Abstract
Background and Purpose
The transmembrane protein LINGO-1 is a negative regulator in the nervous system mainly affecting axonal regeneration, neuronal survival, oligodendrocyte differentiation and myelination. However, the molecular mechanisms regulating its functions are poorly understood. In the present study, we investigated the formation and the role of LINGO-1 cis-dimers in the regulation of its biological activity.
Experimental Approach
LINGO-1 homodimers were identified in both HEK293 and SH-SY5Y cells using co-immunoprecipitation experiments and BRET saturation analysis. We performed a hypothesis-driven screen for identification of small-molecule protein–protein interaction modulators of LINGO-1 using a BRET-based assay, adapted for screening. The compound identified was further assessed for effects on LINGO-1 downstream signalling pathways using Western blotting analysis and AlphaScreen technology.
Key Results
LINGO-1 was present as homodimers in primary neuronal cultures. LINGO-1 interacted homotypically in cis-orientation and LINGO-1 cis-dimers were formed early during LINGO-1 biosynthesis. A BRET-based assay allowed us to identify phenoxybenzamine as the first conformational modulator of LINGO-1 dimers. In HEK-293 cells, phenoxybenzamine was a positive modulator of LINGO-1 function, increasing the LINGO-1-mediated inhibition of EGF receptor signalling and Erk phosphorylation.
Conclusions and Implications
Our data suggest that LINGO-1 forms constitutive cis-dimers at the plasma membrane and that low MW compounds affecting the conformational state of these dimers can regulate LINGO-1 downstream signalling pathways. We propose that targeting the LINGO-1 dimerization interface opens a new pharmacological approach to the modulation of its function and provides a new strategy for drug discovery.
Tables of Links
| LIGANDS |
|---|
| Phenoxybenzamine |
| EGF |
These Tables list key protein targets and ligands in this article which are hyperlinked to corresponding entries in http://www.guidetopharmacology.org, the common portal for data from the IUPHAR/BPS Guide to PHARMACOLOGY (Pawson et al., 2014) and are permanently archived in the Concise Guide to PHARMACOLOGY 2013/14 (a,b,cAlexander et al., 2013a,b,c,,).
Introduction
The transmembrane protein LINGO-1 is a new therapeutic target in several neurodegenerative diseases including Parkinson's disease, chronic glaucoma, amyotrophic lateral sclerosis and multiple sclerosis (Mi, 2008; Rudick et al., 2008; Bessero and Clarke, 2010; McDonald et al., 2011). This single transmembrane protein composed of 620 amino acids displays a large extracellular region consisting of 12 leucine-rich repeat motifs followed by an immunoglobulin domain, and a short cytoplasmic tail. LINGO-1 is selectively expressed in brain and spinal cord on both oligodendrocytes and neurons, and it is not detectable in non-neural tissues (Carim-Todd et al., 2003; Mi et al., 2004; Okafuji and Tanaka, 2005; Barrette et al., 2007; Llorens et al., 2008).
LINGO-1 is a potent negative regulator of neuritic growth and neuronal survival. The combination of four different approaches to reducing endogenous LINGO-1 function in neurons (LINGO-1 RNAi, dominant negative LINGO-1, LINGO-1-Fc and LINGO-1 knockout) demonstrated that the inhibition of LINGO-1 improves survival of dopaminergic neurons (Inoue et al., 2007) and increases neuronal differentiation (Loov et al., 2012), neurite outgrowth (Mi et al., 2004) and axonal sprouting (Ji et al., 2006a).
LINGO-1 also plays a key role in the negative regulation of oligodendrocyte differentiation as well as myelination and remyelination. LINGO-1 functions as a constitutive inhibitor of the differentiation of oligodendrocyte precursor cells (OPCs) into mature oligodendrocytes. All of the strategies for reducing endogenous LINGO-1 function in OPCs invariably result in the observation of morphological changes that are characteristic of highly differentiated and mature cells of the oligodendroglial lineage (Mi et al., 2005; 2009,). By contrast, the over-expression of full-length LINGO-1 had the opposite effects (Mi et al., 2005; Lee et al., 2007; Zhao et al., 2007).
Although LINGO-1 was discovered over 10 years ago (Carim-Todd et al., 2003), the molecular mechanisms involved in the regulation of its functions have remained elusive. Nevertheless, LINGO-1 has been shown to interact homotypically and heterotypically with other membrane proteins. LINGO-1 physically interacts likely in cis (i.e on the same plasma membrane) with the Nogo-A receptor (NgR1) and the p75 neurotrophin receptor (p75NTR; Mi et al., 2004). The formation of the ternary complex LINGO-1/NgR1/p75NTR on neuronal growth cones is essential for the inhibition of neurite regeneration and axonal outgrowth (Mi et al., 2004). The ligands of this complex, Nogo-66, myelin-associated glycoprotein and oligodendrocyte-myelin glycoprotein, are myelin components that each bind to NgR1, using its coreceptors to transduce the activation of the RhoA pathway thereby leading to actin depolymerization and then the collapse or retraction of neurites (Yamashita et al., 2005). LINGO-1 inhibitors (LINGO-1-Fc, LINGO-1-ΔCter) block RhoA activation and reverse the growth inhibitory effects of myelin components (Mi et al., 2004). LINGO-1 also interacts with and mediates the action of TNF receptor orphan Y (TROY). The higher expression of TROY compared with p75NTR in the brain, particularly after birth suggests that TROY can substitute for p75NTR in the LINGO-1/NgR1 complex to activate the RhoA pathway upon interaction with NgR1 ligands (Park et al., 2005; Shao et al., 2005). However, the existence of additional signalling co-factors has been suggested based on the absence of p75 and TROY on LINGO-1 expressing neurons projecting to the spinal cord (Barrette et al., 2007). Zhang and collaborators have identified several candidates co-factors that directly interact with LINGO-1 using the intracellular domain of Lingo-1 as bait, including the serine threonine kinase With No Lysine K (WNK1; Zhang et al., 2009). In addition, LINGO-1 is also able to directly interact with the EGF receptor (EGFR) thereby inhibiting its downstream signalling and leading to the decrease of neuronal survival (Inoue et al., 2007). Until recently, it was considered unlikely that LINGO-1 functioned as a stand-alone receptor because its cytoplasmic domain did not appear to contain any signalling motifs, and no known soluble ligand has yet been identified. However, a recent study demonstrated that LINGO-1 functions both as a ligand and a receptor through trans self-interactions (i.e. between two cells) to inhibit myelination (Jepson et al., 2012). Interestingly, it has been proposed that the secreted glycoprotein myocilin interacts with LINGO-1 and may be considered as a LINGO-1 ligand (Kwon et al., 2014).
The role of dimerization as a step for receptor activation is generally accepted for several classes of receptors. There is some evidence that some RTKs are activated by ligand-induced dimerization (Schlessinger, 2002; Simi and Ibanez, 2010). An increasing number of studies also indicate that some RTKs and GPCRs are present as pre-formed dimers and are activated by a conformational change within the dimers upon agonist addition (Ferre et al., 2014; Maruyama, 2014). Receptor dimerization is recognized as a crucial step in the activation of many plasma membrane receptors (Simi and Ibanez, 2010). Therefore, the aim of the present study was first to explore whether LINGO-1 could interact homotypically in cis- orientation and then, to identify low MW compounds able to bind to these oligomeric complexes, to modulate their conformation and subsequently LINGO-1-mediated signalling. Using the BRET method, we demonstrate that LINGO-1 forms homodimers through cis-orientation in living cells. We report that this constitutive dimerization occurs both in endoplasmic reticulum (ER) and on the plasma membrane. We also demonstrate that a LINGO-1 BRET-based assay may be used as a robust and a mainstreamed platform for screening of LINGO-1 ligands and/or small-molecule protein–protein interaction modulators (SMPPIMs). Finally, the screening of approximately 1000 chemical compounds allowed us to identify phenoxybenzamine as a low MW compound able to enhance LINGO-1-mediated intracellular signalling in HEK-293 cells.
Methods
Antibodies
For Western blot analysis, polyclonal anti-LINGO-1 was purchased from R & D Systems (Abingdon, UK); anti-haemagglutinin-tag (HA) from Roche Diagnostics (Meylan, France), anti-Flag from Sigma Aldrich (St Louis, MO, USA), a living colour anti-YFP antibody from BD Bioscience Clontech; anti Na/K ATPase from Tebu-bio (Le Perray-en-Yvelines, France) and anti-calnexin from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Polyclonal anti-phospho-EGFR (Tyr1173), phospho-Akt (Ser473) and phosphorylated ERK1/2 (pERK1/2) were obtained from Cell Signaling Technology (Danvers, MA, USA). Goat anti-mouse and goat anti-rat-IgG HRP-linked whole antibodies were obtained from Invitrogen Life Technologies.
Plasmid constructs
The human HA-tagged LINGO-1 M1-I620 (LINGO-1-HA) was amplified by PCR from the plasmid corresponding to the clone IMAGE ID (GenBank accession number NM_032808) with specific primers. Primers were designed to introduce the sequence of the HA tag in the C-terminal part of LINGO-1. Then, the PCR product was subcloned into pcDNA3 (Invitrogen) using HindIII and EcoRI restriction sites. To generate LINGO-1 with C-terminal fusions of YFP (LINGO-1-YFP) and Renilla luciferase (RLuc; LINGO-1-RLuc), the sequence of LINGO-1 without the HA tag was amplified from the LINGO-1-HA plasmid. The PCR product was then subcloned in frame into the Sal1/BamH1 site of the pEYFP-N1 vector encoding the YFP variant of green fluorescent protein, and into the pRLuc-N1 vector. The pRLuc-N1 vector was obtained by replacing the sequence of YFP in the peYFP-N1 plasmid using AgeI/BsrGI enzymes with the sequence of RLuc amplified from the pRL-CMV vector (Promega). Human cDNA corresponding to the 5-HT6 receptor without its stop codon was amplified by PCR from the plasmid pRK5 containing the cDNA of the human 5-HT6 receptor (kindly provided by Philippe Marin) with specific primers and subcloned into the pEYFP-N1 and pRLuc-N1 vectors using the EcoRI and KpN1 restriction sites to generate 5-HT6 receptors with C-terminal fusions of YFP (5-HT6-YFP) and RLuc (5HT6-RLuc). A truncated LINGO-1, M1-W582 (LINGO-1-ΔCter), containing the extracellular and the transmembrane domain of LINGO-1, was cloned into the HindIII and Nhe1 sites of the pcDNA3. All constructs were checked by direct DNA sequencing. Plasmid containing the trkB sequence was obtained from Yves-Alan Borde (Bibel et al., 1999; Addgene plasmid 39978).
Animals
All animal care was in accordance with the principles of laboratory animal care of guidelines published by the French Ethical Committee (laboratory agreement n° C 45-234-9) and under the supervision of the authorized investigator (n°75-1331). All experimental procedures in this study complied with the animal welfare guidelines of the European Community and were approved by the local ethical committee CECC0 03. Studies involving animals are reported in accordance with the ARRIVE guidelines for reporting experiments involving animals (Kilkenny et al., 2010; McGrath et al., 2010). A total of 6 animals were used in the experiments described here.
Pregnant Sprague Dawley rats were purchased from Janvier (Le Genest-St-Isle, France). They were housed in light-(12 h dark and 12 h light) and temperature-(21°C) controlled rooms with free access to standard dry food and tap water.
