Abstract
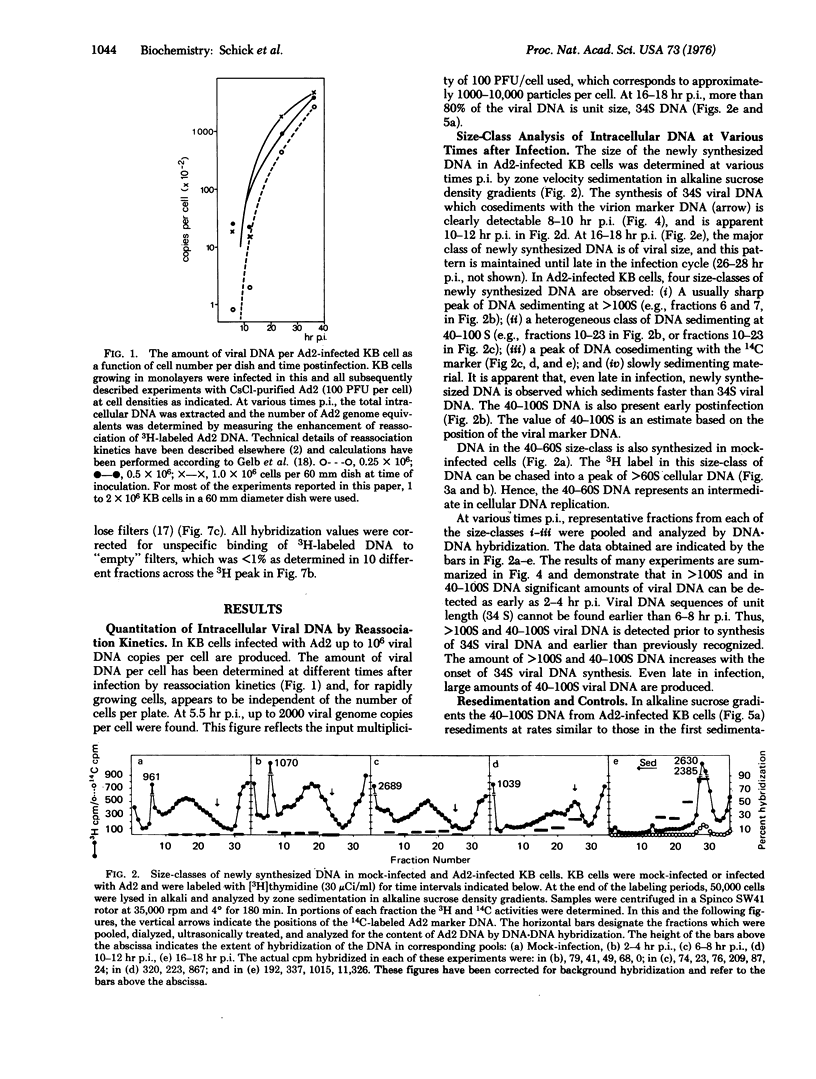
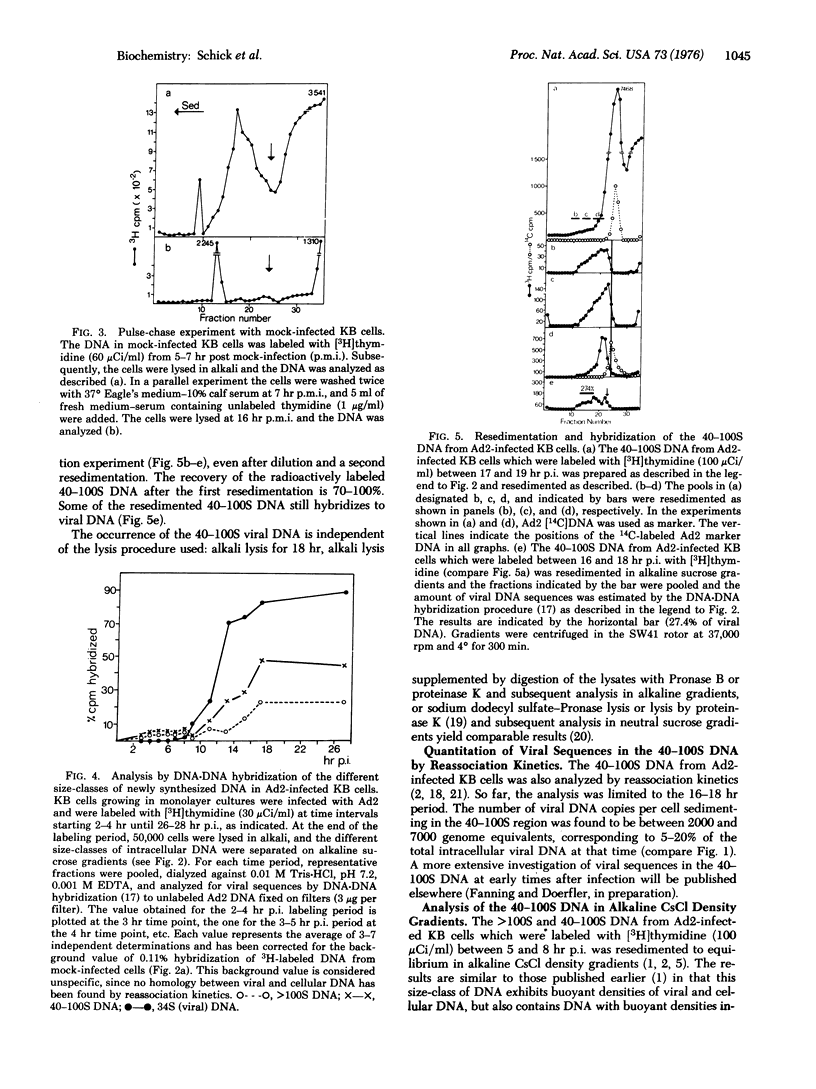
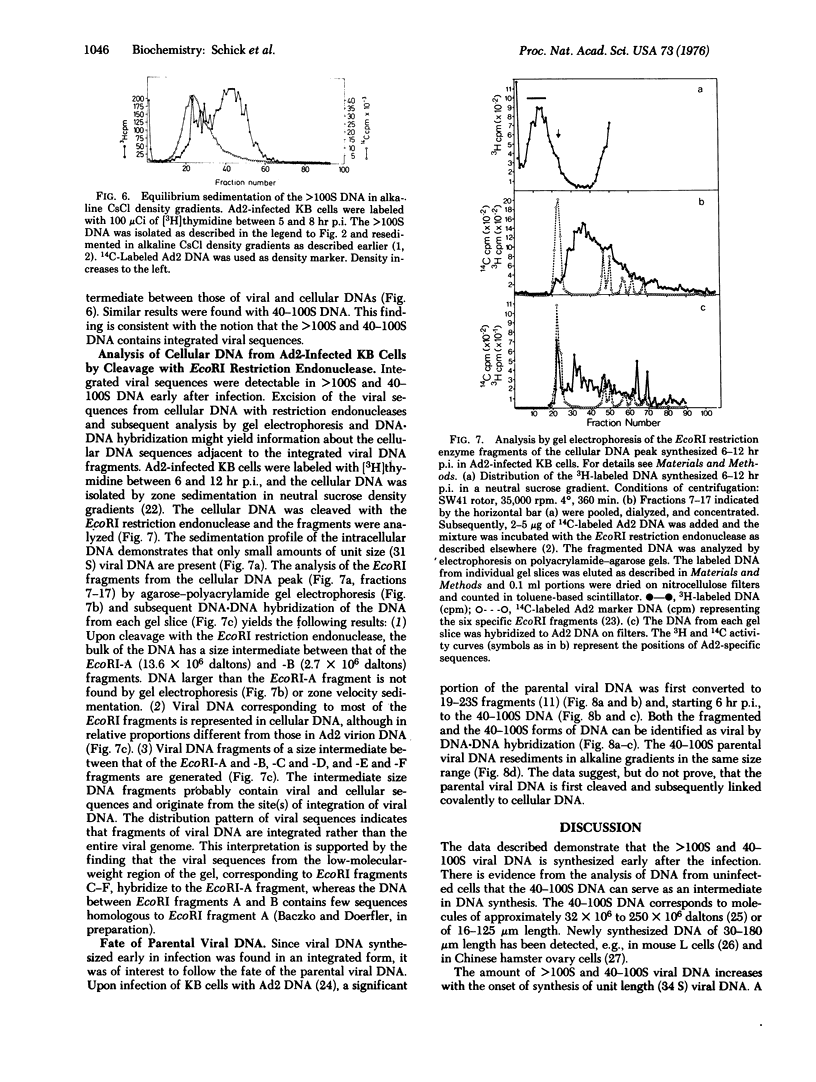
In KB cells productively infected with adenovirus type 2, alkali-stable greater than 100S and 40-100S viral DNAs are synthesized starting 2-4 hr postinfection, i.e., before unit length (34 S) viral DNA is made. The amount of greater than 100S and 40-100S viral DNA increases when 34S viral DNA synthesis begins, and at 16-18 hr postinfection, the 40-100S viral DNA represents 5-20% of the total intracellular viral DNA. The 40-100S viral DNA is synthesized throughout infection. Part of the 40-100S DNA synthesized 5-8 hr postinfection has a density in alkaline CsCl gradients intermediate between those of viral and cellular DNAs. This finding indicates that newly synthesized viral DNA is covalently linked to cellular DNA. Viral sequences can be excised from the cellular DNA of infected cells with the EcoRI restriction endonuclease. Fragments of viral DNA are detected in polyacrylamide-agarose gels by DNA-DNA hybridization, and these fragments correspond in size to most of the known EcoRI fragments of adenovirus 2 DNA. Viral DNA sequences in size-classes between the EcoRI-A and -C fragments are also found and probably represent viral DNA linked to cellular sequences.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bellett A. J. Covalent integration of viral DNA into cell DNA in hamster cells transformed by an avian adenovirus. Virology. 1975 Jun;65(2):427–435. doi: 10.1016/0042-6822(75)90048-3. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Brown D. T., Westphal M., Burlingham B. T., Winterhoff U., Doerfler W. Structure and composition of the adenovirus type 2 core. J Virol. 1975 Aug;16(2):366–387. doi: 10.1128/jvi.16.2.366-387.