Abstract
Background
HIF-1 (hypoxia-inducible factor 1) is a transcriptional activator that functions as a critical regulator of oxygen homeostasis. Recently, a large number of epidemiological studies have investigated the relationship between HIF-1α C1772T/G1790A polymorphisms and cancer susceptibility. However, the results remain inconclusive. Therefore, we performed a meta-analysis on all of the available case-control studies to systematically summarize the possible association.
Methods
A literature search was performed using PubMed and the Web of Science database to obtain relevant published studies. Pooled odds ratios (ORs) and corresponding 95% confidence intervals (CIs) for the relationship between HIF-1α C1772T/G1790A polymorphisms and cancer susceptibility were calculated using fixed- and random-effects models when appropriate. Heterogeneity tests, sensitivity analyses and publication bias assessments were also performed in our meta-analysis.
Results
A total of 40 studies met the inclusion criteria were included in the meta-analysis: 40 studies comprised of 10869 cases and 14289 controls for the HIF-1α C1772T polymorphism and 30 studies comprised of 7117 cases and 10442 controls for the HIF-1α G1790A polymorphism. The results demonstrated that there were significant association between the HIF-1α C1772T polymorphism and cancer susceptibility under four genetic models (TT vs. CC: OR = 1.63, 95% CI = 1.02-2.60; CT + TT vs. CC: OR = 1.15, 95% CI = 1.01-1.34; TT vs. CT + CC: OR = 2.11, 95% CI = 1.32-3.77; T vs. C: OR = 1.21, 95% CI = 1.04-1.41). Similarly, the statistically significant association between the HIF-1α G1790A polymorphism and cancer susceptibility was found to be consistently strong in all of the genetic models. Moreover, increased cancer risk was observed when the data were stratified by cancer type, ethnicity and the source of controls.
Conclusions
This meta-analysis demonstrates that both the C1772T and G1790A polymorphisms in the HIF-1α gene likely contribute to increased cancer susceptibility, especially in the Asian population and in breast cancer, lung cancer, pancreatic cancer and oral cancer. However, further research is necessary to evaluate the relationship between these polymorphisms and cancer risk.
Electronic supplementary material
The online version of this article (doi:10.1186/1471-2407-14-950) contains supplementary material, which is available to authorized users.
Keywords: HIF-1 gene, Polymorphism, Cancer, Susceptibility, Meta-analysis
Background
Human cancer is a major cause of death in the world, and it is estimated that the number of new cases will increase to more than 15 million in the coming decade, creating a substantial worldwide public health burden [1, 2]. Various factors, such as genetic and environmental influences, are associated with cancer prognosis. However, the exact etiology and mechanism of carcinogenesis have not yet been clearly elucidated. In recent years, it has become well-accepted that intrinsic factors, such as host genetic susceptibility, may play important roles in the process of cancer development [3, 4], and an increasing number of studies have focused on the association between genetic factors and cancer susceptibility.
Hypoxia-inducible factor 1 (HIF-1) is a transcriptional activator that functions as a critical regulator of oxygen homeostasis. It is a heterodimer composed of two subunits, HIF-1α and HIF-1β, which dimerize and bind to DNA via the basic helix-loop-helix Per/Arnt/Sim (bHLH-PAS) domain [5, 6]. HIF-1α expression is induced in hypoxic cells, and its level exponentially increase when the cells are exposed to O2 concentration of less than 6%. Under hypoxic condition, HIF-1α ubiquitination decreases dramatically, resulting in an accumulation of the protein, while under normoxic condition, HIF-1α is rapidly degraded through von Hippel-Lindau (VHL)-mediated ubiquitination and proteasomal degradation [7–10]. HIF-1 has also been suggested to play an important role in tumor development, progression and metastasis, and HIF-1 can activate the transcription of more than 60 target genes that are involved in crucial aspects of cancer establishment, including cell survival, glucose metabolism, angiogenesis and invasion [11, 12].
The HIF-1α gene is located on chromosome 14q21-24, and recent studies have shown that there are a total of 35 common single nucleotide polymorphisms (SNPs) throughout the HIF-1α gene in Caucasian and Asian population [13–15]. Two important SNPs in exon 12 of the HIF-1 gene, HIF-1α C1772T (rs11549465) and HIF-1α G1790A (rs11549467), lead to amino acid substitution of proline to serine at position 582 and alanine to threonine at position 588 of the protein, respectively [8, 16, 17]. These two polymorphisms have been demonstrated to be functionally meaningful, resulting in increased transcriptional activity of HIF-1α [14, 18]. Previous studies have shown that the overexpression of HIF-1α is significantly associated with cell proliferation, increased tumor susceptibility, tumor size, lymph node metastasis and prognosis [19, 20].
In recent years, the HIF-1α gene has been a research focus in the scientific community, and many epidemiological studies have been performed to assess the association between HIF-1α C1772T/G1790A polymorphisms and cancer susceptibility. However, the results of the different studies are conflicting. Hence, we performed a meta-analysis of all of the eligible studies to clarify the role of HIF-1α C1772T/G1790A polymorphisms in cancer development.
Methods
Study eligibility and validity assessment
We performed a computerized literature search of the PubMed and Web of Science databases to identify all of the relevant studies of cancer that contained sufficient genotyping data for at least one of the two polymorphisms, HIF-1α C1772T or HIF-1α G1790A. The search strategy was designed by two researchers and included the following keywords: “HIF-1 OR hypoxia-inducible factor-1” and “polymorphism”, and the last search was updated on September 20th, 2013. To obtain all eligible publications, we also manually reviewed the references of the selected articles to identify other potential eligible publications. Articles investigating the association between cancer risk and the HIF-1α polymorphisms were identified with no language restriction.
Inclusion criteria
The studies selected were required to meet the following criteria: 1) evaluate the association between the HIF-1α C1772T and/or HIF-1α G1790A polymorphisms and cancer risk; 2) use a human case-control design; 3) contain sufficient published data for the estimation of an odds ratio (OR) with a 95% confidence interval (CI).
Data extraction
Data were extracted from all of the eligible publications by two investigators (Yan and Chen) independently, according to the inclusion criteria listed above. Disagreements between the two investigators were resolved by discussion until a consensus was reached. The following information was extracted from each of the included publications: the first author’s name, publication data, country of origin, ethnicities of the sample population (categorised as Asians, Caucasians and Mixed), cancer type, source of control group (population- or hospital-based controls), total number of cases and controls, and the number of cases and controls with the HIF-1α C1772T/G1790A polymorphisms.
Statistical methods
The strength of the association between the HIF-1α C1772T/HIF-1α G1790A polymorphisms and cancer risk was measured by ORs with 95% CIs. The statistical significance of the pooled OR was calculated by the Z test, a P < 0.05 was considered to be statistically significant (P-values were two sided). For HIF-1α C1772T polymorphism, we examined the overall ORs and compared the cancer incidence using the allelic model (T versus C), homozygote model (TT versus CC), heterozygote model (TC versus CC), dominant model (TT + TC versus CC), recessive model (TT versus TC + CC). For HIF-1α G1790A polymorphism, we evaluated the risk in the allelic model (A versus G), homozygote model (AA versus GG), heterozygote comparison model (GA versus GG), dominant models (AA + AG versus GG), and recessive model (AA versus AG + GG). Subgroup analyses were also conducted by ethnicity, cancer type (“other cancer groups” means any cancer types with less than two separate publications) and source of controls. Statistical heterogeneity was estimated by a chi-square based Q-test, and when P < 0.05, the heterogeneity was considered to be significant. We combined all of the values from each individual study using the fixed-effect model and the random-effect model. When P > 0.05, the effects were assumed to be homogenous, and the fixed-effect model (the Mantel-Haenszel method) was used [21]. When P < 0.05, the random-effect model (the DerSimonian and Laird method) was more appropriate [22]. The inter-study variance I2 (I2 = 100% × (Q-df)/Q) was used to quantitatively estimate the heterogeneity, and the percentage of I2 was used to describe the extent of the heterogeneity, I2 < 25%, 25-75% and >75% represent low, moderate and high inconsistency, respectively [23, 24]. In addition, we performed sensitivity analyses to evaluate the potential biases of the results in our meta-analyses. The Hardy-Weinberg equilibrium (HWE) of the controls for each study was also calculated using a goodness-of-fit test (chi-square or Fisher’s exact test) and P < 0.05 was considered to be statistically significant. Sensitivity analyses were carried out to assess the stability of the results by conducting analysis of studies with controls in HWE. Finally, the Begg’s funnel plot and Egger’s test were utilised to estimate the publication bias [25]. All analyses were conducted by the software Stata (Version 11; Stata Corporation, College Station, Texas, USA). All P-values were two-sided and a P of < 0.05 was considered to be statistically significant.
Results
Studies selected
Through the literature search and selection, a total of 40 eligible studies met the inclusion criteria and were included in our meta-analysis. One study (Konac et al.) [26] provided data on three types of cancer (cervical cancer, ovarian cancer, and endometrial cancer) and both polymorphisms; therefore, we have grouped them as one in the meta-analyses of all subjects except when stratified by cancer type. Thus, each type of cancer in this study was treated as a separated study in sub-group analyses. Among the 40 eligible studies, 40 studies, representing 10869 cases and 14289 controls, were ultimately analyzed for the HIF-1α C1772T polymorphism [8, 17, 26–63], and 30 studies, representing 7177 cases and 10442 controls, were analyzed for the HIF-1α G1790A polymorphism [8, 17, 26, 29–31, 33–35, 37–43, 45–48, 50, 52–57, 59, 62, 63]. The literature search and study selection procedure are shown in Figure 1. Of the 40 studies on the HIF-1α C1772T polymorphism, 6 studies were conducted on prostate cancer, 6 studies on breast cancer, 3 studies on lung cancer, 4 studies on colorectal cancer, 4 studies on renal cancer, 4 studies on oral cancer and 12 studies on other cancers. Among these eligible studies, 20 were studies on Asians, 16 were studies on Caucasians and 4 studies were performed on a population of mixed ethnicity. The control sources were population-based in 17 studies and hospital-based in 23 studies. For the HIF-1α G1790A polymorphism, 15 of the 30 eligible studies were performed in Asian populations, 13 studies were performed in Caucasian populations and 2 studies were performed in a mixed ethnicity population. Of these studies, 4 studies were conducted on breast cancer, 3 studies on lung cancer, 4 studies on oral cancer, 3 studies on prostate cancer, 3 studies on cervical cancer, 2 studies on pancreatic cancer, 2 studies on colorectal cancer, 4 studies on renal cancer and 7 studies on other cancers. The control sources were population-based in 17 studies and hospital-based in 13 studies. The genotype frequency data of the HIF-1α C1772T and HIF-1α G1790A polymorphisms were extracted from all of these eligible publications. For the HIF-1α C1772T polymorphism, the distributions of the genotypes in the control groups in 11 studies were not in HWE [17, 50, 51, 53, 54, 56–58, 60–62]. For the HIF-1α G1790A polymorphism there was 1 study not in HWE [62]. The main characteristics of the eligible studies in the meta-analysis are listed in Table 1.
