspiel.]
Alterations to the standard genetic code have been found in several organisms and organelle genomes during the past 20 years (for review, see Osawa et al. 1992), resulting in intense debate about their origin and evolution. In an attempt to explain their occurrence, two mutually exclusive models, (1) the “Codon Capture” theory (Osawa and Jukes 1989; Jukes and Osawa 1993) and (2) the “Ambiguous Intermediate” theory (Schultz and Yarus 1994, 1996), have been put forward in recent years. The Codon Capture theory is a neutral theory, which postulates that the reassigned codon completely disappears from the genome under AT or GC pressure. On its reappearance, a tRNA with a different amino acid identity and an anticodon complementary to the codon being reassigned takes over the decoding of the codon. Any decoding ambiguity is excluded. The driving force is the neutral effect of GC/AT pressure. Conversely, the Ambiguous Intermediate theory has no requirement for the disappearance of the codon from the genome. The codon undergoes a transitional stage in which it is ambiguously decoded. Such ambiguity may be one of aminoacylation identity (Schultz and Yarus 1994, 1996), anticodon misreading (Yarus and Schultz 1997), or a competition between two nonambiguous tRNAs (i.e., codon competition). The driving force in this case is likely to be a positive benefit resulting from the reassignment.
An interesting example of a genetic code deviation occurs in Candida yeasts, in which the nuclear CTG codon codes for serine rather than the “universal” leucine (Kawaguchi et al. 1989; Ohama et al. 1993; Santos and Tuite 1995; Sugiyama et al. 1995; Sugita and Nakase 1999). The reassignment of the CTG codon from leucine to serine is mechanistically unusual in that it cannot be accomplished via a single mutation in the anticodon of a serine tRNA (Ser-tRNA). All other known codon reassignments, with the exception of the reassignment of the CUN leucine codon box to threonine in yeast mitochondria (Osawa et al. 1990), may be accomplished via a single mutation of the anticodon of the tRNA concerned. In the Candida spp., a single tRNA with a CAG anticodon, the Ser-tRNACAG, decodes the CTG codon as serine (Yokogawa et al. 1992; Ohama et al. 1993; Santos et al. 1993; Suzuki et al. 1994; Ueda et al. 1994; Sugiyama et al. 1995). Interestingly, this tRNA is charged with serine, but is also mischarged with leucine at a 3% rate in vivo in Candida zeylanoides (Suzuki et al. 1997). Therefore, the CTG codon has the unique property of being “polysemous” (Suzuki et al. 1997), that is, it codes for two amino acids, indicating that the CTG codon in some of the extant Candida species is still ambiguous. This has been taken as providing supporting evidence for an Ambiguous Intermediate mechanism for CTG reassignment (Santos et al. 1996; Knight et al. 2001a,b). However, it questions the origin of the hybrid Ser-tRNACAG. Here we demonstrate that the ancestor of the Ser-tRNACAG was a serine tRNA, based on an analysis of newly identified tRNAs from the Candida albicans genome, tRNAs from other yeast species, and a consideration of tRNA intron sequences. In addition, we date the codon reassignment to ∼170 million years ago using the Ser-tRNACAG sequences of the Candida spp., and show that the ancestor of the Ser-tRNACAG originated some time before the codon reassignment, indicating that CTG evolution should have been driven by a combination of genome GC pressure and ambiguous CTG decoding. In addition, a comparative genomics study was carried out to trace the origin of the 17,000 CTG codons present in the C. albicans genome and to evaluate both the impact of biased AT pressure and serine-CTG misreading on the usage of the CTN codons in C. albicans. Most of the original C. albicans CTG codons mutated to TTA (27.8%) and TTG (25.3%) leucine codons. Remarkably, CTG codons present in the C. albicans genome evolved relatively recently from codons encoding serine or conserved/semiconserved serines but not leucine codons. Only a minor fraction (0.2%; total = 102) of the CTG codons present in C. albicans exist in Saccharomyces cerevisiae, implying that almost all the original CTG codons disappeared from the C. albicans genome. In contradiction to the polysemous nature of the CTG codon, this unexpected observation provides apparent support for a Codon Capture mechanism for CTG reassignment. However, CTG elimination cannot be explained by biased AT pressure, raising the possibility that appearance of the Ser-tRNACAG and consequent serine-CTG misreading was the determining factor driving CTG almost to extinction.
RESULTS
Identity of the Ancestor of the C. albicans Ser-tRNACAG
The tRNAs identified from the C. albicans genome are displayed at http://www.bio.ua.pt/genomica/Lab/Genomedata.html. Pairwise alignments were conducted between the Ser-tRNACAG of C. albicans, Candida cylindracea, Candida tropicalis, and Candida rugosa and the class II tRNAs of S. cerevisiae, Schizosaccharomyces pombe, C. cylindracea, and C. albicans. Without exception, the Ser-tRNACAGs of C. albicans, C. cylindracea, C. tropicalis, and C. rugosa possess a higher mean identity with the serine tRNAs of S. cerevisiae, S. pombe, C. cylindracea, and C. albicans than with the leucine tRNAs, indicating that the ancestral tRNA was a serine tRNA (data not shown). Holmquist et al. (1973) determined that the average divergence for pairs of eukaryotic tRNAs coding for different amino acids is 48%. A somewhat higher mean identity is observed when the Ser-tRNACAG is compared with the leucine tRNAs of S. pombe, S. cerevisiae, C. cylindracea, and C. albicans (59%, 60%, 61%, and 59%, respectively). This is likely indicative of a common origin for serine and leucine tRNAs, consistent with the observation that the isoacceptors are unique in eukaryotes for possessing an extra arm.
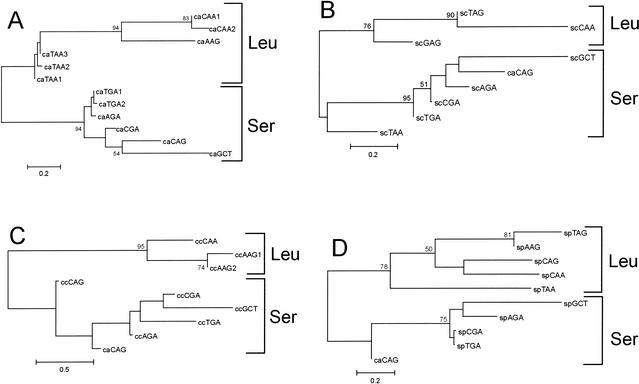
Using neighbor-joining (NJ) analysis, the C. albicans Ser-tRNACAG was compared with the class II tRNAs of C. albicans (Fig. 1A), S. cerevisiae(Fig. 1B), C. cylindracea (Fig. 1C), and S. pombe (Fig. 1D). In all cases the C. albicans Ser-tRNACAG grouped with serine tRNAs rather than leucine tRNAs, with varying degrees of bootstrap support. Thus, the above data indicate that the ancestor of the C. albicans Ser-tRNACAG was a serine tRNA. The close relationship between C. albicans Ser-tRNAAGA and Ser-tRNATGA1 and Ser-tRNATGA2 (Fig. 1A) indicates that these tRNAs have undergone a change in anticodon. Such events make it difficult to predict the specific anticodon of the ancestor of the Ser-tRNACAG.
Figure 1.
Trees of the serine and leucine tRNAs of Candida albicans, Saccharomyces cerevisiae, Candida cylindracea, and Schizosaccharomyces pombe with the C. albicans Ser-tRNACAG. (A) Tree of the serine and leucine tRNAs of C. albicans. (B) Tree of the serine and leucine tRNAs of S. cerevisiae. (C) Tree of the serine and leucine tRNAs of C. cylindracea. (D) Tree of the serine and leucine tRNAs of S. pombe. Trees were constructed using the NJ method and the Kimura 2-parameter distance model (Kimura 1980). The γ shape parameter used was 0.623 for C. albicans, 0.400 for C. cylindracea, 0.833 for S. cerevisiae, and 0.698 for S. pombe. (CACAG) The C. albicans Ser-tRNACAG. The numerals represent the confidence levels from 100 bootstrap replicates. The tRNA sequences used for A are displayed at http://www.bio.ua.pt/genomica/Lab/Genomedata.html. The tRNA sequences used for B, C, and D were obtained from GenBank. The scale represents the average number of substitutions per site.
