Abstract
Selenium-containing compounds and selenized yeast have anti-cancer properties. In order to address possible mechanisms involved in these effects, selenoglycoproteins (SGP) were extracted from selenium-enriched yeast at pH 4.0 and 6.5 (the fractions are called SGP40 and SGP65, respectively), followed by evaluation of their impact on the interactions of lung and breast tumor cells with human brain microvascular endothelial cells (HBMEC). Extracted SGPs, especially SGP40, significantly inhibited adhesion of tumor cells to HBMEC and their transendothelial migration. Because the active component(s) of SGPs are unknown, small selenium-containing compounds (leucyl-valyl-selenomethionyl-arginine [LVSe-MR] and methylseleno adenosine [M-Se-A]), which are normally present in selenized yeast, were introduced as additional treatment groups. Treatment of HBMEC with SGP40, LVSe-MR, and M-Se-A induced changes in gene signatures, which suggested a central involvement of NF-κB-dependent pathway. These observations were confirmed in the subsequent analysis of NF-κB DNA binding activity, quantitative measurements of the expression of selected genes and proteins, and tumor cell adhesion assay with a specific NF-κB inhibitor as the additional treatment factor. These findings indicate that specific organic selenium-containing compounds have the ability to inhibit tumor cell adhesion to brain endothelial cells via downregulation of NF-κB. SGPs appear to be more effective than small selenium-containing compounds, suggesting the role of not only selenium but also the glycoprotein component in the observed protective impact.
Keywords: selenium, brain, tumor cell motility, adhesion, transendothelial migration, NF-κB
1. INTRODUCTION
The role of nutrition in cancer etiology, prevention, and treatment has been a subject of extensive research since the early 1980s [1]. Based on numerous epidemiological and experimental studies it is generally accepted that various dietary agents are biologically active and can efficiently modulate invasiveness and metastatic potential of various tumors [2, 3]. Although the role and significance of conventional cancer therapies are indisputable, therapeutic strategies combining dietary compounds, recognized as cancer chemopreventive agents, are often more effective and result in better survival rates. Moreover, combination treatment can decrease systemic toxicity caused by chemo- or radiotherapy [1, 4].
Selenium (Se) is an essential micronutrient, which functions in human physiology are largely mediated by selenoproteins in which selenium is incorporated as selenocysteine. Due to structural similarity, selenium can also be inserted in place of sulfur in methionine and be incorporated into proteins as selenomethionine [5]. The third group of mammalian selenium-containing proteins is the family of selenium-binding proteins (SBPs), which bind selenium by a mechanism that has yet to be clarified [6].
Compelling evidence indicates that selenium supplementation has significant benefits for human health [7, 8]. Anticarcinogenic properties of selenium, especially at supranutritional levels of supplementation, are well known and usually attributed to its strong antioxidative and anti-inflammatory properties [5, 9, 10]. Indeed, selenium as an essential cofactor for several important enzyme systems, including glutathione peroxidases, selenoprotein P and thioredoxinreductase, possesses the ability to modulate oxidative stress and can affect cancer development in multiple ways [8, 9].
Although selenium and its relation to cancer have been the subject of intense investigations, most reports were concentrated on the primary tumor growth and only few studies explored the role of selenium in later phases of tumor development [10]. Tumor progression is a complex process of overlapping events in which cancer cells invade surrounding tissues, leading to the formation of secondary tumors in organs other than those primarily affected by the disease. Initially, cancer cells become arrested in capillary beds and extravasate into the target organs by interacting with the vascular endothelium [11, 12]. Therefore, tumor cell adhesion to the endothelium and transendothelial migration play key roles in tumor cell invasion. Degradation of extracellular matrix (ECM) by matrix metalloproteinases (MMPs) is also prerequisite for the cells to migrate into target organs. While tumor cells can produce MMPs, during tumor invasion and progression most MMPs are produced by host cells, including endothelial cells [13–15]. It has been demonstrated that a variety of factors, including dietary and environmental agents, can regulate properties of the vascular endothelium and influence tumor progression by modulating the first steps towards extravasation, namely adhesion and transendothelial migration of tumor cells [3, 9, 16].
The role of the unique brain microenvironment in tumor progression is still poorly understood. This lack of knowledge has important health-related implications, since brain metastases affect 200,000 cancer patients in the US annually [11]. We demonstrated previously that feeding mice with selenized yeast markedly decreased growth of brain metastatic tumors [17]. In the present study, we explored potential mechanisms involved in these effects. Our results indicate that the inhibitory effects on NF-κB activation and NF-κB-regulated genes induced by selenoglyproteins are important factors contributing to suppression of the interactions of tumor cells with the brain microvascular endothelium.
2. MATERIALS AND METHODS
2.1. Cell lines and culture conditions
Human brain microvascular endothelial cells (HBMEC; hCMEC/D3 cell line) [18] were cultured in EBM-2 medium supplemented with EBM-2 Bullet-kit containing vascular endothelial growth factor (VEGF), insulin-like growth factor-I (IGF-1), epidermal growth factor (EGF), basic fibroblast growth factor (bFGF), hydrocortisone, ascorbic acid, gentamycin, and fetal bovine serum (FBS) (Lonza, Walkersville, MD). All cell culture plates were coated with 100 μg/mL of rat tail collagen type I (BD Biosciences, San Jose, CA) for 1–3 hours. Cultures were maintained at 37°C in a humidified atmosphere of 5% CO2. The medium was changed every third day. At confluence, the cells were sub-cultured after removal with 0.25% trypsin-EDTA. HBMEC have a doubling time that varies between 27 and 30 h. Prior to each experiment, the cells were maintained in serum-free medium for 12 h in order to increase susceptibility of cells to treatment factors and to promote synchronization of the cell cycle for more uniformed responses.
The human breast cancer cell line MDA-MB231 and human lung adenocarcinoma cell line A549 were purchased from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS, streptomycin (10,000 μg/mL), and penicillin (10,000 U/mL) at 37°C in 5% CO2. MDA-MB231 and A549 cells have doubling times of approximately 23–24 and 20–22 h, respectively.
2.2. Selenium-containing treatment factors
It was reported previously that a large portion of selenium in Se-enriched yeast is present in the protein fractions [19]. The process of selenoglycoproteins (SGPs) extraction from selenium-enriched yeast comprised two stages designed to capture and concentrate as many water-soluble selenoglycoproteins as possible, based upon the main pH ranges of these species. In the first stage, a suspension of spray-dried selenium enriched yeast in 0.3 N HCl (pH 1.5) was stirred and heated at 80°C for 8 hours. The pH of the mixture was maintained by adding small amounts of concentrated HCl during the first hour of the extraction. After 8 hours the mixture was centrifuged and the liquid phase was collected. The pH of the supernatant was adjusted to 1.85 by the addition of 2.0 N NaOH, and the SGPs that have limited solubility at this pH precipitated from the solution and were separated from the liquids by the second centrifugation. The liquids (pH 1.85) were mixed with the solids (pH 1.5) from the first centrifugation, which resulted in change of the pH of the mixture to 1.6. The majority of the SGPs soluble at pH 1.6 were transferred into the liquid phase. In the second stage, the liquid phase from the second centrifugation was transferred to a mixer and the pH adjusted by the addition of 2.0 N NaOH to pH 4.0. The SGPs that have limited solubility at this pH precipitated from the solution and were separated from the liquids (pH 4.0) by third centrifugation and finally freeze-dried to yield a solid SGP fraction at pH 4.0 (SGP40). The liquid phase (pH 4.0) from the third centrifugation was transferred to a mixer and the pH was adjusted by the addition of 2.0 N NaOH to pH 6.5. The SGPs that have limited solubility at this pH precipitated from the solution and were separated from the liquids by the forth centrifugation and then freeze-dried to yield solid SGP fraction of pH 6.5 (SGP65).
Methylselenoadenosine (M-Se-A; C11H15N5O3Se) was chosen as a compound of interest because it was recently identified as a water-soluble component of Se-enriched yeast [19]. Of particular interest is its adenosyl moiety, which may allow this compound to pass with ease into the cell nucleus. If any of the previously reported carcinostatic properties of Se-enriched yeast are due to direct interaction between selenium molecules and DNA, then methylselenoadenosine and similar compounds represent attractive therapeutic compounds. The method for methylselenoadenosine synthesis was developed in the laboratory with all reagents being purchased from Sigma-Aldrich (St. Louis, MO). Briefly, sodium borohydride (227 mg, 6.0 mM, under Argon flow) was placed in a 200 mL round-bottom flask containing 20 mL of anhydrous ethyl alcohol, equipped with a magnetic stirrer and located in an iced, cooling bath. Dimethyldiselenide (190 μL, 376 mg, 2.0 mM) was then added slowly with a syringe. After discoloration to a yellowish solution, solid 5′-chloro-5′-deoxyadenosine (1.143 g, 4.0 mM) was added, along with ethyl alcohol to dissolve the precipitate. The mixture was stirred at room temperature for the following four days. Mass spectrometry was used to monitor the conversion (~75% conversion accomplished after 5 days). The solvents were evaporated and the product (1.522 g) was collected and crystallized from 20 mL of ethyl alcohol. Crystals are highly hygroscopic and were dried over P2O5 to yield 648 mg of white powder. The filtrate was concentrated and dried under high vacuum/P2O5 to yield 667 mg of white-pink solid, which was further purified by reverse phase (C-8) preparative chromatography.