Cell cultures and transfections
HEK-293 cells were grown in DMEM supplemented with 10% (vol/vol) FBS, 1 g·L−1 glucose, 100 U·mL−1 penicillin, 0.1 mg·mL−1 streptomycin and 1 mM glutamine. For BRET experiments and for the generation of stable cell lines, cells were transfected with the calcium phosphate precipitation method. Stably transfected HEK-293 cell lines co-expressing LINGO-1-RLuc and LINGO-1-YFP were maintained in the medium described above with the addition of 2 mg·mL−1 of G418.
Cortical neurons were prepared from rat embryos at embryonic day 18. Neurons were plated on polyornithine coated plastic culture dishes (50 000 cells per cm2). Cultures were grown in neurobasal medium (Invitrogen) supplemented with B27 (Invitrogen) and 2 mM L-glutamine.
Cell fractionation studies
Stably transfected cells co-expressing LINGO-1-RLuc and LINGO-1-YFP were washed with PBS, scraped off and lysed with cold lysis buffer (20 mM HEPES, pH 7,4; 2 mM EDTA, 2 mM EGTA, 6 mM MgCl2; 1 mM PMSF and protease cocktail inhibitor). Cell suspensions were sonicated (on ice, three times for 30 s), and lysates were centrifuged at 1000× g for 5 min. The supernatant was collected and 2 M sucrose was added to achieve a final concentration of 0.2 M. Cell lysates were applied to the top of a discontinuous sucrose step gradient (5 mL per step), made at 0.5, 0.9, 1.2, 1.35, 1.5 and 2.0 M sucrose in lysis buffer. The samples were centrifuged in a Beckman SW28 rotor (27 000× g for 16 h). Fractions were then submitted to fluorescence/luminescence and BRET analysis. The identification of plasma membrane and endoplasmic reticulum (ER)-enriched fractions was achieved by Western blot analysis.
BRET measurements
Forty-eight hours after transfection, HEK-293 cells or cultured cortical neurons were detached with versene (Invitrogen) and resuspended in HBSS saline buffer (Invitrogen). Intact cells or membranes were distributed in 96-well microplates (Optiplate, Perkin Elmer) and incubated for 15 min at 25°C in the absence or presence of the indicated ligands. Coelenterazine H substrate (Molecular Probes) was added at a final concentration of 5 μM, and reading was performed with a Mithras LB 940 Multireader (Berthold, Bad Widbad, Germany), which allows the sequential integration of luminescence signals detected with two filter settings (RLuc filter, 485 ± 10 nm; YFP filter, 530 ± 12 nm). Emission signals at 530 nm were divided by emission signals at 485 nm. The BRET ratio was defined as the difference between the emission ratio obtained with co-transfected RLuc and YFP fusion proteins and that obtained with the RLuc fusion protein alone. The results were expressed in milliBRET units (mBU, with 1 mBU corresponding to the BRET ratio values multiplied by 1000). BRETmax is the maximal BRET signal obtained in milliBRET units and BRET50 represents the ratio of acceptor and donor receptors (acceptor/donor) yielding 50% of the maximum BRET signal. All BRET, luminescence, fluorescence measurements were performed at 21°C using a Mithras LB 940 microplate analyser (Berthold, Bad Widbad, Germany).
Co-immunoprecipitation (IP) assays
HEK-293 cells were co-transfected with C-terminal YFP-fused and HA-tagged proteins. Forty-eight hours after transfection, cells were washed with ice-cold PBS and lysed in buffer containing 50 mM Tris pH 7.5, 150 mM NaCl, 10 mM EDTA, 1% Triton X-100 plus protease cocktail inhibitor on ice for 10 min. The lysates were then centrifuged at 10 000× g for 10 min. The supernatants were incubated with EZview Red anti-HA affinity gel (Sigma) or GFP-Trap (chromoTek, Planegg, Germany) for 3 h at 4°C. The beads were washed five times with lysis buffer and resuspended in 4X Laemmli buffer (200 mM Tris-HCl pH 6.8, 4% SDS, 40% glycerol, 0.02% bromophenol and 0.5 M β-mercaptoethanol).
Western blotting
The cell lysates, immunoprecipitates or membranes from HEK-293 transfected cells or cortical neurons were separated by electrophoresis on SDS/PAGE (8% or 10% gels) and transferred to PVDF membranes (GE Healthcare Life Sciences). Blots containing HA or YFP-tagged proteins were probed with a rat anti-HA antibody (1:5000), or a rabbit anti-BD living colours full-length polyclonal antibody (Clontech). Immunoblots were also probed with a goat anti-LINGO-1 polyclonal antibody (LifeSpan, Biosciences, Seattle, WA, USA). For the analysis of EGF signalling pathways, membranes were incubated with antibodies against phospho-EGFR (Tyr1173), phospho-Akt (Ser473) or pERK1/2. HRP-conjugated rabbit anti-goat, anti-mouse or anti-rat antibodies (1:33 000) were used as secondary antibodies. Immunoreactive bands were detected using the Pico or Dura detection kit. For quantification, blots were analysed using Genetools software (Syngene, Frederick, MD, USA).
AlphaScreen assays
The AlphaScreen SureFire phospho-ERK assay (Perkin Elmer) was also used to quantify pERK1/2. HEK-293 cells were transfected using Lipofectamine 2000 with LINGO-1-YFP (HEK-LINGO-1) or with peYFP-N1 plasmids (control cells). At 24 h after transfection, cells were detached and plated in 12-well plates (2 × 105 cells per well). Then, the medium was replaced with medium without serum 16 h before assays. At 48 h after transfection, cells were washed with HBSS buffer and then incubated with vehicle or EGF (5 ng·mL−1) in the absence or presence of phenoxybenzamine (10 μM or specified concentrations) diluted in HBSS. Cells were lysed 7 min after EGF stimulation in lysis buffer (from Perkin Elmer kit) and pERK1/2 was quantified according to the manufacturer's instructions.
Data analysis
Results are expressed as means ± SEM. All data analyses were carried out with the GraphPad Prism 4 software for Windows (GraphPadSoftware Inc, San Diego, CA, USA). Concentration-response curves were fitted by nonlinear regression and saturation curves by a hyperbolic one-binding site equation. The method provided estimates for EC50 values, BRETmax and BRET50 values and their SEM. Statistical analysis of the difference of the BRETmax values or BRET50 values was determined using Student's t test. Statistical significance of the difference in the detected BRET signals between assays with or without protein competitors was determined using one-way ANOVA using GraphPad Prism software. Statistical significance of the effect of phenoxybenzamine observed in HEK control cells compared to those obtained in HEK-LINGO-1 cells was analysed by two-way ANOVA followed by Newman-Keuls multiple comparisons tests, using GraphPad Prism software.
Materials
Tris-HCl, Tween-20, Triton X100 were obtained from Sigma Aldrich (St Louis, MO, USA). PBS was purchased from Euromedex (Souffelweyersheim, France). Coelenterazine substrate was obtained from Interchim (Montluçon, France). A protease inhibitor cocktail was obtained from Roche (Mannheim, Germany). PVDF membranes, CL-X film were purchased from GE Healthcare (Chalfont St. Giles, UK). Dithiobis-succinimidyl propionate (DSP), SuperSignal West Pico and SuperSignal extended Dura chemiluminescent substrates were purchased from Thermo Fisher Scientific Inc. (Rockford, IL, USA); the plasmids peYFP-N1 was purchased from BD Bioscience Clontech (San Jose, CA, USA). Platinium Taq high fidelity DNA polymerase was obtained from Invitrogen™, Life Technologies (Carlsbad, CA, USA). Age,I, HindIII, KpnI, BsrGI and EcoR1 restriction enzymes were obtained from New England Biolabs France (Evry, France). Recombinant human EGF was obtained from Peprotech (Rocky Hill, CT, USA). Clones IMAGE ID coding for LINGO-1 was from BioValley (Conches, France). We thank M Cazorla for kindly providing the tropomyosin-related kinase B (trkB)-yellow fluorescent protein (YFP) plasmid. All media for cell cultures were obtained from PAA Laboratories (Les Mureaux, France). The Prestwick Chemical Library was purchased from Prestwick Chemical Inc (Illkirch, France).
Results
Self-interaction of LINGO-1 in HEK-293 cells and cortical neurons
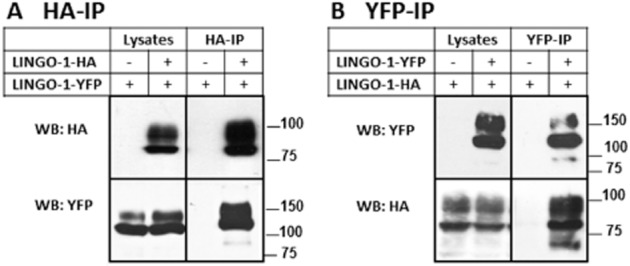
To determine whether LINGO-1 can interact with itself in a cellular system, we performed co-IP analyses of HA-and YFP-tagged LINGO-1 transiently expressed in HEK-293 cells. We tested whether LINGO-1-YFP could be co-immunoprecipitated with the HA-tagged construct by using specific HA antibodies. As a control, the analysis was also carried out with cells expressing LINGO-1-YFP and empty pcDNA3-HA vector only. Under these conditions, YFP-tagged LINGO-1 was specifically co-immunoprecipitated with HA-tagged LINGO-1, indicating an interaction between the two proteins (Figure 1A). To further prove the specificity of our approach, we performed the same experiment in reverse by immunoprecipitating the LINGO-1-YFP with anti-YFP antibodies. As shown in Figure 1B, LINGO-1-HA was specifically co-immunoprecipitated. These results show that LINGO-1 is able to form homotypic interactions in cell lysates.
Figure 1.
Homotypic interactions of LINGO-1 in HEK-293 cells. (A and B) The HA-tagged LINGO-1 was expressed transiently in HEK-293 cells in the presence or absence of YFP-tagged-LINGO-1. In all cases, 1/25 of each lysate was used as an input control (lysates). Cell lysates were immunoprecipitated with the indicated antibodies (IP) and then analysed by 10 or 8% SDS/PAGE and immunoblotted for YFP and HA-tagged constructs. For each experiment, control cells were transfected with empty plasmid controls, pcDNA3.
