Abstract
To investigate geographic structure within U.S. ethnic populations, we analyzed 1705 haplotypes on the basis of 9 short tandem repeat (STR) loci on the Y-chromosome from 9–11 groups each of African-Americans, European-Americans, and Hispanics. There were no significant differences in the distribution of Y-STR haplotypes among African-American groups, whereas European-American and Hispanic groups did exhibit significant geographic heterogeneity. However, the significant heterogeneity resulted from one sample; removal of that sample in each case eliminated the significant heterogeneity. Multidimensional scaling analysis of RST values indicated that African-American groups formed a distinct cluster, whereas there was some intermingling of European-American and Hispanic groups. MtDNA data exist for many of these same groups; estimates of the European-American genetic contribution to the African-American gene pool were 27.5%–33.6% for the Y-STR haplotypes and 9%–15.4% for the mtDNA types. The lack of significant geographic heterogeneity among Y-STR and mtDNA haplotypes in U.S ethnic groups means that forensic DNA databases do not need to be constructed for separate geographic regions of the U.S. Moreover, absence of significant geographic heterogeneity for these two loci means that regional variation in disease susceptibility within ethnic groups is more likely to reflect cultural/environmental factors, rather than any underlying genetic heterogeneity.
The United States harbors an extraordinary amount of genetic diversity, with African, European, Asian, and native American populations (among others) having contributed to the present-day gene pool of the U.S. population. U.S. populations are traditionally classified for official (and other) purposes via ancestry, that is, African-American, Asian-American, European-American, Hispanic, etc., but little work has been done on how patterns of genetic variation correlate with such classifications. Although genetic structure is evident among the source populations that have contributed to U.S. populations (Cavalli-Sforza et al. 1994), the extent to which the several generations of intermarriage and interbreeding between ethnic U.S. populations (i.e., the melting pot) has reduced this genetic structure remains largely unknown. Moreover, the possibility exists for significant geographic structuring within U.S. ethnic groups. For example, the historical record indicates that the slave trade brought ∼400,000 people from a large section of west-central Africa (extending from Senegal to Angola, including coastal and inland regions), and that there were significant differences in the geographic origin of slaves that arrived at the various points of entry into the U.S. (Curtin 1969; Reed 1969). In addition, admixture between African-Americans and European-Americans may have occurred to different extents in different parts of the U.S. (Reed 1969; Chakraborty et al. 1992; Parra et al. 1998), further contributing to geographic structure in the patterns of genetic variation in African-American populations. Similar concerns hold for the other ethnic U.S. populations, in particular Hispanics, as they are defined primarily by cultural criteria and not geographic origin.
The existence of significant genetic differences among geographic subgroups of U.S. populations would have important implications for both the forensic DNA and the disease genetics community. For the forensic DNA community, significant geographic structure in patterns of genetic variation within U.S. populations would then have to be taken into consideration in constructing databases of DNA types for use in determining the probability that unrelated individuals would have matching DNA types. Separate databases would be required for each geographic region. Conversely, the absence of significant geographic structure would mean that databases for each ethnic group need not take into account the geographic origin of individuals.
For the medical community, the question of geographic structure within ethnic U.S. populations influences the interpretation of geographic patterns for susceptibility to various diseases. Although it is well established that disease susceptibility varies across ethnic groups (Gilliland 1997; Keppel et al. 2002), there is also increasing evidence of geographic variation in disease patterns within ethnic groups (Jackson 2000). For example, many types of cancer show striking regional differences in the United States that have persisted for at least 50 yr (Devesa et al. 1999). The existence of significant geographic structure in neutral genetic markers would be consistent with a role for underlying genetic differences in geographic variation for disease susceptibility within ethnic U.S. populations. Conversely, the absence of significant geographic structure would imply that geographic differences in disease susceptibility are instead due to variation in cultural/environmental factors.
To address these issues, we present here an analysis of Y-chromosome haplotypes, on the basis of 9 short-tandem-repeat (STR) or microsatellite loci, for 1705 males from several geographic groups each of African-American, European-American, and Hispanic populations (Figure 1). We also compare the Y-chromosome data to previously published data on mtDNA haplotypes in (largely) the same set of geographic groups (Melton et al. 2001). Y-STR and mtDNA haplotypes are ideal for investigating the genetic structure of human populations, because they behave as (largely) neutral markers, and their rapid rate of evolution and smaller effective population size (due to their haploid, uniparental mode of inheritance) means that they are more sensitive indicators of genetic differences between groups than are autosomal DNA markers. Moreover, comparing patterns of Y-chromosomal and mtDNA variation allows insights into the paternal and maternal history of populations, which may differ, especially in admixed populations.
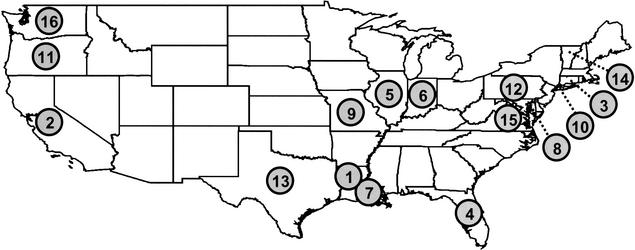
Figure 1.
Map showing sample localities included in this study. (1) Acadian (Lousiana); (2) California; (3) Connecticut; (4) Florida; (5) Illinois; (6) Indiana; (7) Lousiana; (8) Maryland; (9) Missouri; (10) New York City; (11) Oregon; (12) Pennsylvania; (13) Texas; (14) Vermont; (15) Virginia; (16) Washington.
RESULTS
Y-STR Haplotypes
The Y chromosome haplotypes, on the basis of the nine STR loci, exhibit high levels of within-group diversity; average haplotype diversity (H) values range from 0.986–1.000, and the MPSD between haplotypes ranges from 6.76–11.99 (Table 1). On the basis of Mann-Whitney U tests, the range of H and MPSD values is significantly higher in African-American groups than in European-Americans, and H (but not MPSD) values are significantly higher in African-Americans than in Hispanics, whereas MPSD (but not H) values are significantly higher in Hispanics than in European-Americans.
Table 1.