Among proteins characterized in Se-enriched yeast, glyceraldehydes-3-phosphate dehydrogenase (GAPDH) contains approximately 60% of the total selenium in this cellular pool. If dietary selenoprotein fractions were to have biological roles it would likely be via the action of Se-containing peptides generated during the digestive process. LVSe-MR represents a peptide sequence, which would be formed from GAPDH by the action of trypsin. LVSe-MR (C22H44N7O2Se) peptide was custom synthesized, purified (>98%) and supplied by Biomatik, Ltd. (Wilmington, DE).
2.3. Cell cytotoxicity assay
Cell viability was determined by the MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt] assay according to the manufacturer’s protocol (CellTiter 96® AQueous Non-Radioactive Cell Proliferation Kit; Promega, Madison, WI). The assay is based on the ability of metabolically active cells to bioreduce MTS into a formazan product that is soluble in culture medium. Briefly, cells were seeded into individual wells of 96-well plate, allowed to adhere for 24 h, and treated with the tested compounds for 24 h. Then, 20 μl aliquots of the combined MTS/PMS solution were added into each well and plates were incubated at 37°C for 1–2 h. Formazan formation as the indicator of cell viability was measured by SpectraMax 190 Microplate Reader (Molecular Devices, Sunnyvale, CA) at 490 nm.
2.4. Cell adhesion assay
The adhesion of cancer cells to HBMEC was assessed according to the method of Braut-Boucher et al. [20] with modifications. Briefly, HBMEC were grown to confluence on collagen-coated 48-well plates and cancer cells were cultured on 60 mm cell culture dishes (Corning Inc., Corning, NY) and exposed to SGPs (normalized to 5 μM of Se), TNF-α (10 ng/mL), or vehicle for 24 h at 37°C. In experiments with TNF-α co-treatment, cells were first incubated with SGP40 or SGP65 (both at 5 μM of Se), followed by addition of TNF-α (10 ng/mL) for the additional 20 h. TNF-α was used in our experiments as a positive control due to its properties as a strong endothelial cell activator, inducer of cell adhesion molecules and metalloproteinases. The concentration 10 ng/ml was chosen based on previous literature reports [21, 22]. In selected experiments, cultures were also treated for 30 min with SN50 (18 μM; an inhibitor of NF-κB nuclear translocation) or SN50M (18 μM; a negative control for the SN50 peptide with no measurable effect on NF-κB translocation; both from Millipore, Billerica, MA).
Before the adhesion assay the cultures were washed two times with Hank’s balanced salt solution (HBSS). Tumor cells were labeled by incubation with calcein AM (5 μM, Life Technologies, Grand Island, NY) for 30 min at 37°C, followed by two washings with HBSS. Labeled cells were added in the amount of 5.0×105 cells/mL onto endothelial monolayers. Co-cultures were incubated for 20 min at 37°C and then carefully washed three times with HBSS to remove non-adherent tumor cells. The adherence of calcein-labeled cancer cells was quantified by fluorescence measurements (Gemini EM Fluorescence Microplate Reader, Molecular Devices, Sunnyvale, CA) of endothelial monolayers using an excitation of 485 nm and an emission of 530 nm.
To visualize tumor cell adhesion, fluorescent microscopy was performed in co-cultures of HBMEC and calcein-labeled MDA-MB231 or A549 cells. HBMEC were cultured on Collagen Type I Cellware 4-well CultureSlides (BD Biosciences, San Jose, CA). Cells were prepared and treated as described above. Unbound cells were washed away with HBSS and the monolayer was fixed with 4% formaldehyde for 15 min. Images were evaluated under fluorescent microscope (Nikon, Melville, NY) at 20 × magnification.
2.5. Transendothelial cell migration assay
HBMEC were seeded in the amount of 2.0×105 cell/mL on collagen-coated 24-Multiwell Insert System (BD Falcon™ FluoroBlok™, 6.5 mm diameter, 8.0 μm pore size, BD Biosciences, San Jose, CA). The cultures were maintained for 3 days until the cells reached confluency. MDA-MB231 or A549 cells were cultured in 60 mm cell culture dishes (Corning). Both endothelial and cancer cells were treated as for cell adhesion assay. Tumor cells were labeled by incubation with 5 μM calcein AM, suspended in serum-free EBM medium, and placed in the amount of 5.0×105 cells/mL on top of HBMEC in the upper chamber of the transwell system. The 24-Multiwell Insert System used in these experiments contains a proprietary, light-opaque polyethylene terephthalate microporous membrane and was designed for applications where it is desirable to specifically detect fluorescent compounds below the surface of a membrane. After 10 h incubation, fluorescent signal from the lower chamber, including the underside of the membrane, was measured at 485-nm excitation and 530-nm emission wavelengths (Gemini EM Fluorescence Microplate Reader).
2.6. GeneChip Microarray Analysis
HBMEC and MDA-MB231 cells were cultured in 6 and 4 well plates, respectively, and treated with SGP40, LVSe-MR, or M-Se-A at the concentration normalized to 5 μM Se at 37°C in 5% CO2 for 24 h. Then, TNF-α (10 ng/mL) was added for the additional 20 h. Total RNA was isolated and purified using RNeasy Mini Kit (Qiagen, Valencia, CA) following the manufacturer’s protocol.
Microarray gene expression analysis was performed using the Affymetrix GeneChip® Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA). The labeling of RNA samples, hybridization and array scanning were performed following the standard protocols recommended by Affymetrix. Microarray data analysis was conducted as described earlier [21]. GeneSpring GX 12.5 (Silicon Genetics, Redwood, CA) software was employed to normalize the data and to perform statistical and gene expression pattern analyses, where genes that differed from the control with a p ≤ 0.01 and corresponding signal intensity fold change (FC) ≥ 1.2 or FC ≤ −1.2 were deemed as changed. Ingenuity® pathways analysis software (Ingenuity® Systems Inc., Redwood City, CA) was used to dissect the biological pathways and processes in which differentially expressed genes were involved.
2.7. Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared using NE-PER™ Nuclear and Cytoplasmic Extraction Kit (Thermo Scientific, Waltham, MA) following the manufacturer’s protocols and stored at −80°C until used. The double-stranded oligonucleotides (Santa Cruz Biotechnology, Dallas, TX) containing the consensus or mutated sequences of the binding sites for NF-κB were end-labeled with 5′-biotin using Pierce Biotin 3′ End DNA Labeling Kit (Thermo Scientific). Binding reactions were performed in a 20 μl volume using LightShift EMSA Optimization and Control Kit (Thermo Scientific) according to the manufacturer’s instructions. Protein-DNA complexes were analyzed on a 5% polyacrylamide gel (Bio-Rad Laboratories, Hercules, CA) in 0.5 × Tris/boric acid/EDTA buffer (Bio-Rad Laboratories). The binding reactions were transferred to nylon membranes (Thermo Scientific) and fixed by cross-linking with Ultraviolet Crosslinker (UVP, Upland, CA). The biotin-labeled DNA was detected by Chemiluminescent Nucleic Acid Detection Module (Thermo Scientific).
2.8. Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Cells were prepared in the same manner as described above. Total RNA was isolated and purified using RNeasy Mini Kit (Qiagen, Valencia, CA) and reverse transcribed into cDNA using the Reverse Transcription System (Promega, Madison, WI) according to the protocols provided by the manufacturers. IL-1β, IL-6, IL-8 and CCL5 mRNA levels were assessed using a 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) and TaqMan Universal PCR Master Mix. The primers and probes were obtained from Applied Biosystems. The following thermocycling conditions were used: 50°C for 2 min, followed by 95°C for 10 min, 95°C for 15 s, and 60°C for 1 min for 40 cycles. The threshold cycle (CT) from each well was determined using 7500 Software v2.0.5 for Applied Biosystems 7500 and 7500 Fast Real-Time PCR Systems. Expression of mRNA was calculated by the comparative CT method. PCR amplification of GAPDH (housekeeping gene) was performed for each sample to normalize mRNA levels of the target genes.