Sequence Analysis of tRNA Introns
Sequence alignments reveal that the introns of C. albicans Ser-tRNACGA and Candida guilliermondii Ser-tRNACGA are similar to each other and to that of C. cylindracea Ser-tRNACAG (Table 1). This is evidence for a common ancestry of the Ser-tRNACAG and Ser-tRNACGA of the Candida spp. Interestingly, an insertion of A into position 34 of the anticodon of the C. guilliermondii Ser-tRNACGA produces a CAG anticodon and adds an A to the 5′ end of the intron, found in the C. cylindracea Ser-tRNACAG intron. The presence of an intron may have been a predisposing factor for the codon reassignment, because no serine tRNA can change its anticodon to CAG via a single point mutation. An insertion in the anticodon loop would produce a loop larger than the canonical 7 nt, which would likely be detrimental given that the size of the anticodon loop is highly conserved in tRNAs (Sprinzl et al. 1998). However, the presence of an intron would absorb such an anticodon expansion, allowing the loop to maintain its size as the sequence and structure of tRNA introns do not influence splice-site selection by the S. cerevisiae tRNA splicing endonuclease (Reyes and Abelson 1988). Hence, a nucleotide insertion (or deletion) into the intron should not cause any perturbation to the splicing of introns from the anticodon loop, as already noted by Yokogawa et al. (1992).
Table 1.
Introns of ser-tRNACAG and ser-tRNACGA From the Candida spp.
| Anticodon of tRNA | Candida species | Source | Intron |
| Ser(CAG) | C. tropicalis | GenBank accession no. D17535 | tttattctgggat |
| Ser(CAG) | C. lusitaniae | GenBank accession no. D17534 | tttatcagcacc- |
| Ser(CAG) | C. melibiosica | Ohama et al. 1993 | ttacacagcacc- **::::::::::- |
| Ser(CAG) | C. maltosa | GenBank accession no. D26074 | tttttcgtggtaaacgaaggtcaac |
| Ser(CAG) | C. cylindracea | Yokogawa et al. 1992 | atcttcattctcgac |
| Ser(CGA) | C. albicans | Ueda et al. 1994 | -tctt-attcgcgtt |
| Ser(CGA) | C. guilliermondii | Ueda et al. 1994 | -tctt-tt-gagtt -****-:**:::*:: |
| Ser(CGA) | C. zeylanoides | Ueda et al. 1994 | taaatttgagta |
Introns that display similarity were aligned. Bold type indicates identical nucleotides. Symbols displayed below the aligned sequences summarize the alignment: (*) a conserved nucleotide, (:) a nucleotide that is identical with one exception, (.) a position that has two identical nucleotides, and (-) a position with no identical nucleotides.
Date of the CTG Codon Reassignment
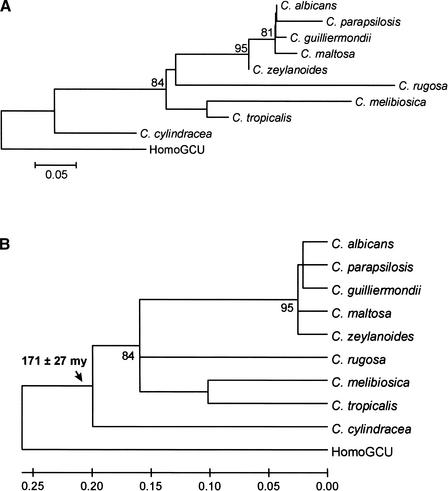
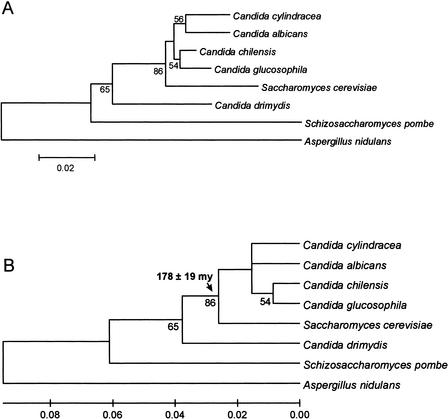
Using the Ser-tRNACAG sequences to construct an NJ tree (Fig. 2), a date of at least 171 ± 27 million years ago was obtained for the emergence of the codon reassignment. Using SSU rRNA sequences to construct an NJ tree, a date of 178 ± 19 million years ago was obtained for the codon reassignment (Fig. 3), which agrees with the date obtained from the Ser-tRNACAG tree. The date is also in agreement with a figure of 150 million years ago, derived using the maximum likelihood method and SSU rRNA sequences (Pesole et al. 1995). The discrepancy is probably due to the use of a limited data set in the previous analysis as C. cylindracea was omitted, which may cause an underestimation of the divergence date. Overall, the evidence points to the codon reassignment having occurred during the Jurassic era. It should be noted that saturation is reached at the base of the tree, and this would lead to an underestimation of the time of divergence of the outgroup, that is, Homo sapiens Ser-tRNAGCT.
Figure 2.
Date of divergence of the CTG codon reassignment using Ser-tRNACAG sequences. (A) Nonlinearized tree; (B) linearized tree. The tree was constructed using the NJ method and the Kimura 2-parameter distance model (Kimura 1980). A γ shape parameter of 1.83 was used. A nucleotide substitution rate of 1.156 × 10−9 substitutions per site per year was used to estimate divergence dates. This value was calculated to be representative of yeast tRNAs, as described in Methods. The standard error of the key divergence time is indicated. Lower numerals represent the confidence level from 100 replicate bootstrap samples. Homo sapiens Ser-tRNAGCT (HomoGCT) was used as an out-group. The scale represents the average number of substitutions per site.
Figure 3.
Date of divergence of the CTG codon reassignment using SSU rRNA sequences. (A) Nonlinearized tree; (B) linearized tree. The tree was constructed using the NJ method and the Kimura 2-parameter distance model (Kimura 1980). A γ shape parameter of 0.146 was used. The standard error of the key divergence time is indicated. Lower numerals represent the confidence level from 100 replicate bootstrap samples. Aspergillus nidulans was used as an out-group. The scale represents the average number of substitutions per site.
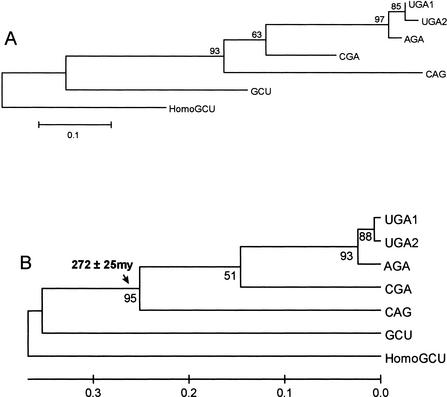
A tree of the C. albicans serine tRNAs indicates that the Ser-tRNACAG originated at least 272 ± 25 million years ago (Fig. 4B), which is considerably more ancient than the date for the codon reassignment. The probability that the codon reassignment and appearance of the Ser-tRNACAG occurred at approximately the same time, that is, within 10 million years of each other, is low, with p < 0.0006. Therefore, a major new feature of this work is that the appearance of the Ser-tRNACAG predates the reassignment event by a significant amount of time. Some caution should be expressed when assigning absolute dates in the absence of a fossil record; however, the dates are valuable in indicating the ancient origin of the tRNA compared with the CTG reassignment event.
Figure 4.
Date of divergence of the Ser-tRNACAG from the serine tRNAs in Candida albicans. (A) Nonlinearized tree; (B) linearized tree. The tree was constructed using the NJ method and the Kimura 2-parameter distance model (Kimura 1980). A γ shape parameter of 0.623 was used. A nucleotide substitution rate of 1.156 × 10−9 substitutions per site per year was used to estimate divergence dates. This value was calculated to be representative of yeast tRNAs, as described in Methods. The standard error of the key divergence time is indicated. Lower numerals represent the confidence level from 100 replicate bootstrap samples. Homo sapiens Ser-tRNAGCT (HomoGCT) was used as an out-group. The scale represents the average number of substitutions per site.
Apparently, all copies of the ancestral tRNA(s) were converted into Ser-tRNACAG, or remaining copies were lost from the genome. Therefore, a close relative of the tRNA is not present in the genomes of C. albicans or C. cylindracea (Fig. 1A,C). Interestingly, considerations of the class II tRNAs of S. cerevisiae do not indicate a close relative either (Fig. 1B), implying that the homolog of the ancestral tRNA was lost from this genome also after the divergence of S. cerevisiae from C. albicans.
The agreement of the SSU rRNA and Ser-tRNACAG phylogenetic trees for the date of the codon reassignment indicates that the Ser-tRNACAG is evolving at a rate comparable to that of the tRNAs used to calibrate the molecular clock. Thus, the reassignment is complete and the other CTN codons, especially the CTA codon, are “safe” from further reassignments, that is, the evolutionary imperative behind the CTG reassignment is not driving further change.
