Lysophosphatidylserines (lyso-PSs) are a class of signaling lipids that regulate (neuro)immunological processes. The metabolism of lyso-PSs remains poorly understood in vivo. Recently, we determined that ABHD12 is a major brain lyso-PS lipase, implicating lyso-PSs in the neurological disease PHARC, which is caused by null mutations in the ABHD12 gene. Here, we couple activity-based profiling with pharmacological and genetic methods to annotate an uncharacterized enzyme ABHD16A as a phosphatidylserine (PS) lipase that generates lyso-PS in mammalian systems. We describe a small-molecule inhibitor of ABHD16A, which depletes lyso-PSs from cells, including PHARC subject-derived lymphoblasts. In mouse macrophages, disruption of ABHD12 and ABHD16A respectively increases and decreases both lyso-PSs and lipopolysaccharide-induced cytokine production. Finally, ABHD16A−/− mice have decreased brain lyso-PSs that run counter to the lyso-PS elevations in ABHD12−/− mice. Our findings illuminate an ABHD16A-ABHD12 axis that dynamically regulates lyso-PS metabolism in vivo, designating these enzymes as potential targets for treating (neuro)immunological disorders.
Lysophospholipids serve as critical small-molecule transmitters that act on both receptors and channels to regulate many facets of mammalian biology and disease1–4. Lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P) represent two well-studied lysophospholipids that produce their biological effects principally through distinct sets of G-protein coupled receptors (GPCRs)1–4. Recently, lysophosphatidylserines (lyso-PSs) have emerged as an additional class of signaling lysophospholipids with activity on Toll-like receptors (TLRs)5 and GPCRs2,6,7 that are highly enriched in immune cells and implicated in human autoimmune disorders8,9. Lyso-PSs have been shown to regulate macrophage activation and clearance of apoptotic cells10, leukemia cell stimulation11, mast cell degranulation12, and chemotactic movements of human glioma cells13.
The enzymes that regulate more established lysophospholipids, such as LPA and S1P, are understood4,14,15; however, the metabolic pathways that generate and terminate lyso-PS signals remain poorly characterized. We recently discovered that the serine hydrolase ABHD12 acts as a major lyso-PS lipase in the mouse brain16. Our interest in ABHD12 stemmed in large part from human genetics studies showing that deleterious mutations in the ABHD12 gene cause the rare autosomal recessive neurological disorder PHARC (Polyneuropathy, Hearing loss, Ataxia, Retinitis pigmentosa, and Cataract)17,18. PHARC is marked by polymodal sensory and motor defects, which are linked to cerebellar atrophy, peripheral neuropathy, early onset of cataract, blindness, and hearing loss, as well as demyelination of sensorimotor neurons. PHARC symptoms progress slowly, first appearing in late childhood or early teens and progressively worsening with age17,18.
ABHD12−/− mice exhibit auditory and motor deficits coupled with elevated brain lyso-PS content and heightened neuroinflammation16, implicating deregulated lyso-PS signaling as a contributory factor to PHARC-like syndromes. Elucidating the enzymes that produce lyso-PSs could thus provide targets for control over these lipid signals to determine their functions in physiology and disease, as well as to potentially treat disorders caused by aberrant lyso-PS activity. While a handful of enzymes have been found to hydrolyze phosphatidylserine (PS) to lyso-PS in vitro19,20, the contribution that these proteins, or others, make to lyso-PS production in vivo remains unknown.
In this paper, we describe the chemical proteomic identification and functional annotation of an uncharacterized integral membrane serine hydrolase ABHD16A [also called HLA-B associated transcript 5 (BAT5)] as a principal PS lipase in mammalian cells and tissues, pharmacological or genetic disruption of which lower lyso-PS and attenuates inflammatory processes that may contribute to PHARC and other (neuro)immunological disorders.
RESULTS
Discovery of ABHD16A as a brain PS lipase
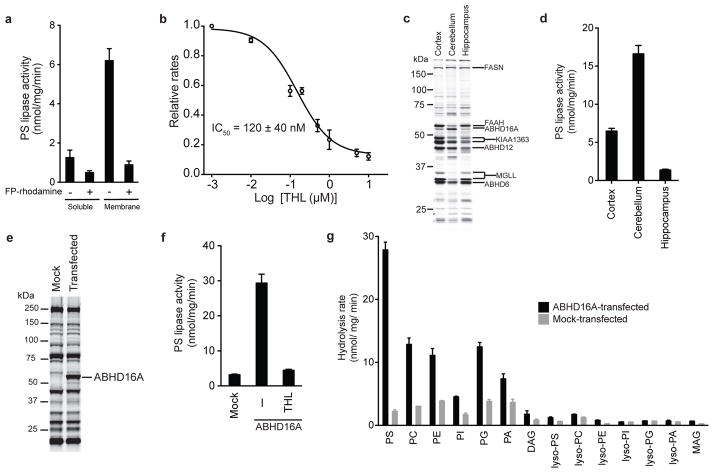
Previous studies have provided biochemical evidence that the brain possesses a PS lipase activity that is distinct in its general properties from other phospholipases (e.g., phosphatidylcholine and phosphatidylinositol lipases) in this tissue21. However, the enzyme responsible for converting PS to lyso-PS in brain has not yet been molecularly characterized. We established an LC-MS assay for monitoring conversion of PS to lyso-PS and determined that the mouse brain proteome possesses an sn-2-selective PS lipase activity that generates lyso-PS and free fatty acid (FFA) products (Supplementary Results, Supplementary Fig. 1). The majority of this activity was membrane-associated (Fig. 1a) and blocked by pre-treatment with serine hydrolase-directed activity-based fluorophosphonate (FP) probes (e.g., FP-rhodamine, FP-biotin22) (Fig. 1a, Supplementary Fig. 1). We also found that the mouse brain PS lipase activity was blocked by the general serine lipase inhibitor tetrahydrolipstatin (THL23; also known as Orlistat) with an IC50 value of 120 ± 40 nM (Fig. 1b).
Figure 1. Identification of ABHD16A as a PS lipase.
(a) PS lipase activity of mouse brain soluble and membrane proteomes after pre-treatment with DMSO or FP-rhodamine (10 μM, 30 min, 37 °C). (b) Concentration-dependent inhibition of brain membrane PS lipase activity by tetrahydrolipstatin (THL). The 95% confidence interval for the reported IC50 value is 40 – 210 nM. (c) ABPP gel of the membrane proteomes (1 mg protein/mL) of the different mouse brain regions treated with FP-rhodamine (2 μM, 30 min, 37 °C). Representative brain serine hydrolases are designated, based on previous assignments24,34. (d) PS lipase activity of membrane proteomes from mouse brain regions. (e) ABPP-gel of membrane proteomes from mock- and ABHD16A-transfected HEK293T cells, showing robust expression and activity of the recombinant mouse ABHD16A enzyme. (f) PS lipase activity of membrane proteomes of mock- and ABHD16A-transfected HEK293T cells. The heightened PS lipase activity of the ABHD16A-transfected proteome was blocked by pre-treatment with THL (20 μM, 30 min, 37 °C) (g) In vitro lipid substrate hydrolysis assays for membrane proteomes of mock and murine ABHD16A transfected HEK293T proteomes. For a, b, d, f, data represent mean values ± s. e. m. for three biological replicates. For g, data represent mean values ± s. e. m. for two biological replicates. See Supplementary Table 1 for complete details of data sets of lipid substrate hydrolysis assays.
THL has previously been shown to inhibit several brain serine hydrolases using competitive activity-based protein profiling (ABPP) assays24,25, including ABHD12 and an uncharacterized 60 kDa membrane enzyme tentatively identified as ABHD16A/BAT5, both of which exhibited IC50 values of ~100 nM24. We have previously demonstrated that ABHD12 does not exhibit PS lipase activity16 and therefore turned our attention to ABHD16A. Consistent with ABHD16A potentially exhibiting PS lipase activity, we found that the cerebellum possessed substantially higher amounts of this enzyme (ABPP analysis; Fig. 1c) and greater PS lipase activity (Fig. 1d) compared to other brain regions. Most other brain serine hydrolases were not enriched in the cerebellum relative to other brain regions (e.g., cortex, hippocampus; Fig. 1c). Large-scale gene expression studies (http://biogps.org) also support the enriched expression of ABHD16A in mouse cerebellum (Supplementary Fig. 1).
We next recombinantly expressed mouse and human ABHD16A by transient transfection in HEK293T cells. Gel-based ABPP confirmed robust expression of ABHD16A proteins in transfected cells compared to mock-transfected counterparts (Fig. 1e and Supplementary Fig. 2). Mouse and human ABHD16A-transfected cell membrane proteomes displayed nearly ten-fold higher PS lipase activity compared to mock-transfected cell membranes, and this heightened activity was blocked by pre-treatment with THL (20 μM, 30 min) (Fig. 1f and Supplementary Fig. 2). We also tested mouse ABHD16A-transfected cell membrane preparations for activity with various mono- and diacyl lipid substrates, which revealed that ABHD16A shows highest specific activity with PS (Km = 40 ± 5 μM and Vmax = 35 ± 3 nmol/ mg/ min for a C18:0/C18:2 PS substrate) and lower activity with other diacylated phospholipids, but negligible activity with lysophospholipids and neutral lipids (Fig. 1g and Supplementary Table 1).