2.9. Enzyme-linked Immunosorbent Assay (ELISA)
The activity of matrix metalloproteinase (MMP)-2 and MMP-9 in cell culture media was detected using SensoLyte® 490 MMP-2 and MMP-9 Assay Kits (AnaSpec, Fremont, CA). Media from the treated cultures were collected and centrifuged. To activate pro-MMPs, the samples were incubated for 2 h at 37°C with 1 mM 4-aminophenylmercuric acetate (APMA). Then, 50 μl aliquots were transferred to 96-well plates and 50 μl of MMP-2 or MMP-9 substrate solutions were added. The plates were incubated at 37°C and fluorescence intensity was measured (Gemini EM Fluorescence Microplate Reader) at 340 nm excitation and 490 nm emission. The background-subtracted results were used for analysis.
Protein levels of IL-6 and CCL-5 in cell culture media were determined using Quantikine Human IL-6 or CCL5/RANTES Immunoassay Kit (R&D Systems, Minneapolis, MN) according to the manufacturer’s recommendations. At the end of the treatment period, 100 μl aliquots of cell culture media were transferred to anti-IL-6 or anti-CCL5 antibody-coated wells and incubated for 2 h. Peroxidase-conjugated secondary polyclonal antibody was added into each well. Following a wash to remove any unbound antibody-enzyme reagents, a substrate solution was added into each well and a reaction was allowed to develop for 20 min. Absorbance was measured at 450 nm using a microplate reader (SpectraMax 190 Microplate Reader, Molecular Devices, Sunnyvale, CA). Results were normalized to total cellular protein.
2.10. Statistical Analysis
Data were analyzed by one-way ANOVA or one-way repeated measured ANOVA using SigmaStat 12.0 (SPSS, Chicago, IL). Statistical probability of p < 0.05 was considered significant. All experiments were repeated at least three times.
3. RESULTS
3.1. Treatment with SGPs does not affect cell viability
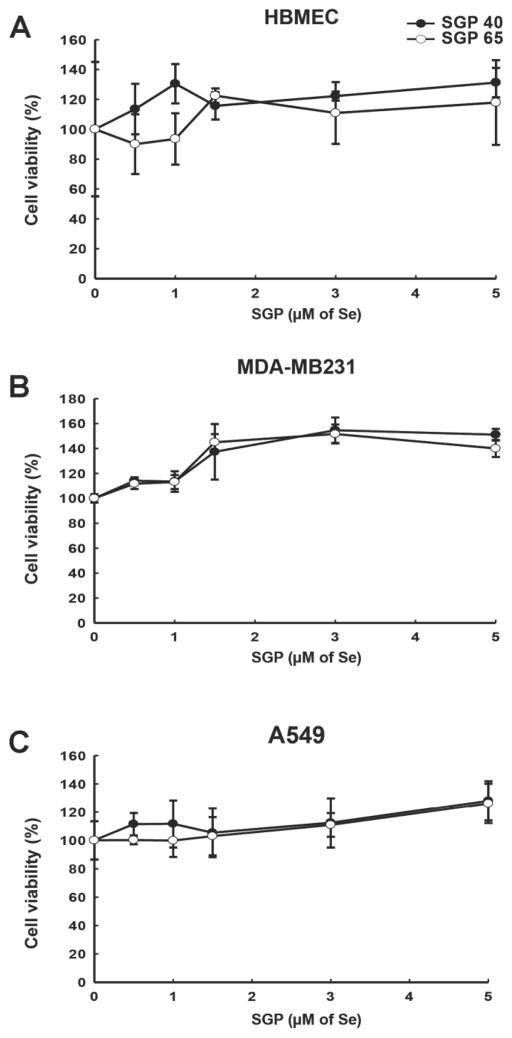
Biological effects of selenium are concentration dependent. Very low (>0.01 μM), as well as high (<10 μM) Se leves result in cell death. At moderate concentration Se possesses antioxidant properties with cancer preventive effects, however at higher levels Se is converted to a strong oxidant, which results in growth inhibition [7]. Therefore, we assessed the impact of SGP40 and SGP65 on viability of cell lines employed in the present study by the MTS assay. Following 24 h incubation with SGP40 or SGP65 at different selenium concentrations, no cytotoxic effects or changes in cell viability were observed (Fig. 1A–C). Based on these results and previous literature reports on the protective effects of Se [22, 23], we selected 5 μM selenium for further experiments.
Fig. 1. Treatment with SGPs does not affect cell viability.
MTS assay was employed to test the impact of SGP40 and SGP65 on viability of the human cell lines used in the present study: (A) HBMEC (human brain microvascular endothelial cells), (B) MDA-MB231 (human breast cancer cells), and (C) A549 (human lung adenocarcinoma cells). Cells were treated with SGP40 or SGP65 at indicated concentrations of Se for 24 h. Values are mean ± SD; n=6 wells per group.
3.2. SGPs attenuate adhesion of tumor cells to human brain endothelial cell monolayers
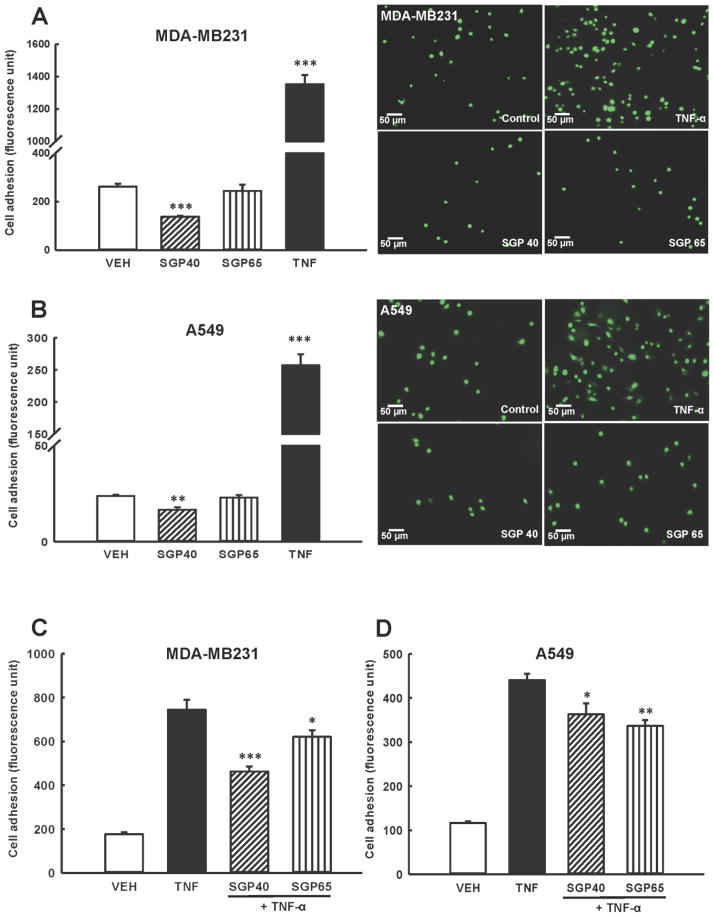
Because adhesion of tumor cells to the vascular endothelium is one of the initial events in tumor progression, we evaluated the impact SGP40 and SGP65 on adhesion of breast and lung tumor cells (MDA-MB231 and A549, respectively) to HBMEC. Adhesion of both types of tumor cells was significantly attenuated in groups treated with SGP40 as compared to vehicle treated control (Fig. 2A and 2B, left panels). In contrast, SGP65 did not cause any significant effects on adhesion of tested cancer cell lines. TNF-α, used as a positive control, effectively increased adhesion of breast and lung tumor cells as compared with vehicle. TNF-α was used as a positive control because it activates HBMEC and promotes tumor cell adhesion and transendothelial migration. Right panels in Fig. 2A and 2B illustrate representative images of calcein-labeled breast and lung tumor cells adhering to HBMEC.
Fig. 2. Adhesion of tumor cells to human brain endothelial cells is attenuated by SGPs.
(A and B) Endothelial and calcein-labeled breast (A) or lung (B) tumor cells were exposed to SGP40, SGP65 (both normalized to 5 μM of Se), TNF-α (10 ng/mL, positive control), or vehicle (VEH) for 24 h. Representative fluorescence images from n=6 experiments are shown on right panels. Left panels show quantitative data from these experiments. (C and D) Endothelial and calcein-labeled tumor cells were exposed to SGP40, SGP65, or vehicle (VEH) as in (A and B). Then, TNF-α (10 ng/mL) or vehicle was added for an additional 20 h incubation. Values are mean ± SEM, n=6–8 wells per group. ***p<0.001, **p<0.01, *p<0.05 compared with vehicle (A and B) or TNF-α (C and D).