Figure 1.

Study flow-chart illustrating the literature search and eligible study selection process.
Table 1.
Characteristics of studies included in the meta-analysis
| First author | Year | Country | Ethnicity | Cancer type | Gene type | Source of controls | Cases | Controls | Case | Control | HWE | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MM | MW | WW | MM | MW | WW | ||||||||||
| Clifford | 2001 | UK | Caucasian | Renal | C1772T | PB | 48 | 143 | 42 | 6 | 0 | 110 | 27 | 6 | 0.02 |
| G1790A | PB | 48 | 144 | 47 | 1 | 0 | 140 | 4 | 0 | 0.87 | |||||
| Tanimoto | 2003 | Japan | Asian | HNSCC | C1772T | PB | 55 | 110 | 45 | 10 | 0 | 98 | 12 | 0 | 0.55 |
| G1790A | PB | 55 | 110 | 51 | 4 | 0 | 101 | 9 | 0 | 0.65 | |||||
| Kuwai | 2004 | Japan | Asian | Colorectal | C1772T | PB | 100 | 100 | 100 | 0 | 0 | 89 | 11 | 0 | 0.56 |
| Ollerenshaw | 2004 | UK | Caucasian | Renal | C1772T | PB | 160 | 162 | 16 | 54 | 90 | 1 | 90 | 71 | 0.001 |
| G1790A | PB | 146 | 288 | 65 | 67 | 14 | 239 | 39 | 10 | 0.001 | |||||
| Ling | 2005 | China | Asian | Esophageal | C1772T | HB | 95 | 104 | 84 | 11 | 0 | 93 | 11 | 0 | 0.57 |
| Chau | 2005 | USA | Caucasian | Prostate | C1772T | PB | 196 | 196 | 161 | 29 | 6 | 179 | 14 | 3 | 0.002 |
| Fransen | 2006 | Sweden | Caucasian | Colorectal | C1772T | PB | 198 | 258 | 167 | 28 | 3 | 213 | 43 | 2 | 0.92 |
| Fransen | 2006 | Sweden | Caucasian | Colorectal | G1790A | PB | 198 | 256 | 189 | 9 | 0 | 247 | 9 | 0 | 0.77 |
| Konac | 2007 | Turkey | Caucasian | Cervical | C1772T | HB | 32 | 107 | 10 | 14 | 8 | 68 | 37 | 2 | 0.23 |
| G1790A | HB | 32 | 107 | 32 | 0 | 0 | 107 | 0 | 0 | 0.99 | |||||
| Caucasian | Ovarian | C1772T | HB | 49 | 107 | 34 | 14 | 1 | 68 | 37 | 2 | 0.23 | |||
| G1790A | HB | 49 | 107 | 47 | 2 | 0 | 107 | 0 | 0 | 0.99 | |||||
| Caucasian | Endometrial | C1772T | HB | 21 | 107 | 4 | 12 | 5 | 68 | 37 | 2 | 0.23 | |||
| G1790A | HB | 21 | 107 | 21 | 0 | 0 | 107 | 0 | 0 | 0.99 | |||||
| Li | 2007 | USA | mixed | Prostate | C1772T | PB | 1041 | 1234 | 818 | 209 | 14 | 995 | 221 | 18 | 0.16 |
| G1790A | PB | 1066 | 1264 | 1053 | 13 | 0 | 1247 | 17 | 0 | 0.81 | |||||
| Orr-Urtreger | 2007 | Israel | Caucasian | Prostate | C1772T | PB | 402 | 300 | 287 | 99 | 16 | 217 | 80 | 3 | 0.14 |
| G1790A | PB | 200 | 300 | 198 | 2 | 0 | 298 | 2 | 0 | 0.95 | |||||
| Apaydin | 2008 | Turkey | Caucasian | Breast | C1772T | PB | 102 | 102 | 79 | 21 | 2 | 68 | 29 | 5 | 0.42 |
| G1790A | PB | 102 | 102 | 102 | 0 | 0 | 98 | 4 | 0 | 0.84 | |||||
| Lee | 2008 | Korea | Asian | Breast | C1772T | PB | 1332 | 1369 | 1207 | 119 | 6 | 1245 | 123 | 1 | 0.25 |
| Kim | 2008 | Korea | Asian | Breast | C1772T | HB | 90 | 102 | 81 | 8 | 1 | 93 | 9 | 0 | 0.64 |
| G1790A | HB | 90 | 102 | 87 | 3 | 0 | 94 | 7 | 1 | 0.06 | |||||
| Nadaoka | 2008 | Japan | Asian | Bladder | C1772T | HB | 219 | 461 | 197 | 21 | 1 | 419 | 42 | 0 | 0.35 |
| G1790A | HB | 219 | 461 | 204 | 13 | 2 | 421 | 40* | - | 0.46 | |||||
| Jacobs | 2008 | USA | mixed | Prostate | C1772T | HB | 1420 | 1450 | 1156 | 252 | 12 | 1138 | 284 | 28 | 0.04 |
| Horree | 2008 | Netherland | Caucasian | Endometrial | C1772T | PB | 58 | 559 | 50 | 5 | 3 | 463 | 84 | 12 | 0.01 |
| Naidu | 2009 | Malaysia | Asian | Breast | C1772T | PB | 410 | 275 | 294 | 100 | 16 | 222 | 50 | 3 | 0.92 |
| G1790A | PB | 410 | 275 | 332 | 72 | 6 | 232 | 41 | 2 | 0.90 | |||||
| Chen | 2009 | Taiwan | Asian | Oral | C1772T | PB | 174 | 347 | 163 | 10 | 1 | 334 | 13 | 0 | 0.72 |
| G1790A | PB | 174 | 347 | 153 | 20 | 1 | 333 | 14 | 0 | 0.70 | |||||
| Konac | 2009 | Turkey | Caucasian | Lung | C1772T | PB | 141 | 156 | 110 | 31 | 0 | 111 | 43 | 2 | 0.34 |
| G1790A | PB | 141 | 156 | 141 | 1 | 0 | 154 | 2 | 0 | 0.94 | |||||
| Morris | 2009 | UK | Caucasian | Renal | C1772T | PB | 332 | 313 | 290 | 39 | 3 | 262 | 46 | 5 | 0.08 |
| G1790A | PB | 325 | 309 | 313 | 10 | 2 | 294 | 15 | 0 | 0.66 | |||||
| Foley | 2009 | Ireland | Caucasian | Prostate | C1772T | PB | 95 | 188 | 65 | 30 | 0 | 175 | 13 | 0 | 0.62 |
| Li | 2009 | China | Asian | Gastric | C1772T | HB | 87 | 106 | 83 | 4 | 0 | 93 | 13 | 0 | 0.50 |
| G1790A | HB | 87 | 106 | 74 | 13 | 0 | 100 | 6 | 0 | 0.76 | |||||
| Munoz- | |||||||||||||||
| Guerra | 2009 | Spain | Caucasian | Oral | C1772T | PB | 70 | 148 | 57 | 6 | 7 | 113 | 27 | 8 | <0.01 |
| G1790A | PB | 64 | 139 | 40 | 21 | 3 | 130 | 9 | 0 | 0.69 | |||||
| Kim | 2010 | Korea | Asian | Cervical | C1772T | HB | 199 | 214 | 177 | 22 | 0 | 187 | 27 | 0 | 0.32 |
| G1790A | HB | 199 | 213 | 187 | 12 | 0 | 200 | 12 | 1 | 0.10 | |||||
| Shieh | 2010 | Taiwan | Asian | Oral | C1772T | HB | 305 | 96 | 282 | 23 | 0 | 89 | 7 | 0 | 0.71 |
| G1790A | HB | 305 | 96 | 281 | 24 | 0 | 89 | 7 | 0 | 0.71 | |||||
| Knechtal | 2010 | Austria | Caucasian | Colorectal | C1772T | PB | 368 | 2156 | 291 | 77** | - | 1773 | 383* | - | >0.05 |
| G1790A | PB | 367 | 2156 | 356 | 11* | - | 2080 | 76* | - | >0.05 | |||||
| Hsiao | 2010 | Taiwan | Asian | Hepatocellul-ar | C1772T | HB | 102 | 347 | 94 | 8 | 0 | 334 | 13 | 0 | 0.72 |
| G1790A | HB | 102 | 347 | 87 | 15 | 0 | 333 | 14 | 0 | 0.70 | |||||
| Xu | 2011 | China | Asian | Glioma | C1772T | HB | 150 | 150 | 121 | 27 | 2 | 135 | 14 | 1 | 0.35 |
| Putra | 2011 | Japan | Asian | Lung | C1772T | PB | 83 | 110 | 74 | 9 | 0 | 98 | 12 | 0 | 0.55 |
| G1790A | PB | 83 | 110 | 72 | 9 | 2 | 101 | 9 | 0 | 0.65 | |||||
| Kang | 2011 | Korea | Asian | Colorectal | C1772T | PB | 50 | 50 | 46 | 4** | - | 38 | 12** | - | <0.01 |
| Wang | 2011 | China | Asian | Pancreatic | C1772T | HB | 263 | 271 | 209 | 54 | 0 | 242 | 29 | 0 | 0.35 |
| G1790A | HB | 263 | 271 | 198 | 65 | 0 | 249 | 22 | 0 | 0.49 | |||||
| Zagouri | 2012 | Greece | Caucasian | Breast | C1772T | HB | 113 | 124 | 98 | 15 | 0 | 107 | 17 | 0 | 0.41 |
| Kuo | 2012 | Taiwan | Asian | Lung | C1772T | HB | 285 | 300 | 153 | 94 | 38 | 216 | 73 | 11 | 0.13 |
| G1790A | HB | 285 | 300 | 150 | 94 | 41 | 215 | 74 | 11 | 0.15 | |||||
| Qin | 2012 | China | Asian | Renal | C1772T | HB | 620 | 623 | 572 | 46 | 2 | 578 | 43 | 2 | 0.22 |
| G1790A | HB | 620 | 623 | 575 | 45 | 0 | 584 | 39 | 0 | 0.42 | |||||
| Li | 2012 | China | Asian | Prostate | C1772T | HB | 662 | 716 | 612 | 48 | 2 | 659 | 57 | 0 | 0.27 |
| G1790A | HB | 662 | 716 | 614 | 47 | 1 | 685 | 31 | 0 | 0.55 | |||||
| Alves | 2012 | Brazil | mixed | Oral | C1772T | PB | 40 | 88 | 0 | 1 | 39 | 0 | 85 | 3 | <0.01 |
| G1790A | PB | 40 | 88 | 2 | 1 | 37 | 81 | 7 | 0 | 0.70 | |||||
| Ruiz-Tovar | 2012 | Spain | Caucasian | Pancreatic | C1772T | PB | 59 | 152 | 47 | 1 | 11 | 116 | 28 | 8 | 0.002 |
| G1790A | PB | 59 | 152 | 54 | 2 | 3 | 142 | 10 | 0 | 0.68 | |||||
| Fu | 2013 | China | Asian | Cervical | C1772T | HB | 518 | 553 | 467 | 49 | 2 | 492 | 60 | 1 | 0.55 |
| G1790A | HB | 509 | 553 | 489 | 20 | 0 | 510 | 42 | 1 | 0.89 | |||||
| Ribeiro | 2013 | Portugal | Caucasian | Breast | C1772T | PB | 96 | 74 | 74 | 21 | 1 | 61 | 7 | 4 | 0.001 |
| G1790A | PB | 96 | 74 | 96 | 0 | 0 | 74 | 0 | 0 | 0.99 | |||||
| Mera- | |||||||||||||||
| Menendez | 2013 | Spain | Caucasian | Glottic | |||||||||||
| larynx | C1772T | HB | 118 | 148 | 85 | 18 | 15 | 113 | 27 | 8 | 0.001 | ||||
| G1790A | HB | 111 | 139 | 107 | 4 | 0 | 130 | 9 | 0 | 0.69 | |||||
| Total | C1772T | 10869 | 14289 | 8994 | 1568 | 307 | 12181 | 1897 | 211 | ||||||
| G1790A | 7117 | 10442 | 6416 | 589 | 112 | 9922 | 494 | 26 | |||||||
W: wide type alleles (1772C or 1790G); M: mutant type alleles (1772 T or 1790A); HWE: Hardy-Weinberg Equilibrium; PB: population based; HB: hospital based.