Discovery of a small-molecule inhibitor of ABHD16A
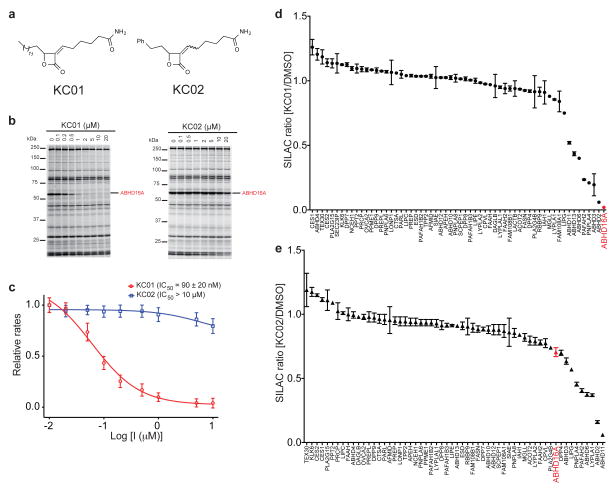
THL inhibits serine hydrolases by covalent reactivity of its electrophilic β-lactone group with active-site serine nucleophiles to form stabilized acyl-enzyme intermediates26. THL can thus serve as a useful scaffold for developing inhibitors with improved activity and/or selectivity for individual serine hydrolases27–29. With this notion in mind, we assayed recombinant human ABHD16A against a focused library of α-methylene-β-lactones or reduced versions of this parent THL-like scaffold30. The compounds (10 μM) were screened by competitive gel-based ABPP and several active ABHD16A inhibitors were identified (Supplementary Fig. 3). Further ABPP experiments designated KC01 (1) (Fig. 2a) as the most potent and selective of the inhibitors for ABHD16A (Fig. 2b and Supplementary Fig. 4). We also generated structural analogues of KC01 (Supplementary Fig. 5), leading to the discovery of an inactive control probe KC02 (2) (Fig. 2a). The synthesis of KC02 consistently yielded a 4:1 mixture of Z/E isomers, which proved difficult to chromatographically separate. We therefore used KC02 as a 4:1 Z/E mixture for our biological experiments. We applied competitive gel-based ABPP to measure IC50 values of ~0.2–0.5 μM and > 10 μM for the inhibition of ABHD16A (human and mouse) by KC01 and KC02, respectively (Fig. 2b and Supplementary Fig. 4). IC50 values were also calculated using a PS substrate assay (KC01 IC50 = 90 ± 20 nM for human ABHD16A; IC50 = 520 ± 70 nM for mouse ABHD16A; KC02 IC50 > 10 μM for both human and mouse ABHD16A; Fig. 2c and Supplementary Fig. 4). We further found that KC01, but not KC02, inhibited the PS lipase activity of brain membrane lysates from two-month old ABHD12+/+ and ABHD12−/− mice (Supplementary Fig. 6).
Figure 2. Identification of an ABHD16A inhibitor and a paired inactive control probe.
(a) Structures of α-methylene-β-lactone probes – the ABHD16A inhibitor KC01 and the inactive control probe KC02. KC02 was synthesized and assayed as a 4:1 mixture of Z:E isomers. (b) Competitive ABPP gels showing the concentration- dependent inhibition of ABHD16A by KC01, but not KC02, in ABHD16A-transfected HEK293T cell proteomes. Proteomes were treated with inhibitors for 30 min at 37 °C, followed by FP-rhodamine (2 μM, 30 min, 37 °C). (c) Concentration-dependent inhibition of the PS lipase activity of the membrane proteome of human ABHD16A-transfected HEK293T cells by KC01 and KC02. Data represent mean values ± s. e. m. for three biological replicates, and the 95% interval for the reported IC50 value of KC01 is 69 – 105 nM. (d, e) ABPP-SILAC analysis of serine hydrolase activities in COLO205 colon cancer cells treated in situ with KC01 (d) or KC02 (e) for 4 h. Data represent mean values ± s.d. for two biological replicates, where each biological replicate value corresponds to the median SILAC ratio for the total quantified peptides observed for each enzyme (minimum of three unique peptides per enzyme required for analysis). See Supplementary Table 3 for complete ABPP-SILAC data sets.
Consistent with the predicted mechanism of covalent inhibition of ABHD16A by KC01, we found that a fluorescent analogue of this inhibitor, termed WHP01, labeled WT-ABHD16A but not the catalytic serine S355A mutant of this enzyme in transfected HEK293T cell lysates (Supplementary Fig. 7). WHP01 also served as a sensitive probe for detecting endogenous ABHD16A activity in mouse brain membrane lysates (Supplementary Fig. 7).
We next tested whether KC01 inhibited ABHD16A in situ. We first confirmed strong ABHD16A activity in the membrane fraction of three human cancer cell lines – COLO205 (colon cancer), K562 (leukemia) and MCF7 (breast cancer) – by the MS method ABPP-MudPIT31 (Supplementary Table 2). We then treated one of these cell lines (K562) with varying concentrations of KC01 for 4 h and analyzed the cell membrane fractions by gel-based ABPP, which confirmed in situ inhibition of ABHD16A with an IC50 value of ~0.3 μM (Supplementary Fig. 8). Very few off-targets were detected for KC01 in this gel-based ABPP experiment. To more thoroughly assess the selectivity of KC01, we performed quantitative MS experiments using the ABPP-SILAC method32. For this study, we investigated the COLO205 cell line, which expresses a rich diversity of serine hydrolases (see Supplementary Table 2). Cells isotopically labeled with light and heavy amino acids were treated with inhibitors (KC01 or KC02, 1 μM) or DMSO for 4 h at 37 °C, lysed, and serine hydrolase activities labeled and enriched with a biotinylated FP probe (FP-biotin33) and analyzed by LC-MS using an LTQ-Orbitrap Velos instrument. Among the 60+ serine hydrolase activities quantified in these experiments, ABHD16A was the most potently inhibited enzyme in KC01-treated cells (98% inhibition; light:heavy ratio of 0.02; Fig. 2d), followed by ABHD2 (94%) and a handful of additional off-targets that were partially inhibited between 50–80% (Fig. 2d and Supplementary Table 3). Most of these enzymes were also inhibited by the control probe KC02, with the exceptions of two partial off-targets of KC01 – ABHD3 and ABHD13 (Fig. 2e and Supplementary Table 3). KC02 also inhibited ABHD11 (94%) and LYPLA1 (63%), but did not substantially inhibit ABHD16A (< 30%; Fig. 2e and Supplementary Table 3). Finally, we found that in situ treatment with KC01 (1 μM, 4 h) but not KC02 (1 μM, 4 h) blocked the PS lipase activity of membrane fractions from COLO205, K562, and MCF7 cell lines (Supplementary Fig. 9).
These data indicate that KC01 and KC02 constitute a suitable pair of active and inactive (control) probes to investigate the function of ABHD16A in cellular systems.
Disruption of ABHD16A impairs cellular PS metabolism
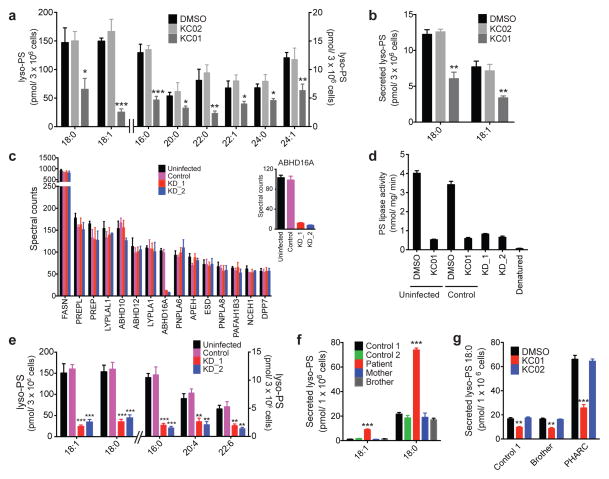
We next performed LC-MS-based lipid profiling of COLO205 cells treated with inhibitors (KC01 or KC02, 1 μM) or DMSO for 4 h and found that KC01-treated cells, but not KC02-treated cells, showed significant reductions in all detected cellular lyso-PSs compared to DMSO-treated control cells (Fig. 3a and Supplementary Table 1). No changes were detected in other lipids, including PSs, PCs, PEs, lyso-PCs, lyso-PEs, or monoacylglycerols (MAGs), in KC01-treated cells (Supplementary Table 1). Secreted lyso-PSs (18:1 and 18:0) were also decreased in COLO205 cells treated with KC01 compared to KC02- or DMSO-treated cells (4 h treatments in serum-free media; Fig. 3b), while other secreted lipids (lyso-PCs, lyso-PEs, MAGs) were unchanged across these treatment groups (Supplementary Table 1). Similar changes in cellular and secreted lyso-PSs were observed in K562 (Supplementary Fig. 10 and Supplementary Table 1) and MCF7 (Supplementary Figure 11 and Supplementary Table 1) cells treated with KC01, but not KC02.
Figure 3. Disruption of ABHD16A reduces the lyso-PS content of human cells.
