Abstract
The discovery of novel classes of non-coding RNAs (ncRNAs) has revolutionized medicine. Long time thought as a mere cellular housekeeper, surprising functions have recently been uncovered. microRNAs (miRNAs), a representative of the class of short ncRNAs, play a fundamental role in the control of DNA and protein biosynthesis and activity as well as pathology. Currently miRNAs are investigated as diagnostic and prognostic markers and potentially therapeutic targets in kidney transplantation for such indolent processes as ischemia-reperfusion injury, humoral rejection or viral infections. It is realistic to believe that monitoring of renal allograft recipients in the future will include genome-wide miRNA profiling of biological fluids. Based on these individual profiles an informed decision on therapeutic consequences will be possible. First success with a specific suppression of miRNAs by antisense oligonucleotides was achieved in experimental studies of reperfusion injury and humoral rejection. Proof of this concept in men comes from studies in such indolent viral infections as Ebola and hepatitis C where anti-miR therapy led to sustained viral clearance.
In this review we summarize the basis of the recent non-coding RNA revolution and its implication for kidney transplantation.
Keywords: acute kidney injury, rejection, biomarker, delayed graft function, ischemic reperfusion injury, miRNAs, renal transplantation
Introduction
Definition and history of miRNAs
MicroRNAs (miRNAs) are short (usually 18-22 nucleotides), endogenous non-coding RNAs that inhibit gene expression post-transcriptionally in a sequence specific mode. miRNAs were discovered about two decades ago by Lee and coworkers while studying the developmental gene lin-14 in the roundworm Caenorhabditis elegans [1]. The investigators observed a mutant strain of the worm with developmental abnormalities such as the inability to form the vulva. The gene lin-4, which was responsible for this mutant phenotype, had two transcripts, a longer (61nt) and a shorter one (21nt). The latter which was not translated, inhibited the lin-14 mRNA in a sequence specific ‘antisense’ manner and thus blocked the translation of the lin-14 mRNA into the lin-14 protein. Without functioning lin-14 protein, the nematode could not progress from the first to the second larval stage and exhibited the above mentioned defects.
This observation was initially not widely appreciated because it was considered to be only of relevance in C. elegans. It took another seven years until another non-coding short RNA with 22 nucleotides, let-7, was identified. Let-7 exhibited sequence specific antisense activity to lin-41 mRNA in the same worm [2]. Subsequently it was discovered that let-7 was not specific to the C. elegans but rather highly conserved in most organisms [3].
This discovery started the torrent of small, non-coding RNA research not only in nematodes and insects but also mammals. Currently, it is believed that the human genome encode over 2000 miRNAs which are abundantly expressed in almost all human cells. The nomenclature of miRNAs by three initial letters referring to the organism and the number behind the miR refers to the sequence of discovery [4]. For example hsa-miR-21 is a human mature miRNA that is encoded by the MIR21 gene and was one of the first to be sequenced in 2001 [5].
Biogenesis and Function of miRNAs
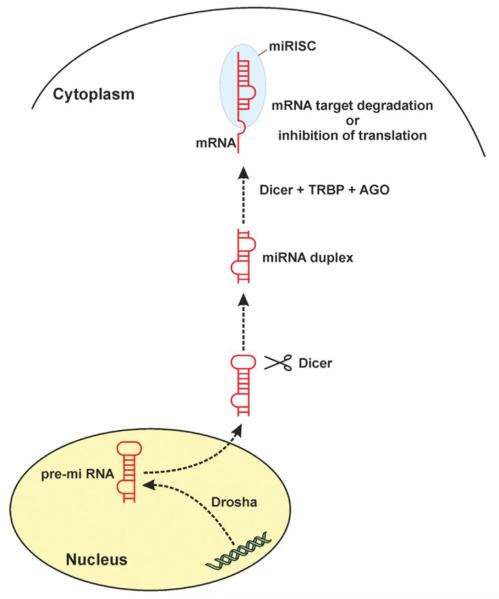
Most microRNAs (miRNAs) are transcribed and further processed into the precursor miRNA (pre-miRNAs) by the RNase III enzyme Drosha in the nucleus (Figure 1). After export to the cytoplasm, Dicer (another RNase III enzyme) cleaves off the loop of the hairpin of the pre-miRNA and generates a double-stranded RNA [6]. The resulting mature miRNA strand is subsequently incorporated into the RNA-induced silencing complex (RISC) responsible for the gene inhibition process coined RNA interference (RNAi). miRNA act similar to siRNAs, the other short and non-coding RNAi molecule, by hybridisation of mRNA binding sites in a sequence specific manner.
Figure 1. Biogenesis and Function of miRNAs.