Sample Sizes, Number of Haplotypes, Haplotype Diversity, and Mean Number of Pairwise Step Differences (MPSD) for 30 U.S. Groups, Based on Y-STR Haplotypes
| Population/Group | Code | Sample size | Number of haplotypes | Haplotype diversity (±SE) | MPSD (±SE) |
| African–American | |||||
| Florida | AFL | 48 | 48 | 1.000 ± 0.004 | 11.08 ± 5.71 |
| Indiana | AIA | 37 | 36 | 0.998 ± 0.007 | 11.99 ± 6.32 |
| Louisiana | ALA | 29 | 27 | 0.995 ± 0.011 | 11.03 ± 6.18 |
| Maryland | AMD | 73 | 72 | 0.999 ± 0.002 | 10.36 ± 5.32 |
| Missouri | AMO | 56 | 52 | 0.997 ± 0.004 | 10.61 ± 6.13 |
| New York | ANY | 150 | 137 | 0.999 ± 0.001 | 11.05 ± 5.90 |
| Oregon | AOR | 47 | 46 | 0.999 ± 0.005 | 11.17 ± 5.84 |
| Pennsylvania | APA | 43 | 39 | 0.994 ± 0.007 | 10.59 ± 5.95 |
| Texas | ATX | 69 | 67 | 0.999 ± 0.003 | 10.51 ± 5.83 |
| Virginia | AVA | 47 | 46 | 0.999 ± 0.005 | 9.56 ± 5.93 |
| TOTAL | 599 | 454 | 0.998 ± 0.003 | 10.76 ± 5.87 | |
| European–American | |||||
| Acadian | ECJ | 22 | 21 | 0.996 ± 0.015 | 7.46 ± 5.04 |
| Florida | EFL | 22 | 20 | 0.987 ± 0.020 | 9.41 ± 4.56 |
| Indiana | EIA | 34 | 31 | 0.995 ± 0.009 | 9.44 ± 5.73 |
| Louisiana | ELA | 31 | 29 | 0.996 ± 0.010 | 7.99 ± 4.00 |
| Maryland | EMO | 64 | 53 | 0.989 ± 0.007 | 8.22 ± 4.66 |
| Missouri | EMO | 59 | 55 | 0.997 ± 0.004 | 9.06 ± 4.73 |
| New York | ENY | 155 | 136 | 0.998 ± 0.001 | 9.42 ± 3.89 |
| Oregon | EOR | 35 | 33 | 0.997 ± 0.008 | 8.14 ± 3.86 |
| Pennsylvania | EPA | 67 | 63 | 0.998 ± 0.003 | 8.9 ± 3.68 |
| Texas | ETX | 78 | 63 | 0.989 ± 0.005 | 6.76 ± 4.29 |
| Virginia | EVA | 61 | 54 | 0.996 ± 0.004 | 7.56 ± 4.30 |
| TOTAL | 628 | 437 | 0.996 ± 0.001 | 8.61 ± 4.38 | |
| Hispanic | |||||
| Connecticut | HCT | 53 | 46 | 0.993 ± 0.006 | 9.79 ± 5.04 |
| Florida | HFL | 23 | 21 | 0.992 ± 0.015 | 8.73 ± 4.49 |
| Louisiana | HLA | 30 | 29 | 0.998 ± 0.009 | 10.45 ± 4.67 |
| Maryland | HMD | 26 | 26 | 1.000 ± 0.011 | 10.89 ± 5.45 |
| New York | HNY | 152 | 119 | 0.994 ± 0.002 | 10.82 ± 5.04 |
| Oregon | HOR | 42 | 38 | 0.994 ± 0.007 | 10.41 ± 4.23 |
| Pennsylvania | HPA | 32 | 28 | 0.986 ± 0.014 | 8.97 ± 4.24 |
| Texas | HTX | 74 | 69 | 0.998 ± 0.003 | 10.63 ± 3.97 |
| Virginia | HVA | 46 | 41 | 0.995 ± 0.006 | 10.68 ± 4.52 |
| TOTAL | 478 | 353 | 0.995 ± 0.001 | 10.54 ± 4.70 |
An analysis of molecular variance (AMOVA) approach was used to assess the degree and significance of between-group differentiation. The AMOVA was based on the RST distance between Y-STR haplotypes; thus, this analysis takes into account both frequency differences among haplotypes as well as relatedness of haplotypes. The results (Table 2) indicate that Y-STR haplotypes differ significantly between African-Americans, European-Americans, and Hispanics; ∼25% of the genetic variance reflects differences between these populations, whereas 1% reflects differences among the regional groups within each population, and 74% reflects the genetic variance within regional groups. When each population is analyzed separately (Table 2), the African-American groups are not significantly different with respect to Y-STR haplotypes (RST = 0.0005, P = 0.39), whereas both European-American and Hispanic groups exhibit significant among-group heterogeneity (European-Americans, RST = 0.018, P < 0.001; Hispanics, RST = 0.026, P < 0.01).
Table 2.
Partition of the Total Genetic Variance Into the Among Population Component (A), The Among Group Within Population Component (B), and the Within Group Component (C), for Y-STR haplotypes and mtDNA SSO-types. Components Are Expressed as Percentages of the Total
| Comparison | Y-STR | MtDNA | ||||
| A | B | C | A | B | C | |
| African-American | — | 0.5 | 99.5 | — | 0.4 | 99.6 |
| European-American | — | 1.8 | 98.2 | — | 0.5 | 99.5 |
| Hispanic | — | 2.6 | 97.4 | — | 1.5 | 98.5 |
| All 3 populations | 24.9 | 0.8 | 74.3 | 18.6 | 0.5 | 80.9 |
To investigate further the cause of the significant heterogeneity among regional groups of European-Americans and Hispanics, each regional group was removed in turn and the AMOVA repeated. Removing the Texas group reduced the heterogeneity among the remaining regional groups to nonsignificant levels for both European-Americans (RST = 0.008, P > 0.05) and Hispanics (RST = 0.009, P > 0.05), whereas removal of any other regional group resulted in RST values that were still statistically significant. Thus, the significant heterogeneity in Y-STR haplotypes among regional groups of European-Americans and Hispanics may be attributed in both cases to the Texas samples.
The AMOVA results are further reinforced by genetic distance (RST) values between each pair of groups (Table 3). Adopting a significance level of P = 0.01, none of the 45 comparisons between pairs of African-American groups are statistically-significant, whereas 6 of 55 comparisons between pairs of European-American groups, and 4 of 36 comparisons between pairs of Hispanic groups are statistically significant. Moreover, four of the six significant comparisons between European-American groups involve the Texas sample, and all four of the significant comparisons between Hispanic groups involve the Texas sample. These results support the genetic distinctiveness of the Texas groups among both the European-Americans and the Hispanics.
Table 3.
Rst Values (Below the Diagonal) and Number of Shared Y-STR Haplotypes (Above the Diagonal) Between Pairs of U.S. Groups. Boldface Rst Values, P < 0.01 Based on 10,000 Permutations
Codes are from Table 1.
With regard to between-population comparisons, all but 1 of the 190 pairwise RST values involving an African-American group and either a European-American or a Hispanic group were statistically significant, whereas 37 of the 99 RST values involving a European-American and an Hispanic group were statistically significant (Table 3). The number of shared haplotypes was also higher for groups from the same population than for groups from different populations (Table 3). The mean number of shared haplotypes was 4.3 between pairs of African-American groups, 4.9 between European-American groups, and 3.6 between Hispanic groups; the mean number of shared haplotypes was 2.5 between African-American and European-American groups, 2.1 between African-American and Hispanic groups, and 3.3 between European-American and Hispanic groups.