In the next series of experiments, tumor cells (MDA-MB231 and A549) and HBMEC were exposed to SGP40 or SGP65 for 24 h, followed by treatment with TNF-α (10 ng/mL) for the additional 20 hours (Fig. 2C and 2D). Co-treatment with TNF-α was used to more closely represent a natural environment during tumor cell invasion. TNF-α effectively increased adhesion of both tumor cell lines; however, incubation of MDA-MB231 cells with SGP40 attenuated this effect by ~40% (Fig. 2C). While SGP65 also protected against endothelial adhesion of MDA-MB231 cells, this effect was less potent than the impact of SGP40. Both SGP40 and SGP65 diminished adhesion of A549 cells to HBMEC, but this protection was less robust than for MDA-MB231 cells (Fig. 2D).
3.3. Transendothelial migration of breast and lung tumor cells is decreased by SGP40
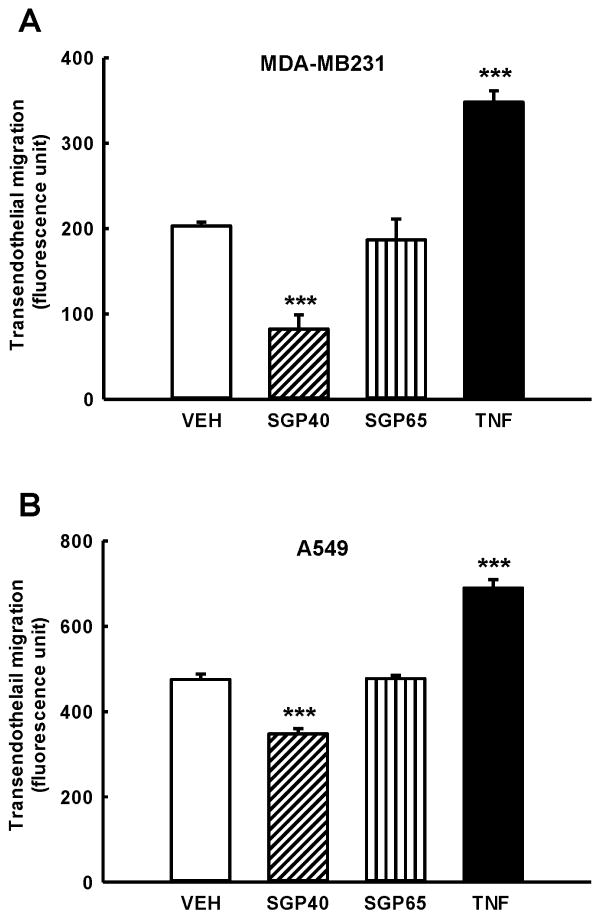
Migration of breast tumor cells (Fig. 3A) and lung tumor cells (Fig. 3B) through HBMEC monolayers was significantly attenuated in groups treated with SGP40 as compared to vehicle treated control (respectively: ~55% and 21% reduction). Exposure of breast and lung tumor cells to SGP65 did not affect transendothelial migration of tumor cells as compared to vehicle treated control. TNF-α, used as a positive control, effectively increased transmigration of breast and lung tumor cells when compared with vehicle (Fig. 3A–B).
Fig. 3. SGP40 decreases transendothelial migration of breast and lung tumor cells.
Brain endothelial cells, breast (A) and lung (B) tumor cells were exposed to SGP40, SGP65 (both normalized to 5 μM of Se), TNF-α (10 ng/mL, positive control), or vehicle (VEH) for 24 h. Values are mean ± SEM, n=6. ***p<0.001 compared with vehicle.
In the experiments in which cells were pretreated with SGP40 or SGP65, followed by exposure to TNF-α, selenoglycoproteins did not affect TNF-α-induced transendothelial migration of breast and lung tumor cells (data not shown). These results indicate that the SGPs were not successful in overcoming the robust effects caused by co-treatment with TNF-α. In addition, SGP40 or SGP65 did not protect against TNF-α-induced activation of MMP-2 and MMP-9 in HBMEC (data not shown).
3.4. NF-κB-dependent genes are downregulated by the exposure to selenium-containing compounds
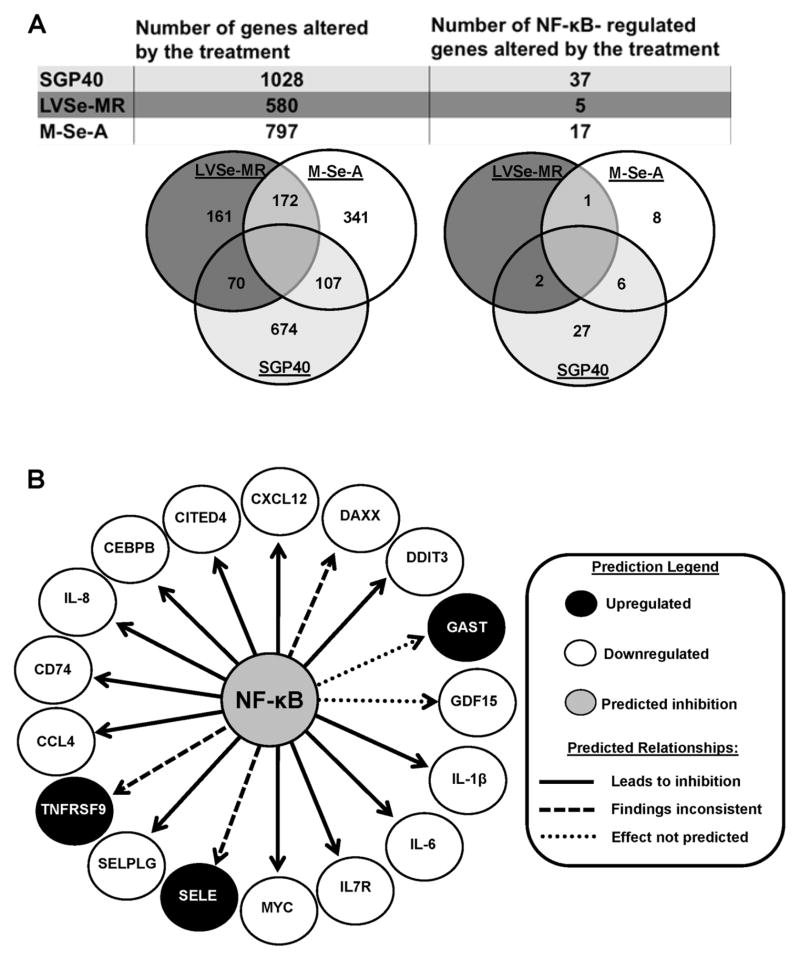
The gene expression profile of HBMEC and MDA-MB231 cells treated with SGP40, LVSe-MR, or M-Se-A in the presence of TNF-α was assessed using microarray technology with the Affymetrix GeneChip® Human Gene 1.0 ST Array. In HBMEC, 1028 genes were significantly altered by treatment with SGP40. The involvement of several pathways were tested; however, the majority of altered genes were related to the NF-κB-dependent pathway. Indeed, among differentially expressed genes 37 (24 downregulated and 23 upregulated) were found to be NF-κB-dependent (Fig. 4A). Exposure to LVSe-MR and M-Se-A caused significant changes in expression of 580 and 797 genes, respectively. Among them, 5 (in cultures exposed to LVSe-MR; 3 downregulated and 2 upregulated) and 17 (in cultures exposed to M-Se-A) genes were found to be dependent on NF-κB (Fig. 4A). For M-Se-A, the vast majority (14 out of 17) genes were downregulated and only 3 genes were upregulated. Classification by function revealed that the treatments altered expression of genes mainly responsible for inflammatory reactions, cellular movement, cell-to-cell signaling, cellular growth and proliferation. Tables 1–3 contain detailed lists of NF-κB-regulated genes in HBMEC, which were markedly altered by the treatments with SGP40, LVSe-MR, and M-Se-A, respectively. Pathway Prediction analysis based on the data obtained from M-Se-A-treated HBMEC is illustrated in Fig. 4B.
Fig. 4. Network and pathway analysis of DNA microarray results.
HBMEC were exposed for 24 h to SGP40, LVSe-MR, or M-Se-A (all normalized to 5 μM Se) or vehicle. Then, TNF-α (10 ng/mL) or vehicle was added for an additional 20 h. Total RNA was isolated and gene microarray analysis was performed. Differentially expressed gene signatures were analyzed using Ingenuity pathways analysis software. (A) Diagram illustrating the number of NF-κB-dependent and independent genes, which were altered by specific treatments. (B) Diagram illustrating the predicted targets under NF-κB regulation in HBMEC following administration of M-Se-A. CXCL12, chemokine ligand 12; DAXX, death-domain associated protein; DDIT3, DNA-damage-inducible transcript 3; GAST, gastrin; GDF15, growth differentiation factor 15; IL-1β, interleukin 1β; IL-6, interleukin 6; IL7R, interleukin 7 receptor; MYC, v-myc myelocytomatosis viral oncogene homolog; SELE, selectin E; SELPLG, selectin P ligand; TNFRSF9, tumor necrosis factor receptor superfamily, member 9; CCL4, chemokine (C-C motif) ligand 4; CD74, CD74 molecule, major histocompatibility complex, class II invariant chain; IL-8, interleukin 8; CEBPB, CCAAT/enhancer binding protein (C/EBP), beta; CITED4, Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain.