Briefly, the primary transcript is transcribed and further processed into the precursor miRNA (pre-RNA) by Drosha, a RNase III enzyme in the nucleus. After export to the cytoplasm, Dicer (another RNase III enzyme) cleaves off the loop of the hairpin and generates a double-stranded RNA. Subsequently the mature miRNA strand binds to a member of the AGO protein family together with Dicer and a dsRNA-binding domain protein such as TRBP building the RNA-induced silencing complex (miRISC)). After mRNA target binding the miRISC complex induce mRNA target degradation or inhibition of protein translation.
miRNAs as well as siRNAs can form a perfect duplex with their targets and therefore direct the cleavage of the target mRNAs at the site of complementarity (mRNA degradation). Or miRNAs can repress protein expression on the translation level through imperfect (semi-complementary) binding to the mRNA target mostly at multiple sites [7]. To which extend mRNA cleavage or translation repression will be guided also depends on into which specific Argonaute (Ago) protein is incorporated in the RISC complex with the miRNA. For example Ago2 is thought to facilitate RISC cleavage activity [8].
RNAi has an universal and important function in organisms in the prevention of translation of parasitic nucleotide sequences (e.g. viruses) and development (see C. elegans and lin-4 above).
RNAi was first described by Andrew Fire and Craig C. Mello in 1998 who subsequently were awarded the Nobel Prize in Physiology or Medicine in the year 2006 [9]. Small RNAs, now called miRNAs, were considered the ‘breakthrough of the year 2002’ in the Science magazine [10]. Current estimates suggest that more than 60% of human protein-coding genes are regulated by miRNAs [11]. It was longer believed that distinct sets of miRNAs exist in different types of cells and organs, but recent research showed that this may not be true since most of the transcribed miRNAs are retained in the nucleus and are not processed to mature miRNA [12]. In fact the situation seems even much more complex as recent reports showed a choreographed interplay between diverse RNA species (long and short non-coding and coding RNAs) and mRNAs [13]. Characteristics of selected ncRNAs are listed in Table 1. Recent data emerged that proved the existence of competing endogenous RNAs (ceRNAs) which act as miRNA sponges and many other small non-coding RNAs such as the lately identified piwi-interacting RNAs (piRNAs) and intron derived miRNAs (miRtrons). In summary, the identification of miRNA seems just to represent the tip of an uncovered iceberg and the prediction of an individual miRNA (or miRNA silencing) effect seems almost impossible.
Table 1. Categories, characteristics and functions of selected ncRNAs.
| Length | Function | ||
|---|---|---|---|
| short ncRNAs | miRNA | −22nt | Posttranscriptional regulation of gene expression |
| siRNA | −21nt | Gene silencing | |
| piRNA | −30nt | Transposon regulation (chromatin) | |
|
| |||
| long ncRNAs | rRNA | +1.9kbases | Protein biosynthesis |
| IncRNA | 200nt | Epigenetics | |
Determination of miRNA and targets
Since miRNAs are short and relatively resistant against cleavage by RNAses, they can be even determined in paraffin embedded tissue. Furthermore, miRNAs are shed in bodily fluids in microvesicles (exosomes) and thus are stable and can be detected fairly reliable in urine and plasma. Basically, the same technical methods/tools used for the identification of mRNA are applicable for the determination and quantification of miRNAs; ranging from multiplexed PCR to microarrays and next generation sequencing. Due to space constraints, technical details and the strengths and weaknesses of each method cannot be described in detail here and thus the reader is referred to recent papers on that topic [14, 15].
The current knowledge on published miRNA sequences, annotations and targets is stored in several databases, examples of the targets are mirdb.org, microrna.org, and mirbase.org which hold up to 24,500 sequences identified from 32 organisms ranging from plants to viruses to vertebrates. However, the many potential target mRNAs of most of the known miRNAs have not been validated experimentally and thus the overlap of the in silico predicted targets among the miRNA databases is low. Bioinformatics solution try to address this important problem such as DIANA Lab Tools (www.microrna.gr), miRWalk [16] and starBase [17]. We created also a webbased information tool that allows the input of multiple miRNAs, combination of prediction algorithms, information of experimentally verified targets and pathway enrichment analysis for providing statistical adjusted, fast and reliable information about miRNA targets and their affected pathways (http://mirway.nephrogene.at/).
The experimental identification of miRNA target mRNA in cells and tissue can be accomplished essentially by three high-throughput approaches:
Coprecipitation of cross-linked miRNAs/argonaute/mRNA complexes and subsequent mRNA sequencing (Argonaute HITS-CLIP - high-throughput sequencing of cross-linking immunoprecipitation) [18].
Genome-wide transcriptomics experiments will uncover the upregulated targets in the absence of the miRNA or the downregulated targets of a overexpressed miRNA
Mass spectrometry yield upregulated proteins in the absence of miRNA or the other way around. This approach however neglects posttranscriptional protein regulation and is only semi-quantitative.
Reporter assays or Western Blot experiments are traditional methods for the verification of direct miRNA targets providing high experimental evidence, but throughput is limited.
