To the editors:
Multiple lines of evidence support a role for microRNA 137 (miR-137) in the etiology of schizophrenia (1–4) and fundamental neuronal processes (5–7). In the largest genome-wide association meta-analysis for schizophrenia,(8) the second most significant association is in MIR137HG, the gene encoding miR-137 (rs1702294, P=3.4×10−19). Prior reports indicated that genes with predicted miR-137 target sites were enriched for smaller GWAS P-values (2), raising the possibility that miR-137 regulates a gene network involved in schizophrenia. To understand more about the role of miR-137, we describe here our characterization of embryonic development, behavior, and gene expression in mice with targeted disruption of the Mir137 transcript (supplementary detailed information can be found at: http://crowley.web.unc.edu/files/2014/03/mir137.ko_.supplement.pdf).
Mir137tm1Mtm mice were created in a project to generate conditional, reporter-tagged targeted mutations in 162 miRNAs (9). These mice were created using a “knockout-first” strategy (10) to produce a knockout at the RNA processing level. We purchased heterozygous (+/−) breeder mice (www.mmrrc.org/catalog/sds.php?mmrrc_id=36301). We confirmed genetic background (~75% C57BL/6J) and backcrossed twice to C57BL/6J to create animals of >95% C57BL/6J ancestry.
To our knowledge, these Mir137 mutant mice have not been characterized previously. Seven heterozygous × heterozygous matings produced 35 living offspring, none of which were homozygous (−/−) for the targeted Mir137 allele (Figure 1A, P < 0.001), suggesting complete embryonic lethality in the presence of knockout of Mir137.
Figure 1.
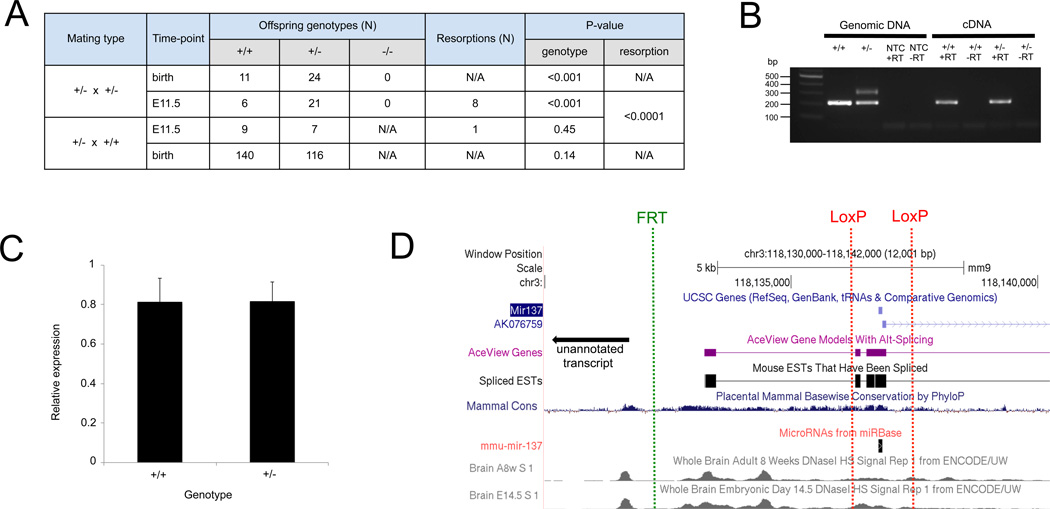
Mir137 knockout mice (−/−) are embryonic lethal. (A) Heterozygous × heterozygous matings failed to yield a live born (−/−) mouse or embryo, and timed matings revealed an excess of resorbed embryos at E11.5. Heterozygous mice did not show a reduction in viability. (B) Verification that the mutant allele does not express Mir137 by allele-specific expression. Representative reverse transcriptase PCR results from adult mouse brain using primers flanking a LoxP insertion site in the Mir137 primary transcript. Amplification from genomic DNA is shown as a positive control for amplification of the mutant and wildtype alleles. The negative control reactions (−RT) confirm the absence of genomic DNA contamination. The absence of an upper band in the (+/−) +RT sample indicates lack of expression from the mutant allele. NTC = no template control. RT = reverse transcriptase. (C) Quantitative PCR analysis of mature miR-137 expression from wildtype and heterozygous brain tissue revealed no significant difference, consistent with the viability and behavioral data. (D) Exogenous gene targeting elements may interfere with endogenous functional elements, preventing conditional mutagenesis. Shown are the positions of the non-native sequences (FRT and LoxP sites that would remain after crossing to Flp) relative to known and predicted transcripts, DNase hypersensitivity sites, and conservation data.
To confirm embryonic lethality and to determine when lethality occurred, we conducted timed matings (six heterozygous × heterozygous and four heterozygous × wildtype). On embryonic day 11.5 (E11.5), all embryos were collected (Figure 1A). Again, no homozygous (−/−) embryos were identified (P < 0.001). We observed significantly more resorbed embryos from heterozygous × heterozygous matings (P < 0.0001), indicating that embryonic lethality must occur after implantation (~E4.5) but before E11.5. Efforts to identify a Mir137 −/− mouse by genotyping resorbed embryos at E11.5 or collecting flushed embryos at E3.5 were unsuccessful, likely due to insufficient and/or impure DNA preparations from small amounts of tissue.