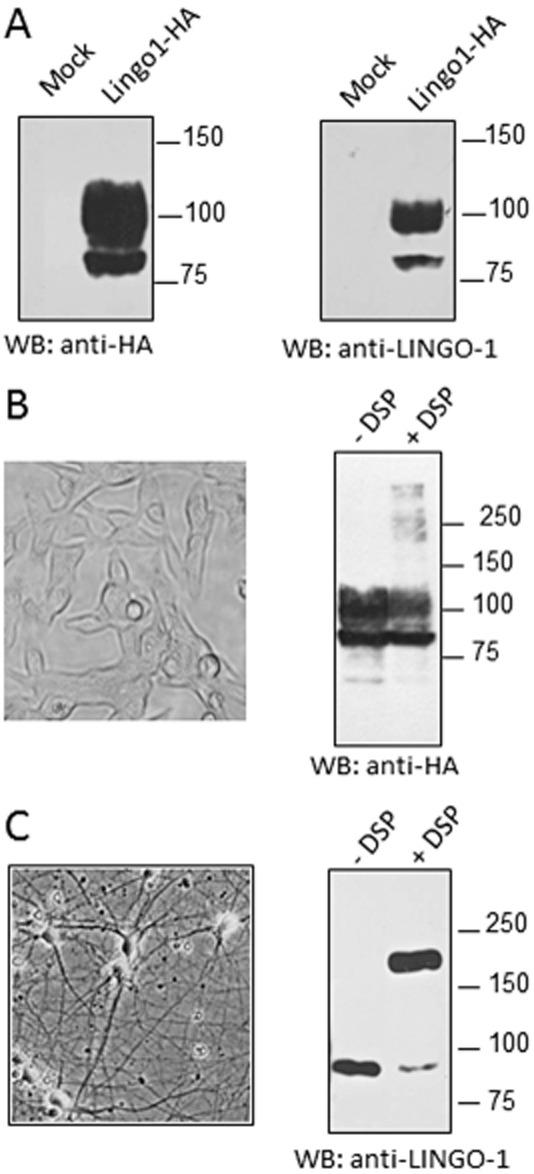
We next examined this self-interaction by testing the formation of complexes using in situ cross-linking in living HEK-293 cells and cortical neurons. For this purpose, we used an anti-LINGO-1 antibody, and its specificity was first evaluated by Western blotting on HEK-293 cell lysates overexpressing LINGO-1-HA and compared with the results obtained with the anti-HA antibody. Both antibodies labelled two prominent immunoreactive species at approximatively 80 and 100 kDa (Figure 2A), corresponding to different states of LINGO-1 glycosylation (Zhong et al., 2009; Supporting Information Fig. S1) and thereby demonstrating the specificity of the anti-LINGO-1 antibody used here. Following chemical cross-linking, cell lysates were subjected to SDS/PAGE under reducing conditions. Western blotting was performed with an anti-LINGO-1 antibody so that oligomeric complexes stabilized by chemical cross-linking could be visualized. This allowed us to detect additional immunoreactive bands at approximately 200 and 250 kDa, corresponding to homotypic interactions of LINGO-1 in living HEK-293 cells (Figure 2B). After the cross-linking of cortical neurons, the specific LINGO-1 antibody labelled two prominent immunoreactive species of 90 and 180 kDa, consistent with monomeric and dimeric forms of the native LINGO-1 protein respectively (Figure 2C). Because chemical cross-linkers have only a limited efficiency, the actual proportions of LINGO-1 monomers, dimers or higher order oligomeric species cannot be determined with this method. Together, these data indicate that LINGO-1 can self-interact in both HEK-293 cells and cortical neurons.
Figure 2.
Detection of LINGO-1 dimers/oligomers in HEK-293 cells and cortical neuron cultures. (A) Lysates from HEK-293 cells expressing pcDNA3-HA (Mock) or HA-tagged LINGO-1 were resolved by 10% SDS-PAGE and analysed by Western blots, with an anti-HA antibody or a specific anti-LINGO-1 antibody. When the cross-linking reagent (DSP) was used, living HEK-293 cells transiently expressing HA-tagged LINGO-1 (B) or cortical neurons 12 days after plating (C) were pretreated with 5 mM DSP for 20 min before lysis. Proteins from cell lysates were then separated by 10% SDS-PAGE and the immunoblots were performed with the indicated antibody.
Constitutive cis-dimerization/ oligomerization of LINGO-1 using BRET analysis
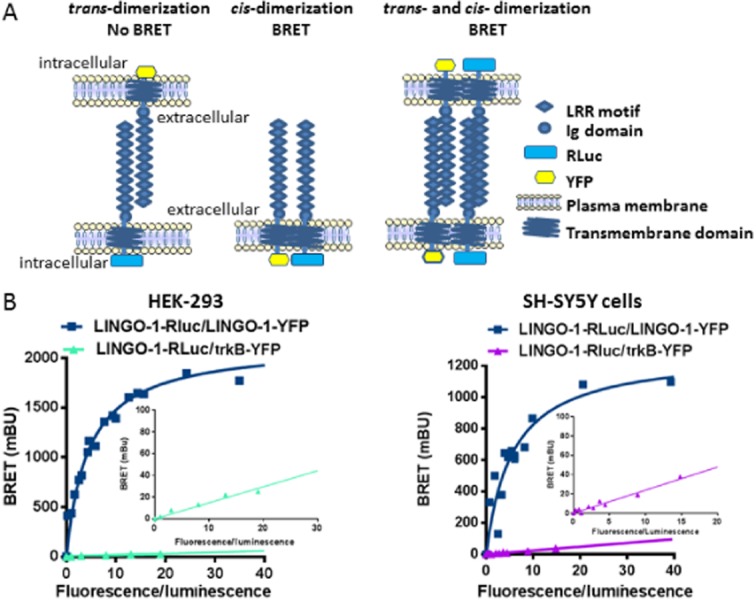
To exclude artefacts generated by the solubilization of membrane-bound receptors, we also studied LINGO-1 self-interaction in intact living cells using BRET analysis. The BRET approach involves the transfer of energy resulting from the degradation of coelenterazine by RLuc, to a YFP, which in turn emits fluorescent light. BRET is strictly dependent on the molecular proximity (10–100 Å) between the energy donor (RLuc) and acceptor (YFP), making it ideal for studying protein–protein interactions in living cells at ‘physiological’ expression levels. The major advantage of this technique over biochemical methods is that protein–protein interactions can be monitored without disrupting the natural environment, which is frequently altered by detergents and membrane preparations. In addition, this method allows for the examination of whether the self-interaction of LINGO-1 that we observed by biochemical approaches occurs in cis-or trans-orientation because a BRET signal is measured only if a cis-interaction takes place (Figure 3A). For this purpose, the homodimerization of LINGO-1 was examined in living HEK-293 and SH-SY5Y cells in suspension using quantitative BRET analysis, with the proper controls required for this approach (James et al., 2006; Bouvier et al., 2007; Bacart et al., 2008; Figure 3B). BRET signals were spontaneously generated by constitutive LINGO-1 dimers in which the C-terminus of one subunit contains YFP (LINGO-1-YFP) and the other contains RLuc (LINGO-1-RLuc). The specificity of this LINGO-1 oligomerization was shown in both cell lines by the very low energy transfer measured in cells co-expressing LINGO-1-Rluc with the trkB receptor, a neurotrophin receptor that it also expressed at the plasma membrane (Figure 3B inset). Data analysis revealed that the BRETmax value obtained in the SH-SY5Y preparation (1288 ± 51 mBU) was lower than that obtained in HEK-293 cells (1725 ± 89 mBU; P < 0.01; n = 4).
Figure 3.
Dimerization of LINGO-1 in HEK-293 cells and neuroblastoma cell line using BRET analysis. (A) Models for LINGO-1 homotypic interaction in trans or cis-orientation and schematic structure of LINGO-1 constructs used in BRET experiments. (B) Adherent HEK-293 cells or SH-SY5Y were transfected transiently with a constant DNA amount of LINGO1-RLuc (25 ng) and increasing amounts (from 0 ng to 2 μg) of LINGO1-YFP, or trkB-YFP), a neurotrophin receptor used as negative controls. The difference between the BRET ratio obtained in cells co-expressing LINGO-1-Rluc and LINGO-1-YFP (BRET ratio varied from 0.8 to 2.45) and the BRET ratio of cells expressing only the LINGO-1-RLuc fusion protein alone (BRET ratio = 0.8) was defined as the BRET value. Results were expressed in milliBRET units (mBU, with 1 mBU corresponding to the BRET values multiplied by 1000). The BRET signal observed between LINGO-1 fusion proteins increases as a hyperbolic curve reaching an asymptote underlying the specificity of the signal. When LINGO-1-Rluc was co-expressed with trkB, a membrane receptor belonging to the LIG protein family, the BRET signal is much lower and increases linearly indicating the occurrence of non-specific and random interactions. This strongly supports that LINGO-1 does not interact with trkB receptors as previously demonstrated by co-IP experiments (Mandai et al., 2009), even if cannot completely exclude the existence of an inappropriate orientation between the donor and the acceptor. Data represent the results of two or three independent experiments read in triplicate.
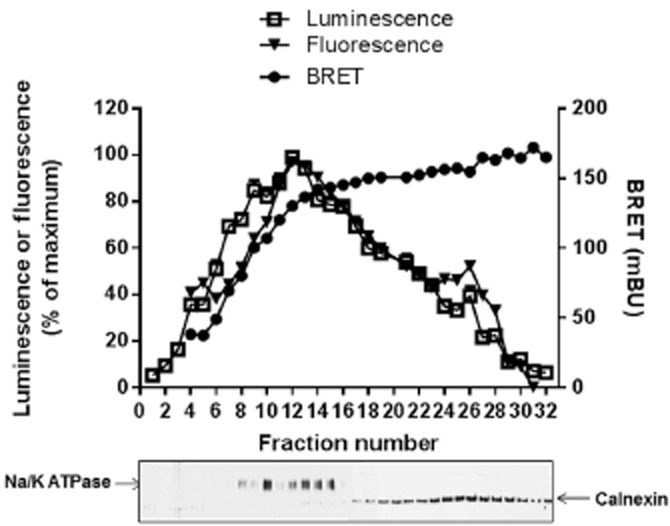
We further analysed the distribution of LINGO-1 dimers by cell fractionation studies. The distributions of the fluorescent and luminescent LINGO-1 both indicated that the vast majority of total LINGO-1 was exported from the ER to the plasma membrane (Figure 4). The BRETmax values were reached not only at the plasma membrane, but also in the ER (Figure 4), indicating that the homo-dimerization of LINGO-1 occurs early during its biosynthesis.
Figure 4.
Constitutive BRET signals between LINGO-1-RLuc and LINGO-1-YFP in plasma membrane and ER. HEK-293 cells stably co-expressing LINGO-1-RLuc and LINGO-1-YFP (basal BRET signal = 175 ± 10 mBu) were lysed and subcellular fractions were resolved on a sucrose gradient. Individual fraction were subjected to fluorescence/luminescence and BRET analysis and resolved by SDS-PAGE and Western blot analysis using anti-NaK-ATPase or anti-calnexin antibodies used as markers of the plasma membrane (fractions 1–15) or the RE (fractions 16–30) respectively.
Validation of BRET-based assays as a screening platform
Because the present study demonstrated that LINGO-1 formed cis-dimers at the plasma membrane, we speculated that the manipulation of this dimerization process may represent an innovative strategy to modulate LINGO-1 function. The identification of proteins and/or molecules interfering with this process may help in further understanding the physiological role of LINGO-1 cis-dimerization. To identify such molecules, we speculated that they may induce conformational changes within LINGO-1 dimers leading to detectable modifications in the measured BRET signals. These modifications may represent changes in the quaternary structure of LINGO-1 (dimerization state of the protein), or its tertiary structure (conformational changes within protomers). To validate the proof of this hypothesis, we tested whether the BRET signal of LINGO-1 dimers could be modulated by low MW compounds as well as by protein partners.