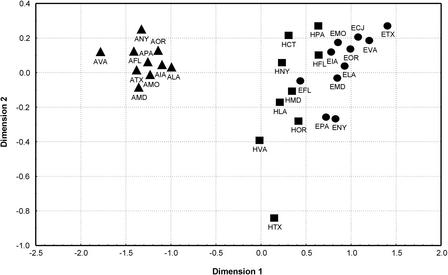
These results suggest that there is some degree of homogeneity among the regional groups within each of the three populations, relative to the comparison of groups from different populations. To further investigate this, a multidimensional scaling (MDS) analysis was carried out (Fig. 2), on the basis of the pairwise RST values (Table 3). The African-American groups are well separated from the European-American and Hispanic groups, whereas there is some overlap between the latter two. The same clustering was obtained from a neighbor-joining tree on the basis of the pairwise RST values (data not shown). Thus, the Y-STR haplotypes indicate closer relationships among European-American and Hispanic groups, and more distant relationships between either of these and African-American groups.
Figure 2.
MDS plot based on RST values for Y-STR haplotypes for U.S. groups. Codes are from Table 1. (▴) African-Americans; (•) European-Americans; (▪) Hispanics.
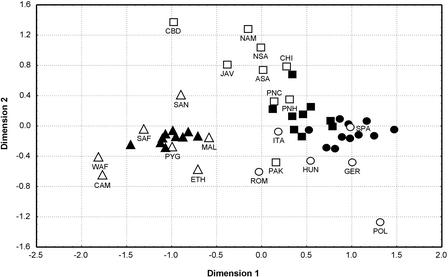
We also compared the U.S. populations to worldwide data for haplotypes for the same nine Y-STR loci. An MDS plot (Fig. 3) shows that sub-Saharan African and African-American groups are clustered together, separate from the other groups. Hispanic groups tend to be associated with populations of Asian and European ancestry, whereas European-American groups tend to be associated with European populations, but there is some intermingling between Asian/Hispanic and European/European-American groups. A neighbor-joining tree shows the same groupings (data not shown).
Figure 3.
MDS plot based on RST values for Y-STR haplotypes, comparing global populations with the U.S. groups. Data for non-U.S. populations, West Africans (WAF), Cameroons (CAM) from this study; Germans (GER), Poles (POL), native South Americans (NSA), Chinese (CHI), Javanese (JAV), and Papua New Guineans from coastal (PNC) and highland (PNH) regions from Kayser et al. (1997, 2000a,b, 2001); Italians (ITA) from Caglia et al. (1997); Hungarians (HUN) and Baranya-Romanies (ROM) from Füredi et al. (1999); South Africans (SAF), Mbuti Pygmies (PYG), Mali (MAL), Ethiopians (ETH), San (SAN), Cambodians (CBD), native Americans (NAM), and Pakistani (PAK) from Seielstad et al. (1999); Spaniards (SPA) from M. Kayser (unpubl.); and Asian-Americans (ASA) from M. Prinz (unpubl.). (▵) Africans; (▴) African-Americans; (○) Europeans; (•) European-Americans; (□) Asian ancestry (including native Americans); (▪) Hispanics.
Comparisons With mtDNA
MtDNA haplotypes were determined previously for many of the groups in this study (Melton et al. 2001) by hybridization of PCR products of the control region with 21 sequence-specific oligonucleotide (SSO) probes directed to 4 locations in the first hypervariable segment of the control region and 4 locations in the second hypervariable segment. The groups analyzed and the associated diversity on the basis of mtDNA SSO-types are shown in Table 4. The AMOVA results on the basis of mtDNA SSO-types are comparable with those for the Y-STR haplotypes (Table 2), with 98%–99% of the total genetic variance shared by the regional groups of African-Americans, European-Americans, and Hispanics. When all three populations are compared and the total genetic variance divided into within group, among group within population, and among population components, the within-group component was lower and the among-population component was higher for the Y-STR haplotypes than for the mtDNA SSO-types (Table 2). Overall, only about 0.5%–0.8% of the total genetic variance is ascribed to differences among regional groups within a population.
Table 4.
Sample Sizes, Number of Haplotypes, and Haplotype Diversity for 27 U.S. Groups, Based on mtDNA SSO-types
| Population/Group | Code | Sample size | Number of haplotypes | Haplotype diversity (±SE) |
| African–American | ||||
| California | ACA | 119 | 62 | 0.975 ± 0.004 |
| Illinois | AIL | 68 | 55 | 0.993 ± 0.002 |
| Louisiana | ALA | 55 | 36 | 0.986 ± 0.008 |
| Maryland | AMD | 38 | 27 | 0.980 ± 0.005 |
| Missouri | AMO | 126 | 64 | 0.977 ± 0.003 |
| Oregon | AOR | 86 | 55 | 0.986 ± 0.002 |
| Pennsylvania | APA | 93 | 64 | 0.988 ± 0.002 |
| Texas | ATX | 85 | 51 | 0.981 ± 0.003 |
| Virginia | AVA | 86 | 58 | 0.984 ± 0.004 |
| Washington | AWA | 49 | 33 | 0.974 ± 0.007 |
| TOTAL | 805 | 251 | 0.983 ± 0.001 | |
| European-American | ||||
| Acadian | ECJ | 58 | 30 | 0.953 ± 0.010 |
| California | ECA | 128 | 72 | 0.972 ± 0.006 |
| Illinois | EIL | 42 | 30 | 0.974 ± 0.009 |
| Louisiana | ELA | 57 | 35 | 0.948 ± 0.014 |
| Maryland | EMD | 38 | 26 | 0.962 ± 0.013 |
| Missouri | EMO | 90 | 51 | 0.970 ± 0.006 |
| Oregon | EOR | 98 | 54 | 0.973 ± 0.005 |
| Pennsylvania | EPA | 105 | 53 | 0.958 ± 0.008 |
| Texas | ETX | 103 | 56 | 0.968 ± 0.006 |
| Vermont | EVT | 94 | 47 | 0.923 ± 0.016 |
| Virginia | EVA | 109 | 56 | 0.973 ± 0.004 |
| TOTAL | 922 | 226 | 0.964 ± 0.002 | |
| Hispanic | ||||
| Louisiana | HLA | 58 | 27 | 0.929 ± 0.014 |
| Oregon | HOR | 74 | 45 | 0.959 ± 0.010 |
| Pennsylvania | HPA | 115 | 40 | 0.925 ± 0.010 |
| Texas | HTX | 60 | 34 | 0.969 ± 0.006 |
| Virginia | HVA | 102 | 52 | 0.967 ± 0.005 |
| Washington | HWA | 52 | 36 | 0.978 ± 0.005 |
| TOTAL | 461 | 153 | 0.967 ± 0.004 | |
Data are from Melton et al. (2001).
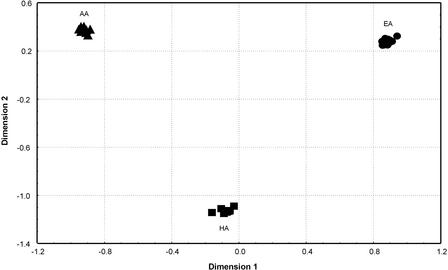
The MDS plot based on mtDNA SSO-types (Fig. 4) is similar to the MDS plot based on Y-STR haplotypes (Fig. 2), in that all of the African-American groups are well separated from the other groups. However, in contrast to the Y-STR MDS plot, the mtDNA plot also separates the Hispanic groups from the European-American groups; Hispanic groups are almost equally distant from European-American and African-American groups with respect to mtDNA (average FST = 0.147 and 0.140, respectively), whereas they are much closer to European-American groups than to African-American groups with respect to Y-STR haplotypes (average RST = 0.097 and 0.241, respectively). A neighbor-joining tree based on the mtDNA SSO-types revealed the same patterns as the MDS plot.