Mixed: Caucasian and African-American; HNSCC: head and neck squamous cell carcinoma.
*Frequency of genotypes “AA + AG”; **Frequency of genotypes “TT + TC”.
Quantitative data synthesis
For the HIF-1α C1772T polymorphism, the overall results from the eligible studies demonstrated a significant association between the HIF-1α C1772T polymorphism and an increased cancer risk in four genetic models (TT vs. CC: OR = 1.63, 95% CI = 1.02-2.60; CT + TT vs. CC: OR = 1.15, 95% CI = 1.01-1.34; TT vs. CT + CC: OR = 2.11, 95% CI = 1.32-3.77; T vs. C: OR = 1.21, 95% CI = 1.04-1.41). In the subgroup analysis by cancer type, the HIF-1α C1772T polymorphism significantly increased the risk of breast cancer in Asians (TT vs. CC: OR = 4.42, 95% CI = 1.60-12.21; TT vs. CT + CC: OR = 4.16, 95% CI = 1.51-11.48; T vs. C: OR = 1.28, 95% CI = 1.05-1.55), other cancers (TT vs.CC: OR = 3.18, 95% CI = 1.90-5.32; TT vs. CT + CC: OR = 3.31, 95% CI = 1.98-5.53; T vs. C: OR = 1.47, 95% CI = 1.10-1.96) and lung cancer (TT vs. CT + CC: OR = 3.27, 95% CI = 1.73-6.17 ). When the data was stratified by ethnicity, the HIF-1α C1772T polymorphism was significantly correlated with an increased cancer risk in Asian population (TT vs. CC: OR = 4.10, 95% CI = 2.49-6.76; CT + TT vs. CC: OR = 1.29, 95% CI = 1.04-1.58; TT vs. CT + CC: OR = 3.67, 95% CI = 2.23-6.02; T vs. C: OR = 1.28, 95% CI = 1.04-1.57) and Caucasian population (TT vs. CT + CC: OR = 1.95, 95% CI = 1.14-3.31). In the analysis stratified by the sources of controls, a significant association was observed in the hospital-based group (CT + TT vs. CC: OR = 1.28, 95% CI = 1.01-1.62; T vs. C: OR = 1.33, 95% CI = 1.04-1.71) and the population-based group (TT vs. CT + CC: OR = 2.01, 95% CI = 1.10-3.71). Sensitivity analyses were carried out to assess the stability of the results by conducting analyses of studies with controls in HWE. The results showed significantly increased cancer risk (TT vs. CC: OR = 2.47, 95% CI = 1.81-3.36; CT + TT vs. CC: OR = 1.25, 95% CI = 1.05-1.49; TT vs. CT + CC: OR = 2.43, 95% CI = 1.41-4.19; T vs. C: OR = 1.27, 95% CI = 1.06-1.52). The other results for the HIF-1α C1772T polymorphism were similar to those when the studies with controls not in HWE were included. The main results of this pooled analysis are shown in Table 2. Figure 2 shows the forest plot of the association between cancer risk and the HIF-1α C1772T polymorphism under the allelic model.
Table 2.
Meta-analysis of the HIF-1α C1772T polymorphism and cancer risk
| Variables | TT vs.CC | CT vs.CC | CT + TT vs.CC | TT vs.CT + CC | T vs.C | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Case/control | I 2 | Phet | OR (95% CI) | Case/control | I 2 | Phet | OR (95% CI) | Case/control | I 2 | Phet | OR (95% CI) | case/control | I 2 | Phet | OR (95% CI) | Case/control | I 2 | Phet | OR (95% CI) | |
| Overall | 40 | 9301/12392 | 67 | <0.001 | 1.63 (1.02-2.60)* | 10562/14078 | 68 | <0.001 | 1.08 (0.92-1.26)* | 10958/14676 | 70 | <0.001 | 1.15 (1.01-1.34)* | 10540/12470 | 71 | <0.001 | 2.11 (1.32-3.37)* | 21738/28578 | 76 | <0.001 | 1.21 (1.04-1.41)* |
| Overall in HWE | 31 | 7429/9947 | 59 | 0.02 | 2.21 (1.27-3.83)* | 8481/11109 | 64 | <0.001 | 1.15 (0.98-1.36)* | 8604/11556 | 70 | <0.001 | 1.20 (1.02-1.41)* | 8275/9350 | 49 | 0.01 | 2.13 (1.28-3.55)* | 17208/22338 | 76 | <0.001 | 1.22 (1.03-1.44)* |
| Cancer type | |||||||||||||||||||||
| Cervical | 3 | 664/750 | 66 | 0.09 | 10.11 (2.55-40.05) | 739/871 | 60 | 0.08 | 0.98 (0.72-1.34) | 749/874 | 80 | 0.01 | 1.32 (0.61-2.87)* | 749/874 | 51 | 0.15 | 8.55 (2.28-32.13) | 2369/1748 | 88 | <0.001 | 1.41 (0.59-3.35)* |
| Breast | 6 | 1859/1809 | 62 | 0.03 | 1.41 (0.34-5.75)* | 2117/2033 | 37 | 0.16 | 1.01 (0.91-1.33) | 2143/2046 | 46 | 0.1 | 1.13 (0.94-1.36) | 2143/2046 | 60 | 0.04 | 1.38 (0.35-5.46)* | 4286/4092 | 56 | 0.04 | 1.09 (0.80-1.48)* |
| Breast in HWE | 5 | 1784/1744 | 55 | 0.08 | 2.30 (1.08-4.91) | 2022/1963 | 35 | 0.19 | 1.07 (0.88-1.29) | 2047/1972 | 56 | 0.06 | 1.12 (0.92-1.35) | 2047/1972 | 49 | 0.12 | 2.27 (1.06-4.82) | 4154/3944 | 65 | 0.02 | 1.09 (0.76-1.55)* |
| Breast in Asian | 3 | 1605/1564 | 0 | 0.93 | 4.42 (1.60-12.21) | 1809/1742 | 36 | 0.21 | 1.14 (0.92-1.41) | 1832/1746 | 51 | 0.13 | 1.22 (0.99-1.49) | 1832/1746 | 0 | 0.91 | 4.16 (1.51-11.48) | 3664/3492 | 55 | 0.11 | 1.28 (1.05-1.55) |
| Lung | 3 | 375/438 | 75 | 0.04 | 1.41 (0.07-30.44)* | 471/553 | 75 | 0.02 | 1.13 (0.59-2.19)* | 509/566 | 86 | 0.01 | 1.19 (0.51-2.76)* | 509/566 | 71 | 0.07 | 3.27 (1.73-6.17) | 1018/1132 | 89 | <0.001 | 1.19 (0.50-2.86)* |
| Colorectal | 4 | 599/2123 | - | - | - | 624/2175 | 79 | 0.03 | 0.24 (0.01-5.51)* | 627/2177 | 71 | 0.02 | 1.12 (0.57-2.18)* | 627/2177 | - | - | - | 1254/4354 | 80 | 0.02 | 0.26 (0.01-6.38)* |
| Prostate | 6 | 3149/3415 | 70 | 0.01 | 1.34 (0.54-3.31)* | 3766/4032 | 86 | <0.001 | 1.34 (0.93-1.92)* | 3816/4084 | 87 | <0.001 | 1.36 (0.95-1.96)* | 3816/4084 | 69 | 0.01 | 1.31 (0.54-3.20)* | 7632/8168 | 87 | <0.001 | 1.35 (0.96-1.89)* |
| Prostate in HWE | 4 | 1814/2067 | 59 | 0.09 | 1.57 (0.89-2.75) | 2168/2417 | 88 | <0.001 | 1.42 (0.84-2.40)* | 2200/2438 | 87 | 0.01 | 1.50 (0.89-2.40)* | 2200/2438 | 61 | 0.08 | 1.55 (0.89-2.72) | 4400/4876 | 85 | <0.001 | 1.44 (0.93-2.21)* |