Table 1.
Down- and upregulation of specific NF-κB-regulated genes in HBMEC cells treated with SGP40.
| Gene symbol | Fold Change | Description |
|---|---|---|
| GDF15 | −1.6 | growth differentiation factor 15 |
| IL7R | −1.4 | interleukin 7 receptor |
| MYC | −1.3 | v-myc myelocytomatosis viral oncogene homolog |
| IL18 | −1.3 | interleukin 18 |
| TFPI2 | −1.3 | tissue factor pathway inhibitor 2 |
| CEBPB | −1.3 | CCAAT/enhancer binding protein (C/EBP), beta |
| ALDH1B1 | −1.3 | aldehyde dehydrogenase 1 family, member B1 |
| SPIB | −1.3 | Spi-B transcription factor |
| DDIT3 | −1.3 | DNA-damage-inducible transcript 3 |
| SCLY | −1.2 | selenocysteine lyase |
| HMOX1 | −1.2 | heme oxygenase (decycling) 1 |
| PPIF | −1.2 | peptidylprolyl isomerase F |
| IL6 | −1.2 | interleukin 6 |
| IL8 | −1.2 | interleukin 8 |
| TNF | 1.2 | tumor necrosis factor |
| CFLAR | 1.2 | CASP8 and FADD-like apoptosis regulator |
| F3 | 1.2 | coagulation factor III (thromboplastin, tissue factor) |
| BCL3 | 1.2 | B-cell CLL/lymphoma 3 |
| VCAM1 | 1.2 | vascular cell adhesion molecule 1 |
| IL24 | 1.2 | interleukin 24 |
| LTA | 1.2 | lymphotoxin alpha (TNF superfamily, member 1) |
| CX3CL1 | 1.2 | chemokine (C-X3-C motif) ligand 1 |
| MMP2 | 1.2 | matrix metallopeptidase 2 |
| IER3 | 1.2 | immediate early response 3 |
| KLF3 | 1.3 | Kruppel-like factor 3 (basic) |
| ABCB1 | 1.3 | ATP-binding cassette, sub-family B (MDR/TAP), member 1 |
| ELL2 | 1.3 | elongation factor, RNA polymerase II, 2 |
| TLR2 | 1.3 | toll-like receptor 2 |
| TMOD2 | 1.3 | tropomodulin 2 (neuronal) |
| CETP | 1.3 | cholesteryl ester transfer protein, plasma |
| MTSS1 | 1.3 | metastasis suppressor 1 |
| LIF | 1.4 | leukemia inhibitory factor |
| IFNE | 1.4 | interferon, epsilon |
| ABCG1 | 1.4 | ATP-binding cassette, sub-family G (WHITE), member 1 |
| ALDH1A3 | 1.5 | aldehyde dehydrogenase 1 family, member A3 |
| SELE | 1.7 | selectin E |
| CCL20 | 1.8 | chemokine (C-C motif) ligand 20 |
Table 3.
Down- and upregulation of specific NF-κB-regulated genes in HBMEC cells treated with M-Se-A
| Gene symbol | Fold Change | Description |
|---|---|---|
| IL7R | −1.3 | interleukin 7 receptor |
| IL1B | −1.3 | interleukin 1, beta |
| CEBPB | −1.3 | CCAAT/enhancer binding protein (C/EBP), beta |
| GDF15 | −1.3 | growth differentiation factor 15 |
| CD74 | −1.3 | CD74 molecule, major histocompatibility complex, class II invariant chain |
| CXCL12 | −1.3 | chemokine (C-X-C motif) ligand 12 |
| IL6 | −1.2 | interleukin 6 |
| SELPLG | −1.2 | selectin P ligand |
| DDIT3 | −1.2 | DNA-damage-inducible transcript 3 |
| CITED4 | −1.2 | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| MYC | −1.2 | v-myc myelocytomatosis viral oncogene homolog |
| DAXX | −1.2 | death-domain associated protein |
| CCL4 | −1.2 | chemokine (C-C motif) ligand 4 |
| IL8 | −1.2 | interleukin 8 |
| GAST | 1.2 | gastrin |
| TNFRSF9 | 1.3 | tumor necrosis factor receptor superfamily, member 9 |
| SELE | 1.3 | selectin E |
In MDA-MB231 cells, 854 genes were significantly altered by the treatment with SGP40. Among these, 26 (16 downregulated and 10 upregulated) genes were found to be NF-κB-dependent (Supplemental Fig. 1). LVSe-MR and M-Se-A caused significant changes in expression of 962 and 942 genes, respectively. Among them, 17 (in LVSe-MR-treated cells; 12 downregulated and 5 upregulated) and 26 (in M-Se-A-treated cells; 19 downregulated and 7 upregulated) genes were found to be dependent on NF-κB (Supplemental Fig. 1). Classification by function revealed that the treatments altered expression of genes mainly involved in inflammatory reactions, cellular movement, cell cycle regulation, cell-to-cell signaling, cellular growth and protein folding. Supplemental Tables 1–3 contain detailed lists of NF-κB-regulated genes, which were markedly altered by SGP40, LVSe-MR, and M-Se-A in MDA-MB231 cells.
3.5. Selenium-containing compounds inhibit NF-κB DNA binding activity and decrease expression of NF-κB-dependent proinflammatory mediators
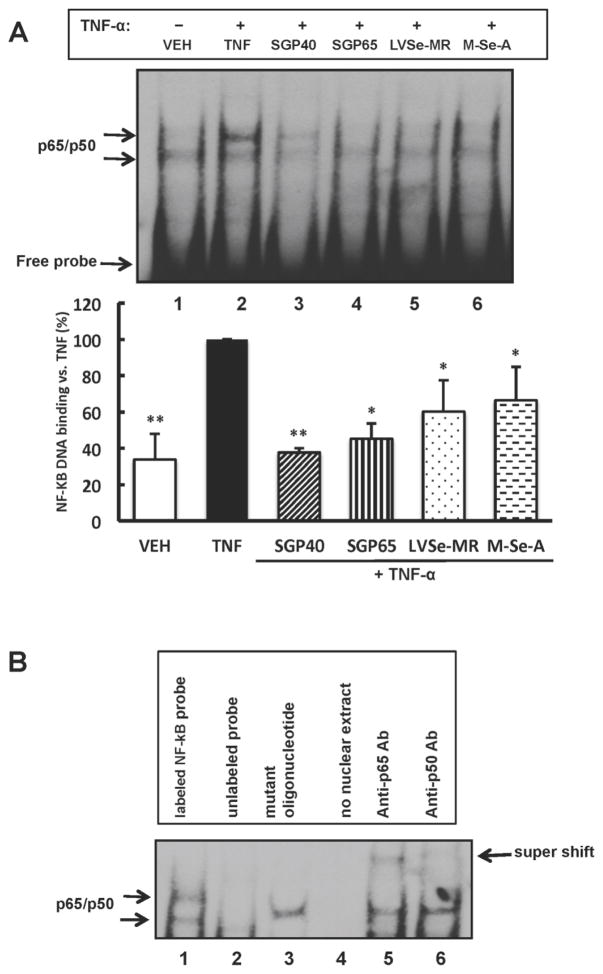
The notion that NF-κB pathway is involved in the protective effects of selenium compounds was further evaluated in the next series of experiments. A low background level of NF-κB DNA binding activity was detected by EMSA in nuclear extracts isolated from control HBMEC (Fig. 5A, lane 1); however, a 20 min exposure to TNF-α (10 ng/mL) markedly enhanced this binding (Fig. 5A, lane 2). Interestingly, a pre-treatment with SGPs or small organic selenium-containing molecules attenuated TNF-α-induced NF-κB binding (Fig. 5A, lanes 3–6). This protective effect was the most pronounced in SGP40 pre-exposed cells (Fig. 5A, lane 3). Specificity of NF-κB binding was demonstrated in experiments with molar excess of unlabeled NF-κB probe, mutated probe, and the super shift analyses with antibodies against NF-κB p50 or p65 (Fig. 5B).
Fig. 5. NF-κB DNA binding activity is inhibited by selenium-containing compounds.
(A) Confluent HBMEC were exposed to SGP40, SGP65, LVSe-MR, M-Se-A (all normalized to 5 μM Se) or vehicle for 24 h. Then TNF-α (10 ng/mL) or vehicle (VEH) was added for the additional 20 min. The image shows representative EMSA blot from n=4 experiments and the bar graph reflects quantitative data from these experiments. Results are mean ± SEM; **p<0.01 and *p<0.05 compared with TNF-α. (B) Specificity of NF-κB DNA binding. Lane 1, HBMEC exposed to TNF-α as in (A) and EMSA was performed as in (A). The assay was modified as follows: Lane 2, co-incubation with an access of unlabeled NF-κB probe; Lane 3, co-incubation with mutated NF-κB probe; Lane 4, no nuclear extract (negative control); Lane 5, co-incubation with anti-p65 antibody (supershift assay); Lane 6, co-incubation with anti-p50 antibody (supershift assay). Shifted bands on Lanes 5 and 6 are identified by arrow.