(a, b) Cellular (a) and secreted (b) concentrations of lyso-PS from COLO205 colon cancer cells treated in situ with inhibitors (KC01 or KC02, 1 μM) or DMSO for 4 h. See Supplementary Table 1 for complete list of quantified lipids. (c) Spectral count values of serine hydrolase activities of the membrane proteomes of the indicated cell models as measured by ABPP-MudPIT. Inset shows a blow-up of the ABHD16A spectral counts. Shown are the 15 most abundant serine hydrolase activities. See Supplementary Table 2 for the complete list of serine hydrolase activities. (d) PS lipase activities of membrane proteomes from the indicated K562 cell lines. Uninfected and control cell line proteomes were pre-treated with DMSO or KC01 (1 μM, 30 min, 37 °C) to establish PS lipase activities for baseline and ABHD16A-inhibited samples. The uninfected proteome was also heat-denatured prior to analysis as an additional control. (e) Cellular concentrations of lyso-PS from the indicated K562 cell lines. (f) Secreted concentrations of lyso-PS from the indicated lymphoblast cell lines (LCLs). (g) Concentrations of secreted C18:0 lyso-PS from the indicated LCLs treated with DMSO or inhibitor (KC01 or KC02; 1 μM; 4 h). Data represent mean values ± s. e. m for three-eight biological replicates. Student’s t-test: * p < 0.05; **p < 0.01; *** p < 0.0001 for experimental versus control groups.
As a complement to our pharmacological studies, we also knocked down ABHD16A expression in K562 cells using RNA-interference technology. We generated two stable shRNA knockdown lines targeting ABHD16A (KD_1 and KD_2) and a control line expressing a scrambled shRNA probe and confirmed near complete (~90%) and selective reductions in ABHD16A in the KD_1 and KD_2 lines compared to uninfected or control lines by ABPP-MudPIT analysis (Fig. 3c and Supplementary Table 2). None of the other 35+ serine hydrolase activities measured in these experiments were affected in the KD_1 and KD_2 lines. ABHD16A knockdown greatly reduced the PS lipase activity of K562 cells to levels observed in uninfected cells treated with KC01 (Fig. 3d). Lipid profiling revealed that cellular lyso-PSs were substantially decreased in the KD_1 and KD_2 lines compared to uninfected or control lines (Fig. 3e). Notably, several PSs were conversely elevated in the KD_1 and KD_2 lines (Supplementary Fig. 12), while other major lipids (PCs, PEs, lyso-PCs, lyso-PEs, and MAGs) were unchanged (Supplementary Table 1). It was not possible to analyze secreted lipids in the shRNA lines, as they showed poor viability when cultured in serum-free media.
ABHD16A-ABHD12 interplay regulates cellular lyso-PS
We next asked whether ABHD16A inhibition altered lyso-PS metabolism in a lymphoblast cell line (LCL) derived from a PHARC subject with compound heterozygotic mutations in the ABHD12 gene18. Five LCLs were investigated, including lines derived from the PHARC subject, their sibling, who possessed a wild-type ABHD12 gene, their mother, who was heterozygotic for ABHD12 gene mutation, and two control subjects with wild-type ABHD12 status. Selective absence of ABHD12 in the PHARC subject-derived LCL was confirmed by gel-based ABPP, as shown previously18, and the membrane lysate from this ABHD12-null LCL also exhibited substantial reductions in lyso-PS lipase activity (Supplementary Fig. 13). The LCL lysate from the ABHD12-heterozygotic mother showed a 50% reduction in lyso-PS lipase activity compared to wild-type LCL controls (Supplementary Fig. 13). The PS lipase activity was comparable across all of the LCL samples and was inhibited by KC01, but not KC02 (Supplementary Fig. 13). These results indicated that ABHD12 and ABHD16A are major lyso-PS and PS lipases, respectively, in the human LCLs.
Targeted LC-MS analysis revealed a dramatic elevation in secreted lyso-PSs in the ABHD12-null LCL compared to control LCLs (Fig. 3f and Supplementary Fig.14). Other secreted and cellular (lyso)phospholipids were unchanged in the ABHD12-null LCL (Supplementary Table 1). Treatment with KC01, but not KC02 (1 μM, 4 h) significantly lowered secreted lyso-PSs of the ABHD12-null LCL, such that they nearly matched the concentrations of secreted lyso-PSs of control LCLs, which were also reduced by KC01 treatment (Fig. 3g). All LCLs also showed decreased cellular lyso-PSs following treatment with KC01, but not KC02 (Supplementary Fig. 14). We confirmed inhibition of ABHD16A in LCLs by KC01, but not KC02 by gel-based ABPP (Supplementary Fig. 14).
Our data indicate that ABHD12 and ABHD16A play complementary roles in the regulation of lyso-PSs in human LCLs, with ABHD16A contributing to the production of both cellular and secreted lyso-PSs and ABHD12 preferentially controlling degradation of secreted lyso-PSs. We have previously shown that ABHD12 is an extracellularly/lumenally-oriented membrane protein34, a localization that is consistent with the enzyme’s role in regulating secreted lyso-PSs. We examined here the membrane orientation of ABHD16A using a protease protection assay35, which revealed that recombinant ABHD16A expressed in HEK293T cells was susceptible to trypsin degradation in the absence of a membrane-solubilizing detergent Triton X-100 (Supplementary Fig. 15). This property is consistent with a cytoplasmic membrane orientation for ABHD16A and contrasted with the behavior of recombinant ABHD12, which showed detergent-dependent sensitivity to trypsin degradation (Supplementary Fig. 15). Also consistent with the respective cytoplasmic and extracellular/lumenal orientations of ABHD16A and ABHD12, only the latter enzyme shows evidence of PNGaseF-sensitive glycosylation (Supplementary Fig. 16 and ref. 34). The localization of ABHD16A on the cytoplasmic face of cellular membranes is consistent with its role as a PS lipase, since PS is largely confined to the inner leaflet of the lipid bilayer in healthy cells36. Although the mechanism(s) that regulate the cellular release of lyso-PS remain unknown, our data argue that, once generated by ABHD16A and secreted from cells, lyso-PS becomes accessible to hydrolytic degradation by ABHD12.
Analysis of ABHD16A/ABHD12-lyso-PS pathway in macrophages
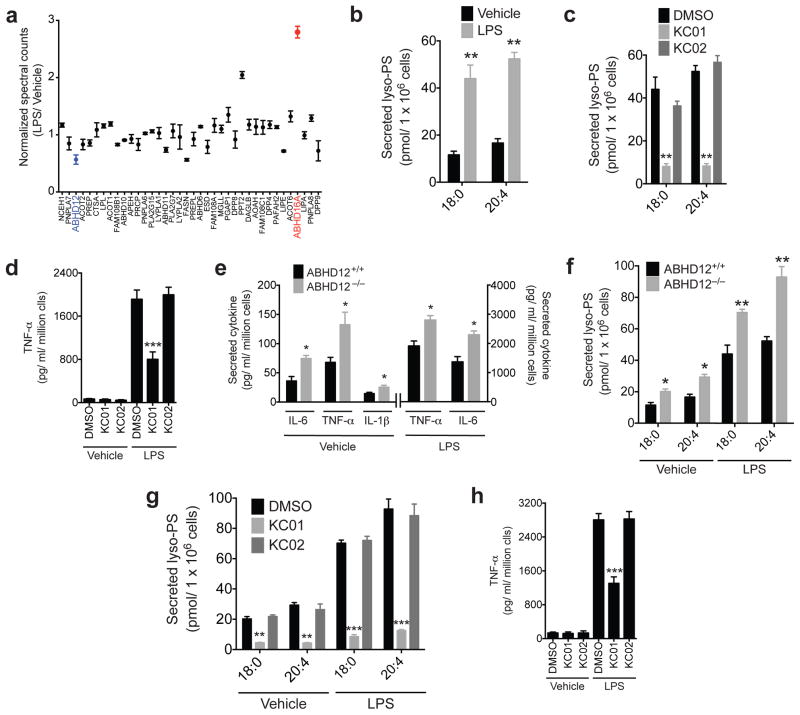
The heightened neuroinflammatory state of ABHD12−/− mice16, combined with the immunoregulatory functions of lyso-PS10,12, motivated us to explore the functions of ABHD16A and ABHD12 in immune cells. A survey of public mRNA expression profiles (http://biogps.org/) revealed that ABHD16A and ABHD12 are differentially regulated by lipopolysaccharide (LPS) treatment of mouse peritoneal macrophages (Supplementary Fig. 17). We verified and extended these findings at the proteomic level by performing a comparative ABPP analysis of thioglycollate-elicited mouse peritoneal macrophages treated with or without LPS for 7 h. Strikingly, among the 40+ serine hydrolases quantified in this study, ABHD16A showed the greatest increase (~three-fold) in activity, while ABHD12 was one of a handful of serine hydrolases that showed reductions (~twofold) in activity (Fig. 4a and Supplementary Table 2). We measured a corresponding elevation in PS lipase activity (Supplementary Fig. 17) and reduction in lyso-PS lipase activity (Supplementary Fig. 17) in LPS-stimulated macrophages, and the former activity was blocked by KC01, but not KC02 (Supplementary Fig. 17). The complementary shift in activities for ABHD16A and ABHD12 in LPS-stimulated macrophages resulted in a significant increase in both cellular (Supplementary Fig. 18) and secreted (Fig. 4b) lyso-PSs. Other cellular and/or secreted (lyso)phospholipids were unchanged by LPS treatment (Supplementary Table 1). Both basal and LPS-induced increases in cellular (Supplementary Fig. 18) and secreted lyso-PSs (Fig. 4c and Supplementary Fig. 19) were reduced by pre-treatment with KC01 (1 μM, 4 h), but not KC02 (1 μM, 4 h).