Figure 4.
MDS plot on the basis of FST values for mtDNA SSO-types for U.S. groups. (▴) African-Americans (AA); (•) European-Americans (EA); (▪) Hispanics (HA).
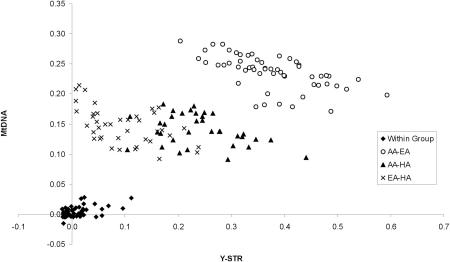
To further compare the relationships among groups on the basis of mtDNA SSO-types versus Y-STR haplotypes, we plotted the FST values for mtDNA versus the RST values for the Y-STR haplotypes for each pair of groups (Fig. 5). Overall, there is a significant relationship between FST and RST (Mantel test, r = 0.78, Z = 6.60, P < 0.0001, on the basis of 10,000 permutations). The plot also indicates that the comparisons involving pairs of groups from the same populations are well separated from comparisons involving pairs of groups from different populations, and that for the latter, comparisons involving one African-American group and one European-American group are well separated from other between-population comparisons. However, there is some overlap in the between-population comparisons involving African-American and Hispanic groups and those involving European-American and Hispanic groups. This overlap is mostly due to the mtDNA distances, which, as noted above, are nearly equal for Hispanic versus European-American groups and Hispanic versus African-American groups.
Figure 5.
Plot of RST values for Y-STR haplotypes vs. FST values for mtDNA SSO-types, for U.S. groups. (AA) African-American; (EA) European-American; (HA) Hispanic.
Admixture Estimates
Estimates of the European genetic contribution to non-European U.S. populations can be obtained for the Y-STR haplotypes and the mtDNA SSO-types, provided that comparable data exist for the parental populations. For the Hispanic populations, such data for appropriate parental populations (in particular, Mexican, Puerto Rican, Cuban, and other Central/South American groups; Chakraborty et al. 1999) do not yet exist; however, for the African-American populations, admixture estimates can be made. For the Y-STR haplotypes, we used the West African and Cameroon populations as the African parental population, and the European-American population (excluding the Acadians, as these are a population isolate, and the Texas group, as this group differed significantly from the other European-American groups) as the European parental population. For the mtDNA SSO-types, we used published data on Yoruban, Mandenka, and Sierra Leone populations (Melton et al. 1997a) as the African parental population, and the published data on European-Americans (Melton et al. 2001), again excluding the Acadians, as the European parental population. We also repeated the analyses using data from European rather than European-American populations and obtained similar results (data not shown). The latter result indicates that the African-American genetic contribution to European-Americans is below the limits of detection with these methods.
The European-American genetic contribution to the African-American gene pool was estimated by use of two methods, one on the basis of a coalescent approach (Bertorelle and Excoffier 1998) and the other on the basis of a genotype assignment test (Paetkau et al. 1995). For the latter method, we first computed the assignment of the parental genotypes to test the ability of the method to distinguish between the European-American and African parental genotypes. For the European-Americans, 9.0% of the mtDNA SSO-types and 8.7% of the Y-STR haplotypes were classified as African, whereas for the Africans, 5.6% of the mtDNA SSO-types and 8.3% of the Y-STR haplotypes were classified as European-American. Thus, in all cases, the level of cross-classification was less than 10%, indicating that the genotype assignments were highly reliable.
For both the coalescent and genotype assignment methods, the estimated European-American genetic contribution to African-Americans (Table 5) was much higher for the Y-chromosome than for mtDNA; ∼27.5%–33.6% of African-American Y-chromosomes were determined to be of European-American ancestry versus only 9.0%–15.4% of African-American mtDNAs. The genotype assignment method gave significantly higher estimates than did the coalescent approach for both Y-STR haplotypes and mtDNA SSO-types (Table 5). A possible explanation for this is that the genotype assignment method assigns a genotype to the population that has the highest expected frequency of that genotype, even if the probability associated with assigning the genotype to the other population is not significantly lower. Thus, the results might be influenced by genotypes that are difficult to classify and, hence, have nearly equal probabilities of arising from either parental population. We therefore used a stricter version of the genotype assignment method, in which genotype assignments were only accepted if the probability associated with the assignment was at least 10 times greater than the probability associated with assigning the genotype to the other population. For the Y-STR haplotypes, 567 of the 598 African-Americans (94.8%) could be assigned under this stricter requirement, and the resulting estimate of the European-American genetic contribution to African-Americans was 32.6%, which is not significantly different from the estimate of 33.6% on the basis of all 598 individuals. For the mtDNA SSO-types, 635 of the 805 African-Americans (78.9%) were assigned under the stricter requirement, and the resulting estimate of the European-American genetic contribution to African-Americans was 11.3%, which is significantly lower than the estimate of 15.4% on the basis of all individuals, but not significantly different from the estimate of 9.0% on the basis of the coalescent approach. Thus, Y-STR haplotypes can be assigned with a higher degree of confidence than mtDNA SSO-types, and mtDNA SSO-types that cannot be assigned with a high degree of confidence appear to be responsible for the difference in admixture estimates between the coalescent approach and the genotype assignment method for mtDNA SSO-types.
Table 5.
Two estimates of the European-American Genetic Contribution to African-Americans (Admixture Estimates), Expressed as the Percent Contribution of European-American Haplotypes, for African-American Y-STR Haplotypes and mtDNA SSO-types
| Group | Y-STR | mtDNA | ||
| 1 | 2 | 1 | 2 | |
| California | N/A | N/A | 18.0 ± 3.3 | 21.0 |
| Florida | 24.6 ± 5.9 | 31.2 | N/A | N/A |
| Indiana | 36.1 ± 7.6 | 37.8 | N/A | N/A |
| Illinois | N/A | N/A | 8.0 ± 4.2 | 13.2 |
| Louisiana | 41.8 ± 8.5 | 44.8 | 3.9 ± 4.1 | 20.0 |
| Maryland | 26.2 ± 4.9 | 31.5 | 21.8 ± 5.6 | 21.1 |
| Missouri | 31.1 ± 5.8 | 33.9 | 5.2 ± 2.9 | 11.9 |
| New York | 25.4 ± 3.8 | 33.3 | N/A | N/A |
| Oregon | 33.8 ± 6.5 | 42.6 | 4.0 ± 3.5 | 10.5 |
| Pennsylvania | 30.2 ± 6.4 | 34.9 | 3.8 ± 3.8 | 15.1 |
| Texas | 25.8 ± 5.1 | 33.3 | 9.0 ± 3.9 | 11.8 |
| Virginia | 11.8 ± 5.9 | 19.6 | 12.4 ± 3.8 | 24.4 |
| Washington | N/A | N/A | 6.0 ± 4.0 | 4.1 |
| All | 27.5 ± 2.5 | 33.6 | 9.0 ± 2.0 | 15.4 |
Methods: (1) coalescent approach (Bertorelle and Excoffier 1998); (2) assignment test (Paetkau et al. 1995). The standard errors for the estimates for the coalescent approach are based on 1000 bootstrap replications.