The Effect of Genome GC Pressure on Codon Usage in C. albicans
Ambiguous CTG decoding provides compelling evidence for its reassignment through a misreading mechanism as proposed by the Ambiguous Intermediate theory (Schultz and Yarus 1994). However, this should have a negative impact on the organism's fitness and therefore prompts the question of whether biased GC pressure reduced CTG usage to a bearable minimum prior to reassignment (Jukes and Osawa 1993). To shed new light on this important question, a comparative study of the effect of GC pressure on the coding component of C. albicans, S. cerevisiae, and S. pombe genomes was carried out. The total GC content is similar in the three species (Table 2); however, C. albicans shows the lowest GC content at the codon third position. A comparative analysis of codon usage carried out for the three species shows that, with very few exceptions, codon usage follows a similar trend, which is represented by the usage of the serine codon family shown in Figure 5B. However, leucine codons do not follow this overall trend, in particular, significant differences in frequency are found for the CTA, CTC, and CTG codons, whose usage is clearly repressed in C. albicans in relation to S. cerevisiae and S. pombe (Fig. 5A). That CTG and CTA usage is repressed whereas TTG usage is favored in C. albicans indicates that AT pressure alone is not the main force driving the evolution of the leucine codons. One alternative explanation is that tRNA selection is responsible for this effect, as is the case in many organisms (Osawa 1995). This is also in line with the observation that S. cerevisiae, S. pombe, and C. albicans decode the CTN codon family with a rather different set of anticodons, that is, GAG and TAG for S. cerevisiae; AAG, TAG, and CAG for S. pombe; and AAG for C. albicans. Because the AAG anticodon in C. albicans decodes the CTC, CTT, and CTA codons using extended wobbling, it is likely that the weak interaction between the AAG anticodon and the CTA/C/T codons, in particular with CTA, is an important factor in reducing CTN usage in C. albicans, as is observed for two-codon sets in highly expressed genes in most organisms (Ikemura 1981a,b; Ohama et al. 1990; Osawa 1995).
Table 2.
Decreased GC Content at the Third Codon Position in Candida Species That Reassigned the CTG Codon From Leucine to Serine
| Candida species | GC content of coding sequences (%) | |||
| Total | 1st codon position | 2nd codon position | 3rd codon position | |
| C. albicans | 36.93 | 44.14 | 37.65 | 29.00 |
| C. dubliniensis | 36.13 | 41.86 | 39.91 | 26.65 |
| C. maltosa | 37.16 | 44.73 | 37.60 | 29.15 |
| C. glabrata | 41.91 | 45.56 | 37.81 | 42.37 |
| S. cerevisiae | 39.71 | 44.59 | 36.58 | 37.96 |
| S. pombe | 39.80 | 48.04 | 38.24 | 33.12 |
All species shown have similar GC content in coding DNA and also at the first and second codon positions. However, GC pressure decreases significantly at the third codon position (N3, bold) in the three species that decode the standard leucine-CTG codon as serine, that is, Candida albicans, Candida dubliniensis, and Candida maltosa. Candida glabrata decodes the CTG codon as a leucine and follows the Saccharomyces crevisiae and Schizosaccharomyces pombe pattern. An exception to the lowering of GC content at the N3 position in Candida species that reassigned the CTG codon is represented by Candida cylindracea. However, the latter uses the CTG codon at a high frequency, which is in sharp contrast to a generalized low CTG usage in the Candida species that reassigned the CTG codon, indicating that other evolutionary forces shape CTG usage in C. cylindracea.
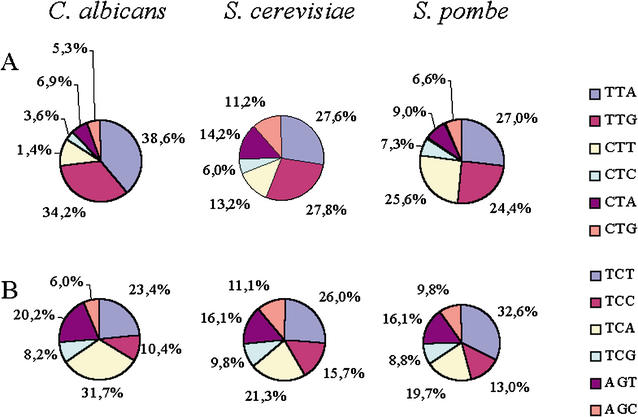
Figure 5.
Usage of leucine and serine codons in Candida albicans, Saccharomyces cerevisiae, and Schizosaccharomyces pombe. (A) Of the six leucine codons, TTA and TTG are the most frequently used by the nuclear genomes of the three yeast species, the exception being CTT in S. pombe. In C. albicans, the usage of CTN codons, in particular CTC and CTA but also CTG codons, is repressed in relation to the same codons in the other two yeasts, whereas usage of the CTT codon is similar between C. albicans and S. cerevisiae. The usage of the CTA codon is also repressed in C. albicans, indicating that other forces apart from genome AT pressure shape CTN usage in this species. (B) The bias in C. albicans CTN usage is not observed for serine codons, whose distribution follows the expected pattern for a genome with low GC content in coding sequences (Table 2). The values indicated are relative to the total synonymous codon count for each genome and independent of amino acid frequency.
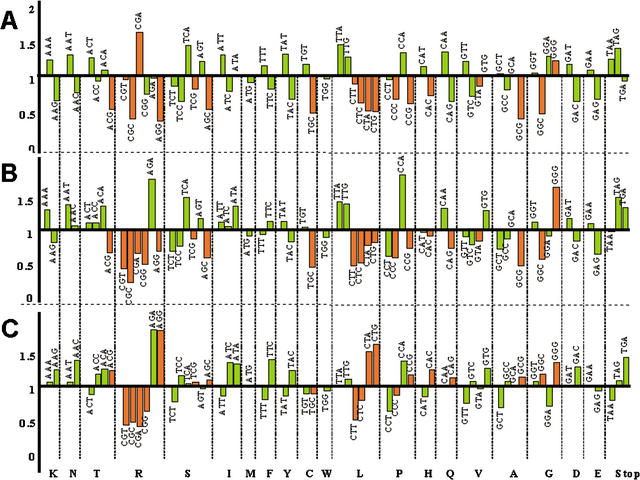
Considering that the three yeast species being studied have similar total GC content and rather conserved tRNA populations (data not shown), the exceptions being higher AT content at N3 and the Leu-tRNA isoacceptors that decode the CTN codon family in C. albicans, specific effects of biased AT pressure at N3 and tRNA selection on CTN usage should be unveiled by determining the relative codon usage frequency for genome pairs, that is, C. albicans versus S. cerevisiae and C. albicans versus S. pombe (Fig. 6A,B). The increase in AT pressure at N3 in C. albicans is clearly visible for frequently used codons in the C. albicans/S. cerevisiae pair, where C. albicans clearly prefers A- and T-ending codons. Interestingly, the Leu-TTG codon, the Val-GTG, and the Gly-GGG codons are exceptions to this rule and imply that translational selection is an important factor modulating usage of these codons in C. albicans. When a similar analysis is carried out for the C. albicans/S. pombe pair, the same trend is observed, but the number of exceptions increases for C-ending and rarely used codons (Fig. 6B), thus further supporting the hypothesis that both AT pressure and tRNA selection modulate codon usage in C. albicans (Fig. 6A,B).
Figure 6.
Candida albicans prefers A- and T-ending codons. Codon usage in C. albicans, Saccharomyces cerevisiae, and Schizosaccharomyces pombe was analyzed by counting the total number of codons for each species. To determine the codon preference between two species, the relative frequency of usage of each codon was divided by the corresponding value for the same codon in the other species. The codon usage ratios obtained for C. albicans/S. cerevisiae (A), C. albicans/S. pombe (B), and S. cerevisiae/S. pombe (C) were plotted as indicated in the graph, with ratio values above 1 (upper part in the graph) indicating the preferred codons in C. albicans in relation to the other two species (A and B) and in S. cerevisiae in relation to S. pombe (C). The C. albicans/S. cerevisiae codon ratios indicate that C. albicans prefers highly used codons (green bars) ending with A and T, with the exception of Leu-TTG and Gly-GGG. For the C. albicans/S. pombe pair, the preference for frequently used A- and T-ending codons is maintained, but four frequently used C-ending codons, Asn-AAC, Thr-ACC, Ile-ATC, Phe-TTC, and also the G-ending codons Leu-TTG, Val-GTG, and Gly-GGG, are preferred. Therefore, the data indicate that in C. albicans the effect of AT pressure is more visible at the third codon position for highly used codons. That the CTG codon is used at low frequency in C. albicans implies that AT pressure was not the main force driving its reassignment to serine. Green and brown bars indicate highly and rarely used codons, respectively.
