Abstract
Bacteriophages are the predominant biological entity on the planet. The recent explosion of sequence information has made estimates of their diversity possible. We describe the genomic comparison of 337 fully sequenced tailed phages isolated on 18 genera and 31 species of bacteria in the Enterobacteriaceae. These phages were largely unambiguously grouped into 56 diverse clusters (32 lytic and 24 temperate) that have syntenic similarity over >50% of the genomes within each cluster, but substantially less sequence similarity between clusters. Most clusters naturally break into sets of more closely related subclusters, 78% of which are correlated with their host genera. The largest groups of related phages are superclusters united by genome synteny to lambda (81 phages) and T7 (51 phages). This study forms a robust framework for understanding diversity and evolutionary relationships of existing tailed phages, for relating newly discovered phages and for determining host/phage relationships.
Keywords: bacteriophage, tailed phage, Caudovirales, Enterobacteriaceae
INTRODUCTION
Experimental study of tailed bacteriophages was seminal in the attainment of our current understanding of many aspects of the nature and function of nucleic acids, genes and proteins (e.g., Luria and Delbruck, 1943; Hershey and Chase, 1952; Cairns et al., 1966). Because of their unique morphology, tailed phage virions can be unambiguously recognized in environmental samples. It is estimated that there are at least 1031 such virions in Earth’s biosphere, making them the most abundant type of biological entity on our planet (Bergh et al., 1989; Wommack and Colwell, 2000; Brussow and Hendrix, 2002; Wilhelm et al., 2002; Hendrix, 2003; Hambly and Suttle, 2005; Suttle, 2005). Their abundance and ability to kill their host bacteria give the tailed phages critical roles in important global and local ecological processes. For example, it has been argued that oceanic phages infect bacterial cells sufficiently often (as many as 1029 phage infections per day) that they release over 1011 kilograms of carbon per day from the biological pool (Suttle, 2007; Brussaard et al., 2008).
In addition to killing their bacterial hosts, temperate phage genomes can carry toxin and other critical virulence factor genes that are important in many human bacterial pathogens (e.g., Casjens, 2003; Canchaya et al., 2004; Casjens and Hendrix, 2005; Chen and Novick, 2009; Oliver et al., 2009; Casas and Maloy, 2011; Boyd, 2012; Fortier and Sekulovic, 2013). Phage also contribute to the diversity of the bacterial community by serving as vectors for transduction of different genetic alleles, such as antibiotic resistance genes, between bacterial cells (e.g., Chen and Novick, 2009; Modi et al., 2013; Bearson et al., 2014; Volkova et al., 2014).
Finally, phages have great medical and nanotechnological potential. Strategies for using tailed phage for detecting bacteria, curing bacterial disease (phage therapy) or decontaminating surfaces have been used for almost 100 years in Russia and Georgia and are currently used to treat agricultural diseases as well as food contamination in the West. In addition, phage virions and related particles are being developed as nanocontainers for specific chemical cargoes that can be delivered to specific targets (e.g., Nam et al., 2006; Lee et al., 2009; Shen et al., 2010; Cardinale et al., 2012; Hyman, 2012; Culpepper et al., 2013; Farkas et al., 2013; Farr et al., 2013; Kale et al., 2013; Qazi et al., 2013).
In spite of their great importance in these varied arenas, a number of factors have inhibited a clear understanding of tailed phage diversity and evolutionary relationships. (i) In the few cases where sufficient numbers of phages have been isolated and studied for a particular host, it is clear that at least tens of very different tailed phage ‘types’ exist that can successfully infect a single bacterial species. For example, hundreds of tailed phages have been isolated that infect the ‘model’ bacteria Escherichia coli, and these are currently placed in 16 different Caudovirales phage genera by the International Committee on Taxonomy of Viruses (http://www.ictvonline.org/virusTaxonomy.asp). The most phages isolated for a single bacterial species is the collection from the Science Education Alliance Phage Hunters program (Hatfull et al., 2006), where over 4700 phages have been isolated and 680 fully sequenced for the Gram-positive host Mycobacterium smegmatis (phagesdb.org). These phages fall into 30 distinct types (called clusters) that have little enough nucleotide sequence similarity to each other to allow the unambiguous assignment of clusters (Hatfull, 2014). Thus, since it has been estimated that there may be at least a billion bacterial species on the planet (Dykhuizen, 1998; Dykhuizen, 2005), the number of tailed phage types is certainly extremely large. This predicted extent of diversity is daunting to an overall understanding of the tailed phages. (ii) Tailed phage virion morphologies are unique and different from other viruses, but many tailed phage which are very similar to one another when viewed with the electron microscope in fact have very different genomes. Thus, nucleotide sequence information (preferably whole genome sequence) is required to understand the relationships among the members of any set of phages being compared. (iii) Viruses would seem to have an evolutionary potential to move between host species, making host species imperfect indicators of phage relatedness; however, we know of no well-documented case of a single tailed phage isolate successfully infecting very distantly related bacterial hosts. Nonetheless, phages are thought to move between related hosts at least in part through physical exchange of tail fiber genes (Haggard-Ljungquist et al., 1992; Sandmeier et al., 1992; Pickard et al., 2008; Casjens and Thuman-Commike, 2011; Jacobs-Sera et al., 2012). Although a few phages are said to have a broad-host-range (e.g., uncharacterized phages BHR1-5 (Jensen et al., 1998) and WHR 8 and 10 (Bielke et al., 2007) which infect hosts from different genera or families within the same order, most are extremely specific for single bacterial species and even serovars/biovars/subspecies. (iv) Finally, evidence has accumulated for a considerable amount of horizontal transfer of genetic material among tailed phages, both within and between largely nonhomologous types. The frequencies of such exchanges in the wild are not known (e.g., George et al., 1983; Haggard-Ljungquist et al., 1992; Sandmeier et al., 1992; Stummeyer et al., 2006; Pickard et al., 2010; Casjens and Thuman-Commike, 2011; Jacobs-Sera et al., 2012), but some measurements have been performed in the laboratory (e.g., De Paepe et al., 2014). Such horizontal exchange complicates whole phage genome comparisons and will make any hierarchical classification scheme nonsensical if it is great enough (Lawrence et al., 2002). However, recent analyses suggest that horizontal exchange does not appear to be so rapid that it destroys the overall relationships within or between ‘phage types’ (Casjens, 2005; Hatfull, 2014). Such types are often referred to as ‘species’ or as ‘genera’ (King et al., 2012), but we refrain from using these terms for phages; to quote Charles Darwin (1859), “…to discuss whether they are rightly called species or varieties, before any definition of these terms has been accepted, is vainly to beat the air”.
In order to further understanding of tailed phage diversity and evolution we examined the genomes of the tailed phages that infect a range of related hosts, the members of the γ-Proteobacteria family Enterobacteriaceae. The fact that hundreds of such phage genomes have been sequenced that include the best characterized ‘model system’ phages (such as λ, T1, T4, T5, T7, N4, P1, P2, P22 and Mu) makes this comparison more informed than comparisons of less well-understood phages that infect less well-understood hosts. In addition, phages have been characterized that infect a number of Enterobacteriaceae genera, allowing a unique opportunity to compare phages that infect related but distinct hosts. The analysis presented forms a framework for comprehending the diversity of phages that infect Enterobacteriaceae, for understanding the relationships of phages that will be isolated in the future, and for comparing these relationships with the relationships among phages that infect other distantly related hosts.
RESULTS AND DISCUSSION
Over three hundred tailed phages that infect Enterobacteriaceae hosts
Recently the pace of tailed phage whole genome sequence determination has accelerated due to increased interest in the potential practical uses of phages and decreased sequencing costs. We searched the extant sequence database at NCBI (Benson et al., 2013; Pruitt et al., 2009) for tailed phages that infect bacteria in the family Enterobacteriaceae. In order to ensure an accurate analysis of the relationships among these phages, this search was limited to phages whose genomes have been completely sequenced. After this initial search we used BLASTn and BLASTp (Altschul et al., 1990) searches with multiple sequences from each phage type (see below) to find relatives that were not present in our initial database (this was necessary because inclusion of specific information about phage hosts has not been rigorously adhered to in GenBank annotations). As of June 1, 2014 our database contained 337 complete or very close to complete genome sequences (a few nearly complete sequences were included to examine specific diversity issues). Space considerations preclude text citations for most individual phages but Table S1 lists the phages in this study with accession numbers and literature references for their isolation and genomic sequencing. Several times this number of prophages (temperate phage genomes integrated into their bacterial host’s chromosome or that exist as plasmids) are present, but are largely unannotated, in Enterobacteriaceae bacterial genome sequences. These were not included in the present analysis unless they have been the focus of a publication. Prophage diversity will be discussed in a subsequent publication.
In categorizing phages we use the term ‘clusters’ for groups of similar phages according to the usage of Hatfull and co-workers (Hatfull et al., 2010; Hatfull et al., 2013) and to avoid confusion with previously used terms such as ‘types’, ‘groups’ or current formal taxonomic classifications. The definition of such a cluster is ≥50% of the nucleotide sequence of the genome is recognizably similar and syntenic (50% homology span length by dot plot analysis at word length 10) to other members of the cluster. We include phages in a cluster whose genome is cumulatively >50% similar by dot plot analysis to other members of the cluster (i.e., some clusters are transitive sets of phages, where A is similar to B, B is similar to C, but C is not very similar to A). This definition creates groups of related phages that correspond quite well to groups such as the T4-like or T7-like phages previously used by workers in the phage research field. We use the term ‘subcluster’ to denote more closely related groups within clusters; this definition is not perfectly quantitative as subclusters are meant to point out relationships within clusters and not connote specific levels of difference. We also use the term ‘supercluster’ to include phages whose encoded gene functions (functional gene order) are largely syntenic and whose parallel proteins show recognizable relationships that may not be detected in the nucleotide sequence. Placing phages into clusters can in a few instances be somewhat ambiguous because of (i) the occasional transitive relationship of phages, (ii) the fact that similarity can vary from near nucleotide sequence identity to barely recognizable relationships among encoded proteins, and (iii) past horizontal exchange of genetic information between phages. In a small number of cases we chose not to merge clusters on the basis of one or a few phages that contain substantial sections representative of two different clusters; we believe it is more informative and useful to retain the clusters defined here and view these few phages as the result of horizontal exchange between clusters (e.g., the lambda and T7 superclusters below).