DISCUSSION
This is, to our knowledge, the first in-depth study of geographic heterogeneity in Y-STR haplotypes in U.S. populations. We found no significant heterogeneity among regional groups of African-Americans, which seems somewhat surprising for two reasons. First, a large number of different African source populations contributed to present-day African-American groups, with about half coming from the area extending from Senegal to Western Nigeria, and the remaining half coming from the area extending from Eastern Nigeria to Angola (Curtin 1969; Reed 1969). However, the amount of genetic heterogeneity among these West and Central African source populations that contributed to African-Americans is not known, as a comprehensive study of genetic variation in these populations has not been carried out. A recent study of Y-SNP haplotype variation in African populations did find significant differences among West African populations (Cruciani et al. 2002), but it is not known to what extent this holds for the more rapidly evolving Y-STR haplotypes. The Y-STR haplotype frequencies in the Cameroon and West African samples analyzed here are at the border of statistical significance (RST = 0.033, P = 0.05), whereas three West African populations analyzed for mtDNA SSO-types (from Sierra Leone, Senegal, and Nigeria) did not differ significantly from one another (Melton et al. 1997a). If the African source populations do not differ significantly in Y-STR haplotype (or mtDNA SSO-type) frequencies, then differences in the contribution of African populations to different African-American populations will not show up in the African-American Y-STR haplotype (or mtDNA SSO-type) distributions.
Second, the amount of admixture of African-Americans with European-Americans is thought to have varied across different geographic regions of the U.S., with generally higher levels of admixture observed in Northern groups (Reed 1969; Chakraborty et al. 1992). However, other studies find a more complex relationship between the amount of admixture and geographic region (Parra et al. 1998), and none of these studies performed statistical tests to determine whether the observed heterogeneity in admixture estimates across groups was statistically significant. Our estimates of the European-American genetic contribution to African-Americans are quite similar across regional geographic groups (Table 5) and do not vary significantly, as discussed in more detail below.
A further complicating factor is migration among geographic regions within the United States. Even with heterogeneity in the founding West African populations and/or the subsequent amount of European-American genetic contribution to African-Americans, migration of African-Americans within the United States may have been extensive enough to eliminate between-group differences in Y-STR haplotype frequencies. In particular, during and following World War I, an estimated one million African-Americans (∼10% of the African-American population) left rural areas in the southern United States for metropolitan areas in the north (Johnson and Campbell 1981; Tanner 1995). The lack of geographic heterogeneity observed in African-American mtDNA and Y-chromosome types may thus reflect this “Great Migration”, the largest internal migration in the history of North America.
In contrast to the African-American groups, the European-American and Hispanic groups do show significant geographic heterogeneity. However, in both cases, this is due to the influence of one group, as removal of that one group reduces the heterogeneity in the remaining groups to statistically insignificant levels. For both European-Americans and Hispanics, it is the Texas group that accounts for the significant geographic heterogeneity. Why this is the case is not obvious; among European-American groups, the Texas group has a low amount of haplotype diversity (but not the lowest) and the lowest MPSD (Table 1), suggesting possibly a lower amount of genetic variation for the Y-chromosome for this group. However, among Hispanic groups, the Texas group does not stand out in terms of either haplotype diversity or MPSD, although this group is quite differentiated in the MDS plot (Fig. 2). Moreover, mtDNA analyses of these same samples do not indicate any differences between these groups and other European-American and Hispanic groups, respectively (Melton et al. 2001). The most likely explanation would appear to be that the significant heterogeneity attributable to the European-American and Hispanic Texans reflects chance rather than any true biological differences; analyses of additional samples from Texas would be required to test this hypothesis.
The overall lack of geographic heterogeneity among European-Americans is not surprising, as European populations exhibit little differentiation with respect to Y-STR haplotypes (Roewer et al. 2001) and mtDNA types (Melton et al. 1997b). However, the striking uniformity among regional groups of Hispanics for both Y-STR haplotypes and mtDNA types (see Fig. 4) is surprising, given that Hispanic does not refer to a defined geographic region, in contrast to European-American and African-American. Instead, the ethnic category, Hispanic, typically can refer to someone of Mexican, Puerto Rican, Cuban, Central/South American, or other Spanish culture ancestry (Chakraborty et al. 1999), and previous analyses have estimated varying degrees of native American, Spanish, and African ancestry in Hispanic populations (Hanis et al. 1991; Merriwether et al. 1997; Chakraborty et al. 1999). As with African-Americans, either a lack of geographic heterogeneity among the source populations and/or extensive migration has resulted in a lack of geographic heterogeneity among Hispanic groups.
A comparison of the Y-STR haplotype analyses with mtDNA analyses of the same samples (Melton et al. 2001) reveals some intriguing similarities and differences. There was no significant heterogeneity among regional groups of African-Americans, European-Americans, and Hispanics with respect to mtDNA, as is (largely) the case with respect to Y-STR haplotypes. Another similarity between the Y-STR and mtDNA analyses is that all of the African-American groups cluster together, well apart from both European-American and Hispanic groups, for both loci. However, a major difference between the Y-STR and mtDNA analyses concerns the relationship of European-American and Hispanic populations. For the mtDNA SSO-types, the Hispanic and European-American groups were completely separated from one another (Fig. 4), whereas for the Y-STR haplotypes, there was some intermingling of Hispanic and European-American groups (Fig. 2). This cannot be attributed simply to a lack of resolution of Y-STR haplotypes, resulting in an inability to distinguish between European-American and Hispanic groups, because, on average, Y-STR haplotype diversity was higher than mtDNA SSO-type diversity (cf. Tables 1 and 4) and, hence, Y-STR haplotypes should provide more information on population relationships. Nor can the failure of Y-STR haplotypes to distinguish between European-American and Hispanic groups be attributed to high rates of parallel mutations in the Y-STR loci leading to a loss of phylogenetic signal, as the Y-STR haplotypes do clearly distinguish between African-American groups and the other groups. Moreover, other studies have indicated that Y-STR haplotypes are informative for studies of human population relationships (Seielstad et al. 1999; Kayser et al. 2001).
Instead, it appears that the paternal and maternal structure of Hispanic groups differ, most likely reflecting a greater contribution of European-American Y-chromosomes than mtDNA haplotypes to the Hispanic gene pool. Although insufficient data from potential source populations among native North American, Central American, and Caribbean populations exist to permit estimates of admixture for Hispanic groups on the basis of Y-STR haplotypes or mtDNA SSO-types, other studies have found a greater contribution of native American mtDNA than nuclear genes to Hispanic populations (Merriwether et al. 1997), which supports our results indicating a greater contribution of European-American males than females to the Hispanic gene pool.