We did not observe reduced viability of heterozygous mice (+/−) after genotyping 256 offspring (P=0.14) collected from birth to three weeks of age, indicating that one copy of Mir137 is sufficient for survival (Figure 1A). We subjected wild-type and heterozygous animals to a battery of behavioral tests (e.g., basic sensorium, activity, social behavior, learning/memory, etc.), and observed no consistent and interpretable differences. These results also suggest compensation from the remaining allele.
We next examined the expression of Mir137 and the levels of mature miR-137 in the brains of heterozygous and wildtype animals (−/− embryos died too early for collection of sufficient tissue). Using allele-specific reverse transcriptase PCR, we confirmed that the targeted allele leads to complete loss of Mir137 expression downstream of the polyA sequence (Figure 1B). However, mature miR-137 levels were not significantly different between heterozygous and wild-type animals (Figure 1C), suggesting that heterozygous mice have compensatory upregulation or decreased degradation of miR-137. These expression data are consistent with the lack of a viability or behavioral phenotype in the heterozygous mice.
To circumvent the embryonic lethality of Mir137 −/− mice, we attempted to make conditional knockouts (9), such that ablation of Mir137 could be limited to a particular tissue. The construct contains a combination of FRT and loxP sites intended to allow conditional mutagenesis by crossing to germline deleter Flp mice (expected to restore the wild-type allele) and then crossing to tissue-specific Cre transgenic mice (expected to generate adult brain-specific knockouts). Crosses between Mir137 +/− mice and an efficient Flp line led to several successfully recombined mice, heterozygous for the rescued allele. Repeated intercrosses of these recombined mice, however, failed to yield any homozygous offspring. Therefore, we were unable to rescue embryonic lethality.
We reasoned that there were two major possibilities for the rescue failure. First, the targeting construct could have inserted into the wrong genomic position or could have sequence errors. We have likely excluded this possibility by confirming the location of the integration site and re-sequencing >95% of the construct without identifying an error. Second, Flp may have been unable to restore proper gene function due to issues inherent with the gene construct (e.g., FRT or LoxP sites in critical part of the gene). We favor this possibility because we were unable to detect any reporter gene (beta-galactosidase) activity in Mir137 +/− embryos or adult tissues, suggesting that the endogenous Mir137 promoter was inadvertently inactivated. Furthermore, bioinformatic analysis of the region (using additional data that became available after we were well into this project), suggests that the exogenous gene targeting elements may indeed interfere with endogenous functional elements, and act to prevent conditional mutagenesis (Figure 1D). The LoxP site upstream of Mir137 is in a putative splice donor sequence and the FRT site lies between highly conserved DNase hypersensitivity sites that are active in embryonic brain.
In conclusion, these results suggest that at least one functional copy of Mir137 is essential for embryonic development. These data are consistent with miR-137 playing important roles in development and perhaps also in neurodevelopmental disorders like schizophrenia. It appears that miR-137’s biological pathway is capable of homeostatic compensation and, while we do not yet understand the functional impact of Mir137 schizophrenia risk variants, it is conceivable that this homeostatic capacity could be affected in some way. It is clear, however, that this genomic region is more complex than previously thought, and the design of targeted mutations in this region should incorporate all available genomic data.
Supplementary Material
Acknowledgements
This work was supported by K01MH094406 (PI Crowley), R21MH099370 (PI Sullivan) and the UNC Intellectual and Developmental Disabilities Research Center, funded by NICHD (U54 HD079124).
Footnotes
Author Contributions
All authors reviewed and approved the final version of the manuscript.
Conflicts of Interest
The authors report no conflicts.
References
- 1.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Erp TG, Guella I, Vawter MP, Turner J, Brown GG, McCarthy G, et al. Schizophrenia miR-137 Locus Risk Genotype Is Associated with Dorsolateral Prefrontal Cortex Hyperactivation. Biol Psychiatry. 2014;75:398–405. doi: 10.1016/j.biopsych.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lett TA, Chakavarty MM, Felsky D, Brandl EJ, Tiwari AK, Goncalves VF, et al. The genome-wide supported microRNA-137 variant predicts phenotypic heterogeneity within schizophrenia. Mol Psychiatry. 2013;18:443–450. doi: 10.1038/mp.2013.17. [DOI] [PubMed] [Google Scholar]
- 5.Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Willemsen MH, Valles A, Kirkels LA, Mastebroek M, Olde Loohuis N, Kos A, et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet. 2011;48:810–818. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- 8.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. In press (Nature) doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park CY, Jeker LT, Carver-Moore K, Oh A, Liu HJ, Cameron R, et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep. 2012;1:385–391. doi: 10.1016/j.celrep.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, et al. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