Relationships among tailed phages that infect Enterobacteriaceae
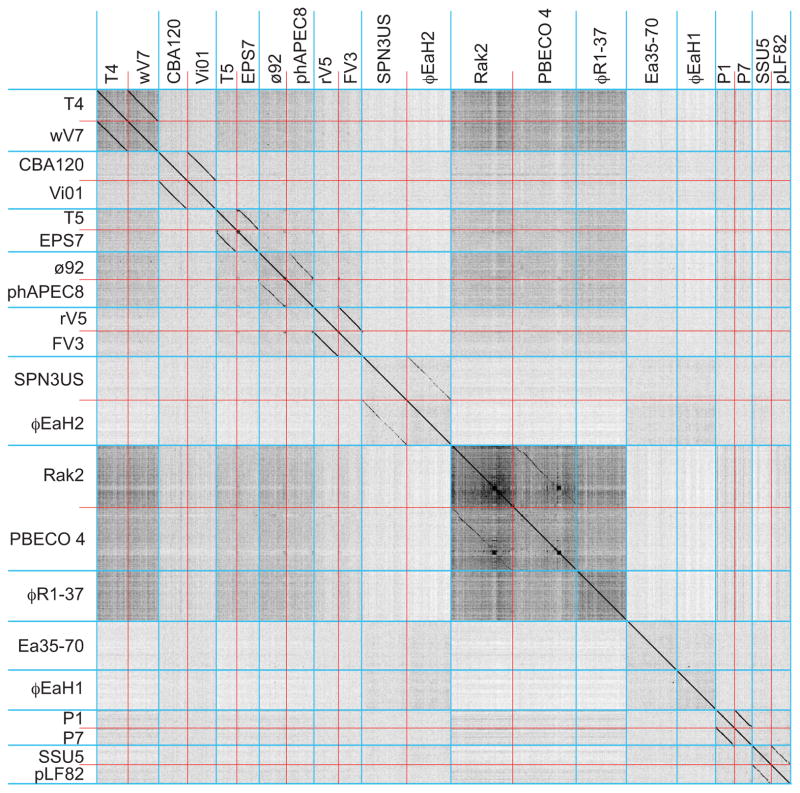
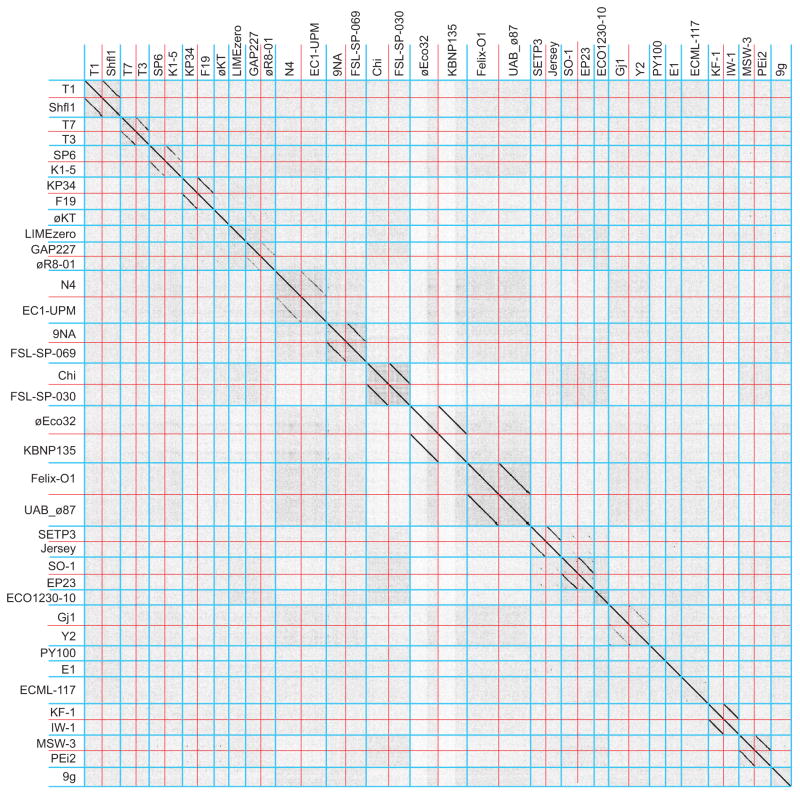
Extensive matrix Gepard dot plot comparisons were made among the 337 phages in our database of Enterobacteriaceae tailed phage genomes. These were examined manually for diagonal lines that indicate similarity and synteny (see METHODS AND MATERIALS for the curation of our phage sequence database and detailed comparison methods). The sequences aggregated convincingly into 56 ‘clusters’ of similar genomes. Table 1 lists these clusters and the number of phages in each cluster, and Table S1 lists all the phages in this study with their cluster designations. In order to demonstrate the existence of these clusters and allow easy visualization of individual phage genomes, Figures 1, 2 and 3 compare representative members from each of the 56 clusters from the larger phages (genomes >90 kbp), small lytic phages (<90 kbp) and small temperate phages (<90 kbp), respectively (comparisons between these three groups show no strong similarities among the clusters; not shown). Figures S1 (and 4, 5, 6 and 7 below) show genome dot plots that include all 337 phages in this study. Nearly all of the clusters with a significant number of members can be unambiguously divided into more highly related subclusters, and in the 56 clusters we recognize 132 subcluster level divisions (including singleton clusters as one subcluster; listed in tables 1 and S1).
Table 1.
Enterobacteriaceae tailed phage clusters
| Cluster1 | Prototype phage | Number of phages | Genome sizes (kbp) | Virion morphology, DNA base modification | Virion chromosome structure4 |
|---|---|---|---|---|---|
| Lytic 1 | T1 | 16 (7) | 43–52 | Siphoviridae8 | Partial CP, TR |
| Lytic 2 | T4 | 37 (10) | 160–181 | Myoviridae8, modified DNA | CP, TR |
| Lytic 3 | Vi01 | 16 (4) | 152–163 | Myoviridae, modified DNA | Circular assembly |
| Lytic 4 | T5 | 7 (2) | 104–123 | Siphoviridae | Long DTR |
| Supercluster | T7 | ||||
| Lytic 5 | T7 | 33 (8) | 37–42 | Podoviridae8 | Short DTR |
| Lytic 6 | SP6 | 10 (4) | 43–46 | Podoviridae | Short DTR |
| Lytic 7 | KP34 | 3 (2) | 43–45 | Podoviridae | Short DTR |
| Lytic 8 | LIMEzero | 1 (1) | 43–44 | Podoviridae | Short DTR6 |
| Lytic 9 | øKT | 1 (1) | 42–43 | Podoviridae2 | Short DTR6 |
| Lytic 10 | GAP227 | 3 (3) | 41–43 | Podoviridae2 | NS, Short DTR6 |
| Lytic 11 | N4 | 11 (5) | 59–78 | Podoviridae | NS, Short DTR |
| Lytic 12 | 9NA | 3 (1) | 52–57 | Siphoviridae | Partial CP, TR7 |
| Lytic 13 | Chi | 9 (3) | 58–61 | Siphoviridae | 12 bp 5′-COS |
| Lytic 14 | øEco32 | 6 (3) | 76–90 | Podoviridae | Short DTR |
| Lytic 15 | Felix-O1 | 9 (2) | 84–89 | Myoviridae | Short DTR |
| Supercluster | SETP3 | ||||
| Lytic 16 | SETP3 | 17 (4) | 41–44 | Siphoviridae | Short DTR |
| Lytic 17 | SO-1 | 9 (2) | 39–46 | Siphoviridae | Circular assembly |
| Lytic 18 | ECO1230-10 | 1 (1) | 41–42 | Myoviridae | Circular assembly |
| Lytic 19 | Gj1 | 3 (3) | 52–57 | Myoviridae | Short DTR |
| Lytic 20 | PY100 | 1 (1) | 50–51 | Myoviridae | Partial CP, TR |
| Supercluster | rV5 | ||||
| Lytic 21 | Ø92 | 2 (1) | 148–149 | Myoviridae | Short DTR |
| Lytic 22 | rv5 | 10 (3) | 136–152 | Myoviridae | Circular assembly |
| Lytic 23 | SPN3US | 3 (3) | 233–244 | Myoviridae | NS |
| Lytic 24 | Rak2 | 3 (3) | 345–359 | Myoviridae | Circular assembly |
| Lytic 25 | øR1-37 | 1 (1) | 262–263 | Myoviridae, modified DNA | NS |
| Lytic 26 | E1 | 1 (1) | 45–46 | Siphoviridae | Circular assembly |
| Lytic 27 | EMCL-117 | 1 (1) | 66–67 | Myoviridae2 | NS |
| Lytic 28 | KF-1 | 2 (1) | 41–42 | Podoviridae2 | Circular assembly |
| Lytic 29 | MSW-3 | 3 (2) | 42–49 | Myoviridae | Circular assembly |
| Lytic 30 | Ea35-70 | 1 (1) | 271–272 | Myoviridae2 | NS |
| Lytic 31 | ϕEaH1 | 1 (1) | 218–219 | Siphoviridae2 | NS |
| Lytic 32 | 9g | 1 (1) | 56–57 | Siphoviridae2 | NS |
| Supercluster | Lambda | ||||
| Temperate 1 | Lambda | 3 (1) | 47–49 | Siphoviridae | 12 bp 5′-COS |
| Temperate 2 | ø80 | 3 (2) | 44–47 | Siphoviridae | 12 bp 5′-COS |
| Temperate 3 | I | 3 (3) | 46–52 | Siphoviridae | 12 bp 5′- & 10 bp 3′-COS |
| Temperate 4 | HK97 | 15 (3) | 36–42 | Siphoviridae | 10 bp-3′ COS |
| Temperate 5 | ES18 | 3 (2) | 46–50 | Siphoviridae | Partial CP, TR |
| Temperate 6 | Gifsy-2 | 9 (6) | 42–523 | Siphoviridae | NS; 5′-COS6 |
| Temperate 7 | BP-4795 | 4 (1) | 54–63 | Siphoviridae | Circular assembly |
| Temperate 8 | SfV | 6 (4) | 37–46 | Myoviridae | 10 bp-3′ COS |
| Temperate 9 | P22 | 16 (2) | 38–44 | Podoviridae | Partial CP, TR |
| Temperate 10 | APSE-1 | 2 (1) | 36–40 | Podoviridae | Circular assembly |
| Temperate 11 | 933W | 11 (1) | 57–66 | Podoviridae | Circular assembly |
| Temperate 12 | HK639 | 1 (1) | 49–50 | Siphoviridae2 | Partial CP, TR |
| Temperate 13 | øES15 | 1 (1) | 49–40 | Siphoviridae2 | Circular assembly |
| Temperate 14 | HS2 | 1 (1) | 58–59 | Siphoviridae2 | NS; Partial CP, TR6 |
| Temperate 15 | ENT47670 | 1 (1) | 47–48 | Myoviridae2 | Circular assembly |
| Temperate 16 | ZF40 | 1 (1) | 48–49 | Myoviridae | Circular assembly |
| Temperate 17 | øEt88 | 1 (1) | 47–48 | Myoviridae2 | NS; Partial CP, TR6 |
| Temperate 18 | ε15 | 7 (4) | 38–45 | Podoviridae | Partial CP, TR |
| Temperate 19 | P1 | 3 (1) | 91–98 | Myoviridae | Partial CP, TR |
| Supercluster | P2 | ||||
| Temperate 20 | P2 | 11 (4) | 29–36 | Myoviridae | 19 bp 5′-COS |
| Temperate 21 | ESSI-2 | 1 (1) | 28–29 | Myoviridae | NS, 3′-COS6 |
| Temperate 22 | Mu | 2 (1) | 34–38 | Myoviridae | Host DNA ends |
| Temperate 235 | SSU5 | 6 (2) | 103–112 | Siphoviridae2 | Circular assembly |
| Temperate 24 | pEP-14 | 1 (1) | 60–61 | Podoviridae2 | NS; Partial CP, TR6 |
See text for supercluster, cluster, and subcluster definitions; numbers in parentheses are total subclusters in each cluster.
No virion electron micrograph has been published for a member of this cluster, but its virion morphology has been reported in either a publication or GenBank (Benson et al., 2013) sequence annotation; tentative morphologies for EMCL-117 and HS2 were deduced here bioinformatically.
CP-1639 prophage is only 39.4 kbp but it appears to contain a large deletion.
CP, the ends of different virion chromosome molecules are circularly permuted relative to one another; TR, terminal repeat or redundancy (molecules with CP and TR are created by headful packaging mechanisms). Partial TR indicates that ends are not randomly distributed across the genome sequence but are restricted to a portion of the genome; DTR, direct terminal sequence repeat that is at the same location in each virion chromosome (note that those of N4 have been reported to be of variable length (Ohmori et al., 1988)). Short DTRs are typically several hundred bps and long DTRs several thousand bps; COS, cohesive single-stranded ends; NS, not yet studied; Circular assembly, since all known tailed phage virion chromosomes are linear DNA molecules, assembly of sequencing runs into a circular nucleotide sequence indicates that it has direct terminal repeats, which could be either DTRs or circularly permuted TRs. To our knowledge, virion DNA structure is uniform within each cluster (with the exception of the N15-like cluster), but the majority of the phages in our panel have not been overtly studied in this regard, so the structures indicated in the table refer to the cases that have been studied.
Temperate nature has not been demonstrated directly (see text).
Virion chromosome structure tentatively deduced from large terminase protein similarity to phages with known chromosome structure (see Casjens and Gilcrease, 2009).
E. Gilcrease and S. Casjens, unpublished results.
We note three cases of apparent reporting errors where T1-like phages øEB49 (Battaglioli et al., 2011) and ESP2949-1 (Lee et al., 2012) were reported to belong to the Myoviridae, and the published electron micrographs of T4-like phage ACG-40 and T7-like phage ACG-91 are apparently switched in figure 2 of (Chibeu et al., 2012).
Figure 1. Dot plot analysis of 21 Enterobacteriaceae tailed phages with genomes larger than 90 kbp.
Blue lines separate phage clusters and red lines separate genomes within the clusters. Dot plot was produced using Gepard (Krumsiek et al., 2007) at a word size setting of 10.
Figure 2. Dot plot analysis of 37 lytic Enterobacteriaceae tailed phages with genomes smaller than 90 kbp.
Blue lines separate clusters and red lines separate genomes within the clusters. Dot plots were produced using Gepard (Krumsiek et al., 2007) at a word size of 10.