A deeper insight into the effect of GC pressure on codon usage can be gained by aligning C. albicans and S. cerevisiae homologous genes and determining the content of A and T at N3 for each homologous codon. That is, for each S. cerevisiae codon in a double alignment, the nucleotide present at the corresponding N3 position in C. albicans can be determined and compared with all other codons for the complete set of homologous genes (Fig. 7). C. albicans prefers A- and T-ending codons without exception, and G- and C-ending codons are repressed. However, leucine codons show a significant deviation to this trend in that there is a clear relative increase in G3. This is mainly achieved by decreasing C3 and T3. A similar trend is observed for the arginine codon family, but in this case there is a relative increase in A3 and the G3 effect is not so visible (Fig. 7). Thus, leucine codons have a slight bias favoring purine- and repressing pyrimidine-ending codons instead of favoring A- and T-ending leucine codons, which contradicts the general trend of an increased frequency of A3 and T3. Considering that this is a rather localized effect, the most likely explanation for it is that the weak interaction between the AAG anticodon and CTA, CTC, and CTT codons and the disappearance of the CAG or TAG anticodons from the C. albicans genome drove a massive conversion of CTN into TTA and TTG codons, which are decoded by a strong interaction with cognate TAA and CAA anticodons, respectively.
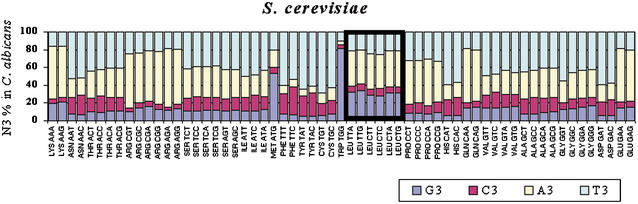
Figure 7.
GC pressure alone at the third codon position does not explain the evolution of leucine codons in Candida albicans. To elucidate the role of GC pressure in the evolution of C. albicans ORFs, an analysis of GC3 pressure (GC pressure at the third codon position) was carried out by comparing the complete ORFs set of both genomes using BLASTP at an E-value of 10−5. For each Saccharomyces cerevisiae codon, the corresponding N3 codon position was identified in C. albicans ortholog genes. The data show that S. cerevisiae codons are represented in C. albicans mainly by A- or T-terminating codons (yellow and light blue bars), in agreement with high AT pressure at the third codon position in the C. albicans genome (N3 = 71% AT; Table 2). However, the leucine codons show a small deviation from the pattern observed for all other codons. That is, they change more frequently than expected into G-terminating codons (blue bar), thus contradicting the high AT pressure at the N3 position observed in C. albicans genes. This increase in G3 is matched by a slight increase in A3, indicating an increase in purines at N3. This is achieved by decreasing the usage frequency of C- and T-ending codons. That is, when compared with all other codons, there is a relative increase in purines at the N3 position in C. albicans genes instead of an increase in AT-ending codons, as would be expected from the relative increase of AT pressure at N3 in the C. albicans genome, thus indicating that other forces apart from GC pressure shaped the evolution of leucine codons in the latter.
Leucine-Encoding CTG Codons Have Disappeared From the C. albicans Genome
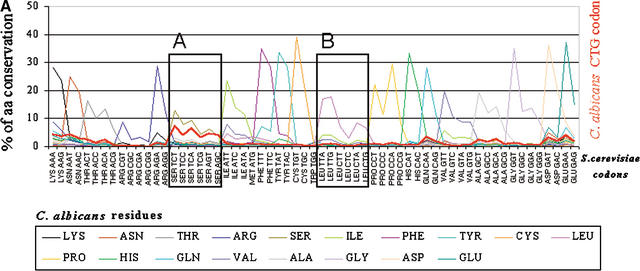
The observation that the Ser-tRNACAG appeared prior to CTG reassignment combined with the unusual evolutionary pattern of the leucine codons raises the interesting possibility that the Ser-tRNACAG itself shaped the evolution of leucine codons, and in particular the CTG codons. That is, the negative pressure imposed on CTG codons by incorporation of serines instead of leucines might have played a major role in the evolution of all six leucine codons. To clarify this question, the complete set of C. albicans and S. cerevisiae homologous genes were aligned as described in Methods, and for each S. cerevisiae amino acid the codons present in the homologous positions in C. albicans genes were computed. By plotting the percentage of the amino acid found in the S. cerevisiae genome for each codon present at homologous positions in the C. albicans genome, an overall picture of amino acid conservation between the two species is obtained. As expected, the level of amino acid conservation between the two species is very high (Fig. 8A,B). If a similar analysis is carried out, but instead of comparing amino acids in S. cerevisiae with codons at homologous positions in C. albicans homologous genes, the CTG codons present in the former are compared with codons present in the latter, an evolutionary pattern for the CTG codon between the two species emerges (red line in Fig. 8A,B). Remarkably, almost all CTG codons present in the S. cerevisiae genome are represented by leucine-TTG and -TTA codons in C. albicans. Only 0.2% of the total number of CTG codons remain conserved at homologous positions (Fig. 8B).
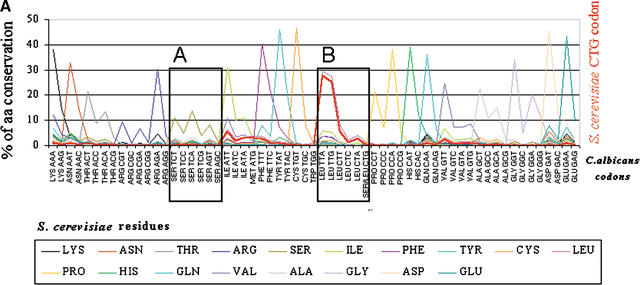
Figure 8.
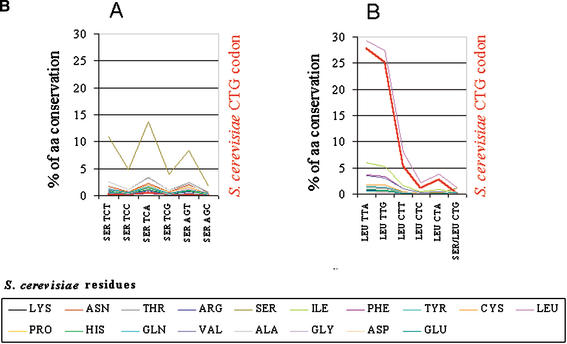
The majority of the CTG codons present in the Saccharomyces cerevisiae genome are encoded by TTG and TTA codons in the Candida albicans genome. (A) To quantify the relative conservation of amino acids and codons between C. albicans and S. cerevisiae, the two genomes were aligned using BLASTP at an E-value of 10−5. For each S. cerevisiae amino acid, the corresponding codons present at homologous positions in C. albicans orthologs were identified, thus providing overall information about amino acid and codon conservation between the two species. To elucidate the mutational pattern of the CTG codon between the two species, its frequency of conversion at each position of the alignment was computed independently of the other leucine codons (thick red line in the graph). As would be expected, the major trend is residue conservation between the two species at each position for each respective codon family. For the CTG and the other leucine codons, two important trends are observed: first, their conversion into leucine TTG and TTA codons and also into conserved amino acids of leucine (Ile, Met, and Phe); second, the avoidance of nonconserved leucine, namely, serine codons. As observed in Figures 6 and 7, the preferential conversion of CTN codons into TTG does not follow the rules imposed by increased AT pressure in C. albicans as TTG is a G-ending codon, thus indicating that translational selection is a strong driving force in the evolution of the CTN codons in C. albicans. (B) Magnification of the previous graph in the regions of serine and leucine codons (boxes A and B, respectively).
The above results only provide a partial picture for the evolution of the CTG codon in that the alignments are not reciprocal and consequently do not show how the 17,000 CTG codons present in C. albicans are represented in the S. cerevisiae genome. For this, the converse comparative analysis was carried out. C. albicans residues in the aligned gene set were fixed, and the corresponding codons present at homologous positions in the S. cerevisiae genome were computed (Fig. 9A,B). The C. albicans CTG codons were then fixed in the alignment, and the codons present in the S. cerevisiae genome were identified. Most strikingly, almost none of the C. albicans CTG codons are represented by leucines in the S. cerevisiae genome; instead, serines appear at homologous positions, indicating that almost all C. albicans CTG codons evolved recently from serine or conserved serine codons (Fig. 9B).
Figure 9.