Figure 4. ABHD16A-ABHD12 interplay regulates secreted lyso-PS and cytokine release from macrophages.
(a) Normalized spectral count ratios for serine hydrolase activities detected in an ABPP-MudPIT analysis of the membrane fractions of thioglycollate-elicited peritoneal macrophages treated with vehicle (PBS) or LPS (5 μg/mL for 7 h). (b) Concentrations of secreted lyso-PS from thioglycollate-elicited peritoneal macrophages, treated with vehicle (PBS) or LPS (5 μg/mL) for 7 h. (c, d) Concentrations of secreted lyso-PS (c) and TNF-α (d) for thioglycollate-elicited peritoneal macrophages treated with DMSO or inhibitor (KC01 or KC02, 1 μM) for 4 h followed by stimulation with LPS (5 μg/mL, 7 h). (e) Concentrations of secreted cytokines from ABHD12+/+ and ABHD12−/− thioglycollate-elicited peritoneal macrophages that were either unstimulated (PBS, vehicle) or stimulated with LPS (5 μg/mL, 7 h). (f) Concentrations of secreted lyso-PS from ABHD12+/+ and ABHD12−/− macrophages treated with vehicle (PBS) or LPS (5 μg/mL) for 7 h. (g, h) Concentrations of secreted lyso-PS (g) and TNF-α (h) from ABHD12−/− macrophages treated with inhibitors (KC01 or KC02, 1 μM) or DMSO for 4 h followed by treatment with vehicle (PBS) or LPS (5 μg/mL) for 7 hours. Data represent mean values ± s. e. m. for four biological replicates. Student’s t-test: * p < 0.05, ** p < 0.0005, *** p < 0.0001 for experimental versus control groups.
LPS, through activation of innate immune receptors, promotes the release of pro-inflammatory cytokines from macrophages37. We found that this LPS-induced cytokine release was significantly blunted by pre-treatment of macrophages with KC01, but not KC02 (Fig. 4d and Supplementary Fig. 20). Conversely, we found that ABHD12−/− macrophages produced greater amounts of cytokines, both basally and in response to LPS stimulation (Fig. 4e). These cytokine changes correlated with heightened secretion of lyso-PSs from ABHD12−/− macrophages (Fig. 4f). The LPS-induced increases in lyso-PSs (Fig. 4g and Supplementary Fig. 21) and cytokine (Fig. 4h and Supplementary Fig. 22) secretion were both blocked by pre-treatment of ABHD12−/− macrophages with KC01 (1 μM, 4 h), but not KC02 (1 μM, 4 h). As was observed in ABHD12-null LCL cells, ABHD12−/− macrophages did not show alterations in cellular lyso-PS content (Supplementary Fig. 23). Finally, we found that C18:0 lyso-PS, which represented one of the most abundant lyso-PS species secreted by macrophages, promoted cytokine release when added exogenously to macrophages (Supplementary Fig. 24).
ABHD16A is a principal PS lipase in vivo
While active in cells, our first-generation ABHD16A inhibitors were not suitable for animal studies, presumably due to their limited bio-distribution and/or stability in vivo. To more directly address the role of ABHD16A in vivo, we established an ABHD16A−/− mouse model (Supplementary Fig. 25). ABHD16A−/− mice were born at a much lower frequency than expected for Mendelian distribution (54 ABHD16A+/+ pups, 87 ABHD16A+/− pups, 10 ABHD16A−/− pups) and were ~30% smaller than ABHD16A+/+ or +/− mice throughout development and life (Supplementary Fig. 26). Despite their smaller size, ABHD16A−/− mice appeared normal in their cage behavior, and we did not observe evidence of increased postnatal lethality in these animals. RT-PCR and ABPP analysis of brain tissue confirmed loss of ABHD16A mRNA expression (Fig. 5a) and protein activity (detected by either the FP-rhodamine (Fig. 5b) or WHP01 (Supplementary Fig. 27) probe) in ABHD16A−/− mice, respectively. We also confirmed the loss of ABHD16A activity by combining ABPP with a quantitative MS-based proteomic analysis using reductive dimethylation38,39 methods (see Supplementary Methods for more details), which revealed complete absence of ABHD16A signals in ABHD16A−/− brains and no changes in the signals for the 40+ other brain serine hydrolases detected in this analysis (Supplementary Fig. 28).
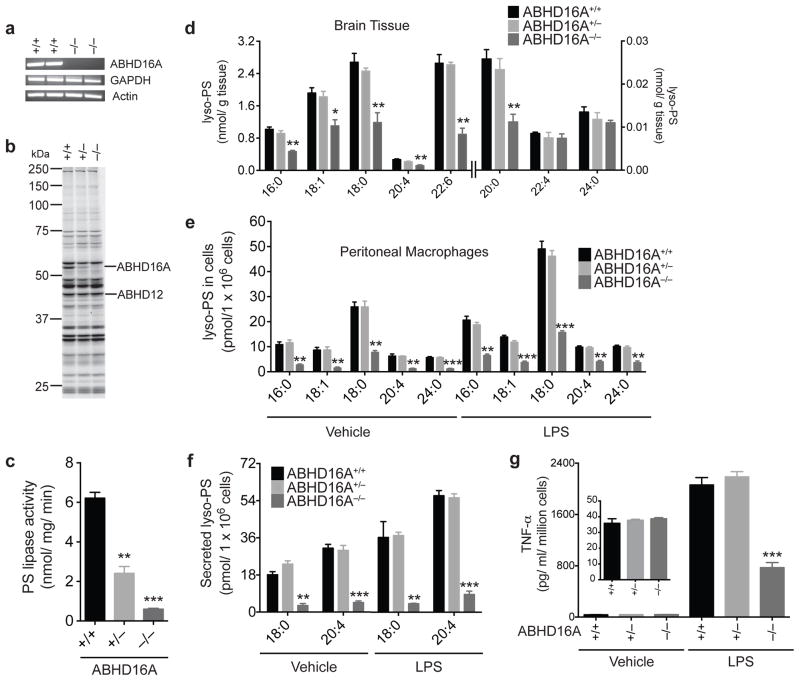
Figure 5. Generation and characterization of ABHD16A−/− mice.
(a, b) Confirmation of absence of ABHD16A mRNA and protein activity in brain tissue from ABHD16A−/− mice using RT-PCR (a) and ABPP (b) analyses, respectively. ABPP gel represents treatment of cerebellar membrane proteomes ABHD16A+/+, +/−, and −/− mice with FP-rhodamine (2 μM, 30 min, 37 °C). (c) PS lipase activity of brain membrane lysates from ABHD16A +/+, +/−, and −/− mice. Data represent mean values ± s. e. m. for three biological replicates. Student’s t-test: ** p < 0.0005, *** p < 0.0001 for ABHD16A−/− versus ABHD16A+/+ groups. (d) Concentrations of lyso-PS from brain tissue of ABHD16A+/+, +/−, and −/− mice. Data represent mean values ± s. e. m. for four biological replicates. Student’s t-test: * p < 0.05, ** p < 0.005, for ABHD16A−/− versus ABHD16A+/+ groups. (eg) Concentrations of cellular (e) and secreted (f) lyso-PS and TNF-α (g) from thioglycollate-elicited peritoneal macrophages derived from ABHD16A+/+, +/−, and −/− mice after treatment with vehicle (PBS) or LPS (5 μg/mL) for 7 h. Data represent mean values ± s. e. m. for four biological replicates Student’s t-test: ** p < 0.0005, *** p < 0.0001, ABHD16A−/− versus ABHD16A+/+ groups.
The PS lipase activity of brain membrane lysates from ABHD16A−/− mice was greatly decreased compared to ABHD16A+/+ and +/− lysates (Fig. 5c). We next evaluated the brain lipid profiles for ABHD16A+/+, +/−, and −/− mice and found that ABHD16A−/− mice exhibited substantial reductions in most of the measured lyso-PSs (Fig. 5d). Similar reductions in lyso-PSs were found in spinal cord of ABHD16A−/− mice (Supplementary Fig. 29), which also exhibited lower PS lipase activity compared to ABHD16A+/+ mice (Supplementary Fig. 29). No changes in CNS lyso-PSs were detected in ABHD16A+/− mice (Fig. 5d and Supplementary Fig. 29). We did not observe changes in PSs or other (lyso)phospholipids in CNS tissues from ABHD16A−/− mice (Supplementary Table 1).
We also examined PS metabolism in thioglycollate-elicited peritoneal macrophages from ABHD16A+/+, +/−, and −/− mice. ABPP analysis with the WHP01 probe confirmed loss of ABHD16A activity in ABHD16A−/− macrophages, which also showed greatly reduced PS lipase activity that was no longer elevated by LPS treatment (Supplementary Fig. 30). ABHD16A−/− macrophages exhibited a corresponding ~80% reduction in cellular (Fig. 5e) and secreted (Fig. 5f) lyso-PSs under either basal or LPS-stimulated conditions. No changes in PSs, free fatty acids, eicasonoids (PGE2, PGD2, LTB4 or TXB2) or other (lyso)phospholipids were detected in ABHD16A−/− macrophages when compared to the ABHD16A+/+ or +/− macrophages (Supplementary Table 1). We also assessed the cytokine profiles of ABHD16A+/+, +/−, and −/− macrophages and found that LPS-induced cytokine release was substantially blunted in ABHD16A−/− macrophages (Fig. 5g and Supplementary Fig. 31). Basal cytokine profiles were unaffected in ABHD16A−/− macrophages (Fig. 5g, inset and Supplementary Fig. 31). To gain further confidence that KC01 produced its pharmacological effects by blocking ABHD16A, we treated macrophages from ABHD16A−/− mice with this inhibitor and KC02. We observed no changes under basal conditions or LPS stimulation in cellular or secreted lyso-PS and other measured lipids, or in secreted proinflammatory cytokines (IL-6 and TNF-α), in either KC01- or KC02-treated ABHD16A−/− macrophages (Supplementary Fig. 32 and Supplementary Table 1).