Figure 3. Dot plot analysis of temperate Enterobacteriaceae tailed phages with genomes smaller than 90 kbp.
Blue lines separate clusters and red lines separate genomes within the clusters. Dot plots were produced using Gepard (Krumsiek et al., 2007) at a word size of 10.
Similarities between phages within the same cluster are clearly seen in these figures, and inter-cluster pairs usually show little nucleic acid sequence similarity. This does not mean that the different clusters are completely unrelated. Distantly related homologues of some proteins are encoded by phages from multiple clusters. For example, the head assembly proteins large terminase, portal protein and major capsid protein (MCP; see below) are the only proteins with homologues encoded by all tailed phages that have been studied. DNA replication and lysis proteins such as DNA polymerases, helicases, holins and endolysins are also encoded by phages in multiple, but not all clusters. Our analysis shows that these genes are usually sufficiently divergent in sequence that they typically do not form detectable diagonal homology lines in inter-cluster nucleotide sequence dot plots. The relationships among clusters within a supercluster that show <50% span length nucleotide sequence similarity are discussed in more detail below.
In order to not lose sight of the immense amount of experimental work that has preceded this purely sequence-based analysis, we prefer that the clusters be referred to by the name of a prototype phage for each cluster, e.g., T4-like, T7-like or P22-like clusters. Towards this end table 1 lists a prototype phage for each cluster that was chosen on the basis of the phage for which the most experimental studies have been performed (i.e., the best understood member), or, in the absence of such information, the first member of the cluster whose genome was completely sequenced.
Within each cluster the phage genome sizes are quite uniform, and with the exception of only a few phages the size range within clusters varies by less than 15% (tables 1 and S1). Within each of the clusters all member phages have the same tail type; 22 clusters have long contractile tails (Myoviridae), 20 have long noncontractile tails (Siphoviridae) and 14 have short tails (Podoviridae). Thirty-two clusters contain 221 lytic phages, and 24 contain 112 temperate phages. No cluster contains both lytic and temperate phages, nor are any lytic and temperate clusters particularly closely related, suggesting that although there have been fairly recent exchanges of a few genes between such phages (George et al., 1983; Casjens and Thuman-Commike, 2011), no whole phage in this panel has recently switched between these two lifestyles. In the course of our analysis we noticed that phage SSU5 (Kim et al., 2012) is syntenic with and homologous to circular plasmids in a number of reported Enterobacteriaceae bacterial genome sequences (see table S1 for examples), and we conclude that SSU5, although it has not been reported as such, is almost certainly a temperate phage with a circular plasmid prophage. Falgenhauer et al. (2014) recently independently reached the same conclusion.
Of the 56 Enterobacteriaceae tailed phage clusters, 18 are singleton clusters that have only one phage member. This large fraction (32%) of singletons indicates that our knowledge of tailed phage diversity is still very far from complete for this family. In addition, the phages analyzed here were isolated on 18 different host Enterobacteriaceae genera, and at present this family currently contains 74 genera (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=543) (Federhen et al., 2012). A complete understanding of tailed phage diversity in the Enterobacteriaceae family will require isolation and characterization of phages that infect the remaining 56 genera in this family.
Space limitations preclude discussion here of all 56 phage clusters identified in this report. Some clusters are quite uniform and assignment of phages to them is straightforward and unambiguous, for example the Felix-O1-, T4-, T5-, Chi-, SETP3- and P1-like clusters. But other cluster relationships are more complex; some clusters have substantial internal diversity and in some cases there are similarities between clusters. We first discuss Felix-O1- and T4-like clusters as examples of ‘simple’ clusters and then proceed to the T7 and lambda superclusters for discussion of complicating issues that arise in attempts to unambiguously classify the tailed phages.
Felix-O1-like cluster
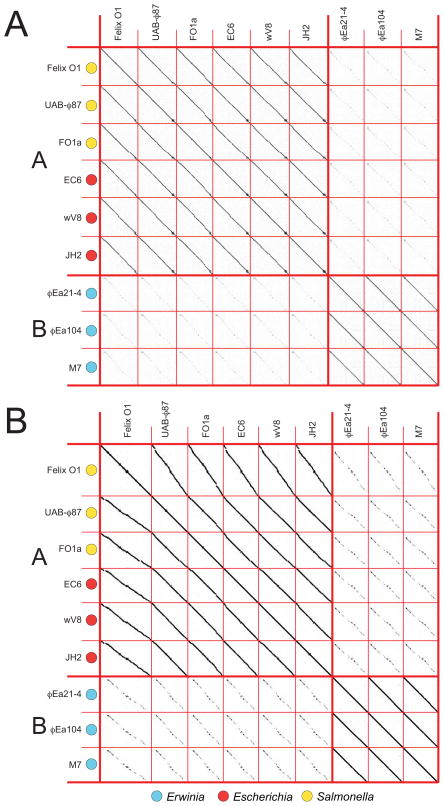
The Felix-O1-like cluster provides a case where cluster and subcluster assignment is straightforward. The first publication of the prototype phage for this cluster, Salmonella phage Felix-O1 (then called phage O1), was in 1943 for use in typing Salmonella serovar Typhi strains (Felix and Callow, 1943; McConnell and Schoelz, 1983). Since then, it has also been used in phage therapy applications (Kuhn et al., 2002; Kuhn, 2007). We identified nine Felix-O1-like phage complete genome sequences that range from 84 to 89 kbp in length; three of these phages infect E. coli (Felix-O1, UAB-φ87, FO1a), three infect Salmonella (EC6, wV8, JH2) and three infect Erwinia (φEa21-4, φEa104, M7); see MATERIALS AND METHODS for reasons ‘phage’ SA1 was not included in this cluster. Whole genome nucleotide sequence (Figure 4A) and encoded gene product amino acid (AA) sequence (Figure 4B) dot plots reveal very substantial syntenic similarity among these nine genomes; no such similarity with any other phage in our database was detected. These nine phages can be grouped into two subclusters based on natural boundaries of phage relatedness as delineated by the strength of dot plot diagonal homology lines. There is particularly close similarity between the six E. coli and S. enterica phages (assigned to subcluster A), and an obvious but more distant relationship between these phages and the three that infect Erwinia (assigned to subcluster B). These relationships are consistent with the closer relationship of Escherichia and Salmonella as compared to the more distant Erwinia (Moran et al., 2005; Gao et al., 2009) and are reflected perfectly in a dot plot of the single protein MCP, where MCP sequences of four additional Salmonella phages and one additional Erwinia phage are available (Figure S1K). In addition to correlating with the hosts, these subclusters closely correlate with GC content. The average content of subcluster A is 38.88±0.04% while B is 43.7± 0.3%.
Figure 4. Whole genome nucleotide (A) and gene product (B) dot plots of known Felix-O1-like phages reveals two subclusters.
Phage genomes and subclusters are separated by thin and thick red lines, respectively. Subcluster names are indicated on the left by red letters, and a key for the phage hosts is shown below. Dot plots were produced using Gepard (Krumsiek et al., 2007) at a word size of 11 for the genome dot plot and 6 for the gene product dot plot. Amino acid sequences for the gene product dot plot consisted of tandem sequences of all the annotated predicted encoded proteins aligned in the order their genes occur in the genome. The reported circular genome sequence assembly of Felix-O1 (Accession No. AF320576) was linearized at bp 16830 in order to align it with the known ends of cluster member phage M7’s linear genome (Born et al., 2011). Other cluster member genomes were oriented to align with these genomes.
The dot plots reveal an overall similarity of all the genomes of the Felix-O1-like cluster phages, except for the first ~13000 bps (as oriented in Figure 4) of the subcluster A genomes which corresponds to ~9000 bp in subcluster B. This region (between ORF64 and ORF111 in Felix-O1) is largely different between subclusters except for a common endolysin and the 20–26 tRNA genes that all of these phages carry. Within each subcluster this region is similar and is predicted to encode a number of small uncharacterized proteins, with the exception of a T4-like nucleotide reductase A protein (NrdA-like gp4 of φEA21-4) and tRNA nucleotidyltransferase/poly(A) polymerase (gp70 of φEA21-4) encoded by the Erwinia phages. The gene products conserved between the subclusters include nucleotide metabolism proteins, structural or phage assembly proteins, a nicotinamide phosphoribosyl transferase (involved in NAD biosynthesis) and many hypotheticals.
Average nucleotide identity (ANI) and conserved gene product (CoreGenes) analyses strongly support the Felix-O1 cluster and subcluster assignments (Table 2). The minimum ANI between phages within each subcluster ranges from 86% (M7 and φEa21-4) to 99.97% (Felix-O1 and FO1a), while the ANI values for phage pairs from the two subclusters lie between 53.8% and 55.2%. CoreGenes analysis shows a similar trend with the fraction of conserved gene products within the subclusters ranging from 82% to 99% and between subclusters from 56% to 62%. We note that 40% conserved proteins cutoff has typically been used to by others to determine groups of ‘related’ phages (Zafar et al., 2002; Mahadevan et al., 2009).
Table 2.
Conserved gene product and average nucleotide identity analysis of the Felix-O1 cluster phages1
| Felix-01 | UAB-φ87 | FO1a | EC6 | wV8 | JH2 | φEa21-4 | φEa104 | M7 | |
|---|---|---|---|---|---|---|---|---|---|
| Felix-01 | 100 (100) | ||||||||
| UAB-φ87 | 82.31 (93.7) | 100 (100) | |||||||
| FO1a | 96.88 (99.97) | 89.84 (93.72) | 100 (100) | ||||||
| EC6 | 83.09 (91.37) | 86.03 (92.4) | 85.94 (91.44) | 100 (100) | |||||
| wV8 | 87.14 (91.98) | 89.29 (91.93) | 83.57 (92.08) | 82.86 (92.48) | 100 (100) | ||||
| JH2 | 86.26 (86.26) | 90.08 (93.03) | 87.5 (91.95) | 86.26 (91.89) | 88.55 (91.18) | 100 (100) | |||
| φEa21-4 | 58.7 (53.82) | 60.17 (53.98) | 56.78 (53.89) | 59.32 (53.98) | 61.02 (53.98) | 59.32 (53.92) | 100 (100) | ||
| φEa104 | 59.32 (53.92) | 61.02 (54.06) | 56.78 (53.99) | 60.17 (53.89) | 61.86 (53.96) | 59.32 (54.00) | 94.92 (98.27) | 100 (100) | |
| M7 | 59.83 (55.06) | 61.54 (55.18) | 59.83 (55.14) | 59.83 (55.00) | 61.54 (54.99) | 60.68 (55.09) | 99.15 (86.36) | 94.87 (86.78) | 100 (100) |
Conserved gene products are presented as the percentage of gene products present in both phages divided by the total number of gene products for the phage in column one. Conserved gene products were analyzed by CoreGenes at a default threshold of 75. ANI (in brackets) was calculated by Kalign. The subcluster B phages are shaded light grey while subcluster A phages are unshaded.