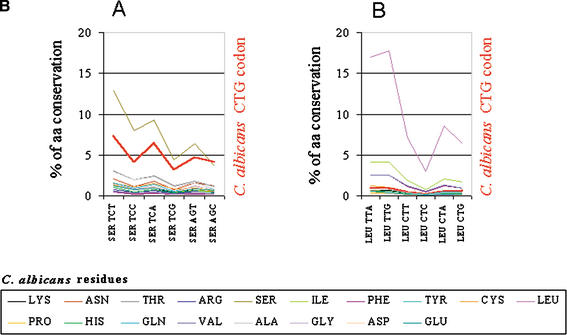
Candida albicans CTG codons display a biased pattern of conversion to serine codons in Saccharomyces cerevisiae. To identify the S. cerevisiae codons that correspond to each amino acid present at the respective position in homologous C. albicans genes, the two genomes were compared as described in Figure 8 with the exception that S. cerevisiae codons were taken as the reference in the alignment. As before, the CTG codon was computed independently. As in Figure 8, the major trend between the two genomes is amino acid conservation. However, the CTG codon shows a major deviation from the other leucine codons. That is, instead of being represented in the S. cerevisiae genome by leucine codons, it is represented by serine codons and codons corresponding to amino acids conserved of serine. (B) Magnification of the previous graph in the regions of serine and leucine codons (boxes A and B, respectively).
Considering that evolution of the CTG codon from serine codons is rather surprising, because of the need for two mutations to convert a serine or conserved serine codon into a CTG codon, a triple alignment between C. albicans, S. cerevisiae, and S. pombe genomes was carried out to remove the background noise inherent in a double genome alignment and consequently improve data quality. The results of the triple alignment are a clear-cut confirmation of the double alignment (Fig. 10). That is, C. albicans CTG codons are represented in the S. cerevisiae and S. pombegenomes by serine and not leucine codons. There is a very low level of conserved leucine codons and an even lower number of conserved CTG codons in the three genomes. That none of the other leucine codons shows this trend highlights the unique evolutionary pathway for CTG codons in the C. albicans genome (Fig. 10). The residual number of conserved CTGs present in the three genomes indicates that they are likely to be located at positions that tolerate leucine or serine residues by C. albicans proteins.
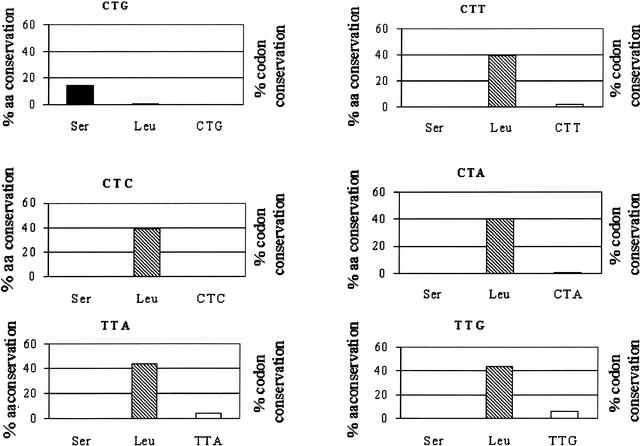
Figure 10.
A high percentage of Candida albicans CTG codons are represented by serine residues in the Saccharomyces cerevisiae and Schizosaccharomyces pombe genomes. To determine the origin of leucine codons present in the C. albicans genome, the percentage of serine and leucine residues present simultaneously in the S. cerevisiae and S. pombe genomes was determined for each of the six leucine codons present in the C. albicans genome. For this, the three genomes were compared using the BLASTP program as in Figures 8 and 9. The conservation of CTN codons between C. albicans and S. cerevisiae/S. pombe is very low and reaches the lowest value for CTG codons (0%). Interestingly, 14% of the C. albicans CTG codons encode serine residues and only 0.7% encode leucine residues in S. cerevisiae and S. pombe. This trend is not observed for any other leucine codon; leucine is always highly conserved in homologous S. cerevisiae/S. pombe genes.
DISCUSSION
The occurrence of genetic code changes raises two important evolutionary questions, namely: How do genetic code alterations evolve? and, Why do they evolve? The reassignment of the leucine-CTG codon to serine in Candida spp. is a paradigm genetic code change whose study has already unveiled several intricate and subtle evolutionary forces involved in the evolution of genetic code changes (Santos et al. 1993, 1996, 1999; Perreau et al. 1999). The present evolutionary study sheds important new light on the evolution of CTG reassignment by showing that a combination of the evolutionary mechanisms postulated by both the Codon Disappearance and Ambiguous Intermediate theories has driven CTG reassignment. That is, increased AT pressure at the third codon position increases usage of A- and T-ending codons; however, the appearance of the Ser-tRNACAG, which decodes the CTG codon as serine, has been the major force repressing CTG, and perhaps CTA/CTC/CTT, and increasing TTR codon usage.
The Evolution of the Ser-tRNACAG
A serine ancestry for the Ser-tRNACAG is consistent with structural probing analyses that demonstrate that the structure of C. albicans Ser-tRNACAG is that of a typical serine tRNA (Perreau et al. 1999). A serine ancestry for the Ser-tRNACAG indicates that the suggested mechanism of tRNA mis-aminoacylation (Schultz and Yarus 1994, 1996) has not occurred in this case, despite the polysemous nature of the CTG codon (discussed further below). Therefore, if codon ambiguity were a part of the mechanism of reassignment, then it was the result of either the decoding ambiguity of the Ser-tRNACAG, mediated via an ambiguous codon–anticodon interaction, or “codon competition” between a cognate leucine and a cognate serine tRNA. Therefore, the ancestor of the Ser-tRNACAG may have initially decoded a serine codon, but also the CTG codon, to a minor extent, via an ambiguously decoding anticodon. The leucine tRNA that originally decoded the CTG codon would have been lost from the genome of C. albicans, consistent with the observation that the S. pombe Leu-tRNACAG bears no similarity to the C. albicans Ser-tRNACAG (Fig. 1D).
The CTG Codon is Polysemous by Default
A problem with proposing a serine ancestry for the Ser-tRNACAG is to explain the presence of m1G37 in the Ser-tRNACAG, which in Candida zeylanoides is responsible for mischarging of the tRNA with leucine (Suzuki et al. 1997), resulting in the polysemous nature of the CTG codon. The proposal has been made that the m1G37 evolved to ameliorate the detrimental effects of reassigning the CTG codon from leucine to serine (Suzuki et al. 1997). In addition, m1G37 could be viewed as a remnant of a putative leucine ancestry of the tRNA. The presence of m1G37 in the Ser-tRNACAG of the Candida spp. is likely an adaptive mutation that occurred after the anticodon of the tRNA had mutated to CAG, because m1G37 is found in tRNAs reading C Y/G N codons from Eubacteria, Archaea, and Eukaryotes (Bjork 1986; Bjork et al. 1987), whereas serine tRNAs possess a modified A37. The Ser-tRNACAG reads a codon (CTG) that belongs to the C Y/G N group. m1G37 appears to have a role in maintaining the fidelity of codon–anticodon interaction (Bjork et al. 1989, 2001; Hagervall et al. 1990, 1993; Li and Bjork 1995; Li et al. 1997; Urbonavicius et al. 2001). We suggest, therefore, that m1G37 is required for the efficient decoding of the CTG codon and that the low level of leucylation is a by-product, not an imperative, for the mutation. m1G37 therefore arose as an adaptive mutation after the reassignment occurred, rather than as a facilitative mutation that allowed the reassignment to occur. The presence of m1G37 is probably mildly detrimental to the yeast, but less detrimental than retaining A37. Further evidence for the above hypothesis is provided by the reassignment of the entire leucine CTN codon box to threonine in yeast mitochondria. This is the only other codon reassignment, apart from the Candida codon reassignment, that involves a change in identity of the 37 nucleotide (N37) in the anticodon loop of the associated tRNA. During the course of the reassignment, A37, typical of threonine tRNAs (Sprinzl et al. 1998), has mutated to m1G37 in the threonine tRNA responsible for decoding the CTN codons.
An explanation for the misacylation of the C. zeylanoides Ser-tRNACAG with leucine, rather than with any of the other 18 amino acids, lies in the observation that, as class II tRNAs, Ser-tRNA and Leu-tRNA are structurally similar. Therefore, the Ser-tRNACAG is most likely to be misrecognized by leucyl-tRNA synthetase, rather than the other 18 aminoacyl-tRNA synthetases that charge class I tRNAs. Experimental evidence supports this assertion; mutations in S. cerevisiae (Soma et al. 1996) and human (Breitschopf et al. 1995) Ser-tRNAs result in misacylation with leucine. Likewise, mutations in S. cerevisiae (Himeno et al. 1997) and human (Breitschopf and Gross 1994) Leu-tRNAs result in misacylation with serine. Hence, we propose that the nature of the polysemous codon results from the process of reassignment, rather than being an integral part of the mechanism.