Discussion
The important roles that PS plays in mammalian biology36,40, along with the more recent emergence of lyso-PS2 as a signaling molecule, has stimulated interest in understanding the enzymatic formation and degradation of these lipids. PS can be metabolized by phospholipases, which hydrolyze the sn-1 or sn-2 acyl chains (PLA1 and PLA2 enzymes, respectively) to generate lyso-PS, and decarboxylases to form phosphatidylethanolamine36,40. At least one phospholipase – PS-PLA1 – has previously been reported to preferentially accept PS as a substrate in vitro19,20. PS-PLA1 is, however, a secreted enzyme and therefore unlikely to have access to bulk PS content in cells19,20, which is mostly localized to the inner leaflet of the membrane bilayer36. Consistent with this conclusion, PS-PLA1 has been shown to more efficiently release lyso-PS from cells undergoing apoptosis19, which results in exposure of PS to the outer leaflet of the bilayer. The intracellular enzymatic pathways that regulate lyso-PS production have, on the other hand, remained a mystery. Here, we provide compelling pharmacological and genetic evidence that ABHD16A is a major enzyme responsible for generating lyso-PSs in mammalian cells and in vivo. That PS lipids were, for the most part, unchanged in ABHD16A-disrupted cells or tissues could indicate alternative metabolic routes (e.g., PS decarboxylation) control bulk PS content in vivo. One exception was the K562 cells, where chronic ABHD16A depletion by RNA-interference resulted in, not only reductions in lyso-PS content, but also elevations in PSs. It therefore remains possible that, in certain cell types or under specific conditions, ABHD16A could coordinately regulate both PS and lyso-PS lipids.
ABHD16A was originally named BAT5, a designation that reflects the location of the ABHD16A gene within the HLA-B region of the human genome, which harbors several other genes that code for proteins that play important roles in immunology (e.g., TNFα)41,42. Our data indicate that ABHD16A, along with its lyso-PS products, and the lyso-PS lipase ABHD12, also function in immunological processes by forming a lipid signaling network that is dynamically regulated by, and in turn contributes to, macrophage inflammatory responses. While we do not yet know the precise receptors involved in transmitting lyso-PS signals in macrophages (or the CNS), previous studies invoke both innate immune receptors5 and GPCRs2,6,7 as candidates. We should also note that, while our data and other studies12 point to an immunostimulatory function for lyso-PSs, other investigations have uncovered an additional role for these lipids as resolving signals in inflammation10, 43.
Several of our findings provide evidence for an interplay between ABHD16A and ABHD12 in the potential regulation of PHARC. First, the ABHD16A inhibitor KC01 reversed the elevated lyso-PS production observed in ABHD12-null cells derived from a PHARC subject. ABHD16A−/− mice also showed lower lyso-PS content in the CNS that ran counter to the heightened lyso-PS profile of ABHD12−/− mice16. We further note that the cerebellum, which was the brain region that exhibits the most dramatic changes in lyso-PSs and microglial activation in ABHD12−/− mice16 and undergoes atrophy in PHARC patients17, is enriched in ABHD16A expression (Fig. 1c). Nonetheless, crosses of ABHD16A−/− and ABHD12−/− mice will be needed to provide a model system that directly addresses whether disruption of ABHD16A can rectify the elevated lyso-PS content and neuroinflammatory and behavioral phenotypes of ABHD12-disrupted animals.
Projecting forward, further studies are needed to elucidate the broader functions of ABHD12 and ABHD16A in mammalian biology. We, for instance, do not understand the basis for the presumed prenatal lethality that results in sub-Mendelian production of ABHD16A−/− mice or the smaller size of these animals compared to their wild-type littermates. Other proteins involved in lysophospholipid signaling, such as LPA and S1P receptors, perform different functions in development and mature organisms3,44, and the latter activities constitute focal points for therapies to treat fibrosis45 and multiple sclerosis3,4. Our findings designate the ABHD16A/ABHD12-lyso-PS pathway as an emerging lysophospholipid signaling network with its own potential therapeutic relevance for PHARC and other (neuro)immunological diseases.
Online Methods
Preparation of mouse tissue proteomes
Mice were anaesthetized with isoflurane and sacrificed by cervical dislocation. CNS tissues (half-brain or spinal cord) were suspended in 0.5 mL of cold PBS and homogenized using a Bullet Blender 24 (Next Advance) using 1 scoop of 0.5 mm diameter glass beads (Next Advance) at a speed setting of 8, for 3 minutes at 4 °C. Thereafter 0.5 mL of cold PBS was added to the homogenates, and mixed by pipetting 8–10 times, and centrifuged at 1400g for 5 minutes at 4 °C to separate tissue debris and glass beads from the proteome. The supernatant (~ 750 μL) was separated and centrifuged at 16,000g for 45 minutes at 4 °C. The supernatant (soluble proteome) was saved and the pellet (membrane proteome) washed with cold PBS (3X), and thereafter re-suspended by pipetting in cold PBS. All protein concentrations were measured using DC Protein Assay Kit (BioRad). All mouse studies were performed following protocols that received approval from The Scripps Research Institute-Institutional Animal Care and Use Committee Office.
Gel-based ABPP analysis
Tissue and cell proteomes (1 mg/mL, 100 μL) were treated with FP-rhodamine (2 μM) for 30 minutes at 37 °C with constant shaking. The reactions were quenched by adding 4X SDS-PAGE loading buffer (40 μL) followed by boiling for 2 minutes. Competitive gel-based ABPP were performed as described previously32. All samples were visualized in-gel using an FMBio II Multiview flatbed fluorescence scanner (Hitachi).
Recombinant expression of ABHD16A
HEK293T cells were grown to 40% confluence in DMEM (25 mL, 15 cm dish) (Caisson Labs) supplemented with L-glutamine (2 mM) (Cellgro), 10% FCS (Gemini) at 37 °C and 5% CO2. The cells were transiently transfected with the full-length cDNA of human or mouse ABHD16A (Open Biosystems) in pCMV-SPORT6 vector using polyethyleneimine “MAX” (MW 40,000) (PEI) (Polysciences Inc.) as the transfection reagent. “Mock” control cells were transfected with an empty vector. 48 hours after transfection, cells were harvested by scraping, washed with PBS (3X), re-suspended in 1 mL of PBS and lysed by sonication.
Substrate hydrolysis assays
20 μg of proteome was incubated with 100 μM lipid substrate (Avanti Polar Lipids) in PBS (100 μL total volume) at 30 °C with constant shaking. After 30 minutes, the reaction was quenched with 350 μL of 2:1 CHCl3: MeOH and an internal standard (0.5 nmol) was added. The mixture was vortexed, and centrifuged at 2800g to separate the aqueous (top) and organic (bottom) phase. The aqueous phase was treated with formic acid (5 μL) and extracted again with 2:1 CHCl3: MeOH. The organic phases were pooled, dried under a stream of N2, and re-solubilized in 150 μL of 2:1 CHCl3: MeOH for MS analysis. A fraction of the organic extract was injected onto an Agilent 6520 quadrupole-time-of-flight (QTOF) LC/MS. LC separation was achieved using a Gemini 5U C-18 column (Phenomenex). The LC solvents were: buffer A: 95:5 = H2O: MeOH + 0.1% NH4OH, and buffer B: 60:35:5 = iPrOH: MeOH: H2O + 0.1% NH4OH. A typical LC run consisted of 15 minutes post-injection: 0.1 mL/min 100% buffer A from for 1.5 minutes, 0.5 mL/min linear gradient to 100% buffer B over 5 minutes, 0.5 mL/min 100% buffer B for 5.5 minutes, and equilibration with 0.5 mL/min 100% buffer A for 3 minutes. All MS analysis was performed using an electrospray ionization source (ESI) in negative ion mode for product formation. The following parameters were used for the ESI-MS analysis: drying gas temperature: 350 °C, drying gas flow rate: 11 mL/ min, fragmentor voltage: 100 V, capillary voltage: 4 kV, and nebulizer pressure: 45 psi. Measuring the area under the peak, and normalizing it to the internal standard quantified the product release for the lipid substrate hydrolysis assays. The substrate hydrolysis rate was corrected by subtracting the non-enzymatic rate of hydrolysis, which was obtained by using heat-denatured (15 minutes at 95 °C) control proteomes. Some products from the substrate hydrolysis assays were detected in positive ion mode for qualitative identification. All the MS parameters for the positive ion mode experiments, and LC separation conditions were the same except for the buffers, which contained 0.1% formic acid instead of 0.1% NH4OH.