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger H., Doerfler W. Intracellular forms of adenovirus DNA. 3. Integration of the DNA of adenovirus type 2 into host DNA in productively infected cells. J Virol. 1974 May;13(5):975–992. doi: 10.1128/jvi.13.5.975-992.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlingham B. T., Doerfler W. Three size-classes of intracellular adenovirus deoxyribonucleic acid. J Virol. 1971 Jun;7(6):707–719. doi: 10.1128/jvi.7.6.707-719.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denhardt D. T. A membrane-filter technique for the detection of complementary DNA. Biochem Biophys Res Commun. 1966 Jun 13;23(5):641–646. doi: 10.1016/0006-291x(66)90447-5. [DOI] [PubMed] [Google Scholar]
- Doerfler W., Burger H., Ortin J., Fanning E., Brown D. T., Mestphal M., Winterhoff U., Weiser B., Schick J. Integration of adenovirus DNA into the cellular genome. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):505–521. doi: 10.1101/sqb.1974.039.01.063. [DOI] [PubMed] [Google Scholar]
- Doerfler W. Integration of the deoxyribonucleic acid of adenovirus type 12 into the deoxyribonucleic acid of baby hamster kidney cells. J Virol. 1970 Nov;6(5):652–666. doi: 10.1128/jvi.6.5.652-666.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerfler W. Integration of viral DNA into the host genome. Curr Top Microbiol Immunol. 1975;71:1–78. doi: 10.1007/978-3-642-66193-8_1. [DOI] [PubMed] [Google Scholar]
- Doerfler W. Nonproductive infection of baby hamster kidney cells (BHK21) with adenovirus type 12. Virology. 1969 Aug;38(4):587–606. doi: 10.1016/0042-6822(69)90179-2. [DOI] [PubMed] [Google Scholar]