As described before, our mechanistic and naive schemes on the regulation of mRNA targets by miRNA may be outdated soon since current papers showed evidence that most mammalian mRNAs are conserved targets of microRNAs [11]. Under what circumstances mRNAs are accessible for a specific miRNAs remains to be determined in the near future.
Use of miRNAs in kidney transplantation
1. Monitoring of the alloimmune response
With the introduction of the solid phase technologies, a major advancement in the prediction and diagnostic of the humoral immunity could be achieved. However especially in the maintenance phase of the transplant life it remains still unclear who of the affected allograft recipient will progress to terminal graft failure with accelerated pace and who will maintain an adequate graft function. The term chronic antibody mediated rejection which is frequently used to describe this clinical course is too imprecise and unspecific. Transcriptomics studies were conducted to address these shortcomings and decipher the molecular basis of the immune response. These studies indeed found associations of transcripts with graft attrition but at the same time ignoring the fundamentals of posttranscriptional regulation of gene expression [19]. Thus it was not surprising that on an individual basis such mRNA profiles in bodily fluids or allograft biopsies did not exhibit sufficient test characteristics (discrimination and calibration) to introduce these tests into clinical routine.
miRNA profiles on the other hand offer novel diagnostic and prognostic avenues to tackle these limitations and together with mRNA profiles allow for a more precise and plausible biological interpretation of findings [20].
1.1. miRNA in allograft biopsies
Wilflingseder and colleagues recently showed that the interplay between selected miRNAs and predicted mRNA targets, when investigated on a genome wide level in 65 biopsies, were able to discriminate and stratify antibody mediated rejection from cellular rejection. However also the overlap of common regulatory pathways such as inflammation and cytokine signaling used in humoral and cellular rejection was elucidated. On the other hand, molecular pathways that were mainly confined to cases of ischemia reperfusion injury included angiogenesis, apoptosis, and transforming growth factor-β signaling as demonstrated by the integrative analysis of miRNA expression and mRNA targets.
In a subsequent paper, the same investigators identified a single miRNA, mir-182-5p, as being a key regulator of postischemic acute kidney injury. This paper will be discussed in more detail in the section on post-transplant acute kidney injury [21].
Investigators from Paris and New York identified a miRNA expression profile that may be used to determine the allograft status. Three miRNAs (miR-142–5p, -155, and -223) that were abundantly expressed in three biopsies with conformed T-cell mediated acute rejection were also found to be activated in PBMCs of these patients [22]. The findings were validated in a test set of nine cases with rejection and 17 controls. The three markers showed complete separation of cases and controls as evidenced by a c-statistics of 1. Based on this study the authors concluded that these three miRNAs may serve as valid biomarker for the alloimmune status of renal allografts. It is of note however, that overfitting likely occurred in this small sample study and clinical utility of this marker set needs to be determined in a larger prospective study.
Sui et al. recently investigated the mechanisms of rejection by integrated protein, mRNA miRNA and long ncRNA in biopsies of three patients with acute rejections and a control group. The main finding was that five transcription factors were activated in rejection biopsies, which correlated to 12 miRNAs and 32 long ncRNAs [23]. The Chinese authors further performed a transcription factor pathway analysis and integrated the identified candidates into the miRNA and mRNA profiles of rejection. The complexity of molecular regulation of alloimmunity can be appreciated from this laborious paper however a clear conclusion remains elusive. Previous the same authors reported an acute rejection signature of 20 miRNAs detected in three acute rejection and three control biopsies [24]. Two miRNAs were confirmed with qRT-PCR, namely miR-320 and miR-324-3p.
Scian correlated miRNA profiles derived from allograft biopsies with IF/TA with urinary profiles and validated findings in an independent data set. The investigators found three miRNAs to be differentially expressed in tissue and urine: miR-142-3p, miR-204 and miR-211. miRNA signatures of allograft fibrosis were also investigated by Ben-Dov and colleagues in a small discovery cohort of eight kidney recipients and validated in 18 patients, ten with fibrosis and eight without [25]. Four miRNAs were found to be upregulated (miR-21, miR-142-5p and miR-142-3p and miR-506) and two suppressed (miR-30 b & c). Discrimination of cases with fibrosis by miRNA profiles is certainly of interest, but what is actually needed is to investigate their contribution to this process over time. If the association can be tested for causality, than an therapeutic miRNA modulation approach against fibrosis might be a great target to be elucidated in greater depth.
A further clinical problem is sometimes the discrimination of acute pyelonephritis from acute cellular rejection of the allograft since overlapping histologic features, and persistent graft dysfunction occur in both clinical entities. Oghumu and colleagues studied the miRNA profiles of biopsies obtained from 11 cases with clear infection vs five with frank rejection [26]. The authors reported a signature of 25 miRNAs that could discriminate both entities, however no external validation was performed in this very small and unmatched case control study and no adjustment for an inflated alpha error was performed.