The Evolution of the CTG Codon
The discovery that the Ser-tRNACAG appeared before CTG reassignment is in agreement with the finding that GC pressure alone cannot explain the evolutionary pattern of the CTN codon family in C. albicans and most likely in the other Candida species that reassigned the CTG codon. This is particularly clear if one considers the relative preference for G- and A-ending codons (N3 position) in C. albicans in relation to S. cerevisiae leucine codons (Fig. 7). That is, C. albicans has a relative preference for purine-ending rather than pyrimidine-ending codons in the leucine codon family. The A3/G3 preference can be explained by the strong directional mutational bias of CTN codons into TTA and TTG codons, and the G3 preference is mainly due to conversion of CTN codons into TTG codons. However, if one considers the higher AT content at N3 in C. albicans codons, one would expect a biased preference for TTA and not TTG as is the case for the six arginine codons (Fig. 7).
Another important discovery emerging from the study presented herein relates to the nature of the 17,000 CTG codons present in extant C. albicans. It is remarkable that almost all CTG codons present in the ancestral yeast species disappeared from the C. albicans genome and that the new codons evolved from serine or conserved serine codons whose conversion into CTG codons require at least two mutations. Alternatively, the conversion of serine codons into CTG codons may have occurred via doublet mutations, such as described for the conversion of serine-AGY codons into serine-TCN codons (Averof et al. 2000). If taken separately, this could be considered as providing strong support for the “Codon Disappearance” theory proposed by Jukes and Osawa (1993); however, the unique evolutionary pattern of the CTN codons in C. albicans and the early appearance of the Ser-tRNACAG in the Candida spp. imply a novel evolutionary mechanism in which the ambiguous decoding of the CTG codon by the newly created Ser-tRNACAG was the main force driving CTG codons to extinction. It is likely that increased AT pressure at the N3 codon position also played an important role. If so, CTG (CTN) evolution has been driven by a combination of the negative pressure imposed by CTG misreading and biased AT pressure at the N3 codon position.
Should the Ambiguous Intermediate Theory Be Reformulated?
The Ambiguous Intermediate theory originally proposed by Schultz and Yarus (1994) postulates that codon reassignment is driven by a mechanism, which requires ambiguous decoding of the codon being reassigned by tRNAs with expanded decoding properties. Briefly, the theory postulates that a mutant tRNA with expanded decoding properties starts decoding a codon belonging to a noncognate amino acid codon family, making it ambiguous. The theory proposes that this ambiguity increases because of additional mutations in the newly created tRNA up to a point at which the mutant tRNA efficiently decodes the ambiguous codon, which can then acquire a new meaning. The implication of this theory is that, contrary to expectation, codon ambiguity provides some sort of selective advantage to drive codon reassignment to completion. That the CTG and TGA codons are ambiguous in C. albicans and Bacillus subtilis, respectively (Lovett et al. 1991; Suzuki et al. 1997) and that reconstruction of the C. albicans CTG reassignment in S. cerevisiae showed that the latter has a selective advantage under some stress conditions provide support for a reassignment mechanism through ambiguous decoding in which the environment may play a significant role (Santos et al. 1999).
A recent study carried out in mitochondria showed a good correlation between codon reassignment and codon disappearance; that is, of the 11 codons that have been reassigned in some lineages, 8 have disappeared in some other lineage (Knight et al. 2001a). However, no significant association between codons predicted to disappear by mutation pressure and reassigned codons was found. Rather, some codons apparently disappear for reasons unrelated to mutational bias, and these are the ones that are more like to be reassigned (Knight et al. 2001a). Our data provide the missing link between codon reassignment and disappearance by showing that codon misreading by mutant tRNAs is an important evolutionary force driving codons to extinction. That most codon reassignments are associated with tRNA mutations, editing, altered tRNA modification, and release factor mutations (Knight et al. 2001a,b) provides strong support for this hypothesis, which unveils a novel mechanism for codon reassignment through codon ambiguity and supports a reformulation of the original Ambiguous Intermediate theory, as follows:
Mutant tRNAs with expanded decoding properties, or mutations in the translational machinery, which create decoding ambiguity, in conjunction with biased GC pressure, impose a negative selective pressure on particular codons, decreasing their usage to very low levels or forcing them to disappear from the genome. Some overall positive advantage must be present in order for this process to occur. These codons can be gradually reassigned through structural change of the translational machinery and loss of the ancestral cognate tRNAs.
The novelty of the reformulated theory relies on the disappearance or reduction of codon usage to a tolerable minimum caused by ambiguous decoding and not GC pressure only. Despite introducing a new mechanism for codon disappearance, the reformulated theory still relies on ambiguous decoding as a mechanism for codon reassignment and, therefore, we consider that the Ambiguous Intermediate theory should maintain its original name.
Conclusions
This study shows that the Ser-tRNACAG evolved from a serine and not a leucine tRNA and that the CTG codons present in extant Candida species are new codons, which have evolved recently. This study also unveils a hidden aspect of Candida biology, that of having a very unstable proteome during the last 272 million years or so. This instability clearly has important consequences at the phenotypic level, and one is urged to unravel its physiological and evolutionary meaning. This instability arises from three distinct mechanisms: (1) initial ambiguous decoding of the CTG codon by the newly created Ser-tRNACAG, forcing its disappearance; (2) evolution of serine or conserved serine codons into CTG codons through an intermediary codon; and (3) ambiguous charging of the Ser-tRNACAG by both the seryl- and leucyl-tRNA synthetases.
METHODS
Genome Data Retrieval and Selection
Sequence data for C. albicans was obtained from the Stanford DNA Sequencing and Technology Centre Web site at http://www-sequence.stanford.edu/group/candida. Sequencing of the C. albicans genome was accomplished with support from the National Institute of Dental and Craniofacial Research and the Burroughs Wellcome Fund.
The sequences are of 10× mean coverage. Complete genome sequence data builds for S. cerevisiae (May 01, 2002) and S. pombe(May 16, 2002) were downloaded from GenBank (ftp://ftp.ncbi.nih.gov).
Sequence data were filtered before any analysis was conducted. All open reading frames (ORFs) were scanned for irregularities in sequence size, start and stop codon positions, and invalid character usage. The total of validated ORFs for each genome is 8467, 6313, and 4659 for C. albicans, S. cerevisiae, and S. pombe, respectively. The C. albicans serine and leucine tRNA genes were identified by homology searching of the database, using the BLASTN program and the S. cerevisiae serine and leucine tRNA gene sequences. Introns were assigned using the tRNAscan-SE 1.1 program (Lowe and Eddy 1997). S. cerevisiae, S. pombe, and C. cylindracea tRNA gene sequences were obtained from GenBank.
RNA Sequence Alignments
The tRNA sequences were manually aligned, excluding introns and ensuring that the stems, anticodons, and invariant nucleotides (U8, A14, G18, G19, A21, G53, U54, U55, and C61) were aligned. The percentage identity of two tRNAs was calculated as the number of identical nucleotides divided by the nucleotide length of the longest of the two tRNA molecules. The 5′-CCA terminus of mature tRNAs was not included in any of the sequence analyses. Small subunit (SSU) rRNA sequences were aligned using the DCSE program (De Rijk and De Wachter 1993), taking into account secondary structure information. The alignments used in this publication are available at http://chuma.cas.usf.edu/∼garey/alignments/alignment.html.
Tree Construction
NJ trees using Kimura 2-parameter distances and γ corrections for site-to-site variation were constructed using the MEGA 2.1 program (Kimura 1980; Kumar et al. 2001). The PAUP 4.0 program (Swofford 1993) was used to calculate the γ shape parameter using maximum likelihood.