First, competitive BRET assays were performed using various HA-tagged proteins. Interestingly, we demonstrated that the truncated construct lacking the intracellular terminal loop of LINGO-1 (LINGO-1-ΔCter) was able to significantly and specifically reduce the basal BRET signal (Figure 5B) in a manner similar to that observed with the full-length LINGO-1 (Figure 5A). Furthermore, the BRET signal was not modified by increasing concentrations of trkB, a neurotrophin receptor used as a negative control (Figure 5C). We then confirmed that LINGO-1 BRET-based assays could be used for screening aimed at identifying LINGO-1 ligands and SMPPIMs by monitoring conformational changes within dimers and/or dimer disruption. To determine the performance (Z′ score) of the assay, we measured the BRET signal in 384-well microplates from cells transiently expressing LINGO-1-RLuc alone (monomer) or co-expressing LINGO-1-RLuc (constant amount) and increasing concentrations of LINGO-1-YFP yielding various BRET levels (from 15 to 60% of BRETmax). Consistent with excellent assay performance, the test runs established a Z′-score within the 0.8–0.9 range, underlying the high quality and suitability of the assay for screening (Figure 6A).
Figure 5.
BRET competition analysis. The basal BRET signal obtained after the transient co-expression of LINGO-1-RLuc and LINGO-1-YFP in HEK-293 cells (yielding to around 50% of the BRETmax) is significantly reduced by the transfection of increasing concentrations (0 to 2600 ng) of both the full length of LINGO-1 (A) and its truncated form (LINGO-1-ΔCter) (B). The specificity of these inhibitions has been demonstrated by the lack of effect of trkB on the basal BRET signal (C). Values were expressed as the percentage of BRET signal measured in the absence of protein competitors (control) from three independent experiments read in triplicate. The expressions of the transfected HA-tagged competitors were confirmed by Western blot analysis (upper panels). The average of basal BRET signal value from three experiments was 915 ± 31 mBU. *P < 0.05; **P < 0.01, ***P < 0.001, compared with no competitor.
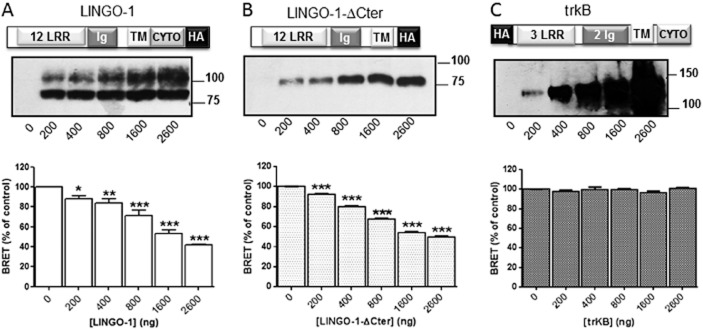
Figure 6.
Validation of the suitability of LINGO-1 BRET-based assay for use in screening. (A) HEK-293 cells were transiently transfected with an equal amount of LINGO-1-RLuc alone or with increasing amounts of LINGO-1-YFP corresponding to various BRET levels (from 15 to 60% of the BRETmax level). Forty-eight hours after transfection, BRET experiments were performed on resuspended cells distributed in white optiplate 384-well plates at a density of 15 000 cells per well. Z′ scores were calculated using the following formula: Z′ = 1-[3 × SD + 3 × SD(monomer)]/(BRET dimers – BRET monomers), where SD = standard deviation. Data shown are representative of two independent experiments. (B) BRET signal reproducibility in a screen searching for ligands/inhibitors of LINGO-1. One hundred and sixty compounds were screened in duplicate on two different groups of cells co-expressing LINGO-1-RLuc and LINGO-1-YFP yielding a BRET ratio of around 1.5. The BRET signals from the duplicate screens were normalized to 1 (1 being the BRET ratio obtained in basal condition).
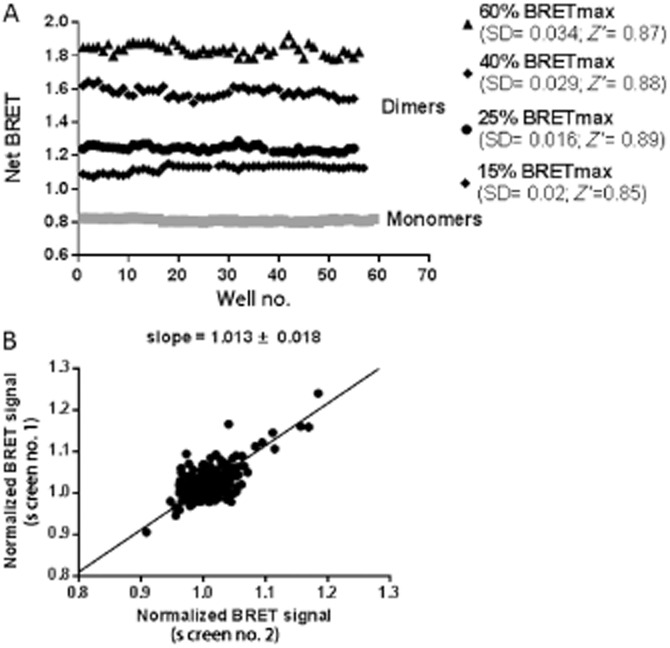
Before starting to screen a large collection of compounds, we tested the signal reproducibility of the assay using 160 compounds belonging to the Prestwick Chemical Library. The screening was performed on a single day with two different transiently transfected cell lines expressing LINGO-1-RLuc and LINGO-1-YFP to reach 50% of BRETmax. As shown in Figure 6B, the LINGO-1 BRET signals obtained by the screening of two different transiently transfected cell lines overlapped well, indicating that the assay is fully reproducible.
Identification of compounds interfering with the LINGO-1 dimerization process
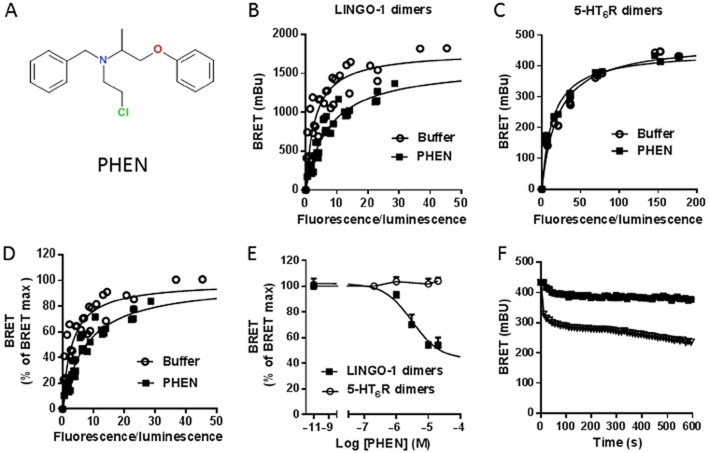
We applied the conditions described above to screen a total of 1263 compounds from the Prestwick chemical library to identify LINGO-1 ligands and/or SMPPIMs of LINGO-1. The primary hit rate of compounds that induced an increase or a decrease of more than three standard deviations of the basal BRET signal was ∼2% allowing the selection of 23 compounds (Table 1). However, some molecules interfering with the absorption properties of the BRET-based assay (fluorescent or coloured compounds, molecules with toxic effects or having quenching effects) may give false positives. To confirm the specificity of these potential LINGO-1 ligands/inhibitors, we tested the effect of these selected compounds on the basal BRET signal produced by the dimerization of an unrelated protein, the 5-HT6 receptor (Table 1). Among the 23 compounds selected in the primary screen, only six were found to be specific for LINGO-1 because they did not modify the BRET signal from 5-HT6 receptor dimers (Table 1). We decided to follow up one compound, phenoxybenzamine (Figure 7A), which produced the strongest decrease (around −40%) of the basal LINGO-1 BRET signal, to test the biological relevance of this hit. Using BRET saturation analysis (Figure 7B), we demonstrated that phenoxybenzamine (10 μM) produced a rightward shift of the BRET saturation curves in cells expressing LINGO-1, whereas it did not affect the BRET signal in cells expressing 5-HT6 receptors (Figure 7C). Because the energy transfer depends on both the relative orientation and the distance between the BRET tags, two parameters (BRET50 and BRETmax) can be modulated upon addition of ligands. The BRETmax level was significantly modified in the presence of phenoxybenzamine (BRETmax = 1629 ± 54 mBU) compared with conditions without phenoxybenzamine (BRETmax = 1805 ± 67 mBU; Figure 7B; P < 0.05). The BRET50 increased to 7.5 ± 1 in the presence of phenoxybenzamine compared with that obtained in its absence (BRET50 = 2.5 ± 0.6; P < 0.01, n = 4; Figure 7B and D). This indicates that phenoxybenzamine leads to a change in the relative affinity between LINGO-1 monomers promoting a dissociation of LINGO-1 performed dimers, mostly upon conformational changes as indicated by the decrease of the BRETmax value. In addition, the phenoxybenzamine-induced decrease of the basal BRET of LINGO-1 dimers was dose dependent with an EC50 = 4 μM, whereas it did not modify the basal BRET of the 5-HT6 receptor dimers (Figure 7E). Finally, although the LINGO-1 BRET signal stayed stable in the presence of buffer, the addition of phenoxybenzamine (20 μM) markedly decreased the BRET signal in a time-dependent manner (Figure 7F). Together, these results indicate that phenoxybenzamine may be considered the first specific SMPPIM of LINGO-1 cis-dimers.
Table 1.
Compounds inducing BRET changes in cells expressing LINGO-1 (primary screen) or 5-HT6 receptors (counterscreen)
| Hits | Screen variation of LINGO-1 BRET signal (% of control) | Counterscreen variation of 5-HT6R BRET signal (% of control) |
|---|---|---|
| BRET signal inhibitors | ||
| Phenoxybenzamine | 57 ± 6 | 100 ± 2 |
| Ethacrynic acid | 87 ± 1 | 100 ± 3 |
| Cyanocobalamin | 84 ± 3 | 100 ± 2 |
| Chicago sky blue | 25 ± 3 | 56 ± 6 |
| Myricetin | 80 ± 4 | 85 ± 4 |
| Amphotericin B | 79 ± 5 | 107 ± 4 |
| BRET signal enhancers | ||
| Bambuterol | 127 ± 9 | 100 ± 2 |
| Clofazimine | 122 ± 4 | 100 ± 1 |
| Pinacidil | 119 ± 3 | 100 ± 2 |
| Nitrofurantoin | 110 ± 2 | 113 ± 2 |
| Balsalazide | 116 ± 2 | 118 ± 2 |
| Perhexiline | 117 ± 5 | 108 ± 3 |
| Nimesulide | 125 ± 4 | 119 ± 2 |
| Fendiline | 125 ± 7 | 118 ± 4 |
| Quinacrine | 127 ± 2 | 140 ± 8 |
| Azapropazone | 134 ± 7 | 129 ± 7 |
| Phenazopyridine | 137 ± 3 | 141 ± 6 |
| Dantrolene | 140 ± 5 | 147 ± 5 |
| Harmalol | 148 ± 10 | 117 ± 2 |
| Riboflavine | 153 ± 10 | 140 ± 5 |
| Nifurtimox | 154 ± 8 | 148 ± 2 |
| Rifampicin | 159 ± 9 | 143 ± 2 |
| Hycanthone | 185 ± 15 | 156 ± 8 |
Cells transiently co-expressing LINGO-1-RLuc and LINGO-1-YFP (screen) or 5-HT6R-RLuc and 5-HT6R-YFP (counterscreen) were resuspended in physiological buffer and distributed in 96-well white plates. Compounds (20 μM in 0.5% of DMSO) from the Prestwick Chemical Library were incubated for 15 min at room temperature. BRET measurement was performed on a Mithras LB940. Results are expressed in percent change of the respective BRET control signal and are the mean ± SEM of two experiments.