1.2. miRNA in urine
The miRNA signatures in tissue may help to elucidate the pathophysiology of transplant related processes such as rejection or fibrosis, but for the utility as biomarkers and screening tool, candidates in bodily fluids such as urine or plasma are needed. Maluf et al. studied miRNA signatures in urinary cell pellets of renal allograft recipients with chronic dysfunction and well-functioning controls in order to identify biomarkers of rejection, tubular atrophy and interstitial fibrosis [27]. The investigator analyzed 191 samples from 125 deceased donor kidney allografts in a discovery, validation and longitudinal study. The investigators identified 22 miRNAs as being able to discriminate between groups. Gene ontologies and pathway analysis identified most of the features as belonging to the inflammation cascade. A subset of this signature was already elevated in a longitudinal study at a time when no morphological detectable changes had occurred. Thus based on these findings the investigators concluded that miRNA panels may be useful in allograft monitoring and perdition of grafts with faster attrition.
The same group found earlier that five miRNAs determined in paired samples of allograft biopsies and urine of 36 patients of the prospective validation set could nicely discriminate fibrosis from allografts with normal biopsy readings [28]. The clinical utility of this combination of markers should be determined by a longitudinal cohort study ideally with sequential biopsies. If the markers were to fulfill the high expectations raised in the discovery case/control study, then a major step forward would have been accomplished in the area of clinical transplantation.
As discussed above, monitoring of humoral alloimmunity has made a great leap forward with the introduction of solid base technologies such as Luminex. However, the monitoring and clear determination of T-cell immunity is still depending solely on invasive allograft biopsies. Thus a urine biomarker of this entity is clearly a clinical need. Lorenzen and coworkers identified miR-210 among two others as valid urinary biomarker of T-cell mediated rejection [29]. Data were confirmed in the urine of 62 patients with acute rejection, 19 control transplant patients without rejection and 13 stable transplant patients with urinary tract infection. Furthermore, the authors showed that urinary miR-210 concentrations were negatively associated with GFR loss in the first year after engraftment. Further bioinformatics and subsequent experimental work on the molecular processes and pathways that are regulated by miR-210 may open further avenues towards improved diagnostic, prognostic and potentially treatment-response guidance.
1.3. miRNA in plasma (blood)
Glowacki and colleagues also investigated allograft fibrosis [21]. The investigators showed increased circulating miR-21 levels to be associated with kidney fibrosis. Furthermore, circulating miR-21 levels were significantly increased in patients with severe fibrosis grades but not with other lesions in allograft biopsies. Based on these data the investigators concluded that miR-21 has a key pathogenic role in kidney fibrosis and may represent a novel fibrosis biomarker in peripheral blood.
Betts and colleagues from Oxford examined miRNAs in the serum of patients with acute rejection [30]. The investigators found two miRNAs in serum (miR-223 and miR-10a) to be different (i.e. suppressed) between the time points at one year and the period of acute rejection but no differences were found to the control group without a history of rejection. Due to the very small sample size of six to eight rejection and four control cases (depending on the measurements of different miRNAs and time points) the data need to be interpreted with great caution as honestly concluded by the authors.
1.4. miRNA in PBMCs
Accordingly, Danger et al. investigated miRNA signatures in PBMCs and allograft biopsies in patients with diagnosed chronic antibody mediated rejection [31]. Ten miRNAs were identified that were associated with cAMR in either PBMCs or tissue and one miRNA (miR-142-5p) was activated in both investigated sources as well as in a rodent model of cAMR. The investigators furthermore showed that this miRNA was not regulated in patients with native renal failure suggesting a specific alloimmune regulatory function. Validation in PBMCs in an independent cohort of patients with cAMR exhibited excellent discrimination (c-statistics of 0.74; p = 0.006). This data led the authors to conclude that this miRNA may be an excellent cAMR biomarker since control patients with acute rejection did not activate miR-142-5p in PBMCs and the suppressed target genes of miR-142-5p belong to the category of immune-regulation.
Earlier the same group from France reported on miR-142-3p as marker of tolerance in PBMCs of such patients [32]. The reported upregulation of miR-142-3p in B-cells of operational tolerant patients was independent of the immunosuppressant regimen and was stable over time. Experimental data showed that this miRNA has roughly 1000 mRNA targets, many associated with the ontologies of immune response in B-cells. Whether miR-142 causes the tolerance phenotype or is just a consequence of it remains to be determined.
In addition to the present overview the reader may be referred to two other recent reviews by Spiegel and Amrouche and colleagues who also nicely summarized the current understanding of miRNAs contribution to immunity and subsequent intragraft events [33],[34]. Besides the discussion on the usefulness as biomarker, the authors present the current knowledge on the molecular mechanisms and potential therapeutic implications of miRNAs.