| Renal | 4 | 1015/1035 | 25 | 0.26 | 0.28 (0.12-1.28) | 1065/1157 | 74 | 0.01 | 0.62 (0.31-1.24)* | 1160/1241 | 70 | 0.02 | 0.62 (0.33-1.18)* | 1160/1241 | 21 | 0.29 | 1.37 (0.92-2.04) | 2320/2482 | 44 | 0.15 | 0.91 (0.73-1.12) |
| Renal in HWE | 2 | 867/847 | 0 | 0.62 | 0.67 (0.21-2.13) | 947/929 | 13 | 0.28 | 0.92 (0.67-1.26) | 952/936 | 29 | 0.24 | 0.90 (0.67-1.22) | 952/936 | 0 | 0.64 | 0.69 (0.22-2.17) | 1904/1872 | 37 | 0.21 | 0.89 (0.67-1.19) |
| Oral | 4 | 549/547 | 0 | 0.46 | 2.01 (0.75-5.41) | 542/668 | 50 | 0.14 | 0.90 (0.55-1.47) | 589/679 | 16 | 0.3 | 1.04 (0.66-1.64) | 589/679 | 93 | <0.001 | 22.82 (0.28-1887.72)* | 1178/1358 | 88 | <0.001 | 2.52 (0.71-8.98)* |
| Oral in HWE | 2 | 446/423 | - | - | - | 478/443 | 0 | 0.5 | 1.28 (0.69-2.38) | 479/443 | 0 | 0.4 | 1.35 (0.73-2.49) | 479/443 | - | - | - | 958/886 | 0 | 0.32 | 1.41 (0.78-2.56) |
| Other | 12 | 1033/2151 | 30 | 0.2 | 3.18 (1.90-5.32) | 1190/2445 | 67 | <0.001 | 1.18 (0.79-1.78)* | 1276/2622 | 60 | <0.001 | 1.34 (0.95-1.87)* | 1276/2622 | 0 | 0.52 | 3.31 (1.98-5.53) | 2434/4940 | 58 | 0.01 | 1.47 (1.10-1.96)* |
| Other in HWE | 9 | 880/1032 | 56 | 0.08 | 5.10 (1.72-15.07) | 1032/1758 | 60 | 0.01 | 1.47 (0.97-2.21)* | 1041/1763 | 64 | 0.01 | 1.52 (0.99-2.34)* | 1041/1763 | 24 | 0.27 | 4.47 (1.53-13.00) | 2082/3526 | 67 | 0.01 | 1.52 (1.02-2.28)* |
| Ethnicity | |||||||||||||||||||||
| Asian | 20 | 5124/5781 | 0 | 0.96 | 4.10 (2.49-6.76) | 5678/6335 | 50 | 0.01 | 1.20 (0.99-1.46)* | 5787/6400 | 75 | <0.001 | 1.29 (1.04-1.58)* | 5787/6400 | 0 | 0.98 | 3.67 (2.23-6.02) | 11574/12800 | 61 | <0.001 | 1.28 (1.04-1.57) |
| Caucasian | 16 | 1791/4247 | 74 | <0.001 | 1.54 (0.72-3.27)* | 2220/4781 | 76 | <0.001 | 0.93 (0.65-1.33)* | 2385/4921 | 59 | 0.01 | 1.07 (0.80-1.43)* | 2385/4921 | 58 | 0.003 | 1.95 (1.14-3.31)* | 4770/9842 | 78 | <0.001 | 1.20 (0.91-1.57) |
| Caucasian in HWE | 9 | 1473/3153 | 76 | <0.001 | 2.28 (0.62-8.35)* | 1738/3152 | 79 | <0.001 | 1.20 (0.99-1.46)* | 1776/3535 | 82 | <0.001 | 1.28 (0.88-1.86)* | 1776/3535 | 69 | 0.002 | 2.08 (0.68-6.37)* | 3552/7070 | 86 | <0.001 | 1.34 (0.86-2.07) |
| Source of control | |||||||||||||||||||||
| HB | 17 | 4608/5249 | 77 | <0.001 | 3.28 (1.29-8.30)* | 5259/6029 | 60 | <0.001 | 1.18 (0.96-1.45)* | 5348/6086 | 72 | <0.001 | 1.28 (1.01-1.62)* | 5348/6086 | 71 | <0.001 | 2.85 (1.24-6.54)* | 10696/12172 | 80 | <0.001 | 1.33 (1.04-1.71)* |
| HB in HWE | 15 | 3340/3962 | 35 | 0.13 | 4.88 (2.96-8.04) | 3748/4467 | 56 | 0.01 | 1.24 (0.99-1.57)* | 3810/4488 | 67 | <0.001 | 1.33 (1.02-1.74)* | 3810/4488 | 4 | 0.4 | 4.23 (2.58-6.93) | 7620/8976 | 74 | <0.001 | 1.38 (1.06-1.80)* |
| PB | 23 | 4693/5303 | 54 | 0.01 | 1.33 (0.76-2.31)* | 5303/7143 | 74 | <0.001 | 0.99 (0.77-1.29)* | 5521/8203 | 70 | <0.001 | 1.10 (0.89-1.36)* | 5521/8203 | 72 | <0.001 | 2.02 (1.10-3.71)* | 11042/16406 | 74 | <0.001 | 1.18 (0.95-1.45)* |
| PB in HWE | 15 | 4089/5985 | 49 | 0.04 | 1.51 (0.74-3.11)* | 4733/6642 | 72 | <0.001 | 1.10 (0.85-1.43)* | 4794/6681 | 72 | <0.001 | 1.17 (0.93-1.48)* | 4794/6681 | 46 | 0.63 | 1.51 (1.01-2.27) | 9588/13362 | 75 | <0.001 | 1.14 (0.89-1.45)* |
HWE: Hardy-Weinberg Equilibrium; PB: population based; HB: hospital based; Phet: P value for heterogeneity. *Random-effects model was used when P value for heterogeneity test <0.05; otherwise, fixed-effects model was used.
Figure 2.
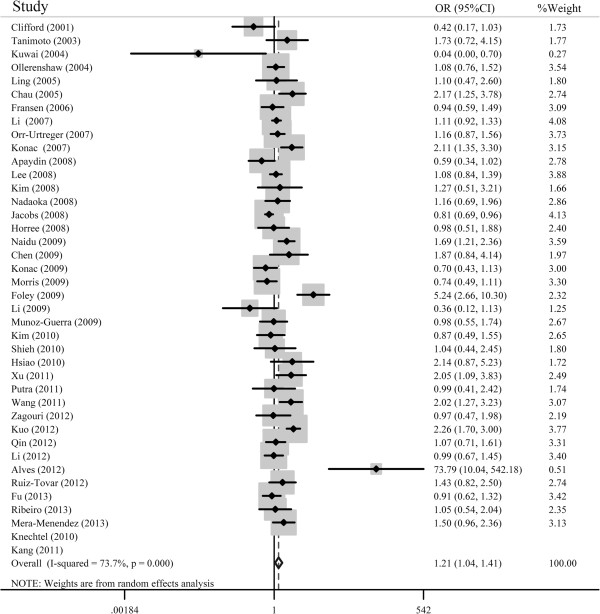
Forest plot of the association between cancer risk and the HIF-1α C1772T polymorphism using the allelicmodel (T vs. C).
For HIF-1α G1790A polymorphism, as shown in Table 3, the association between the HIF-1α G1790A polymorphism and increased cancer risk was significant for the pooled ORs under all of the genetic models (AA vs. GG: OR = 5.11, 95% CI = 2.08-12.56; GA vs. GG: OR = 1.45, 95% CI = 1.05-1.99; AA + AG vs. GG: OR = 1.63, 95% CI = 1.16-2.30; AA vs. GA + GG: OR = 4.41, 95% CI = 1.80-10.84; A vs. G: OR = 1.77, 95% CI = 1.23-2.25). In the subgroup analysis by cancer type, a significant association was observed in lung cancer (AA vs. GG: OR = 5.42, 95% CI = 2.74-10.70; GA vs. GG: OR = 1.72, 95% CI = 1.22-2.41; AA + AG vs. GG: OR = 2.14, 95% CI = 1.56-2.94; AA vs. GA + GG: OR = 4.52, 95% CI = 2.31-8.83; A vs. G: OR = 2.27, 95% CI = 1.74-2.95), pancreatic cancer (AA + AG vs. GG: OR = 3.14, 95% CI = 1.99-2.97; A vs. G: OR = 3.08, 95% CI = 1.98-4.78) and renal cancer (AA vs. GA + GG: OR = 3.09, 95% CI = 1.38-6.92). When the data were stratified by ethnicity, significantly increased cancer risk was observed in Asian population and Caucasian population. When the studies were stratified by the source of controls, a significant association was observed for population-based controls under the homozygote model, the dominant comparison model and the allelic model. Sensitivity analyses were conducted after the removal of the studies with controls not in HWE, the results for the HIF-1α G1790A polymorphism were similar to those when the studies with controls not in HWE were included. Table 3 shows the main results of this pooled analysis for the HIF-1α G1790A polymorphism. Figure 3 shows the forest plot of the association between cancer risk and the HIF-1α G1790A polymorphism under the dominant model.
Table 3.