Figure 7.
Phenoxybenzamine specifically decreases LINGO-1 BRET signals. In HEK-293 cells (A) Chemical structure of phenoxybenzamine (PHEN). (B and C) The effects of phenoxybenzamine (10 μM) were evaluated on LINGO-1 and 5-HT6 receptor dimers at various BRET levels. BRET titration curves were generated in cells expressing a constant amount of either LINGO-1-RLuc (B) or 5-HT6R-RLuc (C) and increasing concentration of YFP-tagged proteins (LINGO-1-YFP or 5HT6R-YFP). (D) Expression of the data shown in B was expressed as percentage of the BRETmax under basal (buffer) and phenoxybenzamine-added conditions. (E) The effect of increasing concentrations of phenoxybenzamine was evaluated on BRET signal after 20 min incubation on HEK-293 cells co-transfected with LINGO-1-RLuc and LINGO-1-YFP (LINGO-1 dimers) or 5-HT6R-RLuc and 5-HT6R-YFP (5HT6R dimers) to reach equivalent BRET signal. Basal BRET signals were 456 ± 15 mBU and 484 ± 2 mBU for LINGO-1 and 5-HT6 receptor dimers respectively. Phenoxybenzamine decreases significantly and specifically the LINGO-1 BRET signal with an EC50 = 4 μM and does not modify the BRET signal resulting from 5-HT6 receptor dimerization. (F) Whereas the LINGO-1 dimers BRET signal stays stable in the presence of buffer, the addition of phenoxybenzamine (10 μM) markedly decreases the basal BRET signal in a time-dependent manner.
Phenoxybenzamine behaves as a positive modulator of LINGO-1 downstream signalling
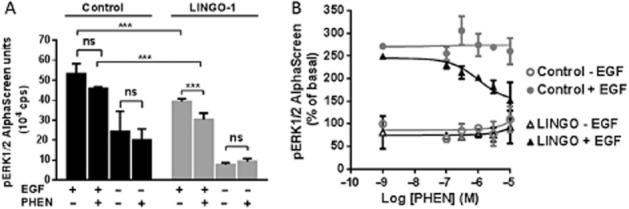
In Cos-7 cells, LINGO-1 activities may be mediated through the inhibition of EGFR downstream signalling (Inoue et al., 2007), a signalling pathway previously documented to be involved in neuronal survival and axonal regeneration as well as oligodendrocyte differentiation (Grimm et al., 2009). Therefore, we decided to explore the effect of phenoxybenzamine on several signalling pathways downstream of EGFR, such as the phosphorylation of EGFR (measured as phosphorylation of Tyr1173), the activation of the MAPK pathway (measured as phosphorylation of ERK 1/2) and the activation of the PI3K pathway (measured as phosphorylation of Akt) in HEK-LINGO-1 cells compared with control cells (Figure 8). To avoid the constitutive activation of these pathways by growth factors contained in the medium, the cells were grown in the absence of serum for 16 h prior to experiments. EGF treatment (5 ng·mL−1) for 7 min induced a strong increase in the phosphorylation of EGFR (+366%; P < 0.0001) and ERK 1/2 (+430%; P < 0.0001), but a lower and non-significant increase in Akt phosphorylation (+15%). After EGF treatment, we observed a significant reduction in EGFR phosphorylation on Tyr1173 of approximately 45% in LINGO-1-transfected cells compared with control cells. Similarly, the levels of Akt and ERK 1/2 phosphorylation were reduced by 35% (P < 0.05) and by 38% (P < 0.01), respectively, in LINGO-1-transfected cells compared with control cells. To elucidate the role of phenoxybenzamine in LINGO-1 function, we first examined its effect in control cells treated with or without EGF treatment. Phenoxybenzamine did not significantly modify either the phosphorylation of EGFR, or the phosphorylation of studied kinases in mock-transfected cells. In contrast, in LINGO-1-transfected cells after EGF treatment, phenoxybenzamine induced a strong and significant decrease of EGFR phosphorylation (73%, P < 0.01) and ERK 1/2 phosphorylation (54%, P < 0.01) but no significant modification of Akt phosphorylation level (Figure 8B–D). To further characterize the effect of phenoxybenzamine on LINGO-1 function, we measured ERK 1/2 activation in HEK-293 cells using the highly sensitive AlphaScreen Phospho-ERK assay. We first verified that this assay gave results similar to those from the Western blot analysis for ERK 1/2 activation (Figure 9A). As expected, EGF-stimulated ERK 1/2 activation was inhibited by 29% after LINGO-1 overexpression compared with control cells. We also confirmed that phenoxybenzamine did not significantly modify the basal ERK 1/2 phosphorylation either in control or in LINGO-1-transfected cells. In contrast, whereas phenoxybenzamine did not significantly inhibit EGF-stimulated ERK activation in control cells, it induced a strong and significant decrease of ERK 1/2 phosphorylation in LINGO-1-transfected cells (−46%, P < 0.001) compared with control cells (−16%; non-significant). A dose-response curve for the effect of phenoxybenzamine was also examined on both basal and EGF-induced ERK1/2 activation. Again, phenoxybenzamine induced a dose-dependent decrease of EGF-induced ERK phosphorylation levels, only in cells expressing LINGO-1, with an EC50 = 2 μM, underlying the specificity of its effect on LINGO-1 (Figure 9B).
Figure 8.
Phenoxybenzamine (PHEN) enhances the LINGO-1 induced inhibition of EGF-stimulated downstream pathways in HEK-293 cells. HEK-293 c ells were transiently transfected with peYFP-N1 control plasmid (control cells) or with YFP-tagged LINGO-1 plasmid (LINGO-1 cells). Forty-eight hours after transfection, cells were stimulated for 7 min with EGF (5 ng·mL−1) in presence of phenoxybenzamine (10 μM) or the vehicle (0.5% DMSO) in HBSS. After cell lysis, EGFR downstream signalling (pEGFR, pERK, pAkt) was analysed by Western blot analysis (A) and were quantified by densitometry (B–D). Equal protein load was verified by analysing the samples with a tubuline antibody. The data shown are the means ± SEM of values from at least three independent experiments. Two-way anova statistical analysis of pEGFR (B) assays was performed as follows: factor cells transfection F(1, 8) = 57.18, P = 0.0001; factor treatment F(3, 8) = 84.5, P < 0.0001; and interaction F(3, 8) = 42.5, P < 0.0001. Then Newman–Keuls multiple comparisons were performed. Two-way anova statistical analysis of pAkt (C) assays was performed as follows: factor cells transfection F(1, 8) = 27.07, P = 0.0008; factor treatment F(3, 8) = 0.9154; and interaction F(3, 8) = 0.2396, P = 0.0415. Then Newman–Keuls multiple comparisons were performed. Two-way anova statistical analysis of pErk (D) assays was performed as followed: factor cells transfection F(1, 8) = 44.35, P = 0.0002; factor treatment F(3, 8) = 52.05, P < 0.0001; and interaction F(3, 8) = 5.114, P = 0.0289. Then, Newman–Keuls multiple comparisons were performed. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus respective control.

Figure 9.
Characterization of the effect of phenoxybenzamine (PHEN) on pERK1/2 signalling. HEK-293 cells were transiently transfected with peYFP-N1 control plasmid (control cells) or with YFP-tagged LINGO-1 plasmid (LINGO-1 cells). Forty-eight hours after transfection, cells were stimulated for 7 min with EGF (5 ng·mL−1) in presence of phenoxybenzamine at 10 μM (A) or at the indicated concentration (B) or the vehicle (0.5% DMSO) in HBSS. After cell lysis, pERK1/2 was quantified using the AlphaScreen assays. Means ± SEM of values from four separate experiments. (A) Two-way anova statistical analysis was performed as follows: factor cells transfection F(1, 25) = 44.17, P < 0.0001; factor treatment F(3, 25) = 148.7, P < 0.0001; and interaction F(3, 25) = 11.94, P = 0.0415. Then, Tukey's multiple comparisons were performed, ***P < 0.001 versus respective control. (B) Tukey's multiple comparisons were performed, ***P < 0.001 versus EGF effect in control cells.
Discussion
Numerous studies have reported negative regulatory functions of LINGO-1 in axonal regeneration, neuronal survival, oligodendrocyte differentiation and myelination (Mi et al., 2008). However, the molecular mechanisms that control these functions are poorly understood.
Within neurons, LINGO-1 has been found to be associated with other membrane proteins typically in a co-receptor function (Mi et al., 2004). Studies based on IP assays after cell solubilization also indicated that LINGO-1 forms homotypic interactions (Jepson et al., 2012; Stein and Walmsley, 2012). However, these earlier studies provided no information on important issues such as the constitutive or induced nature of LINGO-1 homotypic interactions and the putative formation of LINGO-1 cis-dimers in living cells. Using cross-linking experiments and biochemical analyses, we demonstrated for the first time that LINGO-1 formed dimers in both living HEK-293 cells and cortical neurons. Then, in an effort to better characterize LINGO-1 self-interaction in living cells, we took advantage of a biophysical assay based on BRET that was initially developed to monitor the homodimerization of cyanobacteria clock proteins (Xu et al., 1999) and more recently applied to study oligomerization of GPCRs (Ramsay et al., 2002) and activity of RTKs including neurotrophin, insulinotropic or growth factor receptors (Issad et al., 2007; Siddiqui et al., 2013). BRET analysis using the proper controls allowed us to directly demonstrate that LINGO-1 indeed formed cis-dimers/oligomers in living cells. These results are consistent with the model based on the crystal structure of the LINGO-1 ectodomain, showing that it forms a tetrameric structure (Mosyak et al., 2006). By ensuring that the cells were in suspension and present as single cells when the measurements were performed, we minimized the possibility that the observed cis-dimers were secondary to trans-interactions. Moreover, because the known LINGO-1 partners p75NTR, NgR1, TROY (Mi et al., 2004), trkA and trkC (Mandai et al., 2009) are not expressed in HEK-293 cells, our results provide evidence that interactions with these proteins are not necessary for the formation of LINGO-1 dimers. Furthermore, their expression in both cortical neurons and SH-SY5Y cells does not prevent LINGO-1 oligomer formation. Indeed, although the maximal BRET value obtained in SH-SY5Y cells was lower than that observed in HEK-293 cells, this difference does not reflect a lower number of dimers formed in these cells but is more likely to be due to differences in the relative orientation of the dimers. Moreover, protein binding partners of LINGO-1, as well as the lipid composition of membranes, may influence the BRET parameters (Couturier and Jockers, 2003). The detection of BRET signals both at the cell surface and in the ER after cell fractionation on a sucrose gradient indicates that receptors exist as dimers/oligomers in both compartments. Oligomeric assembly within the ER is a general feature of the export system that controls the quality of several plasma membrane proteins such as ion channels (Ma et al., 2001) and GPCRs (White et al., 1998; Bulenger et al., 2005). The decrease in the LINGO-1 BRET signal induced by LINGO-1-ΔCter is likely to reflect a reduction in the formation of wild-type LINGO-1 dimers as a consequence of the formation of heterodimers (LINGO-1/LINGO-1-ΔCter). More importantly, by forming heterodimers with full-length LINGO-1 in the ER, this truncated form prevents LINGO-1 from reaching the plasma membrane (Supporting Information Fig. S2). Thus, we propose that the increased neuronal survival and/or the higher myelination competence previously reported to be induced by LINGO-1-ΔCter overexpression (Mi et al., 2005; Inoue et al., 2007) may reflect an alteration of LINGO-1 maturation and/or the targeting of functional dimers to the cell surface. Therefore, the inhibition of LINGO-1 targeting to the cell surface may represent a new strategy to modulate its functions, particularly because an up-regulation of LINGO-1 expression has been observed in pathological conditions such as in animal models of Parkinson disease (Inoue et al., 2007) or spinal cord injury (Gerin et al., 2011).