2. AKI
2.1. Experimental ischemia reperfusion injury in the non-transplant setting
Bijkerk and colleagues from the Netherlands recently showed that miR-126 protected mice from postischemic acute renal injury [35]. Based on prior knowledge that miR-126 enhances vascular regeneration the investigators overexpressed miR-126 in the hematopoietic compartment of mice and found biochemical evidence of an ameliorated course of acute kidney injury. At the same time, histology of kidneys showed an increased network of peritubular capillaries and novel endothelial cells derived from bone marrow.
Furthermore, this intervention increased the number of circulating hematopoietic stem and progenitor cells by facilitated mobilization from the marrow towards the kidney.
Lorenzen et al. showed very recently that miR-24 antagonism prevents renal reperfusion injury in vivo. The treatment with an antisense oligonucleotide targeting miR-24 before the induction of ischemic reperfusion injury resulted in a significant improved kidney function. The authors conclude that the silencing of miR-24 ameliorates the apoptotic response after the ischemic insult in the kidney via the target genes HO-1 and H2A.X [36]. For the first time this study shows that miRNA inhibition might be a potential therapeutic intervention in the treatment of AKI.
Aguado-Fraile E et al. used a rat model of ischemia reperfusion injury and cultured renal proximal tubule cells to elucidate the effect of mir-127 on the course of injury [37]. The authors identified the kinesin family member 3B as main miRNA target and showed several effects of the activated miRNA via this target such as maintenance of cell-matrix and cell-cell adhesion and regulation of intracellular trafficking.
Sallustio et al. have chosen an interesting approach to study regeneration after renal injury. The researchers investigated whether a distinct set of miRNAs which are expressed in pluripotent renal cortical progenitor cells but no longer in adult kidney tissue play a key role in their self-renewal and differentiation [38]. In particular two of these miRNAs (miR-1915 and miR-1225-5p) have been shown to regulate mRNAs of progenitor cells such as CD133 and PAX2 and TLR2. The authors convincingly showed that suppression of miR-1915 led to activation of CD133 and PAX2 and furthermore that activation of this miRNA led to differentiation of progenitor cells into epithelial-like cells. miR-1225-5p activated TLR2 expression in progenitor cells. Thus it is reasonable to believe that the two identified miRNAs are major players in the repair process after tubular injury.
Cantaluppi and colleagues could show that endothelial progenitor cells were able to protect renal ischemic damage in a rat model by miRNA-dependent reprogramming of resident renal cells [39].
Shapiro and colleagues from Boston reanalyzed previous experimental data with principal component analysis in order to determine whether the identified miRNAs could be used as biomarkers of ischemic injury [40], [41]. In fact the authors could show appropriate discrimination of clusters and thus concluded distinct changes in miR expression may serve as biomarker of injury. Further experimental work however needs to be conducted in order to corroborate these findings.
2.2. Acute renal failure in the non-transplant setting in men
As in most other studies in nephrology, more data are available in native kidney disease than in transplantation. However, the acute kidney injury of patients in the ICU resembles to a big part the postischemic injury after deceased donor transplantation. This is not surprising since the deceased donor usually spends some days in the ICU before organ harvest and suffers from all the events that led to acute renal failure in such a setting. This we believe that acute native kidney failure serves as excellent model and thus the few studies on this topic are presented here.
Ramachandran et al. found a set of miRNAs to be differentially expressed in the urine of patients with acute injury of their native kidneys [42]. Initially, the investigators conducted an omics wide discovery study in only six case equal number of controls using pooled samples. The identified top seven lead miRNAs were then replicated in the urine of 98 patients with documented acute renal injury and 97 controls. Of the seven candidates, four features remained significant and showed excellent discrimination between cases and controls at a cross validated c-statistics of 0.91. However since the positive and negative predictive value depends on the prevalence of the event (acute kidney injury), the utility of these markers need to be tested in a prospective cohort. It is to be expected that test characteristics will decline considerably.