Meta-analysis of the HIF-1α G1790A polymorphism and cancer risk
| Variables | AA vs.GG | GA vs. GG | AA + AG vs.GG | AA vs.GA + GG | A vs.G | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Study | Case/control | I 2 | Phet | OR (95% CI) | Case/control | I 2 | Phet | OR (95% CI) | Case/control | I 2 | Phet | OR (95% CI) | Case/control | I 2 | Phet | OR (95% CI) | Case/control | I 2 | Phet | OR (95% CI) | |
| Overall | 30 | 6538/9948 | 57 | 0.01 | 5.11 (2.08-12.56)* | 7005/10442 | 77 | <0.001 | 1.45 (1.05-1.99)* | 7117/10442 | 83 | <0.001 | 1.63 (1.16-2.30)* | 7117/10442 | 58 | 0.01 | 4.41 (1.80-10.84)* | 14234/20884 | 86 | <0.001 | 1.77 (1.23-2.25)* |
| Overall in HWE | 29 | 6449/9699 | 61 | 0.003 | 5.14 (1.67-15.86)* | 6873/10138 | 69 | <0.001 | 1.35 (1.01-1.81)* | 6971/10154 | 79 | <0.001 | 1.53 (1.10-2.12)* | 6971/10154 | 60 | 0.004 | 4.80 (1.58-14.55)* | 13942/20308 | 85 | <0.001 | 1.70 (1.17-2.46)* |
| Cancer type | |||||||||||||||||||||
| Breast | 4 | 623/501 | 0 | 0.34 | 1.44 (0.38-5.44) | 692/550 | 53 | 0.12 | 1.03 (0.70-1.52) | 698/553 | 60 | 0.08 | 1.05 (0.72-1.53) | 698/553 | 0 | 0.36 | 1.41 (0.37-5.40) | 1396/1466 | 65 | 0.56 | 1.07 (0.76-1.52) |
| Cervical | 3 | 708/819 | 0 | 0.99 | 0.35 (0.04-3.39) | 740/871 | 57 | 0.13 | 0.62 (0.40-0.98) | 740/837 | 51 | 0.15 | 0.60 (0.38-0.94) | 740/837 | 0 | 0.99 | 0.36 (0.04-3.450 | 1480/1746 | 42 | 0.19 | 0.59 (0.38-0.91) |
| Oral | 4 | 517/633 | 75 | 0.02 | 72.11 (2.08-2502.44)* | 542/670 | 70 | 0.02 | 3.17 (1.26-7.92)* | 583/670 | 92 | <0.001 | 7.92 (1.58-39.64)* | 583/670 | 75 | 0.02 | 58.05 (1.70-1985.77)* | 1166/1340 | 96 | 0.01 | 9.66 (1.31-71.15)* |
| Prostate | 3 | 1866/2230 | - | - | - | 1927/2280 | 1 | 0.37 | 1.42 (0.97-2.07) | 1928/2280 | 7 | 0.34 | 1.44 (0.98-2.10) | 1928/2280 | - | - | - | 3856/4560 | 10 | 0.33 | 1.45 (0.99-2.11) |
| Renal | 4 | 1016/1267 | 0 | 0.95 | 5.10 (2.21-11.73) | 1123/1354 | 92 | <0.001 | 1.51 (0.45-5.05)* | 1139/1364 | 92 | <0.001 | 1.58 (0.49-5.04)* | 1139/1364 | 0 | 0.76 | 3.09 (1.38-6.92) | 2278/2728 | 89 | <0.001 | 1.53 (0.60-3.92)* |
| Renal in HWE | 3 | 937/1018 | - | - | - | 991/1076 | 0 | 0.42 | 1.00 (0.69-1.47) | 993/1076 | 0 | 0.6 | 1.04 (0.71-1.52) | 993/1076 | - | - | - | 1986/2152 | 0 | 0.78 | 1.07 (0.74-1.55) |
| Lung | 3 | 405/481 | 0 | 0.87 | 5.42 (2.74-10.70) | 466/555 | 0 | 0.57 | 1.72 (1.22-2.41) | 509/566 | 0 | 0.46 | 2.14 (1.56-2.94) | 509/566 | 0 | 0.79 | 4.52 (2.31-8.83) | 1018/1132 | 0 | 0.48 | 2.27 (1.74-2.95) |
| Colorectal | 2 | 545/2327 | - | - | - | 554/2336 | - | - | - | 554/2336 | 0 | 0.45 | 0.97 (0.57-1.63) | 554/2336 | - | - | - | 1108/4672 | - | - | - |
| Pancreatic | 2 | 255/391 | - | - | - | 319/423 | 82 | 0.02 | 1.61 (0.24-10.76)* | 322/423 | 63 | 0.1 | 3.14 (1.99-4.97) | 322/423 | - | - | - | 644/846 | 0 | 0.42 | 3.08 (1.98-4.78) |
| Other | 7 | 593/1377 | - | - | - | 642/1377 | 74 | <0.001 | 1.53 (0.65-3.59)* | 644/1377 | 72 | <0.001 | 1.57 (0.70-3.53)* | 644/1377 | - | - | - | 1288/2754 | 69 | 0.01 | 1.57 (0.75-3.30)* |
| Ethnicity | |||||||||||||||||||||
| Asian | 15 | 3607/4263 | 13 | 0.33 | 3.50 (2.05-5.98) | 4010/4614 | 74 | <0.001 | 1.44 (1.04-1.99)* | 4063/4630 | 76 | <0.001 | 1.49 (1.07-2.08)* | 4063/4630 | 0 | 0.45 | 3.12 (1.83-5.32) | 8126/9260 | 77 | <0.001 | 1.49 (1.08-2.05)* |
| Caucasian | 13 | 1829/4357 | 0 | 0.69 | 6.63 (3.11-14.12) | 1926/4450 | 81 | <0.001 | 1.36 (0.58-3.19)* | 1948/4460 | 82 | <0.001 | 1.45 (0.69-3.04)* | 1948/4460 | 0 | 0.49 | 4.21 (2.04-8.71) | 3896/8920 | 75 | <0.001 | 1.65 (0.84-3.24)* |
| Caucasian in HWE | 12 | 1750/4108 | 0 | 0.74 | 12.40 (2.19-70.22) | 1794/4172 | 68 | 0.01 | 1.10 (0.48-2.49)* | 1802/4172 | 67 | 0.01 | 1.22 (0.62-2.37)* | 1802/4172 | 0 | 0.79 | 11.37 (2.02-63.93) | 3604/8344 | 68 | 0.01 | 1.65 (1.17-2.32)* |
| Source of control | |||||||||||||||||||||
| HB | 13 | 3197/3945 | 45 | 0.12 | 1.54 (0.35-6.70) | 3510/4234 | 77 | <0.001 | 1.37 (0.92-2.05)* | 3554/4248 | 79 | <0.001 | 1.40 (0.93-2.11)* | 3554/4248 | 35 | 0.19 | 3.13 (1.74-5.62) | 7108/8496 | 79 | <0.001 | 1.38 (0.93-2.05)* |
| PB | 17 | 3133/5705 | 66 | 0.01 | 11.55 (6.62-20.12)* | 3295/5882 | 78 | <0.001 | 1.51 (0.88-2.58)* | 3563/6194 | 85 | <0.001 | 1.90 (1.06-3.39)* | 3563/6194 | 69 | 0.002 | 10.27 (2.42-43.63)* | 6726/11788 | 89 | <0.001 | 2.25 (1.18-4.29)* |
| PB in HWE | 16 | 3054/5456 | 67 | 0.006 | 15.51 (2.53-94.94)* | 3163/5604 | 60 | 0.01 | 1.34 (0.85-2.11)* | 3417/5906 | 81 | <0.001 | 1.71 (0.97-3.03)* | 3417/5906 | 66 | 0.007 | 14.20 (2.38-84.61)* | 6434/11212 | 89 | <0.001 | 2.33 (1.91-2.84)* |
HWE: Hardy-Weinberg Equilibrium; PB: population based; HB: hospital based; Phet: P value for heterogeneity. *Random-effects model was used when P value for heterogeneity test <0.05; otherwise, fixed-effects model was used.
Figure 3.
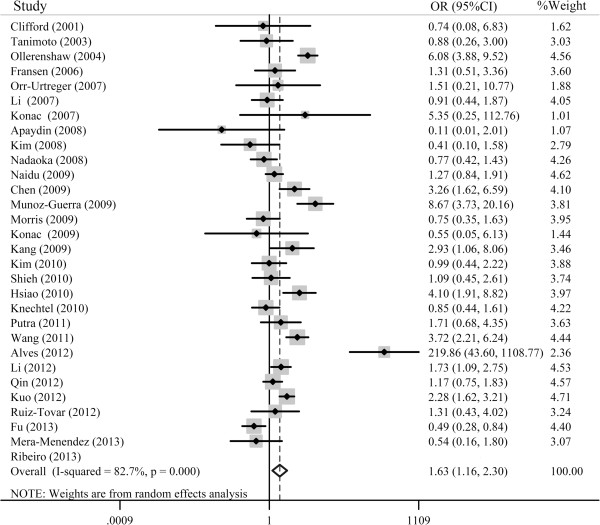
Overall association between the HIF-1α G1790A polymorphism and cancer risk for all subjects using the dominant model (GA + AA vs. GG).
Test of heterogeneity
There was significant heterogeneity observed in the allelic comparison model, the dominant comparison model and the heterozygote comparison model (Tables 2 and 3), and the heterogeneity was effectively decreased or removed in the subgroups stratified by ethnicity, cancer types and source of controls (Tables 2 and 3).
Sensitivity analysis
We performed sensitivity analysis by removing each individual study (including the restudies with controls not in HWE) sequentially for both the HIF-1α C1772T and the HIF-1α G1790A polymorphism (Figure 4 and Additional file 1). The results indicated that the overall significance of the pooled ORs was not altered by any single study in the genetic models for the HIF-1α C1772T/G1790A polymorphisms and cancer susceptibility, suggesting stability and reliability in our overall results.
Figure 4.
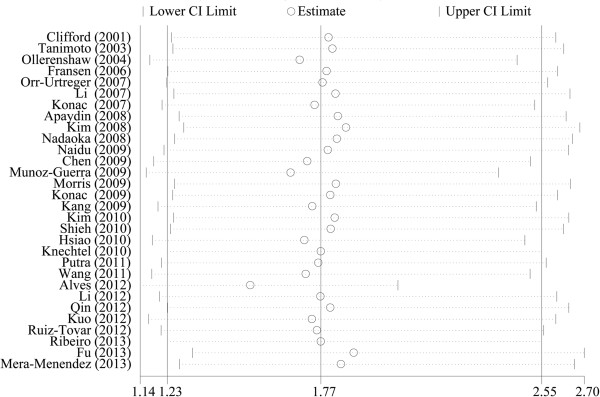
The influence of individual studies on the summary odds ratio (OR) for the HIF-1α G1790A polymorphism.