In situ treatment of human cell lines
The cancer cell lines (COLO205, K562, and MCF7) and LCLs used in this study were cultured in RPMI 1640 (Caisson Labs) supplemented with 10% FCS and 1X penicillin-streptomycin at 37 °C with 5% CO2. 3 x 106 cells were washed with sterile PBS (3X), and treated with KC01 or KC02 (1 μM) or DMSO for 4 h at 37 °C and 5% CO2 in 4 mL of serum free phenol red free RPMI 1640 (Invitrogen). K562 cells could not be cultured without 10% FCS, and were treated using above mentioned compounds in phenol red free RPMI 1640 supplemented with 10% FCS. After treatment, the media was collected for measuring secreted lipids, and the cells were washed with sterile PBS (3X) and harvested for enzyme activity or lipid measurements. The samples were either processed immediately or flash frozen in liquid N2 and stored at −80 °C until further use.
ABPP-MudPIT and ABPP-SILAC sample preparation and analysis
For the ABPP-MudPIT samples, proteomes (1 mg/mL in 1 mL of PBS) were labeled with FP-biotin (5 μM) for 45 minutes at 37 °C with constant shaking. After labeling, the proteomes were denatured and precipitated using 4:1 MeOH: CHCl3, re-suspended in 0.5 mL of 6 M urea in PBS, reduced using tris(2-carboxyethyl)phosphine (TCEP) (10 mM) for 30 minutes at 37 °C, and then alkylated using iodoacetamide (40 mM) for 30 minutes at 25 °C in the dark. The biotinylated proteins were enriched with PBS-washed avidin-agarose beads (100 μL) (Sigma-Aldrich) by shaking at 25 °C for 1.5 hours in PBS with 2% SDS to final volume of 5.5 mL. The beads were washed sequentially with 10 mL PBS with 0.2% SDS (3X), 10 mL PBS (3X), and 10 mL DI H2O (3X). On-bead digestion was performed using sequence-grade trypsin (2 μg) (Promega Catalog) in 2 M urea in PBS with 2 mM CaCl2 for 12 – 14 hours at 37 °C (200 μL). Peptides obtained from this procedure were acidified using formic acid (5%) and stored at -80 °C prior to analysis. All SILAC experiments were performed using the human COLO205 colon cancer cell line generated by twelve passages in either light (100 μg/mL each of L-arginine, and L-lysine) or heavy (100 μg/mL each of [13C615N4] L-arginine and [13C615N2] L-lysine ) SILAC RPMI medium (Thermo Scientific) supplemented with 10% dialyzed FCS. Light and heavy cells were treated with the test compounds or DMSO, respectively for 4 h at 37 °C. Cells were then washed with sterile PBS (3X), harvested, and lysed by sonication in PBS. 2 mg/mL light proteome (0.5 mL) and 2 mg/mL heavy proteome (0.5 mL) were then treated with FP-biotin (10 μM), combined, and thereafter processed similar to the ABPP-MudPIT protocol described above. “Probe-versus-probe” and “probe-versus-no probe” control experiments were performed (Supplementary Table 3) to confirm the full incorporation of heavy-isotopic label and enrichment of active serine hydrolases.
ABPP-reductive dimethylation (ReDiMe)
Mouse half-brain membrane proteomes and ABPP-MudPIT samples were prepared as described above with minor adjustments. The final wash steps and trypsin digestion were performed in 100 mM triethylammonium bicarbonate buffer in preparation for downstream reductive dimethylation labeling. Reductive dimethylation was performed as previously described46. Briefly, either 13C –labeled deuterated formaldehyde (heavy) or formaldehyde (light) was added to each sample (0.15%) followed by addition of sodium cyanoborohydride (22.2 mM). Following a 1 h incubation period at room temperature, the reaction was quenched by addition of NH4OH (0.23%) and formic acid (0.5%). The samples were then combined and analyzed by LC-MS analysis.
Mass spectrometry and data analysis
Mass spectrometry was performed using a LTQ (for spectral counting studies), an LTQ-Orbitrap (for SILAC studies), or an Orbitrap Velos (for ReDiMe studies) following previously described protocols (ThermoFinnigan)31,47. Peptides were eluted using a five-step multidimensional LC-MS protocol in which increasing concentrations of ammonium acetate are injected followed by followed by a gradient of increasing acetonitrile, as previously described48. For all samples, data were collected in data-dependent acquisition mode over a range from 400–1800 m/z. Each full scan was followed by up to 7- or 30- fragmentation events for experiments utilizing the LTQ and Orbitrap or Orbitrap Velos instruments, respectively. Dynamic exclusion was enabled (repeat count of 1, exclusion duration of 20 s) for all experiments. The data were searched using the ProLuCID algorithm, against a mouse or human reverse-concatenated nonredundant (gene-centric) FASTA database that was assembled from the Uniprot database. ProLuCID searches specified static modification of cysteine residues (+57.0215 m/z; iodoacetamide alkylation) and required peptides to contain at least one tryptic terminus. For ReDiMe samples, each data set was independently searched with light and heavy parameter files; for the light search, static modifications on lysine (+ 28.0313 m/z) and N-termini (+ 28.0313 m/z) were specified; for the heavy search, static modifications on lysine (+ 34.06312 m/z) and N-termini (+ 34.06312 m/z) were specified. For SILAC samples, datasets were searched independently with the following parameter files; for the light search, all amino acids were left at default masses; for the heavy search, static modifications on lysine (+8.0142 m/z) and arginine (+10.0082 m/z) were specified. For data collected on the Orbitrap mass spectrometers, precursor-ion mass tolerance was set to 50 ppm. The resulting peptide spectral matches were filtered using DTASelect (version 2.0.47), and only half-tryptic or fully tryptic peptides were accepted for identification. Peptides were restricted to a specified false positive rate of <1%. SILAC and ReDiMe ratios were quantified using in-house CIMAGE software49. Briefly, a 10 minute retention time window was used for peak identification using 10 ppm mass accuracy and requiring a co-elution R2 value greater than 0.8. Peptides detected as singletons, where only the heavy or light isotopically labeled peptide was detected and sequenced, but which passed all other filtering parameters, were given a ratio of 20, which is the maximum SILAC or ReDiMe ratio reported here.
shRNA knockdown studies
ABHD16A MISSION® shRNA bacterial glycerol stocks were purchased, and the lentiviral-based shRNA gene knockdown was performed in K562 cells using manufacturers protocol (Sigma-Aldrich). Briefly, 1 μg shRNA transfer vector, 1 μg of the pCMV-VSVG envelope vector, and 1 μg packaging vector, were transfected using 10 μg of PEI into 2.5 x 105 HEK293T cells cultured in 2 mL DMEM with L-glutamine (2mM) and 10% FCS and 5% CO2 to generate the lentiviral transduction particles (LTP). 1 x 106 K562 cells were infected with the LTP generated from the transfections along with 8 μg/mL polybrene (to enhance infection) cultured in 10 mL RPMI 1640 with 10% FCS and 5% CO2, and thereafter the lentiviral-infected K562 cells were selected on puromycin (2 μg/mL). After six rounds of puromycin selection, K562 cells infected with the LTP generated using the constructs TRCN0000046818 (KD_1) (Sigma-Aldrich) and TRCN0000413301 (KD_2) (Sigma-Aldrich) showed > 90% knockdown of ABHD16A, and were selected for further studies. As a negative control, empty shRNA transfer vector was used.
Generation of ABHD16A−/− mice
ABHD16A+/− mice were obtained from Wellcome Trust Sanger Institute (UK) as a part of the EMMA Mouse Repository (Strain name: B6NTac;B6N-Atm1BrdAbhd16atm1a(EUCOMM)wtsi/Wtsi). ABHD16A+/− mice were generated using the ES clone EPD0605_5_A11, which contains a gene trap-targeting cassette that expresses β-galactosidase. Male ABHD16A+/− mice were initially bred with female ABHD16A+/+ mice to yield a pool of ABHD16A+/− mice. The ABHD16A+/− mice were further bred to eventually yield ABHD16A−/− mice. ABHD16A genotypes were confirmed using PCR amplification of the genomic tail DNA with the following primers: ABHD16A forward 5’-GGCCAGCCTGAGTTCCATAG-3’, ABHD16A reverse 5’-GGGCCTCTTAGGTGGGAAAC-3’, and ABHD16A gene trap reverse 5’-TCGTGGTATCGTTATGCGCC-3’. This PCR amplification strategy yielded a 536-bp product for the WT allele, and a 198-bp product for the gene-trap mutant allele. All mice used in this study were generated from breeding ABHD16A+/− mice, are on the C57Bl/6 genetic background, and had ad libitum access to water and food.
RT-RCR
Total RNA was isolated from half-brains of 10-week old ABHD16A+/+ and ABHD16A−/− mice using RiboZol® (Amresco). SuperScript III® (Life Technologies) was used to synthesize first strand cDNA from the isolated total RNA. PCR amplification of a 206-bp fragment of the ABHD16A cDNA was performed using 5’-TGGAAGCCACACATAGGAACC-3’ and 5’-CCTGTTGAGAAACGTGTCTGC-3’ primers. As controls PCR amplification of a 211-bp fragment of the GAPDH cDNA using 5’-TGGATTTGGACGCATTGGTC-3’ and 5’-TTTGCACTGGTACGTGTTGAT-3’ primers, and a 154-bp fragment of the ACTB cDNA using 5’-GGCTGTATTCCCCTCCATCG-3’ and 5’-CCAGTTGGTAACAATGCCATGT-3’ primers was performed.