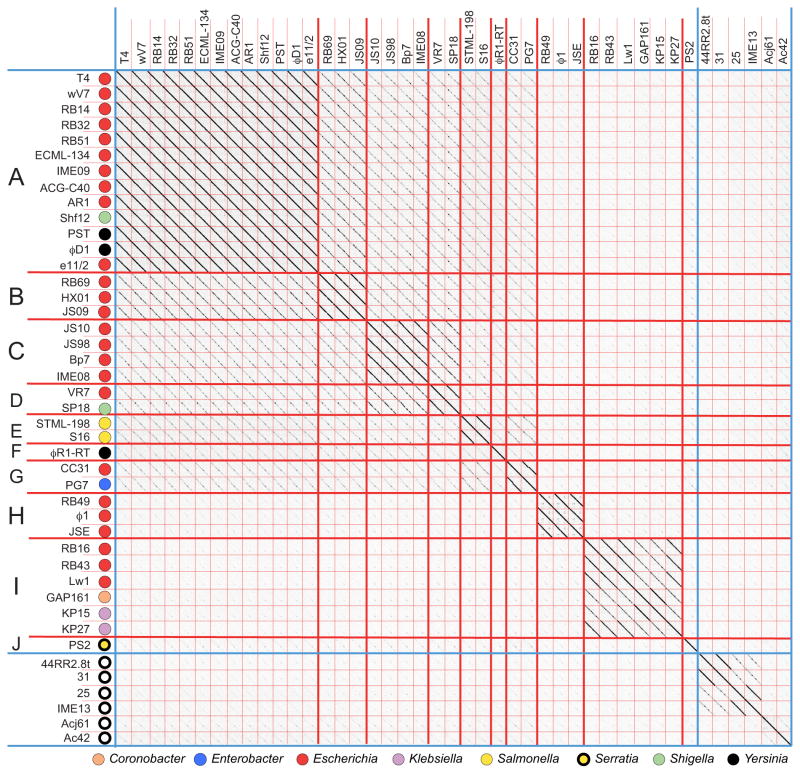
T4-like cluster
The T4-like phage cluster contains 37 lytic Myoviridae phages that infect eight Enterobacteriaceae genera. The prototypic phage for this cluster is the very well studied phage T4, one of the three T-even phages studied by Anderson, Delbruck and Demerec as early as 1944 (Anderson, 1945; Demerec and Fano, 1945; Delbruck, 1946). T4 was pivotal to our understanding of the nature of the genetic code, being the focus of Crick and Brenner’s 1961 experiments (Crick et al., 1961). These 37 phages fall unambiguously into a single cluster by dot plot analysis and all have genome sequences in the 158–181 kbp range. The T4-like phage genomes naturally divide into ten subclusters, which can be grouped into four major subtypes typified by phages T4 (subclusters A–G), RB49 (subcluster H), RB16 (subcluster I) and PS2 (subcluster J)(figure 5). Lavigne et al. (2009) used protein tree comparisons to classify 12 of these phages, and they recognized three of the four major subtypes in our analysis (T4, RB49, and RB16 subtypes; the PS2 sequence was not available at that time). The subdivisions present in an MCP dot plot of the T4-like cluster phages (figure S1B) agree well with the genome-based subcluster divisions, although subcluster A and B MCPs are very similar as are those from subclusters C and D. These subcluster divisions are supported by a CoreGenes analysis in which phages within each subcluster share ≥78% of their gene products (RB43 and KP15 share the least), while inter-subcluster conservation ranges from 40% to 75% (the GAP161/RB69 and PS2/RB16 pairs are most distant at 40%). Krisch and coworkers (Filee et al., 2006; Comeau et al., 2007) have analyzed the nature of this gene product variation among 16 T4-like phages belonging to three subgroups (the “T-evens/Pseudo T-evens”, the “Schizo T-evens” and the “Exo T-evens”). They report two blocks of syntenic ‘core’ genes comprised of 24 viral replication and structural genes, and a more plastic set of ‘accessory’ genes. Although only their nine “T-evens/Pseudo T-evens” are Enterobacteriaceae phages, our analysis of 36 T4-like Enterobacteriaceae phages confirms their analysis in that the core gene products conserved (the 40% of the genome cited above) include the 24 viral replication and structural genes that lie largely in two syntenic blocks.
Figure 5. Whole genome nucleotide dot plot of known T4-like phages reveals ten subclusters.
The dot plot is presented as described in the legend to figure 4. Genomes are aligned with the T4 sequence reported in accession No. AF158101 and compared with a Gepard word size of 12. Open circles indicate phages that infect hosts outside the Enterobacteriaceae family (see text).
The T4-like cluster currently differs from the Felix-O1-like cluster in that, unlike the latter and most of the other clusters in our analysis, it has moderately close relatives that infect other bacterial families. The closest of these infect species in the Moraxellacea, Xanthomonadaceae or Aeromonadaceae families of the γ-Proteobacteria, and six such phages, 44RR2.8t, 25, 31, IME13, Acj61 and Ac42, are shown in figures 5 and S1B. These latter phages are largely syntenic with the T4 cluster members and have core gene similarity throughout their genomes (>40% of their gene products have a homolog encoded by T4 when analyzed by CoreGenes; not shown). Thus, they fall within the T4-like cluster where they form several unique subclusters; none of these phages falls in an Enterobacteriaceae subcluster, and our analysis is consistent with the grouping of the T4-like phages outside the Enterobacteriaceae into “Schizo T-evens” and the “Exo T-evens” by Filee et al. (2006).
The incredible environmental success of the T4-like phages has been observed by many. In fact three of Delbruck’s original seven phages were T4-like phages, most likely due to their successful colonization of the environment (Krish and Comeau, 2008). Other large phages with recognizably similar but less closely related virion assembly genes infect bacteria as distant as the cyanobacteria (Hambly et al., 2001; Hambly and Suttle, 2005, Filee et al., 2005); those that are fully sequenced were analyzed and are do not fall within the T4-like cluster by the rules applied here. Thus, although T4-like phages have been successful in various host families, there is no evidence for recent whole T4-like phages jumping between these diverse host families.
The T7 supercluster
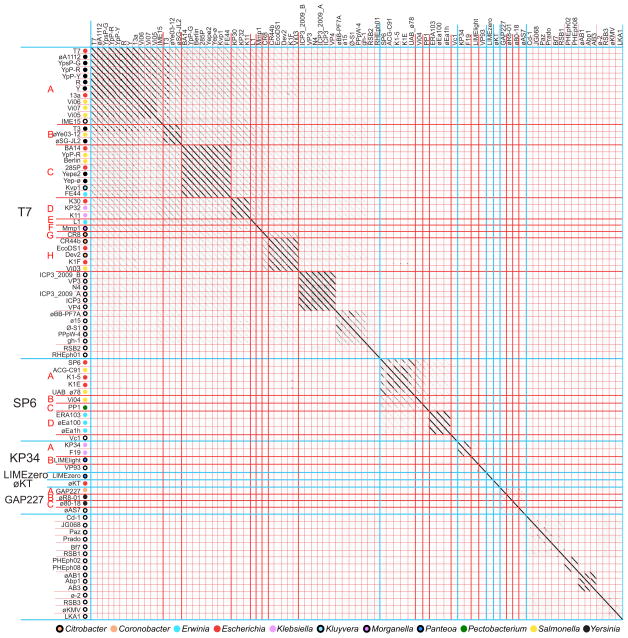
Phage T7, like T4, was officially reported in 1945 as a member of the historical seven ‘T’ phages that infect E. coli B (Demerec and Fano, 1945), although it was the likely phage δ used in the earlier studies of Luria and Delbruck (1943). T7 is one of the best-characterized lytic bacteriophages and its relatives have been identified in abundance around the world. A close relative of T7 was first isolated by d’Herelle in the early 1900’s (d’Herelle, 1926; Hauser et al., 2012) and we identified 51 fully sequenced Enterobacteriaceae phages that have overall gene content and syntenic similarity to T7, which we define as the T7 supercluster.
Phage clusters within the T7 supercluster
Nucleotide dot plot analysis of the 51 Enterobacteriaceae T7-related phages shows the natural formation of six clusters that have rather little overt nucleic acid sequence similarity (Figure 6; Table 1). The T7-like cluster is the largest of these and contains 33 phages that infect 8 different host genera, the SP6-like cluster contains 10 phages from 4 different genera, the KP34-like cluster contains 4 phages from 2 different genera, the GAP227-like contains 4 phages from 3 different genera, and two clusters consist of singleton phages, Panteoa agglomerans phage LIMEzero and E. coli phage øKT. Cluster assignment by inspection of the genome dot plot is unambiguous for all of these phages; the clusters with multiple members are all self-cohesive and each has >50% intra-cluster ANI similarity. MCP and whole gene product AA dot plots were also constructed and the overall pattern of relatedness is similar in all three plots, agreeing perfectly with cluster assignment from the nucleotide dot plots (Figures S1E and S1F, respectively). LIMEzero and øKT were placed in singleton phage clusters due to low nucleotide sequence similarity to other clusters, but whole genome gene product AA dot plots (figure S1F) and CoreGenes analysis (not shown) indicate that they have a T7-like genome architecture as well as some hybrid features, having patches of weak similarity to phages in both the GAP227- and KP34-like clusters (see below). The four clusters with more than one member have considerable substructure and can be split into 17 subclusters (figure 6). The T7-, SP6-, KP34- and GAP227-like clusters naturally form eight, four, two and three subclusters, respectively.
Figure 6. Whole genome nucleotide dot plot of 84 T7 supercluster phages reveals six clusters.
The dot plot is presented as described in the legend to figure 4. Genomes are aligned with the T7 sequence reported in accession No. V01146 and compared with a Gepard word size of 12. Open circles indicate phages that infect hosts outside the Enterobacteriaceae family (see text).
Mosaic module diversity and module shuffling in the T7 supercluster
Nucleotide and AA sequence comparisons clearly suggest that all 51 T7-like supercluster member phages do not all belong in the same cluster. However, comparison of genome content and order argues for their inclusion in a T7 supercluster. This supercluster is largely consistent with previous literature on T7-like phages and consists of phages that have similar gene content and order with obvious divergence and modular shuffling. Historical hallmarks of the T7 phage group include the presence of a T7 type RNA polymerase gene as well as a basic genome organization in which there are three regions harboring the early (the first eleven T7 gene products including host restriction functions, RNA polymerase and DNA ligase), middle (the next 24 T7 gene products including DNA metabolism functions and lysozyme) and late genes (the last 24 T7 gene products, including virion structural and assembly genes), all transcribed from a single strand (Molineux, 2006). CoreGenes and Phamerator analyses indicate that these 51 phages maintain the same strand organization and basic groups of early, middle and late genes, but the order and content of these groups differs somewhat between clusters. Supplementary figure S3 diagrams this basic genome order and gene product content of the T7-like phages by displaying one representative from each cluster.
In spite of their substantial divergence, the genomes of the T7- and SP6-like clusters display remarkable synteny, with the early genes encoding two conserved proteins (the Ocr restriction enzyme inhibitor and RNA polymerase) followed by the middle genes (five conserved genes including primase/helicase, DNA polymerase, exonuclease, endonuclease, and DNA ligase) and finally the late genes (eleven conserved genes including the DNA packaging, virion assembly and lysis proteins). All five middle DNA metabolism genes listed above are present in T7- and SP6-like phages, however the order is dramatically different in the two clusters, and the late genes also have a somewhat different order in the two clusters. The restriction enzyme inhibitor Ocr is the most highly conserved encoded protein of T7 and SP6 (79% AA identity whereas the average for other conserved proteins is 30–52% (Chen and Schneider, 2006)), which is interesting given the wide range of hosts for phages in these clusters. Within each of these two clusters there are differences in gene content with an occasional missing or novel gene, with the early region being the most variable.
The KP34- and GAP227-like clusters, as well as the LIMEzero and øKT singletons, are even more different from T7 in genome content and order. They are more closely related to one another than to the T7- or SP6-like clusters and are somewhat more similar to SP6 than T7 as observed by the AA whole genome plot and whole genome maps (see figures 6 and S1E). These four clusters have groups of putative early, middle and late genes, however the positions of the middle and early regions are switched (the actual expression pattern of these regions in these four clusters has not been examined experimentally). Despite this genome organizational difference, these phages contain a core set of gene products that is similar to T7 and SP6. The genomes begin with the ‘middle’ DNA metabolism genes (including primase, helicase, DNA polymerase, endonuclease and exonuclease), the RNA polymerase ‘early’ gene lies in the middle of the genome, and the conserved virion structural ‘late’ genes reside at the right end. The øKT, KP34, F19 and LIMEzero phages are the only T7-like phages to lack a recognizable DNA ligase gene.
Phages LIMEzero and øKT provide clear examples of how clusters can be related within a supercluster. Phage LIMEzero displays overall genome content and gene order similarity to the KP34- and GAP227-like clusters despite having little nucleotide similarity (see figures 6 and S3), suggesting long evolutionary divergence. E. coli phage øKT has substantial similarity in gene order and content, but its encoded proteins are the most distantly related of the Enterobacteriaceae T7 supercluster phages. In particular, øKT appears to be missing recognizable homologues of a number of genes that are conserved in most of the T7-like phages including DNA ligase and the internal virion proteins, however it does have uncharacterized gene products encoded at the corresponding locations that may have these functions despite the lack of significant AA homology.