Bias diagnostics
A Begg’s funnel plot and Egger’s test were used to assess the publication bias in this meta-analysis. As shown in Figure 5, for the HIF-1α C1772T polymorphism, the funnel plots for the comparison of the five models appear to be basically symmetric. The Egger’s linear regression test did not show any evidence of significant publication bias in five models (TT vs. CC: t = 0.50, P = 0.62; TC vs. CC: t = -0.19, P = 0.85; TT vs. CT + CC: t = 1.11, P = 0.28; T vs. C: t = 1.39, P = 0.17; CT + TT vs. CC: t = 0.59, P = 0.56). For the HIF-1α G1790A polymorphism, no visual publication bias was detected in the funnel plot (Figure 6) and the result showed no significant evidence of a publication bias in five models(AA vs. GG: t = 0.03, P = 0.98; GA vs. GG: t = -0.86, P = 0.40; AA vs. GA + GG: t = 0.33, P = 0.75; AA + AG vs.GG: t = -0.40, P = 0.69; A vs. G: t = -0.41, P = 0.68).
Figure 5.
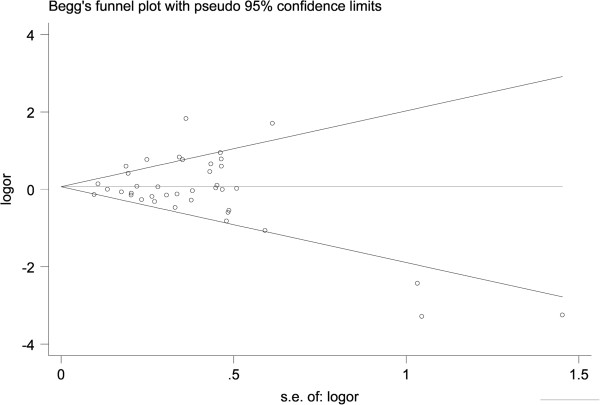
Begg’s funnel plot for evaluating the publication bias of the meta-analysis for the HIF-1α C1772T polymorphism.
Figure 6.
Begg’s funnel plot for evaluating the publication bias of the meta-analysis for the HIF-1α G1790A polymorphism.
Discussion
HIF-1 is a heterodimeric transcription factor and a key regulator of the cellular response to hypoxia [5]. It is composed of HIF-1α and HIF-1β subunits, which are members of the bHLH-PAS transcription factor family. HIF-1α is a unique O2-regulated subunit that determines the function of HIF-1. HIF-1α upregulates the expression of genes whose protein products function to increase O2 availability or to allow metabolic adaptation to O2 deprivation, including VEGF, Epo, NOS2 and others. Most of these aforementioned proteins have been implicated in tumor development and progression [35, 64, 65]. Recent studies have reported that the overexpression of HIF-1α is significantly associated with cell proliferation, tumor susceptibility, tumor size, lymph node metastasis and prognosis [12, 35, 66]. The HIF-1α gene is located on chromosome 14q21-24 and contains a total of 35 common SNPs, according to the dbSNP database (http://www.ncbi.nlm.nih.gov/SNP). Two polymorphisms, C1772T (rs11549465) and G1790A (rs11549467), result in an amino acid substitution of proline to serine and alanine to threonine, respectively, and the present studies show that C1772T (rs11549465) is not in substantial linkage disequilibrium (LD) with G1790A (rs11549467) (R2 = 0.002). Under normoxic condition, the hydroxylation of proline 402 and proline 564 occurs within the oxygen-dependent degradation (ODD) domain of HIF-1α, and HIF-1α is rapidly degraded. The two SNPs examined here are located within the ODD/pVHL binding domain in exon 12 of the HIF-1α gene and may enhance the transcription activity of the HIF-1α gene by causing structural changes, increasing the stability of HIF-1α protein and affecting the expression of downstream target genes [8, 14, 17]. Over the last few years, a great number of studies have been performed to investigate the association between these HIF-1α polymorphisms and cancer risk in different populations. However, the results of these studies remain inconclusive. In a meta-analysis conducted by Zhao et al. in 2009 [67], the HIF-1 C1772T polymorphism was reported to be associated with increased cancer risk, while no significant association was found between the HIF-1α G1790A polymorphism and cancer risk. Additionally, Li et al. reported that the HIF-1α C1772T polymorphism correlates with urinary cancer risk in Caucasian population, and the G1790A polymorphism may increase the risk of prostate cancer [68]. Due to the important role of HIF-1α polymorphisms in the development of cancer and due to the limited statistical power of the previous studies, we conducted a comprehensive literature search and performed a meta-analysis on all of the available case-control studies to systematically evaluate the exact relationship between the C1772T/G1790A polymorphisms in HIF-1α and cancer susceptibility.
Regarding the HIF-1α C1772T polymorphism, our results suggested a significant association in four genetic comparison models, providing convincing evidence that the HIF-1α C1772T polymorphism may be a risk factor in cancer development. When sensitivity analyses were performed, the results were similar to those when the studies with controls not in HWE were included, suggesting that our results were very robust. Moreover, when the data were stratified by cancer type, a significant association was observed between the C1772T polymorphism and breast cancer in Asians. This may be due to the specific genetic variant induced over-expression of HIF-1 under hypoxic condition in breast cancer cells, and the different life style, ethnicity and body composition between Asians and Caucasians, which could contribute to the results. A significant association was also observed in lung cancer. When subgroup analyses were performed according to ethnicity and source of controls, a significant association was found in Asian population, Caucasian population and in hospital-based studies. Zhao et al. [67] reported that the genotype TT was significantly associated with an increased cancer risk in Asians, but the CI was very wide due to the lack of mutant homozygotes in Asians. In our meta-analysis, we also found that the C1772T polymorphism was a risk factor in Asians (Dominant model: OR = 1.29, 95% CI = 1.04-1.58; Allelic model: OR = 1.47, 95% CI = 1.04-1.57). Beyond that, we had not found any significant associations in prostate cancer, renal cancer or oral cancer.
For the HIF-1α G1790A polymorphism, the pooled results from all of the eligible studies suggested that the G1790A polymorphism in HIF-1α is significantly associated with an increased cancer risk in all of the genetic models. We also conducted subgroup analyses based on the cancer type, ethnicities and source of controls. In the subgroup analysis according to cancer type, the results suggested that the HIF-1α G1790A polymorphism significantly increased the risk of lung cancer, renal cancer, oral cancer and pancreatic cancer, but the CI for the oral cancer subgroup was very wide. This may be due to the lack of mutant homozygotes detected, and the association could have been caused by chance. More studies based on large populations should be prusued. The study reported by Putra et al. indicated that even though they did not found any significant differences in genotype for G1790A between lung cancer patients and healthy controls, however, the G1790A variant allele was significantly higher in lung cancer patients, and TP53 LOH and 1p34 LOH were more frequently observed in individuals with the HIF-1α G1790A polymorphism, suggesting that this polymorphism may induce mutations in some tumor suppressor genes involved in lung cancer development [46]. Here, we found a significant association between the G1790A polymorphism and lung cancer risk. When the data were stratified according to ethnicity classification and source of controls, similar to the C1772T polymorphism, significantly increased risks were also found in Asian populations, Caucasian populations and population-based studies. After sensitivity analyses were performed, our results did not vary substantially, which strongly suggests an association between the HIF-1α G1790A polymorphism and increased cancer risk. One important factor that could influence the results is heterogeneity. In our study, significant heterogeneity existed in the analysis of the heterozygote model, the dominant model and the allelic model for the HIF-1α C1772T/G1790A polymorphism. When we performed a subgroup analysis according to cancer type, ethnicity or source of controls, the heterogeneity was reduced significantly or disappeared. The significant heterogeneity may due to the differences in ethnicity or cancer types or even in the selection of the controls. Furthermore, publication bias was not observed in our meta-analysis of the HIF-1α G1790A/C1772T polymorphisms. We also performed a sensitivity analysis to evaluate the sources of heterogeneity. The pooled ORs did not vary substantially, indicating that the results of our meta-analysis are robust and reliable.
To a certain extent, our meta-analysis still includes several limitations that should be interpreted and taken into consideration. First, in the era of GWAS, researchers can obtain the GWAS data for these two SNPs from all cancer studies and conduct a meta-analysis with the GWAS data instead of relying on published data, which may be biased toward positive findings. Second, the lack of observations concerning gene-gene and gene-environment interactions could influence our results. Third, although the total number of studies was not small, there were still not sufficient eligible studies for us to analyze different types of cancers, such as colorectal carcinoma, renal cell carcinoma or glioma; more studies are needed to explore the potential relationship between HIF-1αC1772T/G1790A polymorphisms and cancer susceptibility. Forth, the lack of detailed original data, such as the age and sex of the populations, smoking status, or alcohol consumption in the eligible studies may influence our extended analyses. However, our meta-analysis also has many advantages. First, we searched all possible publications, and the total number of eligible studies was much larger than other previously published meta-analyses; therefore, our results are more convincing. Second, no publication bias was detected in our meta-analysis. Finally, all of the data were extracted from well-selected studies, providing stronger statistical power for our study.
Conclusions
In conclusion, this meta-analysis provides powerful evidence that both the C1772T and G1790A polymorphisms in the HIF-1α gene may contribute to individual susceptibility to cancers. It will be necessary to perform additional research to evaluate the relationship between HIF-1α C1772T/G1790A polymorphisms and cancer risk. Moreover, large sample case-control studies assessing gene-to-gene and gene-to-environment interactions are required to verify these findings.
Authors’ information
Qing Yan, Pin Chen and Songtao Wang are joint first authors.
Electronic supplementary material
Additional file 1: The influence of individual studies on the summary odds ratio (OR) for the HIF-1α C1772T polymorphism. (TIFF 271 KB)
Acknowledgements
This work is supported by the National Natural Science Foundation of China (grant 30901534, 81172694 and 81473013); the Grant for the 135 Key Medical Project of Jiangsu Province (No. XK201117); and the National high technology research and development program 863 (No. 2012AA02A508).
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
QY participated in collection of data and manuscript preparation. PC and QY, SW performed the statistical analysis and participated in the revision. PZ and NL, AG participated in study design and critically revised the manuscript. PZ participated in study design and manuscript preparation. All authors read and approved the final manuscript.
Contributor Information
Qing Yan, Email: yq3880752@hotmail.com.
Pin Chen, Email: chenpin1987@126.com.
Songtao Wang, Email: 986522651@qq.com.
Ning Liu, Email: LIUNING0853@YAHOO.COM.CN.
Peng Zhao, Email: zhaopeng@njmu.edu.cn.