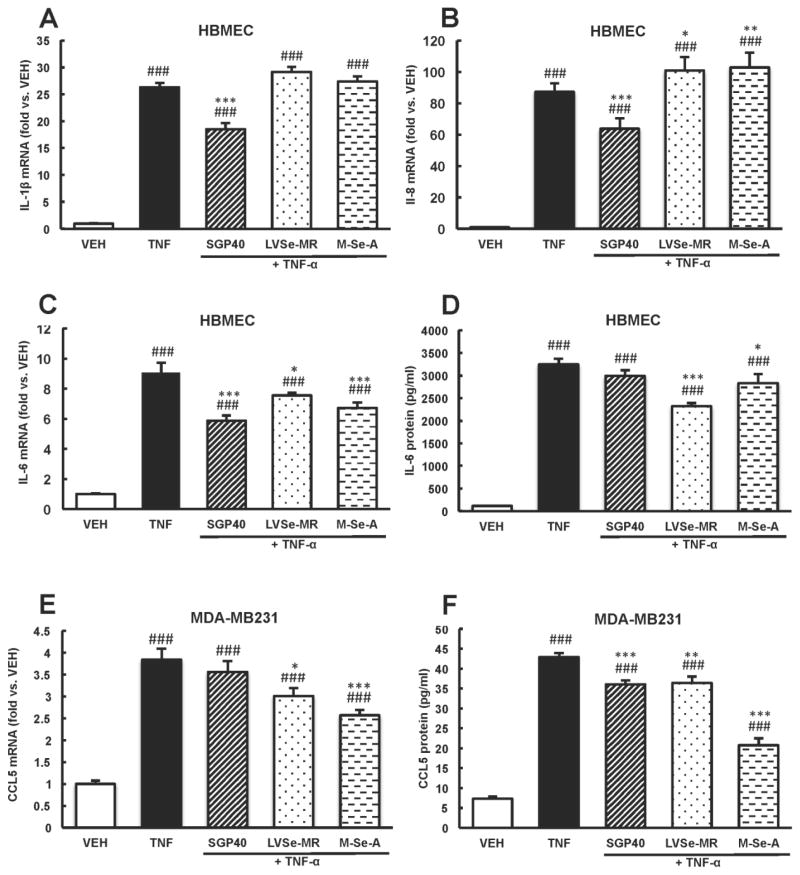
DNA microarray results and the pathway prediction analysis were further validated using qRT-PCR. The candidate genes for these analyses were selected based on the magnitude of responses and their role in tumor cell adhesion. TNF-α-induced upregulation of IL-1β, IL-8, and IL-6 mRNA levels was attenuated by ~30% in cultures pretreated with SGP40 (Fig. 6A–C). While LVSe-MR and M-Se-A diminished mRNA and protein levels of IL-6, no effect of these compounds on IL-1β and IL-8 expression was observed (Fig. 6A–D). In contrast, LVSe-MR and M-Se-A decreased TNF-α-induced CCL5 mRNA (by ~21% and 34%, respectively) and protein levels (by ~18% and 53%, respectively) in MDA-MB231 cells (Fig. 6E–F). SGP40 did not affect TNF-α-induced CCL5 mRNA levels (Fig. 6E); however, it resulted in ~18% reduction of CCL5 protein concentration as compared to TNF-α-treated cultures (Fig. 6F).
Fig. 6. Selenium-containing compounds decrease expression of NF-κB-dependent pro-inflammatory mediators.
HBMEC (A–D) and breast tumor cells (E and F) were incubated with SGP40, LVSe-MR, M-Se-A, TNF-α or vehicle (VEH) as in Fig. 4. mRNA levels of IL-1β (A), IL-8 (B), IL-6 (C), CCL5 (E) were assessed by qRT-PCR. Protein levels of IL-6 (D) and CCL5 (F) were determined by ELISA in cell culture media. Values are mean ± SEM, n= 5–6 wells per group. ***p<0.001, **p<0.01, *p<0.05 compared with TNF-α; ###p<0.001 compared with vehicle.
3.6. SGP40 attenuates tumor cell adhesion via a process involving NF-κB activation
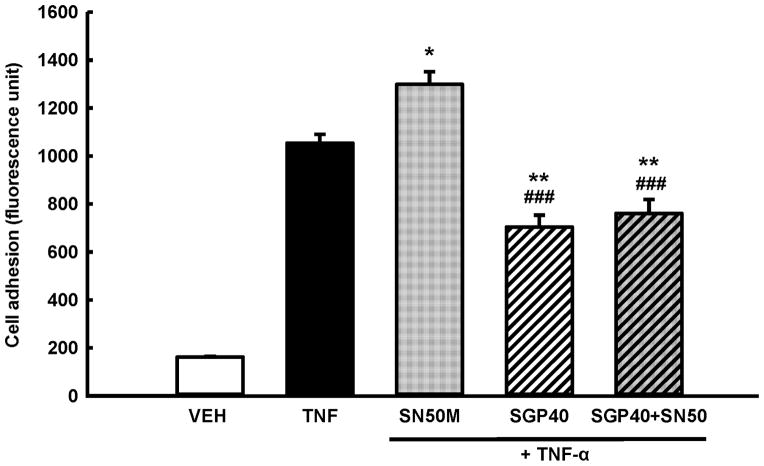
In order to evaluate if the suppression of NF-κB DNA binding activity by SGP40 may contribute for the observed decrease in adhesion of tumor cells to HBMEC, a series of experiments was performed in which additional co-treatment with SN50 (a specific inhibitor of NF-κB nuclear translocation) was employed (Fig. 7). Consistent with the results in Fig. 2 and 3, TNF-α effectively increased adhesion of breast tumor cells and a 24 h incubation with SGP40 attenuated this effect by ~35%. Importantly, treatment with SN50 did not cause any measureable changes in tumor cell adhesion to brain endothelial cell monolayers as compared to treatment with SGP40 alone, indicating the SGP40 acts via NF-κB inhibition. In contrast, pre-treatment with SN50M, a negative control for the SN50 peptide, resulted in increased cell adhesion by ~20% as compared to TNF-α treated cultures.
Fig. 7. SGP40 attenuates tumor cell adhesion to brain endothelial cells via a NF-κB-dependent process.
Endothelial and breast tumor cells were exposed to SGP40 (5 μM of Se) or vehicle for 24 h. Then the cultures were treated for 30 min with 18 μM of SN50 (an inhibitor of NF-κB nuclear translocation), SN50M (a negative control for the SN50 peptide with no measurable effect on NF-κB translocation at 18 μM) or vehicle (VEH). TNF-α (10 ng/mL) or vehicle was added for the additional 20 h incubation. Values are mean ± SEM, n= 8 wells per group. **p<0.01, *p<0.05 compared with TNF-α; ###p<0.001 compared with SN50M.
4. DISCUSSION
Circulating tumor cells are commonly present in the blood of patients with primary solid tumors. While these cells may home and establish metastases at distant sites, there are currently no therapies aimed at preventing the formation of secondary tumors at specific steps during tumor progression [24]. The current study explored nutritional-based intervention specifically targeting adhesion and transendothelial migration of tumor cells. We focused on selenium-containing compounds based on our previous observations that feeding mice with selenium-enriched yeast markedly decreased growth of brain metastatic tumors [17].
The circulatory patterns between the primary tumor and secondary target organs are largely responsible for the formation of organ-specific metastases [25]. Lung and breast cancers represent the most common primary tumors metastasizing to the brain and since the central nervous system does not have lymphatic vessels, the only entry route for tumor cells into the brain is via blood stream [11, 26, 27]. Therefore, the interactions between tumor cells and the brain vascular endothelium are crucial for the formation and growth of brain secondary tumors. These events were mimicked in the present study by co-culture of breast (MDA-MB231) and lung (A549) tumor cells with brain endothelial cells (HBMEC). Because adhesion and transendothelial migration of tumor cells are the critical and rate limiting initial steps responsible for the formation of secondary tumors [28, 29], the main emphasis of the present study was placed on those early events.