CoreGenes analysis confirms these cluster assignments in that at least 49% of gene products are conserved in pairwise comparisons within each cluster, and inter-cluster comparisons are significantly less than this (≤35%, data not shown). For example, the other phages in the T7-like cluster have at least 54% of their gene products in common with T7 whereas phages of the SP6-like cluster share only ~20–27% of their gene products with T7, while having at least 50% of their gene products in common with SP6. Similarly, phages within the GAP227 and KP34-like clusters have >71% and >49% common genes, respectively, whereas ~30% of gene products are conserved between the two clusters. Only 13–19% of GAP227- or KP34-like gene products are in common with T7 and 14–28% with SP6. However, <5% of gene products are conserved when comparing these phages with non-T7-like phages. CoreGenes values also confirm LIMEzero and øKT to be singletons and to be partly ‘hybrid’ phages whose most similar relatives are each other (35% gene product conservation) and phages in the KP34-like and GAP227-like clusters, having almost equal gene product conservation with both clusters (~32–36% each for both LIMEzero and øKT). Gene product conservation is more limited with SP6 (17% and 25% respectively) or T7 (17% and 27% respectively). (The øKT annotation is less complete than the others. Our unpublished analysis indicates it has a typical number of genes for this supercluster, at least 53 genes rather than the published 31, which was taken into account in these calculations by using BLASTn (Altschul et al., 1990) to identify protein homologs). Thus, the GAP227-, KP35-, LIMEzero-, and øKT-like clusters all share about 30% inter-cluster CoreGenes similarity which solidifies the close relationship of these clusters. Due to the transitive nature of many phage genomes and this close relationship, related phages may one day be isolated that will unite these phages into a single cluster.
Comparison of the T7 supercluster with previous analyses
Several investigators have analyzed the relationship of a subset of the T7-related phages by various means, including proteomic phylogenetic tree construction, and these studies agree with the cluster assignments made here. The largest study by Adrianenssens et al. (2011) constructed three single gene product phylogenetic trees of many T7-related phages (including 24 that infect Enterobacteriaceae hosts) that were based on RNA polymerase, DNA polymerase and MCP. The RNA and DNA polymerase trees show relationships very similar to those from our whole genome dot plot analysis, with one branch being the T7 cluster, another branch being the SP6 cluster (phages SP6, ERA103, K1E and K1-5) and a third, more diverse branch harboring phages from the KP34 cluster as well as LIMEzero (no phages of the GAP227 or øKT clusters were analyzed). As in our analysis, LIMEzero is a clear outlier within this latter branch. In the DNA and RNA polymerase trees the SP6-like phages were more related to the T7-like cluster than to the KP34-like cluster, which is supported by gene content and synteny analysis (see figure S3). Their single gene product analysis using the MCP was very similar, except that the SP6 group showed a higher relatedness to the KP34-like phages than the to T7-like cluster. This discrepancy between the DNA/RNA polymerase trees and the MCP tree shows the complex nature of the T7 superfamily phage relationships, with SP6 having some whole genome relationship with both the T7- and KP34-like phages as shown by gene content and synteny (figure S3 and the CoreGenes analysis above).
In another analysis, Chen et al. (2006) classified eight T7-like phages by the relatedness of their T7-like promoters and RNA polymerase. Their analysis also agrees with our whole genome analysis with these eight phages falling into five groups. Three of their five groups are highly related and are comprised of T3 and øYeO3-12 (members of our T7-like cluster, subcluster B), T7 and øA112 (members of our T7-like subcluster A), and K11 (T7-like subcluster D). K1-5 and SP6 form a fourth group (members of our SP6-like cluster) and gh-1 (a Pseudomonas phage) a fifth. Thus, the analyses of Dobbins et al. (2004) and Scholl et al. (2005) agree that the SP6 and K1-5 phages belong to a different group from T7 within this supercluster. The excellent correspondence between these single protein or promoter phylogenetic trees and our more global analyses suggests that there has not been extensive recent exchange of these genes between the clusters of the T7 supercluster.
Host-phage relationships within the T7 supercluster
A striking feature of the T7 supercluster is the variety of hosts on which these phages were isolated (the 51 phages infect eleven different Enterobacteriaceae genera). Of the 19 subclusters, 13 include phages from a single host but eleven of these are low occupancy, containing ≤3 phages. Clearly the current low level of sampling limits our knowledge of host diversity within the T7 subcluster.
The T7 supercluster is a very successful lytic phage type that has managed to parasitize a number of γ-Proteobacteria families and beyond. In addition to Enterobacteriaceae hosts, we identified 32 fully sequenced phages isolated on hosts outside the Enterobacteriaceae that had similar MCP’s (<10−30 BLASTP e-value, ~35% AA identity) and clearly belong to the T7 supercluster by genome nucleotide sequence dot plot analysis (see figure 6). Of these phages, 14 belong to the T7-like cluster, and all but IME15 (Huang et al., 2012) clearly form separate more distantly related singleton subclusters. Stenotrophomonas phage IME15 is remarkable in its similarity to T7, given that its host belongs to a different bacterial family, the Xanthomonadaceae. IME15 shares 73% ANI with T7 and 84% of its gene products by CoreGenes analysis. To our knowledge, no host range studies have been performed on this phage but its tail fiber (gp39) suggests it has a different infection strategy than phage T7. The IME15 tail fiber has 63% identity to the T7 tail fiber over the first 301 AAs of 580 total but lacks the C-terminal region that bears a domain involved in host receptor recognition (Steinbacher et al., 1997; Yu et al., 2000). All of the E. coli and Yersinia phages within the T7 cluster have conservation over the entire T7 tail fiber, but tail fibers from all other phages in this cluster have C-terminal regions that vary by host. Phage IME15 is the best candidate in our panel for a whole phage that has ‘recently’ jumped between hosts in different bacterial families, the Enterobacteriaceae and Xanthomonadacea. The direction of this jump cannot be determined at this time. More T7 supercluster Xanthomonadaceae phages must be studied in order to understand the relationship between the T7 supercluster phages that infect these two host families.
Of the 18 remaining non-Enterobacteriaceae T7 supercluster phages, the Vibrio phage Vc1 and Aeromonas phage øAS7 clearly form singleton subclusters within the SP6-like cluster (46% gene product conservation) and the GAP227-like cluster (58% gene product conservation), respectively. The rest form four clusters within the T7 supercluster that have variable similarity to the GAP227-like, KP34-, øKT- and LIMEzero-like clusters. This trend in related phages that share homology to the GAP227-, KP34-, øKT- and LIMEzero-like clusters is consistent with the high degree of inter-cluster mosaicism between these four clusters that was discussed above. For example, the phages Cd-1, JG068, Paz, Prado and Bf7 phages form a cluster that can be seen by nucleotide dot plot (figure 6), and all five of these phages have between 30–48% CoreGenes gene product conservation with øKT and LIMEzero, GAP227 and KP34 (almost equally with the different clusters). We note that the ‘T7-like’ phages Cd1 and JG068 infect the most distantly related hosts Caulobacter crescentus (α-Proteobacteria) and Burkholderia cenocepacia (β-Proteobacteria), respectively.
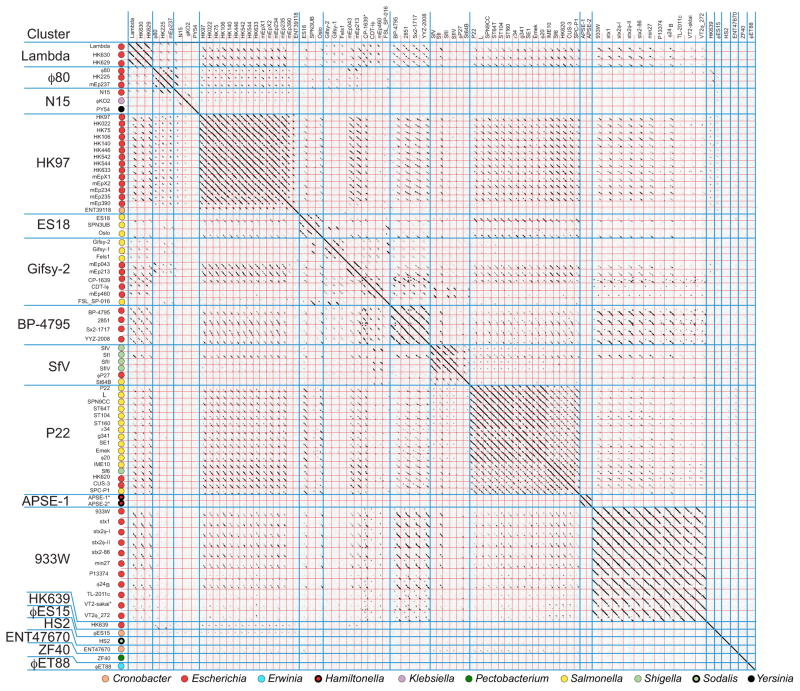
The lambda supercluster
The temperate phage lambda was discovered in 1951 in an E. coli isolate when it was fortuitously induced from its prophage state (Lederberg, 1951). It has served as a very important model system for the study of the nature of genes and the regulation of their expression (e.g., Cairns et al., 1966; Hendrix and Casjens, 2006) and was the first dsDNA virus whose genome was sequenced (Sanger et al., 1982). Many ‘similar’ phages have since been isolated and characterized. These so-called ‘lambdoid’ phages are well-known for their highly mosaic genomes, and the resulting complex genomic relationships pose knotty problems for any attempt to categorize them neatly. The term ‘lambdoid’ has been misused many times in the literature (see Hendrix and Casjens, 2006) and will not be used in the remainder of this discussion, but the ‘lambda supercluster’ that we define here encompasses the phages that are traditionally included in that group. The mosaic relationships among the various members of this group have been anecdotally noted many times (see for example Westmoreland et al., 1969; Simon et al., 1971; Botstein, 1980; Baker et al., 1991; Casjens et al., 1992; Juhala et al., 2000; Ravin et al., 2000; Hendrix, 2002; Casjens, 2005), but the overall extent and complexity of these relationships have not been described.
The lambda supercluster is a cohesive, self-contained yet diverse phage group
We define the lambda supercluster as that group of temperate Enterobacteriaceae phages whose encoded functions are syntenic with the phage lambda genome and whose transcription pattern and gene expression cascade are similar to that of lambda. Analysis of the phages in our database identified 81 phages that have significant syntenic nucleotide or gene product sequence similarity to the other members of the lambda supercluster. Only very rarely do these phages have even short similarities to phages outside this group (and these are largely tail fiber/spike similarities), so these phages naturally form a large transitive, closed set.
One can think of this group of phages as having a menu of multiple different possibilities at each genomic functional position. Listed from left to right across the genome, as it is normally displayed, most of these major functions are as follows: DNA packaging (terminase) – head assembly (portal protein and MCP) – tail assembly – integrase/protelomerase – homologous recombination – early gene transcriptional antiterminator - prophage repressor (CI) - cro repressor – establishment of lysogeny transcriptional activator (CII) – DNA replication – ‘nin region’ genes – late gene transcriptional transcriptional antiterminator – lysis. The phages in the lambda supercluster almost always have these functions encoded by genes arranged in this order. Only the N15- and APSE-1-like clusters have differences in the position of some DNA replication and late antiterminator genes; they were retained in this cluster due to the very close relationship of their other functions to supercluster members. In this supercluster a given function can be performed by divergent homologous proteins (e.g., DnaB-type DNA replication initiation helicases that are only 31% identical in phages Sf6 and ϕEt88) or nonhomologous proteins (e.g., phage lambda Exo/Beta, P22 Arf/Erf homologous recombination proteins and the three types of tails). As we have previously noted (Casjens, 2005), transitive relationships can result in individual pairs of phages within the group that appear unrelated when their nucleic acid sequences are compared; for example, the phage P22 and lambda early regions are rather similar, the lambda and N15 late regions (virion assembly genes) are very similar, but P22 and N15 have only a small number of recognizably homologous genes in common. On the other hand, although phages like lambda and ϕEt88 have no sizeable region of recognizable nucleic acid similarity, they have many syntenic genes that can be recognized as homologous by moderate similarities of their encoded proteins (see for example figure S4). Thus, genome mosaicism among these phages arises from both quantitative and qualitative differences at syntenic functionally equivalent positions on the genomes.