- Doerfler W. The fate of the DNA of adenovirus type 12 in baby hamster kidney cells. Proc Natl Acad Sci U S A. 1968 Jun;60(2):636–643. doi: 10.1073/pnas.60.2.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EAGLE H. Amino acid metabolism in mammalian cell cultures. Science. 1959 Aug 21;130(3373):432–437. doi: 10.1126/science.130.3373.432. [DOI] [PubMed] [Google Scholar]
- EAGLE H. Propagation in a fluid medium of a human epidermoid carcinoma, strain KB. Proc Soc Exp Biol Med. 1955 Jul;89(3):362–364. doi: 10.3181/00379727-89-21811. [DOI] [PubMed] [Google Scholar]
- Gelb L. D., Kohne D. E., Martin M. A. Quantitation of Simian virus 40 sequences in African green monkey, mouse and virus-transformed cell genomes. J Mol Biol. 1971 Apr 14;57(1):129–145. doi: 10.1016/0022-2836(71)90123-9. [DOI] [PubMed] [Google Scholar]
- Graham F. L., van der Eb A. J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973 Apr;52(2):456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- Groneberg J., Brown D. T., Doerfler W. Uptake and fate of the DNA of adenovirus type 2 in KB cells. Virology. 1975 Mar;64(1):115–131. doi: 10.1016/0042-6822(75)90084-7. [DOI] [PubMed] [Google Scholar]
- Gross-Bellard M., Oudet P., Chambon P. Isolation of high-molecular-weight DNA from mammalian cells. Eur J Biochem. 1973 Jul 2;36(1):32–38. doi: 10.1111/j.1432-1033.1973.tb02881.x. [DOI] [PubMed] [Google Scholar]
- Hand R., Tamm I. Initiation of DNA replication in mammalian cells and its inhibition by reovirus infection. J Mol Biol. 1974 Jan 15;82(2):175–183. doi: 10.1016/0022-2836(74)90339-8. [DOI] [PubMed] [Google Scholar]
- Hozier J. C., Taylor J. H. Length distributions of single-stranded DNA in Chinese hamster ovary cells. J Mol Biol. 1975 Apr 5;93(2):181–201. doi: 10.1016/0022-2836(75)90127-8. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Heterogeneity in deoxyribonucleic acids. I. Dependence on composition of the configurational stability of deoxyribonucleic acids. Nature. 1959 May 23;183(4673):1427–1429. doi: 10.1038/1831427a0. [DOI] [PubMed] [Google Scholar]