Tree Calibration and Estimates of Divergence Times
The trees were linearized under the molecular clock assumption using the MEGA 2.1 program and the method of Nei and Kumar (2000) to obtain estimates of divergence times. The tRNA nucleotide substitution rate was estimated by considering tRNAs from C. albicans, S. cerevisiae, and S. pombe, using the respective tRNAs from H. sapiens as out-groups. The tRNAs considered were aspartate, asparagine, cysteine, phenylalanine, histidine, tryptophan, and tyrosine. These tRNAs were chosen because each of the tRNAs specific for these amino acids possesses a single isoacceptor in each genome; thus, the common ancestry of each set of tRNAs was unambiguous. The C. albicans tRNA sequences used are listed at the Web site http://www.ukc.ac.uk/bio/tuite/research/tRNA.htm. The S. cerevisiae, S. pombe, and H. sapiens tRNAs used are listed at the Web site http://rna.wustl.edu/GtRDB/. The tRNA sequences were concatenated into a single sequence for each species. A γ shape parameter of 0.578 was estimated using the maximum likelihood method in PAUP, and a linearized NJ tree was constructed. A nucleotide substitution rate of 1.156 × 10−9 substitutions per site per year was estimated from the NJ tree using 420 million years ago for the divergence of S. cerevisiae from S. pombe (Lum et al. 1996). This tRNA nucleotide substitution rate was used to estimate the divergence times of key genes within the linearized tRNA trees displayed in Figures 2 and 4. The divergence times of key genes within the linearized ribosomal RNA tree (Fig. 3) were estimated by calibrating the rRNA tree using 420 million years ago for the divergence of S. cerevisiae and S. pombe.
To test the statistical significance of the key divergence times, we calculated the standard error of branch lengths of the genes that were used to estimate the respective divergence times. These standard errors were used to estimate the probability that the codon reassignment and origin of the Ser-tRNACAG did not occur at the same time (i.e., within 10 million years of each other), using approximations to the normal distribution.
Codon Usage Analysis
A platform for protein sequence data manipulation and analysis was built on Perl (ActivePerl 5.6.1; http://www.activeperl.com) using BioPerl 1.0 (http://www.bioperl.org) and consists of a series of programs and CGI scripts. The three groups of validated sequences were introduced separately, and each ORF was analyzed according to its codon usage. A range of several statistical values was generated at three different levels of genome comparison: (1) distribution of individual codons, (2) dependencies between groups of synonymous codons, and (3) differences in amino acid usage.
Homolog Alignments
A stand-alone BLAST (Altschul et al. 1997; version 2.2.2, http://www.ncbi.nlm.nih.gov/BLAST/) was used to find S. cerevisiae–C. albicans homologous proteins. All protein pairs that were reciprocal best hits (Rivera et al. 1998) with a BLAST expected value lower than 10−5 were included in this study. In all, 2899 pairs of homologous proteins that matched these criteria were found. The C. albicans genes in this homolog set have a total of 6084 CTGs (40% of the genome total), an average size of 533.68 codons, and an average of 3.52 CTGs per thousand (the genome average is 6.97 CTGs per thousand). The protein homologs were aligned with T-Coffee (Notredame et al. 2000; http://igs-server.cnrs-mrs.fr/∼cnotred/Projects-home-page/t-coffee-home-page.html). N- or C-terminal extensions or insertions in the protein sequence of either species that lacked a mirror residue in the homologous sequence (gaps) were not included in the analysis. The alignments around gaps were analyzed to reduce possible ambiguity. Based on a manual analysis of 30 alignments, a set of sequential rules was determined to formalize our definition of ambiguous regions. We consider ambiguous: (1) all the aligned portions between a gap and a perfect match; (2) all aligned segments of at least five amino acids that surrounded ambiguous regions with at least 20% negative score or; (3) any window of 15 amino acids with one third negative alignment.
All amino acids and their respective codons within S. cerevisiae proteins and genes were compared with their aligned residues in the homologous C. albicans protein and DNA sequences. Codon conservation for each species was determined from the alignments and scored according to the codon or amino acid residue present at the same position in the other species and its respective third-position nucleotide (N3). A triple alignment was also performed between C. albicans, S. cerevisiae, and S. pombeproteins, using the same method already described, to determine the origin of the leucine codons present in modern day C. albicans strains.
WEB SITE REFERENCES
ftp://ftp.ncbi.nih.gov; GenBank.
http://chuma.cas.usf.edu/∼garey/alignments/alignment.html; small subunit rRNA sequences aligned in this paper using the DCSE program.
http://igs-server.cnrs-mrs.fr/∼cnotred/Projects-home-page/t-coffee-home-page.html; T-Coffee.
http://rna.wustl.edu/GtRDB/; the S. cerevisiae, S. pombe, and H. sapiens tRNAs used in this paper.
http://www.activeperl.com; ActivePerl 5.6.1.
http://www.bioperl.org; BioPerl 1.0.
http://www.bio.ua.pt/genomica/Lab/Genomedata.html; tRNAs identified from the C. albicans genome.
http://www.ncbi.nlm.nih.gov/BLAST/; BLAST.
http://www-sequence.stanford.edu/group/candida; Stanford DNA Sequencing and Technology Centre.
http://www.ukc.ac.uk/bio/tuite/research/tRNA.htm; C. albicans tRNA sequences used in this paper.
Acknowledgments
The research reported here was supported by project grants to M.A.S.S. from the Portuguese Foundation for Science and Technology (FCT; Projects POCTI REFs: BME/32938, 32942/99 and BME/39030/01) and the EMBO YIP Programme, and to M.F.T. and M.A.S.S. from the Biotechnological Biological Research Council (BBSRC). G.M. is supported by a postdoctoral research fellowship (SFRH/BPD/7195/2001) from FCT. We thank Nancy Federspiel for her kind permission to use the C. albicans genome data. Finally, we are most thankful to Justin O'Sullivan for his helpful discussions and input into this study.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL msantos@bio.ua.pt; FAX 351234426408.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.811003.
REFERENCES
- 1.Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Averof M., Rokas, A., Wolfe, K.H., and Sharp, P.M. 2000. Evidence for a high frequency of simultaneous double-nucleotide substitutions. Science 287: 1283-1286. [DOI] [PubMed] [Google Scholar]
- 3.Bjork G.R. 1986. Transfer RNA modification in different organisms. Chem. Scripta 26: 91-95. [Google Scholar]
- 4.Bjork G.R., Ericson, J.U., Gustafsson, C.E., Hagervall, T.G., Jonsson, Y.H., and Wikstrom, P.M. 1987. Transfer RNA modification. Annu. Rev. Biochem. 56: 263-287. [DOI] [PubMed] [Google Scholar]
- 5.Bjork G.R., Wikstrom, P.M., and Bystrom, A.S. 1989. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244: 986-989. [DOI] [PubMed] [Google Scholar]
- 6.Bjork G.R., Jacobsson, K., Nilsson, K., Johansson, M.J., Bystrom, A.S., and Persson, O.P. 2001. A primordial tRNA modification required for the evolution of life? EMBO J. 20: 231-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitschopf K. and Gross, H.J. 1994. The exchange of the discriminator base A73 for G is alone sufficient to convert human tRNALeu into a serine-acceptor in vitro. EMBO J. 13: 3166-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breitschopf K., Achsel, T., Busch, K., and Gross, H.J. 1995. Identity elements of human tRNALeu: Structural requirements for converting human tRNASer into a leucine acceptor in vitro. Nucleic Acids Res. 23: 3633-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Rijk P. and De Wachter, R. 1993. DCSE, an interactive tool for sequence alignment and secondary structure research. Comput. Appl. Biosci. 9: 735-740. [DOI] [PubMed] [Google Scholar]
- 10.Hagervall T.G., Ericson, J.U., Esberg, K.B., Li, J.M., and Bjork, G.R. 1990. Role of tRNA modification in translational fidelity. Biochim. Biophys. Acta 1050: 263-266. [DOI] [PubMed] [Google Scholar]
- 11.Hagervall T.G., Tuohy, T.M., Atkins, J.F., and Bjork, G.R. 1993. Deficiency of 1-methylguanosine in tRNA from Salmonella typhimurium induces frameshifting by quadruplet translocation. J. Mol. Biol. 232: 756-765. [DOI] [PubMed] [Google Scholar]
- 12.Himeno H., Yoshida, S., Soma, A., and Nishikawa, K. 1997. Only one nucleotide insertion to the long variable arm confers an efficient serine acceptor activity upon Saccharomyces cerevisiae tRNALeu in vitro. J. Mol. Biol. 268: 704-711. [DOI] [PubMed] [Google Scholar]
- 13.Holmquist R., Jukes, T.H., and Pangburn, S. 1973. Evolution of transfer RNA. J. Mol. Biol. 78: 91-116. [DOI] [PubMed] [Google Scholar]
- 14.Ikemura T. 1981a. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 146: 1-21. [DOI] [PubMed] [Google Scholar]
- 15.___, 1981b. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: A proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 151: 389-409. [DOI] [PubMed] [Google Scholar]
- 16.Jukes T.H. and Osawa, S. 1993. Evolutionary changes in the genetic code. Comp. Biochem. Physiol. 106: 489-494. [DOI] [PubMed] [Google Scholar]
- 17.Kawaguchi Y., Honda, H., Taniguchi-Morimura, J., and Iwasaki, S. 1989. The codon CUG is read as serine in an asporogenic yeast Candida cylindracea. Nature 341: 164-166. [DOI] [PubMed] [Google Scholar]
- 18.Kimura M. 1980. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111-120. [DOI] [PubMed] [Google Scholar]
- 19.Knight R.D., Landweber, L.F., and Yarus, M. 2001a. How mitochondria redefine the code. J. Mol. Evol. 53: 299-313. [DOI] [PubMed] [Google Scholar]
- 20.Knight R.D., Freeland, S.J., and Landweber, L.F. 2001b. Rewriting the keyboard: Evolvability of the genetic code. Nat. Rev. Genet. 2: 49-58. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S., Tamura, K., Jakobsen, I.B., and Nei, M. 2001. MEGA2: Molecular Evolutionary Genetics Analysis software. Bioinformatics 17: 1244-1245. [DOI] [PubMed] [Google Scholar]
- 22.Li J.N. and Bjork, G.R. 1995. 1-Methylguanosine deficiency of tRNA influences cognate codon interaction and metabolism in Salmonella typhimurium. J. Bacteriol. 177: 6593-6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Esberg, B., Curran, J.F., and Bjork, G.R. 1997. Three modified nucleosides present in the anticodon stem and loop influence the in vivo aa-tRNA selection in a tRNA-dependent manner. J. Mol. Biol. 271: 209-221. [DOI] [PubMed] [Google Scholar]
- 24.Lovett P.S., Ambulos, N.P., Mulbry, W., Noguchi, N., and Rogers, E.J. 1991. UGA can be decoded as tryptophan at low efficiency in Bacillus subtilis. J. Bacteriol. 173: 1810-1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowe T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lum P.Y., Edwards, S., and Wright, R. 1996. Molecular, functional and evolutionary characterization of the gene encoding HMG-CoA reductase in the fission yeast, Schizosaccharomyces pombe. Yeast 12: 1107-1124. [DOI] [PubMed] [Google Scholar]
- 27.Nei M. and Kumar, S., 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY.