Clusters within the lambda supercluster
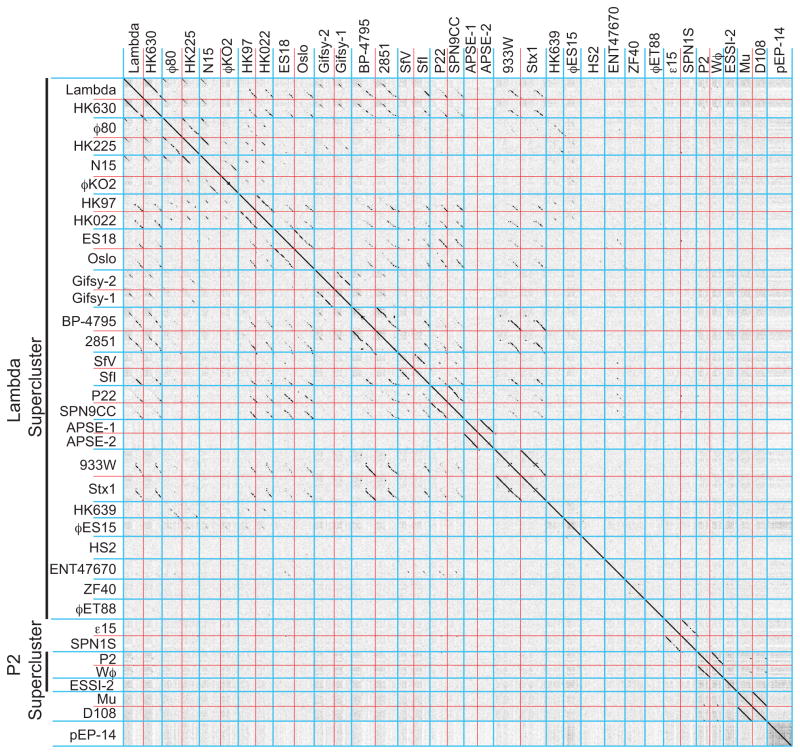
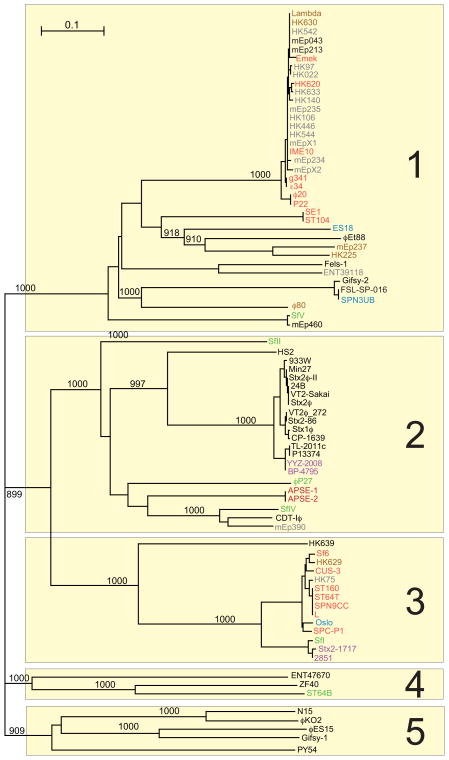
The rules delineated above for cluster membership were applied to determine whether this supercluster of phages could be sensibly divided into discrete groups of more highly related phages. The Gepard genome dot plot in figure 7 shows the complex inter-relationships among these 81 phage genomes (expanded genome dot plots are shown in figures S1N-S). This and more detailed pairwise dot plots created by DNA Strider partition them into 17 different clusters, nearly all of which are quite unambiguously separated (see below for the more complex cases). The prototypes for these seventeen clusters are phages lambda, ϕ80, N15, HK97, ES18, gifsy-2, BP-4795, SfV, P22, APSE-1, 933W, HK639, ϕES15, HS2, ENT47670, ZF40 and ϕET88 (Table 1). There are many partial genome relationships among these clusters; for example, the incomplete diagonal similarity lines when members of the HK97-like genomes are compared with the lambda-, P22-, gifsy-2-, BP-4795- and 933W-like genomes. But these relationships constitute <50% of the genome and so are not sufficient to merge these groups into a single more complex cluster. Because of the many possible combinations of short, exchangeable mosaic module ‘menu selections’ available in the early region, cluster membership within this supercluster tends to be (but is not always) determined by the longer stretches of apparently evolutionarily inseparable virion assembly genes. Thus, with only a few exceptions, each of these clusters has a unique, very different set of virion assembly genes; only the P22-/APSE-1-like and lambda-/ø80-/N15-like cluster groups have convincing similarities between their virion assembly gene clusters (e.g., see the MCP tree in figure S5), but these have very different early regions.
Figure 7. Whole genome nucleotide dot plot of 81 lambda supercluster phages reveals 17 clusters.
The dot plot is presented as described in the legend to figure 4 (Gepard word size of 12). Phage genomes are separated by red lines and clusters by blue lines. Hosts from which the phages were isolated are indicated on the vertical axis. The genomes shown are all oriented according to the standard phage lambda virion chromosome map with the head genes on the left and lysis on the right.
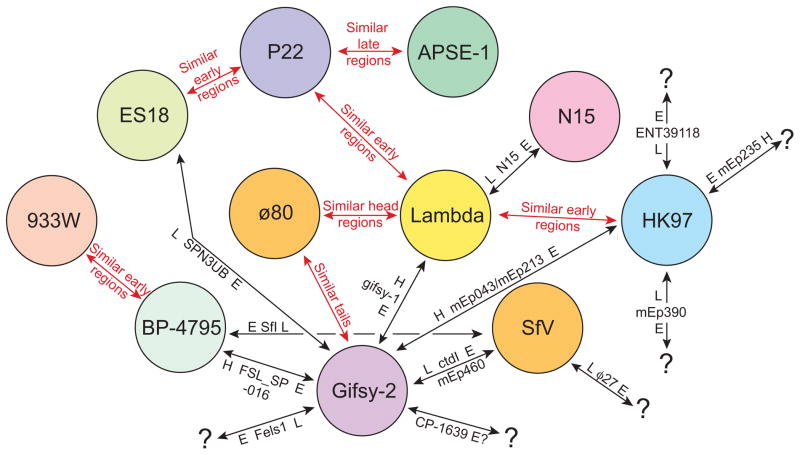
A large majority of the lambda supercluster phages fit unambiguously into individual clusters. These are arranged as contiguous blocks of related phages in figure 7, which appear in the plot as square regions that contain more substantial diagonal similarity lines. However, some phages (for example, SPN3UB and FSL-SP-016; figure S6) contain large blocks of sequence that are similar to two different clusters within the lambda supercluster. We assigned such ‘hybrid’ phages to the cluster with which they have the most in common (typically the virion assembly genes), and rather than obscuring their large differences by merging the affected clusters, we view them as having arisen by recent horizontal transfer of large blocks of genetic material between clusters within the supercluster. The lambda-, ϕ80-, HK97-, BP-4795-, P22-, APSE-1-, and 933W-like clusters are each cohesive and self-contained by virtue of having largely syntenic and similar genomes (long solid lines in a dot plot), while the N15 and gifsy-2 clusters contain more apparent inter-cluster ‘hybrids’ and are less cohesive by this analysis (figure 7). The most difficult of these phages are the two very similar phages mEp043 and mEp213 which have head regions that are similar to gifsy-2, tail regions that are similar to ø80 and early region parts that are most similar to HK97; we arbitrarily assigned these phages to the gifsy-2-like cluster. Figure 8 summarizes the phages from non-singleton clusters that can be viewed as having been formed by inter-cluster hybridization events, including phages that have large novel regions presumably obtained from clusters that have not yet been discovered. Genome mosaicism within clusters with smaller patches of similarity in the early operons is also very frequent and can be seen in the fact that genome dot plots of phages within the clusters show few solid diagonal similarity lines that span the length of the whole genome (see examples in HK97-like cluster subcluster A in figure S1O, S1P and S7).
Figure 8. Relationships among the non-singleton clusters of the lambda supercluster reveals inter-cluster hybridization events.
Circles represent the non-singleton clusters within the lambda supercluster. Red arrows indicated cluster pairs in which most members have significant (usually mosaically related and always divergent) homologies in the indicated regions. Black arrows indicate examples of regions that were rather recently exchanged between these clusters that generated apparently ‘hybrid’ phages (see text). On each black arrow a particular ‘hybrid’ phage(s) is indicated with its genome sections that are closely related to other members of the two clusters connected by the arrow; these sections are labeled L (late region), H (head region) and E (early region), and the source of each region is indicated by the cluster circle nearest to the indicated region (e.g., phage SPN3UB has an early region similar to phages in the gifsy-2-like cluster and a late region similar to phages in the ES18-like cluster. Black question marks (?) indicate that the source of the indicated region is currently unknown.
A CoreGenes analysis of the lambda supercluster supports these cluster assignments. Most clusters share a minimum gene product conservation near or greater than the typically used 40% cutoff for ‘group’ membership (>60% within the lambda-, >68% within φ80-, >36% within N15-, >38% within HK97-, 63% within BP-4795-, >47% within SfV-, 48% within P22-, >83% within APSE-, and >50% within 933W-like clusters). The lowest conservation is within the ES18 cluster (>25% between phages) and within the gifsy-2 cluster (>9% between phages gifsy-1 and mEp043), yet these clusters (except phages mEp043 and mEp213 above) show nucleotide similarity over 50% of the genome by dot plot suggesting proteins that have diverged substantially. For example, in the gifsy-2-like cluster nucleotide similarity between gifsy-2 and Fels-1 exists over the first ~25,000 nucleotides of the ~44,000 nucleotide genomes (figure S1P), however CoreGenes indicates only 8 conserved gene products when using the standard BLASTP similarity score cutoff of 75 (these include integrase, Rz-like phage lysis protein, gpU minor tail protein, gpV major tail protein, gpH tail tape measure, gpK tail assembly, and a gpG tail assembly). This percent conservation increases to 28% if the BLASTP threshold score is decreased to 50, and to 41% if it is decreased to 25 (here we discarded ten poor ‘matches’ with e-values >10−3). Thus, the proteins encoded by phages within this cluster differ to a greater extent than most clusters. This is in part due to the fact that these two clusters have a larger than average number of ‘hybrid’ phages (above); for example, the genome of SPN3UB, a member of the ES18-like cluster, has an ES18-like left half and an FSL-SP-016-like right half (above and figure S6).
Mosaic module diversity and module shuffling
Phages in the lambda supercluster get their huge diversity from the diverse ‘menu choices’ at each chromosomal functional location and from recombinational shuffling of these to generate new combinations. Their virion assembly gene diversity is particularly great, and MCP differences typify this. Neighbor-joining tree analysis shows that the major capsid proteins (MCPs) of these 81 phages fall into 19 types that are more than about 60% different, and in most cases these lambda supercluster MCP types are only extremely distantly related to one another (see figures S1S and S5). These 19 MCP types are represented by phages lambda, HK97, mEp235, ES18, gifsy-2, BP-4795, PY54, SfV, ϕP27, P22, CUS-3, APSE-1, 933W, HK639, ϕES15, HS2, ENT47670, ZF40 and ϕEt88. Only the lambda/ϕ80/N15/gifsy-1, HK97/mEp235, PY54/BP-4795, APSE-1/Sf6 and P22/CUS-3 MCP types have AA sequence similarities that have good bootstrap support in the tree, but as mentioned above, they are all likely to be very distant homologues. Comparison of lambda supercluster genome and MCP dot plots (figures 7 and S1S) shows that there is very good overall correspondence of cluster type with MCP type. Nonetheless, there are a few cases of non-correspondence, mEp235 in the HK97-like cluster B, SfV in the SfV-like cluster, and Sf6 and CUS-3 MCP types in the P22-like cluster, that are discussed in more detail below.
In order to begin to understand the extent of shuffling of other mosaic sections within the lambda supercluster we sought to examine an early protein that has homologues universally encoded by these phages. Some genes that meet this criterion are prophage repressor, Cro repressor, CII transcriptional activator, and late transcriptional antiterminator. We chose the late antiterminator proteins (lambda gene Q protein) that are encoded by the last gene of the early rightward operon; the only exceptions are the N15-like cluster phages, where a homologue occupies a position near the start of their very different early right operons. A neighbor-joining tree of the lambda supercluster ‘Q proteins’ is shown in figure 9. Like the MCPs they span a wide range of diversity, but in this case there are ‘only’ five major sequence types. The branching order of this tree is robust and very different from that of the MCP tree (figure S5). For example, type 1 Q proteins includes the phage lambda Q protein but also include homologues from individual phages in the ϕ80-, ES18-, HK97-, P22-, SfV, gifsy-2 and øEt88-like clusters (e.g., individual phages in the lambda-, P22- and HK97-like clusters carry Q proteins that are ≥97% identical to one another). Similarly, type 3 Q proteins are encoded by phages in the lambda-, HK97-, ES18-, SfV-, BP-4795-, P22-, and HK639-like clusters. Extensive shuffling is also evident from the perspective of the phage clusters; for example, the six phages in the SfV-like cluster carry four of the five different types of Q genes, and the P22-like cluster includes phages with three different types. We conclude that there has been substantial inter-cluster genetic exchange in the lambda supercluster. On the other hand, the frequency of mosaic sectional shuffling is not sufficient to completely randomize Q gene types with respect to cluster membership. For example, all eleven phages in the 933W-like cluster and 12 of the 15 phages in the HK97-like cluster have very closely related Q proteins (figure 9). Perhaps these represent cases where recent population expansion of a cluster has occurred faster than the rate of shuffling (assuming that the current sampling is unbiased and random shuffling is functionally acceptable to the phages).