2.3. Kidney transplantation in men
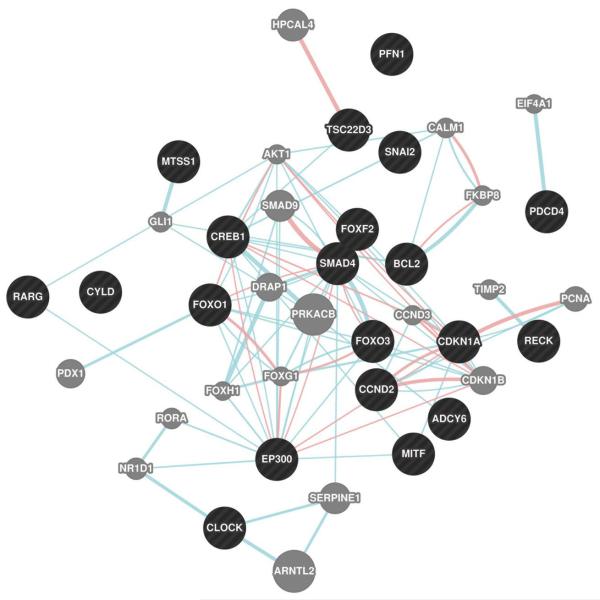
Our knowledge on how miRNAs regulate gene expression in human injured kidney tissue and impact transcript and protein levels is fairly understood. So far the only study that addressed this shortcoming comes from Wilflingseder et al. The investigators prospectively collected donor-baseline, indication as well as protocol allograft biopsies from 166 kidneys. In this cohort, eight patients developed postischemic acute allograft failure, matched allografts without pathology served as control. All analyses were adjusted for baseline miRNA profiles in the donor-baseline biopsies in order to isolate the true evolution of events during acute injury. Independent validation of findings was performed in 42 cases with postischemic acute renal failure and 21 protocol biopsies. The investigators found a specific, molecular AKI signature of twenty mRNAs and two miRNAs (miR-182-5p and miR-21-3p). miR-182-5p was identified as main regulator of this clinically important disorder through mRNA correlation analysis [21]. Further target analysis of miR-182-5p revealed that several AKI associated molecular processes can be potentially regulated by miR-182-5p such as apoptosis (e.g. BCL2) and proliferation (e.g. EP300) (Figure 2). The logical next step is to test for causality by conducting an interventional study. Data are expected to be available in late 2014.
Figure 2.
Protein interaction network of experimentally verified targets of miR-182 (black) and their interaction partners (grey). Targets of miR-182 were extracted from miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/) and are supported by strong experimental evidence (reporter assay or Western Blot). Blue edges: pathway interaction; pink edges: physical interaction. Network was created in Cytocape 2.8.1 with the GeneMANIA plugin (www.genemania.org).
This leads to the ultimate goal of miRNA research – the identification of therapeutic lead candidates. In transplantation no such study exist yet, however in other fields of medicine; antisense against certain miRNAs were successfully tested. These proofs of concept studies in human will be discussed in the next section on viral infections.
3. Viral infection
The most prevalent viral infections after kidney transplantation include HCV, CMV, BK and EBV. As stated above, no therapy studies are available in the transplant setting. However, this would be badly desired since most of the currently used antiviral drugs show low efficiency in viral clearance and use is often limited due to some intrinsic nephrotoxicity.
Proof of concept studies in monkeys on the successful use of RNA antisense treatment in such live threatening viral infection as Ebola make the high potential of such an approach visible [43].
It has also recently been shown in a very elegant study in non-transplant patients, that miRNA inhibition against the HCV genotype 1 miR-122 results in sustained and dose dependent viral response over the observation period of 14 weeks [44]. No renal side effects were reported and this success was also attributed to the liver specificity of miR-122. Very recent data additionally showed that expression of miR-122 in non-hepatic cells affected by the HCV virus facilitates its replication. Thus miR-122 and therapy may not, as previously thought, be tissue specific [45].
Wang and colleagues showed that replication of the human CMV was inhibited by miR-100 and miR-101, two intracellular miRNAs. In this study the investigators showed that the miRNAs do not directly interfere with the virus but rather regulate mTOR which facilitates the reproduction of the virus [46]. Pogglitsch and colleagues could demonstrate in macrophages that sirolimus suppressed CMV replication three to five days after infection [47]. Based on these findings the investigators concluded that their findings may explain the clinically observed anti-CMV activity in renal transplant patients on mTOR-I based immunosuppressive therapy. The reader may be referred to a recent paper by Dölken et al. for a thorough review on CMV induced miRNAs [48].
Tian and colleagues recently identified miR-B1 as potential therapeutic target against BK virus infections [49]. After infection of human renal proximal tubule cells with the BK virus, miR-B1 was strongly activated and inhibited the expression of TAg promoter activity which is required for BK virus replication. Vice versa, miR-B1 silencing enhanced Tag expression in BK virus infected cells. Importantly, renal allograft recipients with polyomavirus nephropathy exhibited significantly higher concentrations of miR-B1 compared to unaffected controls (). Li et al. also found BK virus induced miR-B1 in the blood of renal allograft recipients with BK nephropathy [50]. So far however it remains unclear whether miR-B1 will be a suitable diagnostic marker and/or therapeutic target for renal transplant recipients at risk for BK nephropathy.