- 28.Notredame C., Higgins, D., and Heringa, J. 2000. T-Coffee: A novel method for multiple sequence alignments. J. Mol. Biol. 302: 205-217. [DOI] [PubMed] [Google Scholar]
- 29.Ohama T., Muto, A., and Osawa, S. 1990. Role of GC-biased mutation pressure on synonymous codon choice in Micrococcus luteus, a bacterium with a high genomic GC-content. Nucleic Acids Res. 18: 1565-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohama T., Suzuki, T., Mori, M., Osawa, S., Ueda, T., Watanabe, K., and Nakase, T. 1993. Non-universal decoding of the leucine codon CTG in several Candida species. Nucleic Acids Res. 21: 4039-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osawa S. 1995. Codon usage. In Evolution of the genetic code (ed. S. Osawa), pp. 45–57. Oxford University Press, New York, NY.
- 32.Osawa S. and Jukes, T.H. 1989. Codon reassignment (codon capture) in evolution. J. Mol. Evol. 28: 271-278. [DOI] [PubMed] [Google Scholar]
- 33.Osawa S., Collins, D., Ohama, T., Jukes, T.H., and Watanabe, K. 1990. Evolution of the mitochondrial genetic code. III. Reassignment of CTN codons from leucine to threonine during evolution of yeast mitochondria. J. Mol. Evol. 30: 322-328. [DOI] [PubMed] [Google Scholar]
- 34.Osawa S., Jukes, T.H., Watanabe, K., and Muto, A. 1992. Recent evidence for evolution of the genetic code. Microbiol. Rev. 5: 229-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perreau V.M., Keith, G., Holmes, W.M., Przykorska, A., Santos, M.A., and Tuite, M.F. 1999. The Candida albicans CTG-decoding Ser-tRNA has an atypical anticodon stem–loop structure. Mol. Biol. 293: 1039-1053. [DOI] [PubMed] [Google Scholar]
- 36.Pesole G., Lotti, M., Alberguina, L., and Saccone, C. 1995. Evolutionary origin of nonuniversal CTG Ser codon in some Candida species as inferred from a molecular phylogeny. Genetics 141: 903-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reyes V.M. and Abelson, J. 1988. Substrate recognition and splice site determination in yeast tRNA splicing. Cell 55: 719-730. [DOI] [PubMed] [Google Scholar]
- 38.Rivera M.C., Jain, R., Moore, J.E., and Lake, J.A. 1998. Genomic evidence for two functionally distinct gene classes. Proc. Natl. Acad. Sci. 95: 6239-6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Santos M.A.S. and Tuite, M.F. 1995. The CTG codon is decoded in vivo as serine and not leucine in Candida albicans. Nucleic Acids Res. 23: 1481-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santos M.A.S., Keith, G., and Tuite, M.F. 1993. Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5′-CAG-3′ (leucine) anticodon. EMBO J. 12: 607-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Santos M.A.S., Perreau, V., and Tuite, M.F. 1996. Transfer RNA structural change is a key element in the reassignment of the CUG codon in Candida albicans. EMBO J. 15: 5060-5068. [PMC free article] [PubMed] [Google Scholar]
- 42.Santos M.A.S., Cheesman, C., Costa, V., Moradas-Ferreira, P., and Tuite, M.F. 1999. Selective advantages created by codon ambiguity allowed for evolution of an alternative genetic code in Candida spp. Mol. Microbiol. 31: 937-947. [DOI] [PubMed] [Google Scholar]
- 43.Schultz D.W. and Yarus, M. 1994. Transfer RNA mutation and the malleability of the genetic code. J. Mol. Biol. 235: 1377-1380. [DOI] [PubMed] [Google Scholar]
- 44.___, 1996. On malleability in the genetic code. J. Mol. Evol. 42: 597-601. [DOI] [PubMed] [Google Scholar]
- 45.Soma A., Kumagai, R., Nishikawa, K., and Himeno, H. 1996. The anticodon loop is a major identity determinant of Saccharomyces cerevisiae tRNALeu. J. Mol. Biol. 263: 707-714. [DOI] [PubMed] [Google Scholar]
- 46.Sprinzl M., Horn, C., Brown, M., Ioudovitch, A., and Steinberg, S. 1998. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26: 148-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugita T. and Nakase, T. 1999. Non-universal usage of the leucine CTG codon and the molecular phylogeny of the genus Candida. Syst. Appl. Microbiol. 22: 79-86. [DOI] [PubMed] [Google Scholar]
- 48.Sugiyama H., Ohkuma, M., Masuda, Y., Park, S.M., Ohta, A., and Takagi, M. 1995. In vivo evidence for non-universal usage of the codon CTG in Candida maltosa. Yeast 11: 43-52. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki T., Ueda, T., Yokogawa, T., Nishigawa, K., and Watanabe, K. 1994. Characterization of serine and leucine tRNAs in an asporogenic yeast Candida cylindracea and evolutionary implications of genes for tRNACAGSer responsible for translation of a non-universal genetic code. Nucleic Acids Res. 22: 115-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suzuki T., Ueda, T., and Watanabe, K. 1997. The ‘polysemous’ codon—A codon with multiple amino acid assignment caused by dual specificity of tRNA identity. EMBO J. 16: 1122-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Swofford D.I., 1993. PAUP: Phylogenetic analysis using parsimony, version 3.11. Illinois Natural History Survey, Champaign, IL.
- 52.Ueda T., Suzuki, T., Yokogawa, T., Nishigawa, K., and Watanabe, K. 1994. Unique structure of new serine tRNAs responsible for decoding leucine codon CTG in various Candida species and their putative ancestral tRNA genes. Biochimie 76: 1217-1222. [DOI] [PubMed] [Google Scholar]
- 53.Urbonavicius J., Qian, Q., Durand, J.M., Hagervall, T.G., and Bjork, G.R. 2001. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 20: 4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yarus M. and Schultz, D.W. 1997. Further comments on codon reassignment. Response. J. Mol. Evol. 45: 3-6. [DOI] [PubMed] [Google Scholar]
- 55.Yokogawa T., Suzuki, T., Ueda, T., Mori, M., Ohama, T., Kuchino, Y., Yoshinari, S., Motoki, I., Nishigawa, K., Osawa, S., et al. 1992. Serine tRNA complementary to the nonuniversal serine codon CTG in Candida cylindracea: Evolutionary implications. Proc. Natl. Acad. Sci. 89: 7408-7411. [DOI] [PMC free article] [PubMed] [Google Scholar]