A recent study demonstrated that LINGO-1 was capable of self-association in trans interactions and that such intercellular interactions are involved in the inhibition of OPC differentiation and myelination. It has been proposed that the disruption of such protein–protein interactions may be useful to reduce LINGO-1 function and improve myelination (Jepson et al., 2012). Due to the relatively large surfaces that are typically involved in protein–protein pairing, attempts to modulate such interactions with drug-like, low MW compounds have historically been considered challenging (Arkin and Wells, 2004; Thompson et al., 2012). However, a number of recent successful examples have generated increased interest in this approach (Fry, 2008), and protein–protein interaction disruptors in signalling networks have been recently identified, and their therapeutic properties have been demonstrated in addiction, pain (Ji et al., 2006b; Liu et al., 2008; Pichon et al., 2010) and cancer (Qi et al., 2013). Together, these data suggest that the manipulation of protein–protein interactions represents an innovative strategy to modulate protein functions and therefore that SMPPIMs may be considered as compounds with promising therapeutic applications.
Our observation that LINGO-1 forms cis-homodimers in living cells using a BRET-based homogeneous assay allowed us to suggest that this simple and rapid method could be applied to the screening of LINGO-1 ligands and/or SMPPIMs (Couturier and Deprez, 2012). Because BRET depends strictly on the molecular proximity and the orientation between donor and acceptor, it is theoretically applicable to study conformational changes as a consequence of ligand binding (De Vries et al., 2010; Couturier and Deprez, 2012). Our present data demonstrate that our LINGO-1 BRET-based assay is of high quality and suitable for use in screening (Z′ score > 0.85) and the screening of around 1000 chemical compounds led us to identify one molecule, phenoxybenzamine, that specifically and dose-dependently decreased the LINGO-1 BRET signal. Phenoxybenzamine is an irreversible subtype-nonselective α-adrenoceptor antagonist and has been used to treat hypertension and as a peripheral vasodilator with a long duration of action. Site-directed mutation studies on the α2-adrenoceptor revealed that the covalent binding of phenoxybenzamine to the receptor involves the solvent-exposed cysteine residue in the third transmembrane domain (Frang et al., 2001). Based on the crystal structure of LINGO-1 (Mosyak et al., 2006), all of the cysteine residues in the extracellular domain of the protein are engaged either in disulfide bridges or buried in the protein hydrophobic core, and they are thus unlikely to react with phenoxybenzamine (Supporting Information Fig. S3). We also exclude the role of the transmembrane cysteine C575 in the action of phenoxybenzamine as shown by BRET saturation curves experiments (Supporting Information Fig. S4). Moreover, the reactivity of phenoxybenzamine depends on the formation of a reactive aziridinium intermediate, which in turn depends on steric features of the N-benzyl groups that are released when phenoxybenzamine is bound to adrenoceptors. Consequently, structural and mechanistic considerations eliminate the possibility of covalent binding of phenoxybenzamine to LINGO-1, consistent with our experimental findings (Supporting Information Fig. S5). We therefore propose that phenoxybenzamine induces conformational changes within the LINGO-1 dimer, promoting a dissociation of preformed LINGO-1 dimers.
To evaluate whether phenoxybenzamine modulated biological processes downstream of LINGO-1, we tested its effect on EGFR signalling in HEK-293 cells overexpressing LINGO-1 compared with control cells. The EGF-induced activation of EGFR occurs through receptor dimerization, which induces the activation of several transduction pathways, including MAPK (Ras, Raf, MEK, Erk) and the PI3K pathways (Ras, PI3K, Akt; Jorissen et al., 2003). A previous study demonstrated that the overexpression of LINGO-1 inhibits signalling downstream of EGFR and that some LINGO-1 inhibitors (LINGO-1-Fc, LINGO-1-ΔCter or anti-LINGO-1 antibody) can suppress the inhibitory effects of LINGO-1 on EGFR activity and PI3K/Akt signalling (Inoue et al., 2007). Our data demonstrated that the overexpression of LINGO-1 not only significantly inhibited the EGF-induced MAPK pathway (Akt phosphorylation), as previously reported (Inoue et al., 2007), but also the EGF-induced activation of ERK. Although the mechanisms by which LINGO-1 mediates inhibition of the ERK pathway should be further characterized, It is possible that WNK1 is involved, as this kinase is known to physically interact with LINGO-1 (Zhang et al., 2009) and to activate downstream cascades in response to EGF (Xu et al., 2004; Huang et al., 2008).
We also demonstrated that phenoxybenzamine acts as a positive modulator of LINGO-1 by increasing the inhibitory action of LINGO-1 on EGFR downstream signalling. Phenoxybenzamine is a known blocker of α-adrenoceptors (Frang et al., 2001). However, several arguments lead us to conclude that its binding to α-adrenoceptors is not involved in its inhibitory effect on EGFR downstream signalling. First, α1 adrenoceptors are not endogenously expressed in HEK-293 cells (Theroux et al., 1996). Second, even if α2 adrenoceptors were endogenously expressed in HEK-293 cells (Cooper et al., 1997), phenoxybenzamine does not modify the EGF-induced MAPK/ERK activation when tested in control cells (transfected with empty vector, peYFP-N1). Because EGFR itself plays a crucial role in the regulation of cell proliferation and is involved in many aggressive human cancers, novel strategies to inhibit EGFR activation could represent novel potential therapeutic approaches. Interestingly, phenoxybenzamine has recently been identified as a low MW inhibitor of glioblastoma cell growth (Wang et al., 2012).
Given the emerging role of LINGO-1 as a negative regulator of several degenerative diseases, targeting the functions of LINGO-1 is intensively investigated for the treatment of neurodegenerative and demyelinating diseases. Here, we have demonstrated that LINGO-1 can form dimers or higher order oligomers in living cells under physiological conditions. We took advantage of this property to use a BRET-based assay for the screening of ligands and/or SMPPIMs of LINGO-1. The identification of the first chemical compound able to promote conformational changes within the dimers of LINGO-1 allowed us to propose that this method will be particularly useful to identify innovative pharmacological inhibitors or activators of LINGO-1. Such compounds will serve as tools to decipher the role of LINGO-1 dimerization in its downstream signalling and its relevance to physiological processes.
Acknowledgments
We thank Professor Tobias Hevor and Philippe Moreau (Neurobiology Laboratory, Orleans) for their contribution to neuronal cultures. We thank David Gosset (CBM, Orleans) for his technical help to obtain clonal cell lines. We also thank Béatrice Vallee and Michel Doudeau (CBM, Orléans) for all discussions of the results and for their technical advices
This work was supported by a Trampoline grant (15058) from the AFM (Association Française contre les Myopathies), by the CNRS structure of valorization and transfer (03247-02), the Ligue contre le Cancer (Loiret and Cher comities) and a FEDER grant (N° 5789/39115) from Region Centre. This work has been published within the LABEX ANR-10-LABX-0034_Medalis and received a financial support from French government managed by ‘Agence National de la Recherche’ under ‘Programme d'investissement d'avenir’.
Glossary
- DSP
Dithiobis[succinimidyl] propionate
- EGFR
EGF receptor
- ER
endoplasmic reticulum
- HA
haemagglutinin-tag
- IP
immunoprecipitation
- OPCs
oligodendrocyte precursor cells
- RLuc
Renilla luciferase
- SMPPIM
small-molecule protein–protein interaction modulator
- trkB
tropomyosin-related kinase B
- YFP
yellow fluorescent protein
Author contributions
L.C. performed the BRET experiments, drug screening, Western blot analysis and AlphaScreen experiments. M.-L.de T., I.H. and F.G. generated the plasmid constructs. J.F performed the functional assays on primary culture. E.K.and D. R. performed the 3D modelling of LINGO-1 and contributed to writing the manuscript. E.T., J.P. and H.B. contributed to scientific discussion and to writing the manuscript. S.M.-L. designed experiments and wrote the manuscript.
Conflict of interest
All authors declare no competing financial interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article at the publisher's web-site:
Figure S1 Deglycosylation of LINGO-1-HA with peptide-N-glycosidase (PNGase F). Lysates from HEK-293 cells transiently transfected with the LINGO-HA were incubated in buffer B (50 mM sodium citrate, pH 7.5, 1% Triton, cocktail of protease inhibitor) with or without PNGase F for 2 h at 37°C. The reaction was stopped by adding loading sample buffer. Samples were separated using 8% SDS-PAGE and analysed by immunoblotting with the anti-BD living colours antibody (BD Biosciences). Treatment of membranes with PNGase F reduced the 80 kDa and 100 kDa mature forms to about 60 kDa, which represented the deglycosylated forms of LINGO-HA. These data demonstrate that recombinant LINGO-1 expressed in HEK-293 cells is N-glycosylated.
Figure S2 Effect of LINGO-1ΔCter on the distribution and dimerization of wild-type LINGO-1. HEK-293 cells were transiently transfected with LINGO-1-RLuc and LINGO-1-YFP with or without LINGO-1ΔCter. Forty-eight hours after transfection, cells were lysed and subcellular fractions were resolved on a sucrose gradient. Individual fractions were subjected to luminescence and BRET analysis.
Figure S3 Solvent accessibility of cysteine residues in human LINGO-1 and human α2-adrenoceptor. The protein backbone is represented with grey ribbon. Only cysteine residues are shown, their side chain being depicted using CPK-coloured ball and sticks. A. The three-dimensional structure of LINGO1 ecto-domain (residues 1–476) was solved by X-ray crystallography (PDB: 2ID5) and revealed a tetrameric organization (doi: 10.1074/jbc.M607314200). Each protomer contains 13 cysteine residues, 10 engaged in disulfide bridges and the remaining three burying their side chain into the core structure of the leucine-rich repeats. B. The three-dimensional structure of α2-adrenoceptors was modelled by homology with human α2-adrenoceptors (PDB: 3P0G). Details of modelling: The template file was chosen because of sequence similarity between the protein (25.8% identity) as well as shape similarity between phenoxybenzamine and the ligand bound to transmembrane cavity of the receptor. The model was built using MOE v2011.10 (Chemical Computing Group Inc) based on sequence alignment guided by GPRC constraints. The 3D structure of phenoxybenzamine was generated with Corina v3.1 (Molecular Network GmbH). Model of complex between phenoxybenzamine and the α2-adrenoceptor was obtained from covalent docking of phenoxybenzamine into the transmembrane cavity (defined in a radius of 10 Å centred on OE1 atom of ASP3.32) using GOLD v5.2 (The Cambridge Crystallographic Data Centre). The best scored pose selected with ASP scoring function closely reproduces the binding mode of ligand bound to the template receptor.