Aihua Gu, Email: aihuagu@njmu.edu.cn.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59(4):225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Pharoah PD, Dunning AM, Ponder BA, Easton DF. Association studies for finding cancer-susceptibility genetic variants. Nat Rev Cancer. 2004;4(11):850–860. doi: 10.1038/nrc1476. [DOI] [PubMed] [Google Scholar]
- 4.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411(6835):366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 5.Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92(12):5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Semenza GL. HIF-1 and mechanisms of hypoxia sensing. Curr Opin Cell Biol. 2001;13(2):167–171. doi: 10.1016/S0955-0674(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 7.Sutter CH, Laughner E, Semenza GL. Hypoxia-inducible factor 1alpha protein expression is controlled by oxygen-regulated ubiquitination that is disrupted by deletions and missense mutations. Proc Natl Acad Sci U S A. 2000;97(9):4748–4753. doi: 10.1073/pnas.080072497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanimoto K, Yoshiga K, Eguchi H, Kaneyasu M, Ukon K, Kumazaki T, Oue N, Yasui W, Imai K, Nakachi K, Poellinger L, Nishiyama M. Hypoxia-inducible factor-1alpha polymorphisms associated with enhanced transactivation capacity, implying clinical significance. Carcinogenesis. 2003;24(11):1779–1783. doi: 10.1093/carcin/bgg132. [DOI] [PubMed] [Google Scholar]
- 9.Bardos JI, Ashcroft M. Negative and positive regulation of HIF-1: a complex network. Biochim Biophys Acta. 2005;1755(2):107–120. doi: 10.1016/j.bbcan.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Ravi R, Mookerjee B, Bhujwalla ZM, Sutter CH, Artemov D, Zeng Q, Dillehay LE, Madan A, Semenza GL, Bedi A. Regulation of tumor angiogenesis by p53-induced degradation of hypoxia-inducible factor 1alpha. Genes Dev. 2000;14(1):34–44. [PMC free article] [PubMed] [Google Scholar]
- 11.Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Hypoxia-inducible factor-1alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60(15):4010–4015. [PubMed] [Google Scholar]
- 12.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3(10):721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 13.Rannala B. Finding genes influencing susceptibility to complex diseases in the post-genome era. Am J Pharmacogenomics. 2001;1(3):203–221. doi: 10.2165/00129785-200101030-00005. [DOI] [PubMed] [Google Scholar]
- 14.Yamada N, Horikawa Y, Oda N, Iizuka K, Shihara N, Kishi S, Takeda J. Genetic variation in the hypoxia-inducible factor-1alpha gene is associated with type 2 diabetes in Japanese. J Clin Endocrinol Metab. 2005;90(10):5841–5847. doi: 10.1210/jc.2005-0991. [DOI] [PubMed] [Google Scholar]
- 15.Semenza GL, Rue EA, Iyer NV, Pang MG, Kearns WG. Assignment of the hypoxia-inducible factor 1alpha gene to a region of conserved synteny on mouse chromosome 12 and human chromosome 14q. Genomics. 1996;34(3):437–439. doi: 10.1006/geno.1996.0311. [DOI] [PubMed] [Google Scholar]
- 16.Fu XS, Choi E, Bubley GJ, Balk SP. Identification of hypoxia-inducible factor-1alpha (HIF-1alpha) polymorphism as a mutation in prostate cancer that prevents normoxia-induced degradation. Prostate. 2005;63(3):215–221. doi: 10.1002/pros.20190. [DOI] [PubMed] [Google Scholar]
- 17.Clifford SC, Astuti D, Hooper L, Maxwell PH, Ratcliffe PJ, Maher ER. The pVHL-associated SCF ubiquitin ligase complex: molecular genetic analysis of elongin B and C, Rbx1 and HIF-1alpha in renal cell carcinoma. Oncogene. 2001;20(36):5067–5074. doi: 10.1038/sj.onc.1204602. [DOI] [PubMed] [Google Scholar]
- 18.Smaldone MC, Maranchie JK. Clinical implications of hypoxia inducible factor in renal cell carcinoma. Urol Oncol. 2009;27(3):238–245. doi: 10.1016/j.urolonc.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Yoshiba S, Ito D, Nagumo T, Shirota T, Hatori M, Shintani S. Hypoxia induces resistance to 5-fluorouracil in oral cancer cells via G(1) phase cell cycle arrest. Oral Oncol. 2009;45(2):109–115. doi: 10.1016/j.oraloncology.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 20.Lin PY, Yu CH, Wang JT, Chen HH, Cheng SJ, Kuo MY, Chiang CP. Expression of hypoxia-inducible factor-1 alpha is significantly associated with the progression and prognosis of oral squamous cell carcinomas in Taiwan. J Oral Pathol Med. 2008;37(1):18–25. doi: 10.1111/j.1600-0714.2007.00571.x. [DOI] [PubMed] [Google Scholar]
- 21.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 22.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 23.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21(11):1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konac E, Onen HI, Metindir J, Alp E, Biri AA, Ekmekci A. An investigation of relationships between hypoxia-inducible factor-1 alpha gene polymorphisms and ovarian, cervical and endometrial cancers. Cancer Detect Prev. 2007;31(2):102–109. doi: 10.1016/j.cdp.2007.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Kuwai T, Kitadai Y, Tanaka S, Kuroda T, Ochiumi T, Matsumura S, Oue N, Yasui W, Kaneyasu M, Tanimoto K, Nishyama M, Chayama K. Single nucleotide polymorphism in the hypoxia-inducible factor-1alpha gene in colorectal carcinoma. Oncol Rep. 2004;12(5):1033–1037. [PubMed] [Google Scholar]
- 28.Ling TS, Shi RH, Zhang GX, Zhu H, Yu LZ, Ding XF. Common single nucleotide polymorphism of hypoxia-inducible factor-1alpha and its impact on the clinicopathological features of esophageal squamous cell carcinoma. Chin J Dig Dis. 2005;6(4):155–158. doi: 10.1111/j.1443-9573.2005.00223.x. [DOI] [PubMed] [Google Scholar]
- 29.Fransen K, Fenech M, Fredrikson M, Dabrosin C, Soderkvist P. Association between ulcerative growth and hypoxia inducible factor-1alpha polymorphisms in colorectal cancer patients. Mol Carcinog. 2006;45(11):833–840. doi: 10.1002/mc.20209. [DOI] [PubMed] [Google Scholar]
- 30.Orr-Urtreger A, Bar-Shira A, Matzkin H, Mabjeesh NJ. The homozygous P582S mutation in the oxygen-dependent degradation domain of HIF-1 alpha is associated with increased risk for prostate cancer. Prostate. 2007;67(1):8–13. doi: 10.1002/pros.20433. [DOI] [PubMed] [Google Scholar]
- 31.Li P, Cao Q, Shao PF, Cai HZ, Zhou H, Chen JW, Qin C, Zhang ZD, Ju XB, Yin CJ. Genetic polymorphisms in HIF1A are associated with prostate cancer risk in a Chinese population. Asian J Androl. 2012;14(6):864–869. doi: 10.1038/aja.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee JY, Choi JY, Lee KM, Park SK, Han SH, Noh DY, Ahn SH, Kim DH, Hong YC, Ha E, Yoo KY, Ambrosone CB, Kang D. Rare variant of hypoxia-inducible factor-1alpha (HIF-1A) and breast cancer risk in Korean women. Clin Chim Acta. 2008;389(1–2):167–170. doi: 10.1016/j.cca.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Kim HO, Jo YH, Lee J, Lee SS, Yoon KS. The C1772T genetic polymorphism in human HIF-1alpha gene associates with expression of HIF-1alpha protein in breast cancer. Oncol Rep. 2008;20(5):1181–1187. [PubMed] [Google Scholar]
- 34.Apaydin I, Konac E, Onen HI, Akbaba M, Tekin E, Ekmekci A. Single nucleotide polymorphisms in the hypoxia-inducible factor-1alpha (HIF-1alpha) gene in human sporadic breast cancer. Arch Med Res. 2008;39(3):338–345. doi: 10.1016/j.arcmed.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Chen MK, Chiou HL, Su SC, Chung TT, Tseng HC, Tsai HT, Yang SF. The association between hypoxia inducible factor-1alpha gene polymorphisms and increased susceptibility to oral cancer. Oral Oncol. 2009;45(12):e222–e226. doi: 10.1016/j.oraloncology.2009.07.015. [DOI] [PubMed] [Google Scholar]
- 36.Foley R, Marignol L, Thomas AZ, Cullen IM, Perry AS, Tewari P, O’Grady A, Kay E, Dunne B, Loftus B, Waston WR, Fitzpatirck JM, Woodson K, Lehman T, Hollywood D, Lynch TH, Lawler M. The HIF-1alpha C1772T polymorphism may be associated with susceptibility to clinically localised prostate cancer but not with elevated expression of hypoxic biomarkers. Cancer Biol Ther. 2009;8(2):118–124. doi: 10.4161/cbt.8.2.7086. [DOI] [PubMed] [Google Scholar]
- 37.Konac E, Dogan I, Onen HI, Yurdakul AS, Ozturk C, Varol A, Ekmecki A. Genetic variations in the hypoxia-inducible factor-1alpha gene and lung cancer. Exp Biol Med (Maywood) 2009;234(9):1109–1116. doi: 10.3181/0902-RM-49. [DOI] [PubMed] [Google Scholar]
- 38.Morris MR, Hughes DJ, Tian YM, Ricketts CJ, Lau KW, Gentle D, Shuib S, Serrano-Fernandez P, Lubinski J, Wiesener MS, Pugh CW, Latif F, Ratcliffe PJ, Maher ER. Mutation analysis of hypoxia-inducible factors HIF1A and HIF2A in renal cell carcinoma. Anticancer Res. 2009;29(11):4337–4343. [PubMed] [Google Scholar]