The current study provides evidence that specific SGPs extracted from selenium-enriched yeast possess the ability to attenuate adhesion of tumor cells to human brain endothelial cells. These effects were most prominent in cultures treated with SGP40 both alone and in co-treatment with TNF-α. SGP40 also attenuated transendothelial migration of tumor cells, suggesting that the diminished transmigration resulted from the decreased number of adhering tumor cells to endothelial monolayers. These observations are consistent with the notion that tumor cell adhesion and transendothelial migration are two, closely associated albeit distinct, steps in tumor progression [24, 25]. Further experiments revealed that TNF-α-induced MMP-2 and MMP-9 activities, which have been shown to correlate with enhanced tumor progression [15, 24, 30], were not affected by SGP40 or SGP65, indicating an alternative mechanism of diminished affectivity of these compounds in protection against transendothelial migration of tumor cells.
NF-κB is a transcription factor and a master regulator of pro-inflammatory reactions due to its role in the induction of a variety of genes involved in vascular inflammation, including cell adhesion molecules, cytokines, and chemokines [31]. Therefore, it was important that exposure to SGP40 and small selenium-containing organic compounds, such as LVSe-MR and M-Se-A, reduced DNA binding activity of NF-κB in HBMEC. The most robust effects were observed in cells treated with SGP40, which is consistent with strong protection against tumor cell adhesion and transendothelial migration by this compound. Additional experiments in which an inhibitor of NF-κB nuclear translocation was employed, supported the hypothesis that inhibition of NF-κB binding was, at least partially, responsible for decreased interactions between tumor and endothelial cells. Indeed, NF-κB inhibition was linked to reduced expression of numerous pro-metastatic genes, including chemokine receptor CXCR4 [32–34]. Thus, targeting NF-κB activity appears to be a promising strategy aimed to improve the efficacy of anticancer therapies and prevent metastasis formation [35–38]. Established chemopreventive agents frequently act via NF-κB signaling pathways, making selenium-containing compounds attractive candidates for anti-cancer and anti-metastatic therapeutic strategies [32, 37].
Proinflammatory cytokines and chemokines are the major mediators of communication between cells in the inflammatory tumor microenvironment and their overexpression is associated with cancer progression [39]. Consistent with the results on NF-κB activation, supplementation with SGP40 or selenium-containing small compounds protected against expression of NF-κB-dependent IL-1β, IL-6, IL-8 and CCL5 genes. Compelling evidence indicates the role of these molecules in cancer progression. For example, 3-methylcholanthrene-induced skin carcinogenesis was delayed in IL-1β-deficient mice compared to wild-type animals [40]. It was also demonstrated that IL-1β enhances tumor cell adhesion to the vascular surface [41, 42]. IL-6 and several IL-6-related signaling pathways have been identified as important in transmitting signals associated with tumor cell invasion. For example, it has been shown that IL-6 enhances glioblastoma cell transendothelial migration in a dose-dependent manner [43]. Additionally, overexpression of IL-6 in specific organs, including brain, attracts circulating tumor cells and promotes metastatic progression [44]. IL-8 is known to be a potent mediator of angiogenesis and has been reported to activate CXCR1 and CXCR2 receptors on endothelial cells [45], which contribute to increased vascular permeability [45, 46]. Furthermore, several clinical studies have observed overexpression of IL-8 in advanced stages of cancer, suggesting that IL-8 suppression may serve as treatment strategies for aggressive and metastatic tumors [47]. CCL5 signaling has also been linked to cancer progression, as CCL5 levels in the plasma of breast cancer patients have been correlated with the severity of the disease [48, 49]. It has also been suggested that a variety of CCL5 analogues and antagonists of the CCR5 receptor may be utilized in treatment of metastatic disease [49, 50]. Importantly, cancer cell extravasation was identified as the crucial metastatic step affected by CCL5/CCR5 inhibition [50].
In summary, the present study indicates that specific selenoglycoproteins extracted from selenium-enriched yeast have the ability to attenuate adhesion of breast and lung tumor cells to brain endothelial cells via a process involving inhibition of NF-κB pathway. While small organic selenium-containing compounds, LVSe-MR and M-Se-A, may be involved in the observed effects, the most robust protection was observed in cells treated with SGP40, indicating not only the role of selenium but also the importance of the glycoprotein component. Impairment of tumor cell homing is a highly desirable strategy; therefore selenium-containing compounds may constitute valuable options for adjuvant therapies aiming at the inhibition of early steps in cancer progression.
Supplementary Material
Table 2.
Down- and upregulation of specific NF-κB-regulated genes in HBMEC cells treated with LVSe-MR
| Gene symbol | Fold Change | Description |
|---|---|---|
| IL7R | −1.3 | interleukin 7 receptor |
| ALDH1B1 | −1.2 | aldehyde dehydrogenase 1 family, member B1 |
| IL1B | −1.2 | interleukin 1, beta |
| SELE | 1.3 | selectin E |
| IFNE | 1.3 | interferon, epsilon |
Acknowledgments
FUNDING SOURCES
The authors would like to thank Dr. Luc Bertrand for his valuable advice. The authors also would like to thank the other members of the laboratory for their crucial assistance and constructive discussions. The study was supported by the grants from the National Institutes of Health CA133257 and Alltech Nutrigenomics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Taylor PR, Greenwald P. Nutritional interventions in cancer prevention. J Clin Oncol. 2005;23:333–45. doi: 10.1200/JCO.2005.06.190. [DOI] [PubMed] [Google Scholar]
- 2.Meadows GG. Diet, nutrients, phytochemicals, and cancer metastasis suppressor genes. Cancer Metastasis Rev. 2012;31:441–54. doi: 10.1007/s10555-012-9369-5. [DOI] [PubMed] [Google Scholar]
- 3.Chambers AF. Influence of diet on metastasis and tumor dormancy. Clin Exp Metastasis. 2009;26:61–6. doi: 10.1007/s10585-008-9164-4. [DOI] [PubMed] [Google Scholar]
- 4.Sarkar FH, Li Y. Using chemopreventive agents to enhance the efficacy of cancer therapy. Cancer Res. 2006;66:3347–50. doi: 10.1158/0008-5472.CAN-05-4526. [DOI] [PubMed] [Google Scholar]
- 5.Patrick L. Selenium biochemistry and cancer: a review of the literature. Altern Med Rev. 2004;9:239–58. [PubMed] [Google Scholar]
- 6.Fang W, Goldberg ML, Pohl NM, Bi X, Tong C, Xiong B, et al. Functional and physical interaction between the selenium-binding protein 1 (SBP1) and the glutathione peroxidase 1 selenoprotein. Carcinogenesis. 2010;31:1360–6. doi: 10.1093/carcin/bgq114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selenius M, Rundlof AK, Olm E, Fernandes AP, Bjornstedt M. Selenium and the selenoprotein thioredoxin reductase in the prevention, treatment and diagnostics of cancer. Antioxid Redox Signal. 2010;12:867–80. doi: 10.1089/ars.2009.2884. [DOI] [PubMed] [Google Scholar]
- 8.Rayman MP. The importance of selenium to human health. Lancet. 2000;356:233–41. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 9.Chen YC, Prabhu KS, Mastro AM. Is selenium a potential treatment for cancer metastasis? Nutrients. 2013;5:1149–68. doi: 10.3390/nu5041149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen YC, Prabhu KS, Das A, Mastro AM. Dietary selenium supplementation modifies breast tumor growth and metastasis. Int J Cancer. 2013;133:2054–64. doi: 10.1002/ijc.28224. [DOI] [PubMed] [Google Scholar]
- 11.Eichler AF, Chung E, Kodack DP, Loeffler JS, Fukumura D, Jain RK. The biology of brain metastases-translation to new therapies. Nat Rev Clin Oncol. 2011;8:344–56. doi: 10.1038/nrclinonc.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Zijl F, Krupitza G, Mikulits W. Initial steps of metastasis: cell invasion and endothelial transmigration. Mutat Res. 2011;728:23–34. doi: 10.1016/j.mrrev.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sternlicht MD, Lochter A, Sympson CJ, Huey B, Rougier JP, Gray JW, et al. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell. 1999;98:137–46. doi: 10.1016/s0092-8674(00)81009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eum SY, Lee YW, Hennig B, Toborek M. Interplay between epidermal growth factor receptor and Janus kinase 3 regulates polychlorinated biphenyl-induced matrix metalloproteinase-3 expression and transendothelial migration of tumor cells. Mol Cancer Res. 2006;4:361–70. doi: 10.1158/1541-7786.MCR-05-0119. [DOI] [PubMed] [Google Scholar]