Harvesting and treating thioglycollate-elicited macrophages
Peritoneal macrophages were elicited by thioglycollate treatment and harvested from mice according to the standard protocols (http://www.lipidmaps.org/). After removal of the RBC lysis buffer, the macrophages were re-suspended in RPMI 1640 with 10% FCS, 1X penicillin-streptomycin and 5% CO2. Cell counts measured, and 2 x106 cells were plated. Macrophages were allowed to adhere to the plates for 2 h at 37 °C and 5% CO2. The macrophages were washed with sterile PBS (3X) to remove cellular debris and treated with KC01 or KC02 (1 μM) or DMSO (control) for 4 h in 4.5 mL of serum free phenol red free RPMI 1640. Macrophages were then stimulated with lipopolysaccharide (5 μg/mL) for 7 hours in sterile PBS. Secreted metabolites and cytokines were measured from collected cell media (4 and 0.5 mL, respectively). Macrophages were harvested by scraping and washed with sterile cold PBS (3X) before further use. All samples were either used immediately or flash frozen in liquid N2, and stored at – 80 °C until further use.
Cytokine measurements
Cytokine analyses were performed on media harvested from mouse peritoneal macrophages (described above) using mouse inflammatory cytokine single analyte ELISA kits (R&D Systems).
Sample preparation and targeted LC-MS metabolite profiling
ABHD16A+/+, ABHD16A+/−, and ABHD16A−/− littermates of 10 weeks age were anaesthetized by isoflurane and sacrificed by decapitation. CNS tissues (brain; spinal cord) were harvested, weighed, and immediately submerged in liquid N2. Tissues were then Dounce-homogenized in 8 mL of 2:1:1 CHCl3: MeOH: PBS containing the internal standard mix (1 nmol 17:1 FFA (Sigma-Aldrich), and 100 pmol each of 17:1 lyso-PS, 17:0–20:4 PS, 17:1 lyso-PC, 17:0–20:4 PC, 17:0–20:4 PE (Avanti Polar Lipids) and 50 pmol each of 2-AG-d5, PGE2-d9 (Cayman Chemical)). Homogenates were centrifuged at 2800g for 10 minutes to separate the two phases. The organic phase (bottom) was removed, 200 μL of formic acid was added to acidify the aqueous homogenate (to enhance extraction of phospholipids), and CHCl3 was added to make up 8 mL volume. The mixture was vortexed, and separated using centrifugation described above. Both the organic extracts were pooled, and dried under a stream of N2. The metabolomes were re-solubilized in 120 μL of 2:1 CHCl3: MeOH, and 10 μL were used for the targeted LC/MS analysis. For cellular and secreted metabolomics studies, serum free phenol red free RPMI 1640 was used to prevent any promiscuous contributions from serum lipids. For the cellular metabolomics studies, 3 x 106 cells were used for all cancer cell lines and LCLs, and 2 x106 cells were used for the peritoneal macrophages. 4 mL of the serum free phenol red free RPMI 1640 was harvested from all cultures to measure secreted metabolites (with the exception of the K562 leukemia cell line which need 10% FCS complementation for culturing and did not suit secreted metabolic studies protocol). For cellular metabolomics samples, the cells were washed with PBS (3X), and suspended in 1 mL PBS. 3 mL of 2:1 CHCl3: MeOH with the internal standard mix was added, and the mixture was vigorously vortexed. The two phases were separated by centrifugation at 2800g for 10 minutes. The organic phase (bottom) was collected, and the aqueous phase was re-extracted after acidification by 50 μL of formic acid. The organic phases were pooled, and dried under a stream of N2. The metabolome was re-solubilized using 120 μL of 2:1 CHCl3: MeOH, and 10 μL were used for the targeted LC-MS analysis. For the secreted metabolomic profiles, 4 mL of the serum free phenol red free RPMI 1640 harvested from the cells, was processed by addition of 12 mL of 2:1 CHCl3: MeOH with the internal standard mix, and the mixture was vigorously vortexed. The two phases were separated by centrifugation at 2800g for 10 minutes, and the organic phase (bottom) was collected. The aqueous phase was re-extracted after acidification with 200 μL of formic acid. The organic phases were pooled, and dried using a stream of N2. Thereafter the secreted metabolome was re-solubilized using 120 μL of 2:1 CHCl3: MeOH, and 20 μL were used for the targeted LC-MS analysis.
Metabolites analyzed in this study were quantified using LC/MS-based multiple reaction monitoring (MRM) methods (Agilent Technologies 6460 Triple Quad). The solvents and LC separation conditions were the same as those described in the substrate hydrolysis assays section. MS analysis was performed using ESI with the following parameters: drying gas temperature = 350 °C, drying gas flow = 9 L/min, nebulizer pressure = 45 psi, sheath gas temperature = 375 °C, sheath gas flow = 12 L/min, fragmentor voltage = 100 V, and capillary voltage = 3.5 kV. A typical run consisted of 33 minutes, with the following solvent run sequence post injection: 0.1 ml/min 0% buffer B for 5 minutes, 0.4 ml/min linear gradient of buffer B from 0 – 100% over 15 minutes, 0.5 ml/min of 100% buffer B for 8 minutes, and re-equilibration with 0% buffer B for 5 minutes. Lipid species were quantified by measuring areas under the curve in comparison to the appropriate unnatural internal standard and then normalizing to with wet tissue weight or number of cells.
Synthesis and characterization of ABHD16A inhibitors and probes
See Supplementary Note for information on the synthesis and characterization of β-lactones.
Statistical analysis
Statistical analyses were performed using the GraphPad Prism 6 (for Mac OS X) software. All data are shown as mean values ± s.e.m. Student’s t-test (two-tailed) was used to study statistically significant differences between study groups. A p-value of < 0.05 was considered statistically significant for this study.
Supplementary Material
Acknowledgments
We are grateful to K. Masuda, A. Viader and K-L. Hsu for their helpful discussions, guidance and technical expertise in harvesting tissues, G. Simon and J. Blankman for helpful discussions, M. Niphakis for advice on the SAR studies on the β-lactone scaffolds, O. Ulanovskaya for advice on generating the shRNA knockdown cell lines, B. Correia for the guidance on using CIMAGE, C. Joslyn, T. Takei for technical assistance, and M. Lau for contributions to initial synthetic studies. This work was supported by the US National Institutes of Health (DA033760) and the 9th Irving S. Sigal Postdoctoral Fellowship, American Chemical Society (SSK), a Hewitt Foundation for Medical Research Fellowship (WHP), and the US National Science Foundation (AH, CHE-0111522 and CHE-1048717).
Footnotes
Author Contributions.
SSK and BFC conceived the project, and designed the experiments. SSK performed the biochemical and cell biological experiments. KC and AH synthesized and chemically characterized all β-lactone compounds, D-H C, and TDD generated the LCL lines from the patient and controls in this study, WHP assisted with chemical and biochemical studies, and MMD performed the ABPP-reductive dimethylation experiments. SSK and BFC wrote the manuscript.
Competing financial interests.
The authors declare competing financial interests. BFC is a founder and advisor to Abide Therapeutics, a biotechnology company interested in developing serine hydrolase inhibitors and therapeutics.
Additional information.
Supplementary information and chemical compound information is available with the online version of this paper.