Conclusions
The discovery of miRNA has revolutionized cellular biology in the past decade. The unrevealing of its function introduced a whole new level of gene and protein transcription regulation by interference with coding mRNA. Research has focused on different expression profiles in health and disease. In the field of transplantation several miRNAs have been identified that are associated with allograft rejection, ischemia-reperfusion injury or fibrosis. Additionally to their disease specific expression profiles, miRNAs are readily accessible for analysis from plasma or urine specimen (as miRNA is shed in microvesicles into bodily fluids) and therefore represent a group of promising candidate molecular biomarkers that may substantially change clinical decision-making by providing further insight into specific pathophysiological mechanisms in individual patients. Furthermore, biotechnologically engineered miRNA modulators can mimic or bind and thereby enhance or silence endogenous miRNAs offering novel treatment strategies. In humans miRNA inhibition for HCV specific miRNA – 122 has been successfully used in the treatment of HCV infection resulting in sustained virological response. Besides this general proof of concept, antisense miRNA is tested in animal models for ischemia reperfusion injury in the transplant setting. Today we are on the verge of implementing many of these new technologies into clinical routine.
Acknowledgments
Sources of support: This study was supported by grants from the Austrian Science Fund (FWF P-25726) and the European Union, grant MC-IOF#328613
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Reinhart BJ, Slack FJ, Basson M, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403(6772):901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 3.Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408(6808):86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V, Bartel B, Bartel DP, et al. A uniform system for microRNA annotation. RNA. 2003;9(3):277–9. doi: 10.1261/rna.2183803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294(5543):853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 8.Meister G, Landthaler M, Patkaniowska A, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15(2):185–97. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Fire A, Xu S, Montgomery MK, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391(6669):806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 10.Kennedy D. Breakthrough of the year. Science. 2002;298(5602):2283. doi: 10.1126/science.298.5602.2283. [DOI] [PubMed] [Google Scholar]
- 11.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee EJ, Baek M, Gusev Y, et al. Systematic evaluation of microRNA processing patterns in tissues, cell lines, and tumors. RNA. 2008;14(1):35–42. doi: 10.1261/rna.804508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam S, de Borja R, Tsao MS, McPherson JD. Robust global microRNA expression profiling using next-generation sequencing technologies. Laboratory investigation; a journal of technical methods and pathology. 2014;94(3):350–8. doi: 10.1038/labinvest.2013.157. [DOI] [PubMed] [Google Scholar]
- 15.van Rooij E. The art of microRNA research. Circ Res. 2011;108(2):219–34. doi: 10.1161/CIRCRESAHA.110.227496. [DOI] [PubMed] [Google Scholar]
- 16.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–47. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460(7254):479–86. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halloran PF, Reeve JP, Pereira AB, Hidalgo LG, Famulski KS. Antibody-mediated rejection, T cell-mediated rejection, and the injury-repair response: new insights from the Genome Canada studies of kidney transplant biopsies. Kidney international. 2014;85(2):258–64. doi: 10.1038/ki.2013.300. [DOI] [PubMed] [Google Scholar]
- 20.Wilflingseder J, Regele H, Perco P, et al. miRNA profiling discriminates types of rejection and injury in human renal allografts. Transplantation. 2013;95(6):835–41. doi: 10.1097/TP.0b013e318280b385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilflingseder J, Sunzenauer J, Toronyi E, et al. Molecular pathogenesis of post-transplant Acute Kidney Injury: assessment of whole-genome mRNA and miRNA profiles. PloS one. 2014 doi: 10.1371/journal.pone.0104164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anglicheau D, Sharma VK, Ding R, et al. MicroRNA expression profiles predictive of human renal allograft status. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(13):5330–5. doi: 10.1073/pnas.0813121106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sui W, Lin H, Peng W, et al. Molecular dysfunctions in acute rejection after renal transplantation revealed by integrated analysis of transcription factor, microRNA and long noncoding RNA. Genomics. 2013;102(4):310–22. doi: 10.1016/j.ygeno.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Sui W, Dai Y, Huang Y, et al. Microarray analysis of MicroRNA expression in acute rejection after renal transplantation. Transpl Immunol. 2008;19(1):81–5. doi: 10.1016/j.trim.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Dov IZ, Muthukumar T, Morozov P, et al. MicroRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation. 2012;94(11):1086–94. doi: 10.1097/TP.0b013e3182751efd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oghumu S, Bracewell A, Nori U, et al. Acute pyelonephritis in renal allografts: a new role for microRNAs? Transplantation. 2014;97(5):559–68. doi: 10.1097/01.TP.0000441322.95539.b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maluf DG, Dumur CI, Suh JL, et al. The urine microRNA profile may help monitor post-transplant renal graft function. Kidney international. 2014;85(2):439–49. doi: 10.1038/ki.2013.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scian MJ, Maluf DG, David KG, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. American journal of transplantation. 2011;11(10):2110–22. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenzen JM, Thum T. Circulating and urinary microRNAs in kidney disease. Clin J Am Soc Nephrol. 2012;7(9):1528–33. doi: 10.2215/CJN.01170212. [DOI] [PubMed] [Google Scholar]