Figure 9. A neighbor-joining tree of the lambda supercluster Q proteins.
A CLUSTAL tree of the different lambda supercluster Q protein types shows bootstrap values (out of 1000 trials; values less than 900 and those on very short branches are not shown). The five major Q protein sequence types are numbered on the right. Four of the five major types have weak but recognizable sequence similarity to phage lambda Q protein, but type 4 does not. None of the four ‘type 4’ phages carry a gene with Q homology, so the type 4 putative late operon activator proteins were chosen because they are each encoded by a gene that lie between recognizable nin region genes and recognizable terminase genes (like the other true Q homologues).
Relationship of the lambda supercluster to phages that infect other bacterial families
In contrast to the T4 and T7 superclusters (above), we found no phage that infects other bacterial families that can legitimately be included in the lambda supercluster as defined here. Pseudomonas phage D3 and its close relatives are perhaps most closely related; many of D3’s encoded proteins are very weakly similar to those of phages in the lambda supercluster, and its genome has considerable functional synteny to these phages (Kropinski, 2000). Its transcriptional pattern is predicted to be similar to lambda; however, its lysis genes are not syntenic with the lambda supercluster. We also note that the ε15-like Enterobacteriaceae cluster has a number of functions that are syntenic with lambda supercluster phages; however, their lysis genes are also not syntenic with the lambda supercluster, and only a few ε15-like cluster encoded proteins have clear homology with the lambda supercluster phage proteins (see also Kropinski et al., 2007). In neither case does the evidence support extensive recent horizontal exchange with the lambda supercluster phages.
Individual proteins that have convincing sequence similarity to proteins encoded by lambda supercluster phages are nonetheless found in other types of phages. Many of these are the tail fiber/spike exchanges mentioned above, but there are others; for example the ES18, HK639 and SfV MCPs are 69%, 41% and 62% identical to the MCPs of Pseudomonas (γ-Proteobacteria family Pseudomonaceae) phage ϕ297, Burkholderia (family Rhizobiaceae, class α-Proteobacteria) phage P106B, and Aggregatibacter (γ-Proteobacteria family Pasteurellaceae) phage Aabϕ01, respectively. Thus, there is occasional genetic exchange of apparently rather short regions between very disparate phages, but such exchange appears to be much less frequent than exchange between clusters within the lambda supercluster.
Major capsid protein sequence as predictor of cluster membership
Can any one tailed phage-encoded protein be used as a universal indicator of cluster and/or subcluster membership? Recent studies have shown that the tail tape measure protein (TMP) type correlates well with clusters for Siphoviridae and Myoviridae phages that infect Mycobacteria smegmatis (Smith et al., 2013), but the short tails of the Podoviridae have no TMP. In fact, there is no known tail protein that is common to all types of tails and, as mentioned above, phage DNA replication/metabolism and lysis mechanisms are sufficiently diverse that homologous proteins are not universally utilized. However, all tailed phages assemble heads by similar underlying mechanisms (Casjens and Hendrix, 1988), and three genes, those that encode the large terminase subunit (TerL), portal protein and MCP, appear to be universally encoded by every tailed phage genome (homologues of phage SPP1 gp7 ‘procapsid assembly protein’, head decoration proteins, scaffolding proteins and procapsid proteases, are often but not universally encoded by tailed phage genomes; head-tail joining or connecting proteins are likely present in all tailed phage virions but are too diverse to be recognizable in all of them at present). MCPs form the icosahedral shell of the tailed phage head, portal proteins form the hole at one vertex of this shell through which DNA is packaged and ejected, and TerL is the ATPase that pumps DNA into the capsid during packaging. After DNA is packaged, tails attach to the portal vertex. These three proteins have been studied in a sufficient number of diverse phages that we are confident that homologues are encoded by all tailed phages that have been studied and almost certainly by all tailed phages. Their protein products can be recognized by AA similarity in nearly all tailed phages (these genes have diverged to the point that they cannot be recognized as having similar nucleotide sequence in most inter-cluster comparisons, and in some cases even the AA sequences are not recognizably similar). Our previous analysis has suggested that terL genes have undergone sufficient horizontal transfer between phage groups to disrupt correlation between terminase sequence type and cluster relationships (Casjens and Thuman-Commike, 2011). However, either MCP or portal protein could be suitable for cluster determination if horizontal exchange has not obscured the relationships.
The correlation between MCP type and cluster membership was examined for the Enterobacteriaceae phages. Although they have diverged to the point that they do not always have easily recognizably similar AA sequence similarities, very divergent tailed phage MCPs have been examined and found to have similar protein folds (Parent et al., 2012; Shen et al., 2012; Zhang et al., 2013; Rizzo et al., 2014; Grose et al., 2014 and references therein). Thus, although they are amazingly diverse, all tailed phage MCPs are almost certainly very ancient homologues. Figure S2A, B and C show MCP dot plots for the phages shown in figures 1, 2 and 3, respectively, and there is a very good correlation between MCP types and genome clusters within the Enterobacteriaceae tailed phages in these comparisons. Our less comprehensive analysis indicates that portal proteins appear to have largely co-evolved with MCPs (not shown; see also Casjens and Thuman-Commike (2011)), so it is very likely that portal protein sequence correlates equally well with clusters.
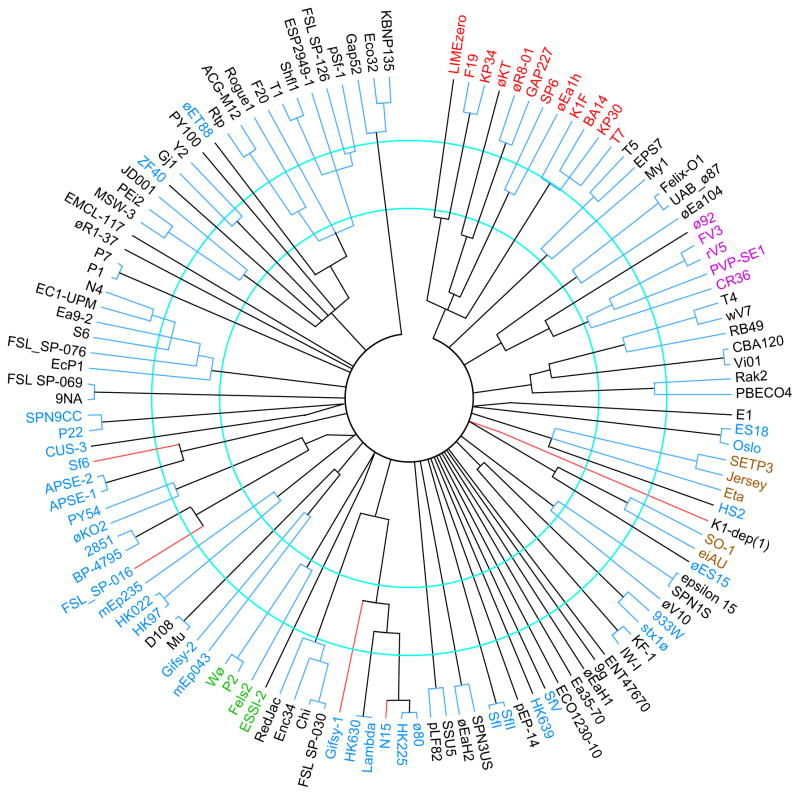
Determination of MCP sequence type of a newly isolated Enterobacteriaceae phage will therefore give a good preliminary indication of the cluster to which the phage belongs; however, closer examination shows that there are a small number of clear exceptions to this rule. Figure S1 shows MCP dot plots of all the phages in our panel, and figure 10 shows an MCP neighbor-joining tree that includes representatives of all the major sequence types of MCPs identified by this analysis (126 of the 337 MCPs are shown, the other 211 were not included because they are closely related to one of the shown MCPs). There are very different ranges of MCP sequence variation within clusters (see contiguous blue intra-cluster branch lengths in figure 10). The tree has 51 branches whose members are less than about 25% identical to one other and 67 whose members are less than about 50% identical to one another. Neither value corresponds perfectly with the 56 phage clusters since there are a few cases of MCPs in different clusters being rather closely related to one another and of MCPs within a cluster being very different from one another.
Figure 10. Neighbor-joining tree of representative Enterobacteriaceae tailed phage major capsid proteins.
A representative set of MCP types was aligned and a tree constructed by ClustalX; representative MCPs from all clusters and many subclusters are shown. The phages that encode the MCPs are indicated outside of the tree. The members of the following superclusters are indicated by label color as follows: T7, red; lambda, blue; rV5, magenta; SETP3, brown, P2, green. Nodes with bootstrap values less than 985 (out of 1000 trials) were collapsed; all the nodes shown thus have >985 bootstrap support, except slanted lines near the center which denote convincing similarities in the 25–30% identity range in pairwise alignments that fail to show this in CLUSTAL multiple alignments (probably because an MCP in another branch has weak similarity). The red branch lines indicate MCPs that are very different from others in their cluster and were most likely obtained by horizontal transfer (see text), and contiguous blue lines connect members of the same cluster. The inner and outer blue circles indicate the locations of approximately 50% and 75% AA sequence identity. An apparent frameshift in the phage Stx1ø MCP gene was ‘fixed’ for purposes of comparison.
The MCPs of some clusters are recognizably related to one another, for example the MCPs of T1-, Gj1-, PY100-, ZF40-, øET88- and MW-3-like clusters are all rather different, but reside in the same major branch in figure 10, and in particular the MCPs of the lambda- and ø80-like clusters are quite similar (~93% AA identity). On the other hand, there are several cases of very different MCPs encoded by phages in the same cluster as follows: Sf6 and CUS-3 types MCP are encoded by P22-like cluster phages but are on very different branches, and N15, SfV, FSL-016, mEp235 and K1-dep(1) type MCPs are very different from other members of their clusters (figure 10). Thus, there appears to have been fairly recent horizontal exchange between clusters or very long divergence of MCPs within clusters in these cases. For example, N15 and FSL-SP-016 MCPs appear to be recent horizontal acquisition of a virion assembly genes from BP-4795- and lambda-like Enterobacteriaceae phages, respectively, and mEp235 and K1-dep(1) MCPs are unique sequence types that appear to be head gene acquisitions from as yet uncharacterized clusters. On the other hand we have argued from their unique structures that the P22, Sf6 and CUS-3 MCPs have diverged within the P22-like group of phages (Parent et al., 2012; Parent et al., 2014). Thus, only twelve of the 337 phages (the three ø80-like phages, N15, gifsy-1, FSL-SP-016, Sf6, HK620, CUS-3, SCP-P1, mEp235 and K1-dep(1)) have MCPs that do not correlate robustly with the phage’s cluster. It is interesting to note that eleven of these twelve phages (all but K1-dep(1)) reside in the lambda supercluster - are these more likely to exchange MCPs than other phages? The phages in our panel have a 96.4% chance of being assigned to the correct cluster by their MCP sequence. This is remarkably similar to the reported 97.6% correlation between Siphoviridae and Myoviridae mycobacteriophage TMP sequence and cluster (Smith et al., 2013). The correlation between subcluster membership and MCP type is also strong but not universal. One example of MCP exchange between subclusters within a cluster is in phage øEB49, a subcluster B T1-like phage, which appears to have obtained its MCP from a subcluster C T1-like phage (Figures S1A).