- McDongall J. K., Dunn A. R., Gallimore P. H. Recent studies on the characteristics of adenovirus-infected and -transformed cells. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):591–600. doi: 10.1101/sqb.1974.039.01.073. [DOI] [PubMed] [Google Scholar]
- Ortin J., Doerfler W. Transcription of the genome of adenovirus type 12. I. Viral mRNA in abortively infected and transformed cells. J Virol. 1975 Jan;15(1):27–35. doi: 10.1128/jvi.15.1.27-35.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson U., Mulder C., Deluis H., Sharp P. A. Cleavage of adenovirus type 2 DNA into six unique fragments by endonuclease R-RI. Proc Natl Acad Sci U S A. 1973 Jan;70(1):200–204. doi: 10.1073/pnas.70.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwirth B., Tjia S., Westphal M., Doerfler W. Incomplete particles of adenovirus. II. Kinetics of formation and polypeptide composition of adenovirus type 2. Virology. 1974 Aug;60(2):431–437. doi: 10.1016/0042-6822(74)90337-7. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Botchan M., Gallimore P., Ozanne B., Pettersson U., Williams J., Sharp P. A. Viral DNA sequences in cells transformed by simian virus 40, adenovirus type 2 and adenovirus type 5. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 1):615–632. doi: 10.1101/sqb.1974.039.01.075. [DOI] [PubMed] [Google Scholar]
- Zur Hausen H., Sokol F. Fate of adenovirus type 12 genomes in nonpermissive cells. J Virol. 1969 Sep;4(3):256–263. doi: 10.1128/jvi.4.3.256-263.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]