Sufficient information does exist, however, to permit estimates of the European-American genetic contribution to African-Americans. Previous studies based on nuclear loci have generally found ∼20% European genetic contribution to African-American populations (Reed 1969; Chakraborty et al. 1992; Parra et al. 1998; Destro-Bisol et al. 1999; Collins-Schramm et al. 2002), in agreement with our estimate (averaged for mtDNA and the Y-chromosome) of 18%–24%. Our results indicate substantially higher contribution of European-American Y-chromosome (27.5%–33.6%) than mtDNA (9.0%–15.4%) to African-Americans, also in agreement with previous studies (Parra et al. 1998, 2001). Presumably, this disparity in admixture estimates for the Y-chromosome versus mtDNA reflects the greater genetic contribution of European-American men than women to African-Americans during the slavery period. However, there is currently an increasing trend toward more marriages between African-American men and European-American women; census data indicate that in 1960 there were 25,000 marriages involving African-American men and European-American women and 26,000 marriages involving African-American women and European-American men, whereas in 1992, there were 163,000 marriages involving African-American men and European-American women and 83,000 marriages involving African-American women and European-American men (source, U.S. Census Bureau, http://www.census.gov/population/socdemo/race/interractab1.txt). In our study, on the basis of self-reported ancestry, the offspring of marriages between African-Americans and European-Americans would generally be assigned as African-Americans rather than European-Americans. Hence, if this trend continues, the disparity between mtDNA and Y-chromosome-based estimates of the European genetic contribution to African-Americans may eventually diminish or even reverse direction.
A question of some interest is the extent to which the European-American genetic contribution to African-Americans has varied among African-American groups from different geographic regions. Previous studies suggest that, in general, the amount of European-American ancestry is higher for African-American groups in the north than in the south (Reed 1969), although other studies have found that variation among northern and southern groups was as great as the variation between groups (Parra et al. 1998). For our data on Y-STR haplotypes, estimates of the European-American genetic contribution to African-Americans (on the basis of the coalescent approach) did not differ significantly among the geographic groups of African-Americans (χ2 = 12.33, df = 9, P > 0.10). However, for mtDNA haplotypes there was significant heterogeneity among the admixture estimates (on the basis of the coalescent approach) for different geographic groups (χ2 = 31.02, df = 9, P < 0.01). This heterogeneity is due to the higher admixture estimates for Maryland (21.8%) and California (18.0%), as removal of these two groups reduces the heterogeneity to nonsignificant levels for the remaining eight groups (χ2 = 8.98, df = 7, P > 0.2). Moreover, we do not detect any significant differences in admixture estimates for either Y-STR haplotypes or mtDNA SSO-types when comparing northern versus southern populations (analysis not shown). Thus, our results support the view that the dynamics of the European-American genetic contribution to African-Americans is more complicated than a simple north–south division would suggest (Parra et al. 1998, 2001).
In conclusion, our results indicate a lack of substantial geographic structuring of Y-STR haplotypes among regional groups of African-Americans, European-Americans, and Hispanics. For both African-Americans and Hispanics we find evidence of a much higher genetic contribution of European-American males than European-American females. We also do not find any geographic heterogeneity for the European-American genetic contribution to African-Americans experienced by the different African-American groups examined. Analyses of mtDNA SSO-types are largely concordant in indicating a lack of substantial geographic structuring, which is also in agreement with studies of autosomal STR loci (Budowle et al. 2001). These results have important implications for the forensic DNA community, as they argue against the necessity for incorporating geographic structure into forensic databases of Y-STR/mtDNA haplotypes by allowing pooling of data from geographic subpopulations of U.S. ethnic groups.
They also have important implications for understanding regional variation in disease susceptibility, as a lack of regional variation in (presumably) neutral DNA markers, such as the Y-chromosome and mtDNA, suggests that any regional variation in disease susceptibility is caused by environmental/cultural factors, rather than underlying genetic heterogeneity. To be sure, more loci need to be evaluated before one can conclude that there is no genetic heterogeneity among regional groups of U.S. populations. In fact, recently, Kittles et al. (2002) suggested that there was significant population stratification in an African-American population, on the basis of 10 autosomal genetic markers. However, this conclusion was based on differences in allele frequencies across the loci in a single population; it remains to be seen if there is significant geographic heterogeneity with respect to any of these markers. MtDNA and the Y chromosome, by virtue of their haploid and uniparental mode of inheritance, should be more sensitive indicators of population structure than autosomal markers. Thus, the lack of significant geographic heterogeneity for mtDNA and the Y chromosome in U.S. populations leads us to predict that neutral autosomal markers also will not exhibit significant geographic heterogeneity. Still, much more remains to be done on the genetic structure of U.S. populations, including more thorough geographic sampling, as well as more thorough genetic characterization of the source populations that have contributed to the rich diversity of U.S. populations.
METHODS
DNA Samples
All U.S. samples used here (Fig. 1) were provided to us by U.S. crime laboratories, with the exception of the Louisiana and the Acadian samples (provided by M.A. Batzer). Samples were selected by the crime laboratories to be representative for the respective geographic region. Ancestry of each individual is self-reported as African-American, European-American, or Hispanic. Most of the samples were studied previously for mtDNA diversity (Melton et al. 2001). Additional samples used for Y chromosome analysis came from Connecticut, Florida, Indiana, and New York, whereas samples from California, Illinois, and Washington, which were analyzed previously for mtDNA, could not be analyzed for Y-chromosomal markers. We use the term “population” to refer to the composite African-American, European-American, and Hispanic groups, and “group” to refer to the geographic subgroups within these populations. In addition to the U.S. groups, 54 samples from Cameroon and 79 samples from West Africa (including 22 from Ghana, 6 from Guinea, 16 from the Ivory Coast, 7 from Senegal, and 27 from Sierra Leone) were analyzed for the purpose of estimating the European-American genetic contribution to African-Americans (admixture estimates); these DNA samples have been described elsewhere (Zimmerman et al. 1992, 1996).
Y-STR Typing
Nine Y-STR loci (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, and DYS385a/b) were amplified via the PCR and genotyped as described elsewhere (http://www.ystr.org/usa). Alleles are designated by the number of repeats (Kayser et al. 1997). The Y-chromosomal data are accessible via the Y-chromosomal short tandem repeat Haplotype Reference Database (YHRD) for U.S. populations (http://www.ystr.org/usa), which is described in more detail elsewhere (Kayser et al. 2002), and are also available from the authors.