References
- 1.Rivera R, Chun J. Biological effects of lysophospholipids. Reviews of physiology, biochemistry and pharmacology. 2008;160:25–46. doi: 10.1007/112_0507. [DOI] [PubMed] [Google Scholar]
- 2.Makide K, et al. Novel lysophosphoplipid receptors; their structure and function. J Lipid Res. 2014 doi: 10.1194/jlr.R046920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosen H, Stevens RC, Hanson M, Roberts E, Oldstone MB. Sphingosine-1-phosphate and its receptors: structure, signaling, and influence. Annu Rev Biochem. 2013;82:637–662. doi: 10.1146/annurev-biochem-062411-130916. [DOI] [PubMed] [Google Scholar]
- 4.Lynch KR, Macdonald TL. Sphingosine 1-phosphate chemical biology. Biochimica et biophysica acta. 2008;1781:508–512. doi: 10.1016/j.bbalip.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van der Kleij D, et al. A novel host-parasite lipid cross-talk. Schistosomal lyso-phosphatidylserine activates toll-like receptor 2 and affects immune polarization. J Biol Chem. 2002;277:48122–48129. doi: 10.1074/jbc.M206941200. [DOI] [PubMed] [Google Scholar]
- 6.Inoue A, et al. TGFalpha shedding assay: an accurate and versatile method for detecting GPCR activation. Nat Methods. 2012;9:1021–1029. doi: 10.1038/nmeth.2172. [DOI] [PubMed] [Google Scholar]
- 7.Sugita K, Yamamura C, Tabata K, Fujita N. Expression of orphan G-protein coupled receptor GPR174 in CHO cells induced morphological changes and proliferation delay via increasing intracellular cAMP. Biochemical and biophysical research communications. 2013;430:190–195. doi: 10.1016/j.bbrc.2012.11.046. [DOI] [PubMed] [Google Scholar]
- 8.Chu X, et al. An X chromosome-wide association analysis identifies variants in GPR174 as a risk factor for Graves' disease. Journal of medical genetics. 2013;50:479–485. doi: 10.1136/jmedgenet-2013-101595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szymanski K, et al. rs3827440, a nonsynonymous single nucleotide polymorphism within GPR174 gene in X chromosome, is associated with Graves' disease in Polish Caucasian population. Tissue antigens. 2014;83:41–44. doi: 10.1111/tan.12259. [DOI] [PubMed] [Google Scholar]
- 10.Frasch SC, Bratton DL. Emerging roles for lysophosphatidylserine in resolution of inflammation. Progress in lipid research. 2012;51:199–207. doi: 10.1016/j.plipres.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park KS, Lee HY, Kim MK, Shin EH, Bae YS. Lysophosphatidylserine stimulates leukemic cells but not normal leukocytes. Biochemical and biophysical research communications. 2005;333:353–358. doi: 10.1016/j.bbrc.2005.05.109. [DOI] [PubMed] [Google Scholar]
- 12.Lloret S, Moreno JJ. Ca2+ influx, phosphoinositide hydrolysis, and histamine release induced by lysophosphatidylserine in mast cells. Journal of cellular physiology. 1995;165:89–95. doi: 10.1002/jcp.1041650112. [DOI] [PubMed] [Google Scholar]
- 13.Lee SY, et al. Lysophosphatidylserine stimulates chemotactic migration in U87 human glioma cells. Biochemical and biophysical research communications. 2008;374:147–151. doi: 10.1016/j.bbrc.2008.06.117. [DOI] [PubMed] [Google Scholar]
- 14.Knowlden S, Georas SN. The autotaxin-LPA axis emerges as a novel regulator of lymphocyte homing and inflammation. Journal of immunology. 2014;192:851–857. doi: 10.4049/jimmunol.1302831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Billich A, Baumruker T. Sphingolipid metabolizing enzymes as novel therapeutic targets. Sub-cellular biochemistry. 2008;49:487–522. doi: 10.1007/978-1-4020-8831-5_19. [DOI] [PubMed] [Google Scholar]
- 16.Blankman JL, JZL, Trauger SA, Siuzdak G, Cravatt BF. ABHD12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease PHARC. Proc Natl Acad Sci U S A. 2013;110:1500–1505. doi: 10.1073/pnas.1217121110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiskerstrand T, et al. Mutations in ABHD12 cause the neurodegenerative disease PHARC: An inborn error of endocannabinoid metabolism. American journal of human genetics. 2010;87:410–417. doi: 10.1016/j.ajhg.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen DH, et al. Two novel mutations in ABHD12: expansion of the mutation spectrum in PHARC and assessment of their functional effects. Human mutation. 2013;34:1672–1678. doi: 10.1002/humu.22437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hosono H, et al. Phosphatidylserine-specific phospholipase A1 stimulates histamine release from rat peritoneal mast cells through production of 2-acyl-1-lysophosphatidylserine. J Biol Chem. 2001;276:29664–29670. doi: 10.1074/jbc.M104597200. [DOI] [PubMed] [Google Scholar]
- 20.Aoki J, Nagai Y, Hosono H, Inoue K, Arai H. Structure and function of phosphatidylserine-specific phospholipase A1. Biochimica et biophysica acta. 2002;1582:26–32. doi: 10.1016/s1388-1981(02)00134-8. [DOI] [PubMed] [Google Scholar]
- 21.Kelleher JA, Sun GY. Enzymic hydrolysis of arachidonoyl-phospholipids by rat brain synaptosomes. Neurochemistry international. 1985;7:825–831. doi: 10.1016/0197-0186(85)90038-5. [DOI] [PubMed] [Google Scholar]
- 22.Simon GM, Cravatt BF. Activity-based proteomics of enzyme superfamilies: serine hydrolases as a case study. J Biol Chem. 2010;285:11051–11055. doi: 10.1074/jbc.R109.097600. R109.097600 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Borgstrom B. Mode of action of tetrahydrolipstatin: a derivative of the naturally occurring lipase inhibitor lipstatin. Biochimica et biophysica acta. 1988;962:308–316. doi: 10.1016/0005-2760(88)90260-3. [DOI] [PubMed] [Google Scholar]
- 24.Hoover HS, Blankman JL, Niessen S, Cravatt BF. Selectivity of inhibitors of endocannabinoid biosynthesis evaluated by activity-based protein profiling. Bioorganic & medicinal chemistry letters. 2008;18:5838–5841. doi: 10.1016/j.bmcl.2008.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Baggelaar MP, et al. Development of an activity-based probe and in silico design reveal highly selective inhibitors for diacylglycerol lipase-alpha in brain. Angew Chem Int Ed Engl. 2013;52:12081–12085. doi: 10.1002/anie.201306295. [DOI] [PubMed] [Google Scholar]
- 26.Pemble CWt, Johnson LC, Kridel SJ, Lowther WT. Crystal structure of the thioesterase domain of human fatty acid synthase inhibited by Orlistat. Nature structural & molecular biology. 2007;14:704–709. doi: 10.1038/nsmb1265. [DOI] [PubMed] [Google Scholar]
- 27.Dekker FJ, et al. Small-molecule inhibition of APT1 affects Ras localization and signaling. Nat Chem Biol. 2010;6:449–456. doi: 10.1038/nchembio.362. [DOI] [PubMed] [Google Scholar]
- 28.Ortar G, et al. Tetrahydrolipstatin analogues as modulators of endocannabinoid 2-arachidonoylglycerol metabolism. J Med Chem. 2008;51:6970–6979. doi: 10.1021/jm800978m. [DOI] [PubMed] [Google Scholar]
- 29.Ngai MH, et al. Click-based synthesis and proteomic profiling of lipstatin analogues. Chemical communications. 2010;46:8335–8337. doi: 10.1039/c0cc01276a. [DOI] [PubMed] [Google Scholar]
- 30.Camara K, Kamat S, Lasota CC, Cravatt BF, Howell AR. Combining cross-metathesis and activity-based protein profiling: new beta-lactone motifs for targeting serine hydrolases. manuscript submitted. 2014 doi: 10.1016/j.bmcl.2014.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jessani N, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- 32.Adibekian A, et al. Click-generated triazole ureas as ultrapotent in vivo-active serine hydrolase inhibitors. Nat Chem Biol. 2011;7:469–478. doi: 10.1038/nchembio.579. nchembio.579 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci USA. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blankman JL, Simon GS, Cravatt BF. A Comprehensive Profile of Brain Enzymes that Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hua X, Sakai J, Ho YK, Goldstein JL, Brown MS. Hairpin orientation of sterol regulatory element-binding protein-2 in cell membranes as determined by protease protection. J Biol Chem. 1995;270:29422–29427. doi: 10.1074/jbc.270.49.29422. [DOI] [PubMed] [Google Scholar]
- 36.Vance JE, Tasseva G. Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochimica et biophysica acta. 2013;1831:543–554. doi: 10.1016/j.bbalip.2012.08.016. [DOI] [PubMed] [Google Scholar]
- 37.Rossol M, et al. LPS-induced cytokine production in human monocytes and macrophages. Critical reviews in immunology. 2011;31:379–446. doi: 10.1615/critrevimmunol.v31.i5.20. [DOI] [PubMed] [Google Scholar]
- 38.Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nature protocols. 2009;4:484–494. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- 39.Wilson-Grady JT, Haas W, Gygi SP. Quantitative comparison of the fasted and re-fed mouse liver phosphoproteomes using lower pH reductive dimethylation. Methods. 2013;61:277–286. doi: 10.1016/j.ymeth.2013.03.031. [DOI] [PubMed] [Google Scholar]
- 40.Kim HY, Huang BX, Spector AA. Phosphatidylserine in the brain: Metabolism and function. Progress in lipid research. 2014 doi: 10.1016/j.plipres.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spies T, Blanck G, Bresnahan M, Sands J, Strominger JL. A new cluster of genes within the human major histocompatibility complex. Science. 1989;243:214–217. doi: 10.1126/science.2911734. [DOI] [PubMed] [Google Scholar]
- 42.Spies T, Bresnahan M, Strominger JL. Human major histocompatibility complex contains a minimum of 19 genes between the complement cluster and HLA-B. Proc Natl Acad Sci U S A. 1989;86:8955–8958. doi: 10.1073/pnas.86.22.8955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frasch SC, et al. Neutrophils regulate tissue Neutrophilia in inflammation via the oxidant-modified lipid lysophosphatidylserine. J Biol Chem. 2013;288:4583–4593. doi: 10.1074/jbc.M112.438507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ishii I, Fukushima N, Ye X, Chun J. Lysophospholipid receptors: signaling and biology. Annu Rev Biochem. 2004;73:321–354. doi: 10.1146/annurev.biochem.73.011303.073731. [DOI] [PubMed] [Google Scholar]
- 45.Swaney JS, et al. A novel, orally active LPA(1) receptor antagonist inhibits lung fibrosis in the mouse bleomycin model. British journal of pharmacology. 2010;160:1699–1713. doi: 10.1111/j.1476-5381.2010.00828.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Inloes JM, et al. The hereditary spastic paraplegia-related enzyme DDHD2 is a principal brain triglyceride lipase. Proc Natl Acad Sci U S A. 2014;111:14924–14929. doi: 10.1073/pnas.1413706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsu KL, et al. DAGLβ inhibition perturbs a lipid network involved in macrophage inflammatory responses. Nat Chem Biol. 2012;8:999–1007. doi: 10.1038/nchembio.1105. nchembio.1105 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 49.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. nature09472 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.