- 30.Betts G, Shankar S, Sherston S, Friend P, Wood KJ. Examination of serum miRNA levels in kidney transplant recipients with acute rejection. Transplantation. 2014;97(4):e28–30. doi: 10.1097/01.TP.0000441098.68212.de. [DOI] [PubMed] [Google Scholar]
- 31.Danger R, Paul C, Giral M, et al. Expression of miR-142-5p in peripheral blood mononuclear cells from renal transplant patients with chronic antibody-mediated rejection. PloS one. 2013;8(4):e60702. doi: 10.1371/journal.pone.0060702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Danger R, Pallier A, Giral M, et al. Upregulation of miR-142-3p in peripheral blood mononuclear cells of operationally tolerant patients with a renal transplant. Journal of the American Society of Nephrology: JASN. 2012;23(4):597–606. doi: 10.1681/ASN.2011060543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spiegel JC, Lorenzen JM, Thum T. Role of microRNAs in immunity and organ transplantation. Expert Rev Mol Med. 2011;13:e37. doi: 10.1017/S1462399411002080. [DOI] [PubMed] [Google Scholar]
- 34.Amrouche L, Rabant M, Anglicheau D. MicroRNAs as biomarkers of graft outcome. Transplant Rev. 2014;28:111–118. doi: 10.1016/j.trre.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 35.Bijkerk R, van Solingen C, de Boer HC, et al. Hematopoietic MicroRNA-126 Protects against Renal Ischemia/Reperfusion Injury by Promoting Vascular Integrity. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2013060640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lorenzen JM, Kaucsar T, Schauerte C, et al. MicroRNA-24 Antagonism Prevents Renal Ischemia Reperfusion Injury. J Am Soc Nephrol. 2014 doi: 10.1681/ASN.2013121329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguado-Fraile E, Ramos E, Saenz-Morales D, et al. miR-127 protects proximal tubule cells against ischemia/reperfusion: identification of kinesin family member 3B as miR-127 target. PloS one. 2012;7(9):e44305. doi: 10.1371/journal.pone.0044305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sallustio F, Serino G, Costantino V, et al. miR-1915 and miR-1225-5p regulate the expression of CD133, PAX2 and TLR2 in adult renal progenitor cells. PloS one. 2013;8(7):e68296. doi: 10.1371/journal.pone.0068296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cantaluppi V, Gatti S, Medica D, et al. Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia-reperfusion injury by microRNA-dependent reprogramming of resident renal cells. Kidney international. 2012;82(4):412–27. doi: 10.1038/ki.2012.105. [DOI] [PubMed] [Google Scholar]
- 40.Shapiro MD, Bagley J, Latz J, et al. MicroRNA expression data reveals a signature of kidney damage following ischemia reperfusion injury. PloS one. 2011;6(8):e23011. doi: 10.1371/journal.pone.0023011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Godwin JG, Ge X, Stephan K, et al. Identification of a microRNA signature of renal ischemia reperfusion injury. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(32):14339–44. doi: 10.1073/pnas.0912701107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ramachandran K, Saikumar J, Bijol V, et al. Human miRNome profiling identifies microRNAs differentially present in the urine after kidney injury. Clin Chem. 2013;59(12):1742–52. doi: 10.1373/clinchem.2013.210245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitka M. Experimental RNA therapy shows promise against Ebola virus in monkey studies. JAMA. 2010;304(1):31. doi: 10.1001/jama.2010.868. [DOI] [PubMed] [Google Scholar]
- 44.Janssen HL, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368(18):1685–94. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 45.Fukuhara T, Kambara H, Shiokawa M, et al. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with hepatitis C virus. J Virol. 2012;86(15):7918–33. doi: 10.1128/JVI.00567-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang FZ, Weber F, Croce C, et al. Human cytomegalovirus infection alters the expression of cellular microRNA species that affect its replication. J Virol. 2008;82(18):9065–74. doi: 10.1128/JVI.00961-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poglitsch M, Weichhart T, Hecking M, et al. CMV late phase-induced mTOR activation is essential for efficient virus replication in polarized human macrophages. American journal of transplantation. 2012;12(6):1458–68. doi: 10.1111/j.1600-6143.2012.04002.x. [DOI] [PubMed] [Google Scholar]
- 48.Dolken L, Pfeffer S, Koszinowski UH. Cytomegalovirus microRNAs. Virus Genes. 2009;38(3):355–64. doi: 10.1007/s11262-009-0347-0. [DOI] [PubMed] [Google Scholar]
- 49.Tian YC, Li YJ, Chen HC, et al. Polyomavirus BK-encoded microRNA suppresses autoregulation of viral replication. Biochem Biophys Res Commun. 2014;447(3):543–9. doi: 10.1016/j.bbrc.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 50.Li JY, McNicholas K, Yong TY, et al. BK Virus Encoded MicroRNAs Are Present in Blood of Renal Transplant Recipients With BK Viral Nephropathy. American journal of transplantation. 2014;14(5):1183–90. doi: 10.1111/ajt.12694. [DOI] [PubMed] [Google Scholar]