Statistical Analyses
The number of haplotypes, haplotype diversity, and the mean number of pairwise step differences (MPSD), which takes into account the difference in the number of repeats between the two alleles compared at each locus, were calculated using Arlequin 2.0 (http://lgb.unige.ch/arlequin) (Schneider et al. 2000). Nonparametric Mann-Whitney tests of the differences in haplotype diversity and MPSD among populations were performed with Statistica (Statsoft). Decomposition of the total genetic variance into within-group and among-group components was done via the AMOVA procedure in Arlequin 2.0; RST values (Slatkin 1995), which are analogous to FST values but are based on a stepwise mutation model, were also computed with Arlequin 2.0. The statistical significance of the variance components and the RST values were assessed by permutation tests with 10,000 permutations. Population relationships on the basis of the RST values were determined by use of neighbor-joining trees constructed with programs in PHYLIP 3.5 (http://evolution.genetics.washington.edu/phylip.html), (Felsenstein 1993), and using multidimensional scaling analysis (MDS) as implemented in Statistica. The Mantel test (Smouse and Long 1992) was used, as implemented in Arlequin 2.0 (http://lgb.unige.ch/arlequin), to test the statistical significance of the correlation between genetic distances on the basis of Y-STR and mtDNA haplotypes. The European-American genetic contribution to African-Americans was estimated by two different methods. The first method is based on a coalescent approach that incorporates both allele frequencies as well as the molecular distance among alleles (Bertorelle and Excoffier 1998), and is implemented in the program ADMIX (http://www.unife.it/genetica/Giorgio/Giorgio_soft.html#ADMIX). The second method is an assignment test (Paetkau et al. 1995), in which the probability that an individual comes from each of several populations is calculated on the basis of genotype frequencies, and then the individual is assigned to the population associated with the highest probability. Assignments of African-Americans to either African or European ancestry was done via this method with the program Doh (http://www.biology.ualberta.ca/jbrzusto/Doh.php).
For all statistical analyses, alleles at DYS389II were considered excluding variation at DYS389I. For DYS385, which is a duplicated Y-STR locus, the allele locus assignment was performed so that for each individual, the smaller allele was assigned to one locus (DYS385a) and the longer to the other (DYS385b). This procedure may result in incorrect genotypes (Kittler et al. 2003); we therefore repeated relevant analyses without DYS385a/b, and in no case did the conclusions change.
WEB SITE REFERENCES
http://www.ystr.org/usa; Y-chromosome STR haplotype reference database (YHRD) for US populations.
http://www.unife.it/genetica/Giorgio/Giorgio_soft.html#ADMIX; ADMIX software.
http://lgb.unige.ch/arlequin; Arelquin software.
http://www.biology.ualberta.ca/jbrzusto/Doh.php; Doh software.
http://evolution.genetics.washington.edu/phylip.html; PHYLIP software.
http://www.census.gov/population/socdemo/race/interractab1.txt; U.S. Census Bureau, “Interracial Tables, (Table) 1. Race of Wife by Race of Husband: 1960, 1970, 1980, 1991, and 1992”; published 10 June 1998.
Acknowledgments
We thank the following colleagues for providing blood and/or DNA samples: Bruce Budowle, Thomas Grant, Deborah Grippando, Barbara Llewellyn, Teresa M. Long, Miguel Lorente, Keith McKenney, Tamyra Moretti, Joanne B. Sgueglia, Mohammad A. Tahir, Chris Tomsey, and Cecilia H. von Beroldingen. Daniel Corach, Sandor Füredi, and Mark Seielstad are gratefully acknowledged for providing electronic access to published data. This research was supported by the Louisiana Board of Regents Millennium Trust Health Excellence Fund HEF (2000-05)-05, (2000-05)-01, and (2001-06)-02 (MAB), and awards NIJ98-LB-VX-005 (MS) and 2001-IJ-CX-K004 (M.A.B) from the Office of Justice Programs, National Institute of Justice, Department of Justice, and by funds from the Max Planck Society (M.S.). Points of view in this document are those of the authors and do not necessarily represent the official position of the U.S. Department of Justice.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL kayser@eva.mpg.de; FAX 49-341-9952555.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.463003.
REFERENCES
- 1.Bertorelle G. and Excoffier, L. 1998. Inferring admixture proportions from molecular data. Mol. Biol. Evol. 15: 1298-1311. [DOI] [PubMed] [Google Scholar]
- 2.Budowle B., Shea, B., Niezgoda, S., and Chakraborty, R. 2001. CODIS STR loci data from 41 sample populations. J. Forensic Sci. 46: 453-489. [PubMed] [Google Scholar]
- 3.Caglia A., Novelletto, A., Dobosz, M., Malaspina, P., Ciminelli, B., and Pascali, V. 1997. Y-chromosome STR loci in Sardinia and continental Italy reveal islander-specific haplotypes. Eur. J. Hum. Genet. 5: 288-292. [PubMed] [Google Scholar]
- 4.Cavalli-Sforza L.L., Menozzi, P., and Piazza, A., 1994. The history and geography of human genes. Princeton University Press, Princeton, NJ.
- 5.Chakraborty B.M., Fernandez-Esquer, M.E., and Chakraborty, R. 1999. Is being Hispanic a risk factor for non-insulin dependent diabetes mellitus (NIDDM)? Ethn. Dis. 9: 278-283. [PubMed] [Google Scholar]
- 6.Chakraborty R., Kamboh, M., Nwankwo, M., and Ferrell, R. 1992. Caucasian genes in American blacks: New data. Am. J. Hum. Genet. 50: 145-155. [PMC free article] [PubMed] [Google Scholar]
- 7.Collins-Schramm H., Phillips, C., Operario, D., Lee, J.L., Hanson, R., Knowler, W., Cooper, R., Li, H., and Seldin, M. 2002. Ethnic-difference markers for use in mapping by admixture linkage disequilibrium. Am. J. Hum. Genet. 70: 737-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruciani F., Santolamazza, P., Shen, P., Macaulay, V., Moral, P., Olckers, A., Modiano, D., Holmes, S., Destro-Bisol, G., Coia, V., et al. 2002. A back migration from Asia to Sub-Saharan Africa is supported by high-resolution analysis of human Y-chromosome haplotypes. Am. J. Hum. Genet. 70: 1197-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Curtin P., 1969. The Atlantic slave trade. University of Wisconsin Press, Madison, WI.
- 10.Destro-Bisol G., Maviglia, R., Caglia, A., Boschi, I., Spedini, G., Pascali, V., Clark, A., and Tishkoff, S. 1999. Estimating European admixture in African Americans by using microsatellites and a microsatellite haplotype (CD4/Alu). Hum. Genet. 104: 149-157. [DOI] [PubMed] [Google Scholar]
- 11.Devesa S., Grauman, D., Blot, W., Pennello, G., Hoover, R., and Fraumeni, J., 1999. Atlas of cancer mortality in the United States, 1950–94. US Government Printing Office, Washington, DC.
- 12.Felsenstein J., 1993. PHYLIP (Phylogeny Inference Package) version 3.5c. Department of Genetics, University of Washington, Seattle, WA.