- 39.Li K, Zhang Y, Dan Z, Wang Y, Ren ZC. Association of the hypoxia inducible factor-1alpha gene polymorphisms with gastric cancer in Tibetans. Biochem Genet. 2009;47(9–10):625–634. doi: 10.1007/s10528-009-9254-2. [DOI] [PubMed] [Google Scholar]
- 40.Naidu R, Har YC, Taib NA. Associations between hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms and risk of developing breast cancer. Neoplasma. 2009;56(5):441–447. doi: 10.4149/neo_2009_05_441. [DOI] [PubMed] [Google Scholar]
- 41.Kim YH, Park IA, Park WY, Kim JW, Kim SC, Park NH, Song YS, Kang SB. Hypoxia-inducible factor 1alpha polymorphisms and early-stage cervical cancer. Int J Gynecol Cancer. 2011;21(1):2–7. doi: 10.1097/IGC.0b013e318204f6e6. [DOI] [PubMed] [Google Scholar]
- 42.Hsiao PC, Chen MK, Su SC, Ueng KC, Chen YC, Hsieh YH, Liu YF, Tsai HT, Yang SF. Hypoxia inducible factor-1alpha gene polymorphism G1790A and its interaction with tobacco and alcohol consumptions increase susceptibility to hepatocellular carcinoma. J Surg Oncol. 2010;102(2):163–169. doi: 10.1002/jso.21539. [DOI] [PubMed] [Google Scholar]
- 43.Shieh TM, Chang KW, Tu HF, Shih YH, Ko SY, Chen YC, Liu CJ. Association between the polymorphisms in exon 12 of hypoxia-inducible factor-1alpha and the clinicopathological features of oral squamous cell carcinoma. Oral Oncol. 2010;46(9):e47–e53. doi: 10.1016/j.oraloncology.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 44.Xu G, Wang M, Xie W, Bai X. Hypoxia-inducible factor-1 alpha C1772T gene polymorphism and glioma risk: a hospital-based case-control study from China. Genet Test Mol Biomark. 2011;15(6):461–464. doi: 10.1089/gtmb.2010.0265. [DOI] [PubMed] [Google Scholar]
- 45.Wang X, Liu Y, Ren H, Yuan Z, Li S, Sheng J, Zhao T, Chen Y, Liu F, Wang F, Huang H, Hao J. Polymorphisms in the hypoxia-inducible factor-1alpha gene confer susceptibility to pancreatic cancer. Cancer Biol Ther. 2011;12(5):383–387. doi: 10.4161/cbt.12.5.15982. [DOI] [PubMed] [Google Scholar]
- 46.Putra AC, Tanimoto K, Arifin M, Hiyama K. Hypoxia-inducible factor-1alpha polymorphisms are associated with genetic aberrations in lung cancer. Respirology. 2011;16(5):796–802. doi: 10.1111/j.1440-1843.2011.01972.x. [DOI] [PubMed] [Google Scholar]
- 47.Kuo WH, Shih CM, Lin CW, Cheng WE, Chen SC, Chen W, Lee YL. Association of hypoxia inducible factor-1alpha polymorphisms with susceptibility to non-small-cell lung cancer. Transl Res. 2012;159(1):42–50. doi: 10.1016/j.trsl.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 48.Qin C, Cao Q, Ju X, Wang M, Meng X, Zhu J, Yan F, Li P, Ding Q, Chen J, Gu M, Zhang W, Yin C, Zhang Z. The polymorphisms in the VHL and HIF1A genes are associated with the prognosis but not the development of renal cell carcinoma. Ann Oncol. 2012;23(4):981–989. doi: 10.1093/annonc/mdr325. [DOI] [PubMed] [Google Scholar]
- 49.Zagouri F, Sergentanis TN, Gazouli M, Tsigginou A, Dimitrakakis C, Papaspyrou I, Eleutherakis-Papaiakovou E, Chrysikos D, Theodoropoulos G, Zografos GC, Antsaklis A, Dimopoulos AM, Papadimitriou CA. HSP90, HSPA8, HIF-1 alpha and HSP70-2 polymorphisms in breast cancer: a case-control study. Mol Biol Rep. 2012;39(12):10873–10879. doi: 10.1007/s11033-012-1984-2. [DOI] [PubMed] [Google Scholar]
- 50.Ribeiro AL, Gaspar JF, Pereira T, Ribeiro V. Lack of relevance of HIF-1alpha polymorphisms in breast cancer in a Portuguese population. Anticancer Res. 2013;33(6):2549–2555. [PubMed] [Google Scholar]
- 51.Kang MJ, Jung SA, Jung JM, Kim SE, Jung HK, Kim TH, Shim KN, Yi SY, Yoo K, Moon IH. Associations between single nucleotide polymorphisms of MMP2, VEGF, and HIF1A genes and the risk of developing colorectal cancer. Anticancer Res. 2011;31(2):575–584. [PubMed] [Google Scholar]
- 52.Knechtel G, Szkandera J, Stotz M, Hofmann G, Langsenlehner U, Krippl P, Samonigg H, Renner W, Langner C, Dehchamani D, Gerger A. Single nucleotide polymorphisms in the hypoxia-inducible factor-1 gene and colorectal cancer risk. Mol Carcinog. 2010;49(9):805–809. doi: 10.1002/mc.20655. [DOI] [PubMed] [Google Scholar]
- 53.Alves LR, Fraga CA, Oliveira MV, Sousa AA, Jorge AS, Marques-Silva L, Santos SH, Jones KM, de Paula AM, Guimaraes AL. High HIF-1alpha expression genotypes increase odds ratio of oral cancer. Head Neck Oncol. 2012;4(5):87. [Google Scholar]
- 54.Munoz-Guerra MF, Fernandez-Contreras ME, Moreno AL, Martin ID, Herraez B, Gamallo C. Polymorphisms in the hypoxia inducible factor 1-alpha and the impact on the prognosis of early stages of oral cancer. Ann Surg Oncol. 2009;16(8):2351–2358. doi: 10.1245/s10434-009-0503-8. [DOI] [PubMed] [Google Scholar]
- 55.Fu SL, Miao J, Ding B, Wang XL, Cheng WJ, Dai HH, Han SP. A polymorphism in the 3’ untranslated region of Hypoxia-Inducible Factor-1 alpha confers an increased risk of cervical cancer in aChinese population. Neoplasma. 2013;61(1):63–69. doi: 10.4149/neo_2014_002. [DOI] [PubMed] [Google Scholar]
- 56.Mera-Menendez F, Hinojar-Gutierrez A, Guijarro Rojas M, de Gregorio JG, Mera-Menendez E, Sanchez JJ, Quintanilla M, Cerezo L, Gamallo C. Polymorphisms in HIF-1alpha affect presence of lymph node metastasis and can influence tumor size in squamous-cell carcinoma of the glottic larynx. Clin Transl Oncol. 2013;15(5):358–363. doi: 10.1007/s12094-012-0930-z. [DOI] [PubMed] [Google Scholar]
- 57.Ruiz-Tovar J, Fernandez-Contreras ME, Martin-Perez E, Gamallo C. Association of thymidylate synthase and hypoxia inducible factor-1alpha DNA polymorphisms with pancreatic cancer. Tumori. 2012;98(3):364–369. doi: 10.1177/030089161209800314. [DOI] [PubMed] [Google Scholar]
- 58.Horree N, Groot AJ, van Hattem WA, Heintz AP, Vooijs M, van Diest PJ. HIF-1A gene mutations associated with higher microvessel density in endometrial carcinomas. Histopathology. 2008;52(5):637–639. doi: 10.1111/j.1365-2559.2008.02991.x. [DOI] [PubMed] [Google Scholar]
- 59.Nadaoka J, Horikawa Y, Saito M, Kumazawa T, Inoue T, Narita S, Yuasa T, Satoh S, Nishiyama H, Ogawa O, Tsuchiya N, Habuchi T. Prognostic significance of HIF-1 alpha polymorphisms in transitional cell carcinoma of the bladder. Int J Cancer. 2008;122(6):1297–1302. doi: 10.1002/ijc.23256. [DOI] [PubMed] [Google Scholar]
- 60.Jacobs EJ, Hsing AW, Bain EB, Stevens VL, Wang Y, Chen J, Chanock SJ, Zheng SL, Xu J, Thun MJ, Calle EE, Rodriguez C. Polymorphisms in angiogenesis-related genes and prostate cancer. Cancer Epidemiol Biomarkers Prev. 2008;17(4):972–977. doi: 10.1158/1055-9965.EPI-07-2787. [DOI] [PubMed] [Google Scholar]
- 61.Chau CH, Permenter MG, Steinberg SM, Retter AS, Dahut WL, Price DK, Figg WD. Polymorphism in the hypoxia-inducible factor 1alpha gene may confer susceptibility to androgen-independent prostate cancer. Cancer Biol Ther. 2005;4(11):1222–1225. doi: 10.4161/cbt.4.11.2091. [DOI] [PubMed] [Google Scholar]
- 62.Ollerenshaw M, Page T, Hammonds J, Demaine A. Polymorphisms in the hypoxia inducible factor-1alpha gene (HIF1A) are associated with the renal cell carcinoma phenotype. Cancer Genet Cytogenet. 2004;153(2):122–126. doi: 10.1016/j.cancergencyto.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 63.Li H, Bubley GJ, Balk SP, Gaziano JM, Pollak M, Stampfer MJ, Ma J. Hypoxia-inducible factor-1alpha (HIF-1alpha) gene polymorphisms, circulating insulin-like growth factor binding protein (IGFBP)-3 levels and prostate cancer. Prostate. 2007;67(12):1354–1361. doi: 10.1002/pros.20589. [DOI] [PubMed] [Google Scholar]
- 64.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of hypoxia-inducible factor 1alpha in common human cancers and their metastases. Cancer Res. 1999;59(22):5830–5835. [PubMed] [Google Scholar]
- 65.Bardos JI, Ashcroft M. Hypoxia-inducible factor-1 and oncogenic signalling. Bioessays. 2004;26(3):262–269. doi: 10.1002/bies.20002. [DOI] [PubMed] [Google Scholar]
- 66.Vaupel P. The role of hypoxia-induced factors in tumor progression. Oncologist. 2004;9(Suppl 5):10–17. doi: 10.1634/theoncologist.9-90005-10. [DOI] [PubMed] [Google Scholar]
- 67.Zhao T, Lv J, Zhao J, Nzekebaloudou M. Hypoxia-inducible factor-1alpha gene polymorphisms and cancer risk: a meta-analysis. J Exp Clin Cancer Res. 2009;28:159. doi: 10.1186/1756-9966-28-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li D, Liu J, Zhang W, Ren J, Yan L, Liu H, Xu Z. Association between HIF1A P582S and A588T polymorphisms and the risk of urinary cancers: a meta-analysis. PLoS One. 2013;8(5):e63445. doi: 10.1371/journal.pone.0063445. [DOI] [PMC free article] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/14/950/prepub
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: The influence of individual studies on the summary odds ratio (OR) for the HIF-1α C1772T polymorphism. (TIFF 271 KB)