- 15.Nabeshima K, Inoue T, Shimao Y, Sameshima T. Matrix metalloproteinases in tumor invasion: role for cell migration. Pathol Int. 2002;52:255–64. doi: 10.1046/j.1440-1827.2002.01343.x. [DOI] [PubMed] [Google Scholar]
- 16.Sipos E, Chen L, Andras IE, Wrobel J, Zhang B, Pu H, et al. Proinflammatory adhesion molecules facilitate polychlorinated biphenyl-mediated enhancement of brain metastasis formation. Toxicol Sci. 2012;126:362–71. doi: 10.1093/toxsci/kfr349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wrobel JK, Seelbach MJ, Chen L, Power RF, Toborek M. Supplementation with selenium-enriched yeast attenuates brain metastatic growth. Nutr Cancer. 2013;65:563–70. doi: 10.1080/01635581.2013.775315. [DOI] [PubMed] [Google Scholar]
- 18.Weksler BB, Subileau EA, Perriere N, Charneau P, Holloway K, Leveque M, et al. Blood-brain barrier-specific properties of a human adult brain endothelial cell line. FASEB J. 2005;19:1872–4. doi: 10.1096/fj.04-3458fje. [DOI] [PubMed] [Google Scholar]
- 19.Bierla K, Bianga J, Ouerdane L, Szpunar J, Yiannikouris A, Lobinski R. A comparative study of the Se/S substitution in methionine and cysteine in Se-enriched yeast using an inductively coupled plasma mass spectrometry (ICP MS)-assisted proteomics approach. J Proteomics. 2013;87:26–39. doi: 10.1016/j.jprot.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Braut-Boucher F, Pichon J, Rat P, Adolphe M, Aubery M, Font J. A non-isotopic, highly sensitive, fluorimetric, cell-cell adhesion microplate assay using calcein AM-labeled lymphocytes. J Immunol Methods. 1995;178:41–51. doi: 10.1016/0022-1759(94)00239-s. [DOI] [PubMed] [Google Scholar]
- 21.Xiao R, Power RF, Mallonee D, Crowdus C, Brennan KM, Ao T, et al. A comparative transcriptomic study of vitamin E and an algae-based antioxidant as antioxidative agents: investigation of replacing vitamin E with the algae-based antioxidant in broiler diets. Poult Sci. 2011;90:136–46. doi: 10.3382/ps.2010-01018. [DOI] [PubMed] [Google Scholar]
- 22.Hazane-Puch F, Champelovier P, Arnaud J, Garrel C, Ballester B, Faure P, et al. Long-term selenium supplementation in HaCaT cells: importance of chemical form for antagonist (protective versus toxic) activities. Biol Trace Elem Res. 2013;154:288–98. doi: 10.1007/s12011-013-9709-5. [DOI] [PubMed] [Google Scholar]
- 23.Park JS, Ryu JY, Jeon HK, Cho YJ, Park YA, Choi JJ, et al. The effects of selenium on tumor growth in epithelial ovarian carcinoma. J Gynecol Oncol. 2012;23:190–6. doi: 10.3802/jgo.2012.23.3.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labelle M, Hynes RO. The initial hours of metastasis: the importance of cooperative host-tumor cell interactions during hematogenous dissemination. Cancer Discov. 2012;2:1091–9. doi: 10.1158/2159-8290.CD-12-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–72. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 26.Lassman AB, DeAngelis LM. Brain metastases. Neurol Clin. 2003;21:1–23. vii. doi: 10.1016/s0733-8619(02)00035-x. [DOI] [PubMed] [Google Scholar]
- 27.Wolff G, Toborek M. Targeting the therapeutic effects of exercise on redox-sensitive mechanisms in the vascular endothelium during tumor progression. IUBMB Life. 2013;65:565–71. doi: 10.1002/iub.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen LM, Kuo CH, Lai TY, Lin YM, Su CC, Hsu HH, et al. RANKL increases migration of human lung cancer cells through intercellular adhesion molecule-1 up-regulation. J Cell Biochem. 2011;112:933–41. doi: 10.1002/jcb.23009. [DOI] [PubMed] [Google Scholar]
- 29.Schluter K, Gassmann P, Enns A, Korb T, Hemping-Bovenkerk A, Holzen J, et al. Organ-specific metastatic tumor cell adhesion and extravasation of colon carcinoma cells with different metastatic potential. Am J Pathol. 2006;169:1064–73. doi: 10.2353/ajpath.2006.050566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo J. Role of matrix metalloproteinase-2 in ethanol-induced invasion by breast cancer cells. J Gastroenterol Hepatol. 2006;21 (Suppl 3):S65–8. doi: 10.1111/j.1440-1746.2006.04578.x. [DOI] [PubMed] [Google Scholar]
- 31.Csiszar A, Wang M, Lakatta EG, Ungvari Z. Inflammation and endothelial dysfunction during aging: role of NF-kappaB. J Appl Physiol (1985) 2008;105:1333–41. doi: 10.1152/japplphysiol.90470.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aggarwal BB. Nuclear factor-kappaB: the enemy within. Cancer Cell. 2004;6:203–8. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 33.Fujioka S, Sclabas GM, Schmidt C, Frederick WA, Dong QG, Abbruzzese JL, et al. Function of nuclear factor kappaB in pancreatic cancer metastasis. Clinical cancer research: an official journal of the American Association for Cancer Research. 2003;9:346–54. [PubMed] [Google Scholar]
- 34.Helbig G, Christopherson KW, 2nd, Bhat-Nakshatri P, Kumar S, Kishimoto H, Miller KD, et al. NF-kappaB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J Biol Chem. 2003;278:21631–8. doi: 10.1074/jbc.M300609200. [DOI] [PubMed] [Google Scholar]
- 35.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaturvedi MM, Sung B, Yadav VR, Kannappan R, Aggarwal BB. NF-kappaB addiction and its role in cancer: ‘one size does not fit all’. Oncogene. 2011;30:1615–30. doi: 10.1038/onc.2010.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–6. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 38.Nakanishi C, Toi M. Nuclear factor-kappaB inhibitors as sensitizers to anticancer drugs. Nat Rev Cancer. 2005;5:297–309. doi: 10.1038/nrc1588. [DOI] [PubMed] [Google Scholar]
- 39.Candido J, Hagemann T. Cancer-related inflammation. J Clin Immunol. 2013;33 (Suppl 1):S79–84. doi: 10.1007/s10875-012-9847-0. [DOI] [PubMed] [Google Scholar]
- 40.Krelin Y, Voronov E, Dotan S, Elkabets M, Reich E, Fogel M, et al. Interleukin-1beta-driven inflammation promotes the development and invasiveness of chemical carcinogen-induced tumors. Cancer Res. 2007;67:1062–71. doi: 10.1158/0008-5472.CAN-06-2956. [DOI] [PubMed] [Google Scholar]
- 41.Lauri D, Bertomeu MC, Orr FW, Bastida E, Sauder D, Buchanan MR. Interleukin-1 increases tumor cell adhesion to endothelial cells through an RGD dependent mechanism: in vitro and in vivo studies. Clin Exp Metastasis. 1990;8:27–32. doi: 10.1007/BF00155590. [DOI] [PubMed] [Google Scholar]
- 42.Giavazzi R, Garofalo A, Bani MR, Abbate M, Ghezzi P, Boraschi D, et al. Interleukin 1-induced augmentation of experimental metastases from a human melanoma in nude mice. Cancer Res. 1990;50:4771–5. [PubMed] [Google Scholar]
- 43.Liu Q, Li G, Li R, Shen J, He Q, Deng L, et al. IL-6 promotion of glioblastoma cell invasion and angiogenesis in U251 and T98G cell lines. J Neurooncol. 2010;100:165–76. doi: 10.1007/s11060-010-0158-0. [DOI] [PubMed] [Google Scholar]
- 44.Ara T, Declerck YA. Interleukin-6 in bone metastasis and cancer progression. Eur J Cancer. 2010;46:1223–31. doi: 10.1016/j.ejca.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schraufstatter IU, Chung J, Burger M. IL-8 activates endothelial cell CXCR1 and CXCR2 through Rho and Rac signaling pathways. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1094–103. doi: 10.1152/ajplung.2001.280.6.L1094. [DOI] [PubMed] [Google Scholar]
- 46.Brat DJ, Bellail AC, Van Meir EG. The role of interleukin-8 and its receptors in gliomagenesis and tumoral angiogenesis. Neuro Oncol. 2005;7:122–33. doi: 10.1215/S1152851704001061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Waugh DJ, Wilson C. The interleukin-8 pathway in cancer. Clin Cancer Res. 2008;14:6735–41. doi: 10.1158/1078-0432.CCR-07-4843. [DOI] [PubMed] [Google Scholar]
- 48.Niwa Y, Akamatsu H, Niwa H, Sumi H, Ozaki Y, Abe A. Correlation of tissue and plasma RANTES levels with disease course in patients with breast or cervical cancer. Clin Cancer Res. 2001;7:285–9. [PubMed] [Google Scholar]
- 49.Velasco-Velazquez M, Jiao X, De La Fuente M, Pestell TG, Ertel A, Lisanti MP, et al. CCR5 antagonist blocks metastasis of basal breast cancer cells. Cancer Res. 2012;72:3839–50. doi: 10.1158/0008-5472.CAN-11-3917. [DOI] [PubMed] [Google Scholar]
- 50.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.