Figure S4 BRET analysis on LINGO1C575A. TheCys575 residue is located is the transmembrane domain of LINGO-1. In order to investigate its potential role in the action of phenoxybenzamine, we performed two constructions containing the mutated LINGO fused to Renilla luciferase (LINGO-1-C575A-RLuc) or with yellow fluorescent protein (LINGO-1-C575AYFP). BRET saturation curves analysis demonstrates that the mutation does modify neither the ability of LINGO-1 to form dimers (A) nor the ability of phenoxybenzamine to decrease BRET signal (B).
Figure S5 Phenoxybenzamine binds LINGO-1 in a reversible manner. BRET experiments were performed in adherent HEK-293 cells transiently transfected with LINGO-1-RLuc alone or with LINGO-1-RLuc and LINGO-1-YFP. BRET signal was determined in the presence of phenoxybenzamine immediately before and immediately after extensive washing of the cell culture. After the washing, the BRET signal reached a value not significantly different from the control value. Data are the mean ± SEM of two independent experiments. ***P < 0.001.
References
- Alexander SPH, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, et al. The Concise Guide to PHARMACOLOGY 2013/14: Catalytic Receptors. Br J Pharmacol. 2013a;170:1676–1705. doi: 10.1111/bph.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, et al. The Concise Guide to PHARMACOLOGY 2013/14: Enzymes. Br J Pharmacol. 2013b;170:1797–1867. doi: 10.1111/bph.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander SPH, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, et al. The Concise Guide to PHARMACOLOGY 2013/14: G Protein-Coupled Receptors. Br J Pharmacol. 2013c;170:1459–1581. doi: 10.1111/bph.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkin MR, Wells JA. Small-molecule inhibitors of protein–protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- Bacart J, Corbel C, Jockers R, Bach S, Couturier C. The BRET technology and its application to screening assays. Biotechnol J. 2008;3:311–324. doi: 10.1002/biot.200700222. [DOI] [PubMed] [Google Scholar]
- Barrette B, Vallieres N, Dube M, Lacroix S. Expression profile of receptors for myelin-associated inhibitors of axonal regeneration in the intact and injured mouse central nervous system. Mol Cell Neurosci. 2007;34:519–538. doi: 10.1016/j.mcn.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Bessero AC, Clarke PG. Neuroprotection for optic nerve disorders. Curr Opin Neurol. 2010;23:10–15. doi: 10.1097/WCO.0b013e3283344461. [DOI] [PubMed] [Google Scholar]
- Bibel M, Hoppe E, Barde YA. Biochemical and functional interactions between the neurotrophin receptors trk and p75NTR. EMBO J. 1999;18:616–622. doi: 10.1093/emboj/18.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier M, Heveker N, Jockers R, Marullo S, Milligan G. BRET analysis of GPCR oligomerization: newer does not mean better. Nat Methods. 2007;4:3–4. doi: 10.1038/nmeth0107-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulenger S, Marullo S, Bouvier M. Emerging role of homo- and heterodimerization in G-protein-coupled receptor biosynthesis and maturation. Trends Pharmacol Sci. 2005;26:131–137. doi: 10.1016/j.tips.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Carim-Todd L, Escarceller M, Estivill X, Sumoy L. LRRN6A/LERN1 (leucine-rich repeat neuronal protein 1), a novel gene with enriched expression in limbic system and neocortex. Eur J Neurosci. 2003;18:3167–3182. doi: 10.1111/j.1460-9568.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- Cooper J, Hill SJ, Alexander SP. An endogenous A2B adenosine receptor coupled to cyclic AMP generation in human embryonic kidney (HEK 293) cells. Br J Pharmacol. 1997;122:546–550. doi: 10.1038/sj.bjp.0701401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier C, Deprez B. Setting up a bioluminescence resonance energy transfer high throughput screening assay to search for protein/protein interaction inhibitors in mammalian cells. Front Endocrinol (Lausanne) 2012;3:1–13. doi: 10.3389/fendo.2012.00100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier C, Jockers R. Activation of the leptin receptor by a ligand-induced conformational change of constitutive receptor dimers. J Biol Chem. 2003;278:26604–26611. doi: 10.1074/jbc.M302002200. [DOI] [PubMed] [Google Scholar]
- De Vries L, Finana F, Cachoux F, Vacher B, Sokoloff P, Cussac D. Cellular BRET assay suggests a conformational rearrangement of preformed TrkB/Shc complexes following BDNF-dependent activation. Cell Signal. 2010;22:158–165. doi: 10.1016/j.cellsig.2009.09.027. [DOI] [PubMed] [Google Scholar]
- Ferre S, Casado V, Devi LA, Filizola M, Jockers R, Lohse MJ, et al. G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol Rev. 2014;66:413–434. doi: 10.1124/pr.113.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frang H, Cockcroft V, Karskela T, Scheinin M, Marjamaki A. Phenoxybenzamine binding reveals the helical orientation of the third transmembrane domain of adrenergic receptors. J Biol Chem. 2001;276:31279–31284. doi: 10.1074/jbc.M104167200. [DOI] [PubMed] [Google Scholar]
- Fry DC. Drug-like inhibitors of protein–protein interactions: a structural examination of effective protein mimicry. Curr Protein Pept Sci. 2008;9:240–247. doi: 10.2174/138920308784533989. [DOI] [PubMed] [Google Scholar]
- Gerin CG, Madueke IC, Perkins T, Hill S, Smith K, Haley B, et al. Combination strategies for repair, plasticity, and regeneration using regulation of gene expression during the chronic phase after spinal cord injury. Synapse. 2011;65:1255–1281. doi: 10.1002/syn.20903. [DOI] [PubMed] [Google Scholar]
- Grimm I, Messemer N, Stanke M, Gachet C, Zimmermann H. Coordinate pathways for nucleotide and EGF signaling in cultured adult neural progenitor cells. J Cell Sci. 2009;122(Pt 14):2524–2533. doi: 10.1242/jcs.044891. [DOI] [PubMed] [Google Scholar]
- Huang CL, Kuo E, Toto RD. WNK kinases and essential hypertension. Curr Opin Nephrol Hypertens. 2008;17:133–137. doi: 10.1097/MNH.0b013e3282f4e4fd. [DOI] [PubMed] [Google Scholar]
- Inoue H, Lin L, Lee X, Shao Z, Mendes S, Snodgrass-Belt P, et al. Inhibition of the leucine-rich repeat protein LINGO-1 enhances survival, structure, and function of dopaminergic neurons in Parkinson's disease models. Proc Natl Acad Sci U S A. 2007;104:14430–14435. doi: 10.1073/pnas.0700901104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issad T, Blanquart C, Gonzalez-Yanes C. The use of bioluminescence resonance energy transfer for the study of therapeutic targets: application to tyrosine kinase receptors. Expert Opin Ther Targets. 2007;11:541–556. doi: 10.1517/14728222.11.4.541. [DOI] [PubMed] [Google Scholar]
- James JR, Oliveira MI, Carmo AM, Iaboni A, Davis SJ. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nat Methods. 2006;3:1001–1006. doi: 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
- Jepson S, Vought B, Gross CH, Gan L, Austen D, Frantz JD, et al. LINGO-1, a transmembrane signaling protein, inhibits oligodendrocyte differentiation and myelination through intercellular self-interactions. J Biol Chem. 2012;287:22184–22195. doi: 10.1074/jbc.M112.366179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji B, Li M, Wu WT, Yick LW, Lee X, Shao Z, et al. LINGO-1 antagonist promotes functional recovery and axonal sprouting after spinal cord injury. Mol Cell Neurosci. 2006a;33:311–320. doi: 10.1016/j.mcn.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Ji SP, Zhang Y, Van Cleemput J, Jiang W, Liao M, Li L, et al. Disruption of PTEN coupling with 5-HT2C receptors suppresses behavioral responses induced by drugs of abuse. Nat Med. 2006b;12:324–329. doi: 10.1038/nm1349. [DOI] [PubMed] [Google Scholar]
- Jorissen RN, Walker F, Pouliot N, Garrett TP, Ward CW, Burgess AW. Epidermal growth factor receptor: mechanisms of activation and signalling. Exp Cell Res. 2003;284:31–53. doi: 10.1016/s0014-4827(02)00098-8. [DOI] [PubMed] [Google Scholar]
- Kilkenny C, Browne W, Cuthill IC, Emerson M, Altman DG. NC3Rs Reporting Guidelines Working Group. Br J Pharmacol. 2010;160:1577–1579. doi: 10.1111/j.1476-5381.2010.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HS, Nakaya N, Abu-Asab M, Kim HS, Tomarev SI. Myocilin is involved in NgR1/Lingo-1-mediated oligodendrocyte differentiation and myelination of the optic nerve. J Neurosci. 2014;34:5539–5551. doi: 10.1523/JNEUROSCI.4731-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee X, Yang Z, Shao Z, Rosenberg SS, Levesque M, Pepinsky RB, et al. NGF regulates the expression of axonal LINGO-1 to inhibit oligodendrocyte differentiation and myelination. J Neurosci. 2007;27:220–225. doi: 10.1523/JNEUROSCI.4175-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XJ, Gingrich JR, Vargas-Caballero M, Dong YN, Sengar A, Beggs S, et al. Treatment of inflammatory and neuropathic pain by uncoupling Src from the NMDA receptor complex. Nat Med. 2008;14:1325–1332. doi: 10.1038/nm.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens F, Gil V, Iraola S, Carim-Todd L, Marti E, Estivill X, et al. Developmental analysis of Lingo-1/Lern1 protein expression in the mouse brain: interaction of its intracellular domain with Myt1l. Dev Neurobiol. 2008;68:521–541. doi: 10.1002/dneu.20607. [DOI] [PubMed] [Google Scholar]
- Loov C, Fernqvist M, Walmsley A, Marklund N, Erlandsson A. Neutralization of LINGO-1 during in vitro differentiation of neural stem cells results in proliferation of immature neurons. PLoS ONE. 2012;7:e29771. doi: 10.1371/journal.pone.0029771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Zerangue N, Lin YF, Collins A, Yu M, Jan YN, et al. Role of ER export signals in controlling surface potassium channel numbers. Science. 2001;291:316–319. doi: 10.1126/science.291.5502.316. [DOI] [PubMed] [Google Scholar]
- Mandai K, Guo T, St Hillaire C, Meabon JS, Kanning KC, Bothwell M, et al. LIG family receptor tyrosine kinase-associated proteins modulate growth factor signals during neural development. Neuron. 2009;63:614–627. doi: 10.1016/j.neuron.2009.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama IN. Mechanisms of activation of receptor tyrosine kinases: monomers or dimers. Cells. 2014;3:304–330. doi: 10.3390/cells3020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald CL, Bandtlow C, Reindl M. Targeting the Nogo receptor complex in diseases of the central nervous system. Curr Med Chem. 2011;18:234–244. doi: 10.2174/092986711794088326. [DOI] [PubMed] [Google Scholar]