- 13.Füredi S., Woller, J., Padar, Z., and Angyal, M. 1999. Y-STR haplotyping in two Hungarian populations. Int. J. Legal Med. 113: 38-42. [DOI] [PubMed] [Google Scholar]
- 14.Gilliland F. 1997. Ethnic differences in cancer incidence: A marker for inherited susceptibility? Environ. Health Persp. 105: 897-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanis C.L., Hewett-Emett, D., Bertin, T.K., and Schull, W.J. 1991. Origins of U.S. Hispanics: implications for diabetes. Diabetes Care 14: 618-627. [DOI] [PubMed] [Google Scholar]
- 16.Jackson F. 2000. Anthropological measurement: The mismeasure of African Americans. Ann. Am. Acad. Pol. Soc. Sci. 568: 154-171. [Google Scholar]
- 17.Johnson D. and Campbell, R., 1981. Black migration in America: A social demographic history. Duke University Press, Durham, NC.
- 18.Kayser M., Caglia, A., Corach, D., Fretwell, N., Gehrig, C., Graziosi, G., Heidorn, F., Herrmann, S., Herzog, B., Hidding, M., et al. 1997. Evaluation of Y-chromosomal STRs: A multicenter study. Int. J. Legal Med. 110: 125-133., 141–149. [DOI] [PubMed] [Google Scholar]
- 19.Kayser M., Brauer, S., Weiss, G., Underhill, P.A., Roewer, L., Schiefenhövel, W., and Stoneking, M. 2000a. Melanesian origin of Polynesian Y chromosomes. Curr. Biol. 10: 1237-1246. [DOI] [PubMed] [Google Scholar]
- 20.Kayser M., Roewer, L., Hedman, M., Henke, L., Henke, J., Brauer, S., Krüger, C., Krawczak, M., Nagy, M., Dobosz, T., et al. 2000b. Characteristics and frequency of germline mutations at microsatellite loci from the human Y chromosome, as revealed by direct observation in father/son pairs. Am. J. Hum. Genet. 66: 1580-1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kayser M., Krawczak, M., Excoffier, L., Dieltjes, P., Corach, D., Pascali, V., Gehrig, C., Bernini, L., Jespersen, J., Bakker, E., et al. 2001. An extensive analysis of Y-chromosomal microsatellite haplotypes in globally dispersed human populations. Am. J. Hum. Genet. 68: 990-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kayser M., Brauer, S., Willuweit, S., Schädlich, H., Batzer, M.A., Zawacki, J., Prinz, M., Roewer, L., and Stoneking, M. 2002. Online Y-chromosomal short tandem repeat haplotype reference database (YHRD) for U.S. populations. J. Forensic Sci. 47: 513-519. [PubMed] [Google Scholar]
- 23.Keppel K., Pearcy, J., and Wagener, D., 2002. Trends in racial and ethnic-specific rates for the health status indicators: United States, 1990–98. Healthy people statistical notes, no. 23. National Center for Health Statistics, Hyattsville, MD. [PubMed]
- 24.Kittler, R., Erler, A., Brauer, S., Stoneking, M., and Kayser, M. 2003. Apparent intra-chromosomal exchange on the human Y chromosome explained by population history. Eur. J. Hum. Genet. 4: (in press). [DOI] [PubMed]
- 25.Kittles, R.A., Chen, W., Panguluri, R.K., Ahaghotu, C., Jackson, A., Adebamowo, C.A., Griffin, R., Williams, T., Ukoli, F., Adams-Campbell, L., et al. 2002. CYP3A4-V and prostate cancer in African Americans: Causal or confounding association because of population stratification? Hum. Genet. online DOI 10.1007/s00439-002-0731-5. [DOI] [PubMed]
- 26.Melton T., Ginther, C., Sensabaugh, G., Soodyall, H., and Stoneking, M. 1997a. Extent of heterogeneity in mitochondrial DNA of sub-Saharan African populations. J. Forensic Sci. 42: 582-592. [PubMed] [Google Scholar]
- 27.Melton T., Wilson, M., Batzer, M., and Stoneking, M. 1997b. Extent of heterogeneity in mitochondrial DNA of European populations. J. Forensic Sci. 42: 437-446. [PubMed] [Google Scholar]
- 28.Melton T., Clifford, S., Kayser, M., Nasidze, I., Batzer, M., and Stoneking, M. 2001. Diversity and heterogeneity in mitochondrial DNA of North American populations. J. Forensic Sci. 46: 46-52. [PubMed] [Google Scholar]
- 29.Merriwether D., Huston, S., Iyengar, S., Hamman, R., Norris, J., Shetterly, S., Kamboh, M., and Ferrell, R. 1997. Mitochondrial versus nuclear admixture estimates demonstrate a past history of directional mating. Am. J. Phys. Anthropol. 102: 153-159. [DOI] [PubMed] [Google Scholar]
- 30.Paetkau D., Calvert, W., Stirling, I., and Strobeck, C. 1995. Microsatellite analysis of population structure in Canadian polar bears. Mol. Ecol. 4: 347-354. [DOI] [PubMed] [Google Scholar]
- 31.Parra E., Marcini, A., Akey, J., Martinson, J., Batzer, M., Cooper, R., Forrester, T., Allison, D., Deka, R., Ferrell, R., et al. 1998. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 63: 1839-1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parra E., Kittles, R., Argyropoulos, G., Pfaff, C., Hiester, K., Bonilla, C., Sylvester, N., Parrish-Gause, D., Garvey, W., Jin, L., et al. 2001. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am. J. Phys. Anthropol. 114: 18-29. [DOI] [PubMed] [Google Scholar]
- 33.Reed T.E. 1969. Caucasian genes in American Negroes. Science 165: 762-768. [DOI] [PubMed] [Google Scholar]
- 34.Roewer L., Krawczak, M., Willuweit, S., Nagy, M., Alves, C., Amorim, A., Anslinger, K., Augustin, C., Betz, A., Bosch, E., et al. 2001. Online reference database of European Y-chromosomal short tandem repeat (STR) haplotypes. Forensic Sci. Int. 118: 106-113. [DOI] [PubMed] [Google Scholar]
- 35.Schneider S., Roessli, D., and Excoffier, L., 2000. Arlequin ver 2.000: A software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland.
- 36.Seielstad M., Bekele, E., Ibrahim, M., Toure, A., and Traore, M. 1999. A view of modern human origins from Y chromosome microsatellite variation. Genome Res. 9: 558-567. [PMC free article] [PubMed] [Google Scholar]
- 37.Slatkin M. 1995. A measure of population subdivision based on microsatellite allele frequencies. Genetics 139: 457-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smouse P. and Long, J. 1992. Matrix correlation analysis in anthropology and genetics. Yearb. Phys. Anthropol. 35: 187-231. [Google Scholar]
- 39.Tanner H., 1995. The settling of North America. Macmillan, New York, NY.
- 40.Zimmerman P., Dadzie, K., De Sole, G., Remme, J., Alley, E., and Unnasch, T. 1992. Onchocerca volvulus DNA probe classification correlates with epidemiologic patterns of blindness. J. Infect. Dis. 165: 964-968. [DOI] [PubMed] [Google Scholar]
- 41.Zimmerman P., Steiner, L., Titanji, V., Nde, P., Bradley, J., Pogonka, T., and Begovich, A. 1996. Three new DPB1 alleles identified in a Bantu-speaking population from central Cameroon. Tissue Antigens 47: 293-299. [DOI] [PubMed] [Google Scholar]