- McGrath J, Drummond G, Kilkenny C, Wainwright C. Guidelines for reporting experiments involving animals: the ARRIVE guidelines. Br J Pharmacol. 2010;160:1573–1576. doi: 10.1111/j.1476-5381.2010.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S. Troy/Taj and its role in CNS axon regeneration. Cytokine Growth Factor Rev. 2008;19:245–251. doi: 10.1016/j.cytogfr.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Mi S, Lee X, Shao Z, Thill G, Ji B, Relton J, et al. LINGO-1 is a component of the Nogo-66 receptor/p75 signaling complex. Nat Neurosci. 2004;7:221–228. doi: 10.1038/nn1188. [DOI] [PubMed] [Google Scholar]
- Mi S, Miller RH, Lee X, Scott ML, Shulag-Morskaya S, Shao Z, et al. LINGO-1 negatively regulates myelination by oligodendrocytes. Nat Neurosci. 2005;8:745–751. doi: 10.1038/nn1460. [DOI] [PubMed] [Google Scholar]
- Mi S, Sandrock A, Miller RH. LINGO-1 and its role in CNS repair. Int J Biochem Cell Biol. 2008;40:1971–1978. doi: 10.1016/j.biocel.2008.03.018. [DOI] [PubMed] [Google Scholar]
- Mi S, Miller RH, Tang W, Lee X, Hu B, Wu W, et al. Promotion of central nervous system remyelination by induced differentiation of oligodendrocyte precursor cells. Ann Neurol. 2009;65:304–315. doi: 10.1002/ana.21581. [DOI] [PubMed] [Google Scholar]
- Mosyak L, Wood A, Dwyer B, Buddha M, Johnson M, Aulabaugh A, et al. The structure of the Lingo-1 ectodomain, a module implicated in central nervous system repair inhibition. J Biol Chem. 2006;281:36378–36390. doi: 10.1074/jbc.M607314200. [DOI] [PubMed] [Google Scholar]
- Okafuji T, Tanaka H. Expression pattern of LINGO-1 in the developing nervous system of the chick embryo. Gene Expr Patterns. 2005;6:57–62. doi: 10.1016/j.modgep.2005.04.016. [DOI] [PubMed] [Google Scholar]
- Park JB, Yiu G, Kaneko S, Wang J, Chang J, He XL, et al. A TNF receptor family member, TROY, is a coreceptor with Nogo receptor in mediating the inhibitory activity of myelin inhibitors. Neuron. 2005;45:345–351. doi: 10.1016/j.neuron.2004.12.040. [DOI] [PubMed] [Google Scholar]
- Pawson AJ, Sharman JL, Benson HE, Faccenda E, Alexander SP, Buneman OP, et al. NC-IUPHAR. The IUPHAR/BPS Guide to PHARMACOLOGY: an expert-driven knowledge base of drug targets and their ligands. Nucl Acids Res. 2014;42(Database Issue):D1098–D1106. doi: 10.1093/nar/gkt1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichon X, Wattiez AS, Becamel C, Ehrlich I, Bockaert J, Eschalier A, et al. Disrupting 5-HT(2A) receptor/PDZ protein interactions reduces hyperalgesia and enhances SSRI efficacy in neuropathic pain. Mol Ther. 2010;18:1462–1470. doi: 10.1038/mt.2010.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Q, He K, Liu X, Pham C, Meyerkord C, Fu H, et al. Disrupting the PIKE-A/Akt interaction inhibits glioblastoma cell survival, migration, invasion and colony formation. Oncogene. 2013;32:1030–1040. doi: 10.1038/onc.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsay D, Kellett E, McVey M, Rees S, Milligan G. Homo- and hetero-oligomeric interactions between G-protein-coupled receptors in living cells monitored by two variants of bioluminescence resonance energy transfer (BRET): hetero-oligomers between receptor subtypes form more efficiently than between less closely related sequences. Biochem J. 2002;365(Pt 2):429–440. doi: 10.1042/BJ20020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudick RA, Mi S, Sandrock AW., Jr LINGO-1 antagonists as therapy for multiple sclerosis: in vitro and in vivo evidence. Expert Opin Biol Ther. 2008;8:1561–1570. doi: 10.1517/14712598.8.10.1561. [DOI] [PubMed] [Google Scholar]
- Schlessinger J. Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell. 2002;110:669–672. doi: 10.1016/s0092-8674(02)00966-2. [DOI] [PubMed] [Google Scholar]
- Shao Z, Browning JL, Lee X, Scott ML, Shulga-Morskaya S, Allaire N, et al. TAJ/TROY, an orphan TNF receptor family member, binds Nogo-66 receptor 1 and regulates axonal regeneration. Neuron. 2005;45:353–359. doi: 10.1016/j.neuron.2004.12.050. [DOI] [PubMed] [Google Scholar]
- Siddiqui S, Cong WN, Daimon CM, Martin B, Maudsley S. BRET biosensor analysis of receptor tyrosine kinase functionality. Front Endocrinol (Lausanne) 2013;4:46. doi: 10.3389/fendo.2013.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simi A, Ibanez CF. Assembly and activation of neurotrophic factor receptor complexes. Dev Neurobiol. 2010;70:323–331. doi: 10.1002/dneu.20773. [DOI] [PubMed] [Google Scholar]
- Stein T, Walmsley AR. The leucine-rich repeats of LINGO-1 are not required for self-interaction or interaction with the amyloid precursor protein. Neurosci Lett. 2012;509:9–12. doi: 10.1016/j.neulet.2011.11.029. [DOI] [PubMed] [Google Scholar]
- Theroux TL, Esbenshade TA, Peavy RD, Minneman KP. Coupling efficiencies of human alpha 1-adrenergic receptor subtypes: titration of receptor density and responsiveness with inducible and repressible expression vectors. Mol Pharmacol. 1996;50:1376–1387. [PubMed] [Google Scholar]
- Thompson AD, Dugan A, Gestwicki JE, Mapp AK. Fine-Tuning multiprotein complexes using small molecules. ACS Chem Biol. 2012;7:1311–1320. doi: 10.1021/cb300255p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Zhao H, Cui K, Yao L, Ren M, Hao A, et al. Identification of novel small-molecule inhibitors of glioblastoma cell growth and invasion by high-throughput screening. Biosci Trends. 2012;6:192–200. doi: 10.5582/bst.2012.v6.4.192. [DOI] [PubMed] [Google Scholar]
- White JH, Wise A, Main MJ, Green A, Fraser NJ, Disney GH, et al. Heterodimerization is required for the formation of a functional GABA(B) receptor. Nature. 1998;396:679–682. doi: 10.1038/25354. [DOI] [PubMed] [Google Scholar]
- Xu BE, Stippec S, Lenertz L, Lee BH, Zhang W, Lee YK, et al. WNK1 activates ERK5 by an MEKK2/3-dependent mechanism. J Biol Chem. 2004;279:7826–7831. doi: 10.1074/jbc.M313465200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Piston DW, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc Natl Acad Sci U S A. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita T, Fujitani M, Yamagishi S, Hata K, Mimura F. Multiple signals regulate axon regeneration through the Nogo receptor complex. Mol Neurobiol. 2005;32:105–111. doi: 10.1385/MN:32:2:105. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xu X, Zhang Y, Zhou J, Yu Z, He C. LINGO-1 interacts with WNK1 to regulate nogo-induced inhibition of neurite extension. J Biol Chem. 2009;284:15717–15728. doi: 10.1074/jbc.M808751200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao XH, Jin WL, Ju G. An in vitro study on the involvement of LINGO-1 and Rho GTPases in Nogo-A regulated differentiation of oligodendrocyte precursor cells. Mol Cell Neurosci. 2007;36:260–269. doi: 10.1016/j.mcn.2007.07.008. [DOI] [PubMed] [Google Scholar]
- Zhong X, Pocas J, Liu Y, Wu PW, Mosyak L, Somers W, et al. Swift residue-screening identifies key N-glycosylated asparagines sufficient for surface expression of neuroglycoprotein Lingo-1. FEBS Lett. 2009;583:1034–1038. doi: 10.1016/j.febslet.2009.02.034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Deglycosylation of LINGO-1-HA with peptide-N-glycosidase (PNGase F). Lysates from HEK-293 cells transiently transfected with the LINGO-HA were incubated in buffer B (50 mM sodium citrate, pH 7.5, 1% Triton, cocktail of protease inhibitor) with or without PNGase F for 2 h at 37°C. The reaction was stopped by adding loading sample buffer. Samples were separated using 8% SDS-PAGE and analysed by immunoblotting with the anti-BD living colours antibody (BD Biosciences). Treatment of membranes with PNGase F reduced the 80 kDa and 100 kDa mature forms to about 60 kDa, which represented the deglycosylated forms of LINGO-HA. These data demonstrate that recombinant LINGO-1 expressed in HEK-293 cells is N-glycosylated.
Figure S2 Effect of LINGO-1ΔCter on the distribution and dimerization of wild-type LINGO-1. HEK-293 cells were transiently transfected with LINGO-1-RLuc and LINGO-1-YFP with or without LINGO-1ΔCter. Forty-eight hours after transfection, cells were lysed and subcellular fractions were resolved on a sucrose gradient. Individual fractions were subjected to luminescence and BRET analysis.
Figure S3 Solvent accessibility of cysteine residues in human LINGO-1 and human α2-adrenoceptor. The protein backbone is represented with grey ribbon. Only cysteine residues are shown, their side chain being depicted using CPK-coloured ball and sticks. A. The three-dimensional structure of LINGO1 ecto-domain (residues 1–476) was solved by X-ray crystallography (PDB: 2ID5) and revealed a tetrameric organization (doi: 10.1074/jbc.M607314200). Each protomer contains 13 cysteine residues, 10 engaged in disulfide bridges and the remaining three burying their side chain into the core structure of the leucine-rich repeats. B. The three-dimensional structure of α2-adrenoceptors was modelled by homology with human α2-adrenoceptors (PDB: 3P0G). Details of modelling: The template file was chosen because of sequence similarity between the protein (25.8% identity) as well as shape similarity between phenoxybenzamine and the ligand bound to transmembrane cavity of the receptor. The model was built using MOE v2011.10 (Chemical Computing Group Inc) based on sequence alignment guided by GPRC constraints. The 3D structure of phenoxybenzamine was generated with Corina v3.1 (Molecular Network GmbH). Model of complex between phenoxybenzamine and the α2-adrenoceptor was obtained from covalent docking of phenoxybenzamine into the transmembrane cavity (defined in a radius of 10 Å centred on OE1 atom of ASP3.32) using GOLD v5.2 (The Cambridge Crystallographic Data Centre). The best scored pose selected with ASP scoring function closely reproduces the binding mode of ligand bound to the template receptor.
Figure S4 BRET analysis on LINGO1C575A. TheCys575 residue is located is the transmembrane domain of LINGO-1. In order to investigate its potential role in the action of phenoxybenzamine, we performed two constructions containing the mutated LINGO fused to Renilla luciferase (LINGO-1-C575A-RLuc) or with yellow fluorescent protein (LINGO-1-C575AYFP). BRET saturation curves analysis demonstrates that the mutation does modify neither the ability of LINGO-1 to form dimers (A) nor the ability of phenoxybenzamine to decrease BRET signal (B).
Figure S5 Phenoxybenzamine binds LINGO-1 in a reversible manner. BRET experiments were performed in adherent HEK-293 cells transiently transfected with LINGO-1-RLuc alone or with LINGO-1-RLuc and LINGO-1-YFP. BRET signal was determined in the presence of phenoxybenzamine immediately before and immediately after extensive washing of the cell culture. After the washing, the BRET signal reached a value not significantly different from the control value. Data are the mean ± SEM of two independent experiments. ***P < 0.001.