The MCPs of the different Enterobacteriaceae tailed phage clusters are in general not more like each other than they are like phages that infect other bacterial families. The canonical MCPs (those of the majority of cluster members) of 17 of the 56 clusters have no known relatives with >35% AA sequence identity, and in only six clusters (ø92-, rV5-, lambda-, ø80-, øES15- and HS2-like) are the canonical MCPs more closely related to other Enterobacteriaceae clusters than they are to known phages that infect other bacterial families. Table 2 shows that 35 of the canonical cluster MCP types and the two exceptional MCPs of K1-dep(1) and mEp325 (above) have closer relatives (with >35% identity) among known phages that infect other bacterial families than among known Enterobacteriaceae phages. Indeed, eleven of the MCP types in table 3 have such ‘outside’ relatives that are >65% identical, with the highest similarities being 82% identity between the N4-like cluster and Achromobacter phage JWDelta (Wittmann et al., 2014) and between the T7-like cluster and Stenotrophomonas phage IME15 (Huang et al., 2012), and 78% between the T4-like cluster and Acinetobacter phage Ac42 (Petrov et al., 2010). In these three cases and some of the other ‘outside’ phages with closely related MCPs there is a degree of overall genome synteny between the Enterobacteriaceae phages and the ‘outside’ phages, but in all cases we are aware of except IME15 (above), such outside phages fall well outside of the clusters as defined here or form novel subclusters within them (see discussions of T4- and T7-clusters above). These relationships point out possible examples of past (but not recent) movement of whole phages between host families or long co-divergence of host and phage, rather than horizontal transfer of virion assembly genes among phages. We also note that nine of the 11 MCP types with >65% ‘outside’ relatives are from lytic phages, suggesting lytic phages are more likely to have relatives from distant hosts.
Table 3.
Database matches for Enterobacteriaceae tailed phages
| Cluster1 | Phage MCP | Best matches to other Enterobacteriaceae tailed phages (cluster-% identity)2 | Best matches outside the Enterobacteriaceae tailed phages (host genus/phage-% identity)2 |
|---|---|---|---|
| T1-like | T1 | øET88-46% | Xanthomonas/DiDDI-46% |
| T4-like | T4 | CBA120-37% | Acinetobacter/AC42-78%; Vibrio/KVP40-60% |
| CBA120-like | Vi01 | T4-37% | Delftia/øW-14-54% |
| T5-like | T5 | – | Vibrio/pVp-1-60% |
| T7-like | T7 | – | Streptophomonas/IME15-82%; Pseudomonas/gh1-73% |
| SP6-like | SP6 | – | Vibrio/Vc1-44% |
| LIMEzero-like | LIMEzero | F19 – 48% | Ralstonia/RSB3-52% |
| KP34-like | KP34 | LIMEzero-47% | Vibrio/VP93-74% |
| øKT-like | øKT | GAP227-47% | Caulobacter/Cd1-50% |
| GAP227-like | GAP227 | øKT-47% | Aeromonas/øAS7-82% |
| N4-like | N4 | – | Achromobacter/JWDelta-86%Pseudomonas/LUZ7-57% |
| 9NA-like | 9NA | – | – |
| Chi-like | Chi | – | Burkholderia/AH2-47% |
| øEco32-like | øEco32 | – | – |
| Felix-O1-like | Felix-O1 | – | – |
| SETP3-like | SETP3 | – | Burkholderia/BcepGomr-37% |
| SETP3-like | K1-dep(1) | – | Synechococcus/S-CBS4-56% |
| SO-1-like | SO-1 | – | – |
| Eco1230-10-like | Eco1230-10 | – | Pseudomonas/PPpW-3-68% |
| Gj1-like | Gj1 | – | Shewanella/Spp001-56% |
| PY100-like | PY100 | – | – |
| ø92-like | ø92 | rV5-43% | – |
| rV5-like | rV5 | ø92-43% | Vibrio/1189-B1-35% |
| SPN3US-like | SPN3US | – | – |
| Rak2-like | Rak2 | – | – |
| øR1-37-like | øR1-37 | – | – |
| E1-like | E1 | – | – |
| EMCL-117-like | EMCL-117 | – | Pseudomonas/F8-67%; Burkholderia/BcepF1-61% |
| KF-1-like | K1 | – | Vibrio/VPMS1-69% |
| MSW-3-like | MSW-3 | – | Iodobacter/øPLPE-68% |
| Ea35-70-like | EA35-70 | – | – |
| øEaH1-like | øEaH1 | – | – |
| 9g-like | 9g | Eta-37% | Sponge associated α-proteobacterium/øJL001-51% |
| Lambda-like | Lambda | N15-90%; ø80-87% | Xylella/Sano-35% |
| ø80-like | ø80 | N15-97%; lambda-89% | – |
| HK97-like | HK97 | – | Xanthomonas/øL7-45% |
| HK97-like | mEp235 | – | Pseudomonas/D3-48% |
| N15-like | PY54 | – | Burkholderia/fE125-50% |
| N15-like | N15 | ø80-97%; lambda-90% | Xylella/Sano-35% |
| ES18-like | ES18 | – | Pseudomonas/øi297-69% |
| Gifsy-2-like | gifsy-2 | – | Pseudomonas/F10-55% |
| BP-4795-like | BP-4795 | – | Pseudomonas/PAJU2-48% |
| SfV-like | SfI | – | Burkholderia/Bcep176-41% |
| SfV-like | SfV | – | Aggregatibacter/Aabø01-62% |
| P22-like | P22 | – | – |
| P22-like | CUS-3 | – | – |
| P22-like | Sf6 | APSE-1-80% | – |
| APSE-1-like | APSE-1 | – | – |
| 933W-like | 933W | – | – |
| HK639-like | HK639 | – | Rhizobium/P106B-41% |
| øES15-like | øES15 | SO-1-38% | – |
| HS2-like | HS2 | SO-1-40% | – |
| ENT47670-like | ENT47670 | – | – |
| ZF40-like | ZF40 | – | – |
| øEt88-like | øET88 | T1-46% | Xanthomonas/DiBBI-53% |
| ε15 | ε15 | – | Bordetella/BPP-1-46%; Burkholderia/BcepC6B-44%; |
| P2-like | P2 | – | Rasltonia RSA1-57%; Burkholderia/KL3-56% |
| ESSI-2-like | ESSI-2 | – | Aeromonas/øiO18P-55%; Vibrio/K139-52% |
| P1-like | P1 | – | – |
| Mu-like | Mu | – | Pseudomonas/DMS3-51%; Rhodobacter/RC1-47%; |
| SSU5-like | AAU5 | – | Xanthomonas/Xp15-36%; Pseudomonas/YuA-36% |
| pPEP-14-like | pPEP-14 | – | Sinorhizobium/PBC5-66%; Burkhoderia/Bcep22-65% |
Unusual MCPs in a cluster (see text) are inset in the column.
Only matches >35% AA sequence identity over most of the protein are shown (as identified by BLASTp); ‘–’ indicates no such matches were found.
Relationships between host taxonomy and phage clusters
The large number of genome sequences of phages that infect hosts in 18 genera and 31 species in the Enterobacteriaceae family (genera listed in Table 4) offers a unique opportunity to examine the relationship between host similarity and phage similarity. Because of their importance as bacterial pathogens and as bacterial model systems, 240 of the 337 phages in our panel were isolated on E. coli/Shigella or S. enterica hosts, and these phages reside in 40 of the 56 clusters. Is our knowledge of diversity closer to complete in these rather closely related genera? Of the 40 clusters that contain phages that infect these two species, 17 contain phages that infect both species. The contents of these clusters range from 29 and 16 Escherichia and Salmonella phages in the T4- and SETP3-like clusters, respectively, to 8 clusters that contain only one phage that infects either of these species. This fraction of singleton clusters (20%) is lower than that of all the Enterobacteriaceae hosts (32%), but nonetheless suggests that even for the well-studied E. coli and S. enterica hosts, tailed phage cluster discovery is rather far from complete.
Table 4.
Host genera of sequenced Enterobacteriaceae tailed phages
| Prototype phage | Cluster |
Escherichia/ Shigella |
Salmonella | Yersinia | Erwinia | Cronobacter | Klebsiella | Edwardsiella | Pectobacterium | Enterobacter | Citrobacter | Serratia | Hamiltonella | Pantoea | Sodalis | Dickeya | Kluyvera | Morganella | Providencia |
Single host Subcluster1 |
Multiple host subcluster |
Number of subclusters |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T1 | Lytic1 | 12 | 1 | 1 | 1 | 1 | 5 | 2 | 7 | |||||||||||||
| T4 | Lytic2 | 27 | 2 | 3 | 1 | 2 | 1 | 1 | 7 | 3 | 10 | |||||||||||
| Vi01 | Lytic3 | 4 | 9 | 1 | 1 | 1 | 2 | 2 | 4 | |||||||||||||
| T5 | Lytic4 | 2 | 3 | 1 | 1 | 1 | 1 | 2 | ||||||||||||||
| T7 | Lytic5 | 7 | 5 | 1 | 2 | 1 | 2 | 3 | 1 | 1 | 3 | 5 | 8 | |||||||||
| SP6 | Lytic6 | 3 | 3 | 3 | 1 | 3 | 1 | 4 | ||||||||||||||
| KP34 | Lytic7 | 2 | 1 | 2 | 0 | 2 | ||||||||||||||||
| LIMEzero | Lytic8 | 1 | 1* | 0 | 1 | |||||||||||||||||
| øKT | Lytic9 | 1 | 1* | 0 | 1 | |||||||||||||||||
| GAP227 | Lytic10 | 2 | 1 | 3 | 0 | 3 | ||||||||||||||||
| N4 | Lytic11 | 6 | 2 | 2 | 1 | 5 | 0 | 5 | ||||||||||||||
| 9NA | Lytic12 | 3 | 1 | 0 | 1 | |||||||||||||||||
| Chi | Lytic13 | 7 | 1 | 1 | 3 | 0 | 3 | |||||||||||||||
| øEco32 | Lytic14 | 4 | 1 | 1 | 3 | 0 | 3 | |||||||||||||||
| Felix-O1 | Lytic15 | 3 | 3 | 3 | 1 | 1 | 2 | |||||||||||||||
| SETP3 | Lytic16 | 5 | 11 | 1 | 4 | 0 | 4 | |||||||||||||||
| SO-1 | Lytic17 | 4 | 4 | 1 | 1 | 1 | 2 | |||||||||||||||
| Eco1230-10 | Lytic18 | 1 | 1* | 0 | 1 | |||||||||||||||||
| Gj1 | Lytic19 | 1 | 1 | 1 | 3 | 0 | 3 | |||||||||||||||
| PY100 | Lytic20 | 1 | 1* | 0 | 1 | |||||||||||||||||
| ø92 | Lytic21 | 2 | 1 | 0 | 1 | |||||||||||||||||
| rv5 | Lytic22 | 4 | 2 | 3 | 1 | 1 | 2 | 3 | ||||||||||||||
| SPN3US | Lytic23 | 1 | 1 | 1 | 3 | 0 | 3 | |||||||||||||||
| Rak2 | Lytic24 | 1 | 1 | 1 | 3 | 0 | 3 | |||||||||||||||
| øR1-37 | Lytic25 | 1 | 1* | 0 | 1 | |||||||||||||||||
| E1 | Lytic26 | 1 | 1* | 0 | 1 | |||||||||||||||||
| EMCL-117 | Lytic27 | 1 | 1* | 0 | 1 | |||||||||||||||||
| KF-1 | Lytic28 | 2 | 1 | 0 | 1 | |||||||||||||||||
| MSW-3 | Lytic29 | 1 | 2 | 2 | 0 | 2 | ||||||||||||||||
| Ea35-70 | Lytic30 | 1 | 1* | 0 | 1 | |||||||||||||||||
| øEaH1 | Lytic31 | 1 | 1* | 0 | 1 | |||||||||||||||||
| 9g | Lytic32 | 1 | 1* | 0 | 1 | |||||||||||||||||
| lambda | Temperate1 | 3 | 1 | 0 | 1 | |||||||||||||||||
| ø80 | Temperate2 | 3 | 2 | 0 | 2 | |||||||||||||||||
| N15 | Temperate3 | 1 | 1 | 1 | 3 | 0 | 3 | |||||||||||||||
| HK97 | Temperate4 | 14 | 1 | 3 | 0 | 3 | ||||||||||||||||
| ES18 | Temperate5 | 3 | 2 | 0 | 2 | |||||||||||||||||
| gifsy-2 | Temperate6 | 5 | 4 | 6 | 0 | 6 | ||||||||||||||||
| BP-4795 | Temperate7 | 4 | 1 | 0 | 1 | |||||||||||||||||
| SfV | Temperate8 | 5 | 1 | 4 | 0 | 4 | ||||||||||||||||
| P22 | Temperate9 | 13 | 3 | 1 | 1 | 2 | ||||||||||||||||
| APSE-1 | Temperate10 | 2 | 1 | 0 | 1 | |||||||||||||||||
| 933W | Temperate11 | 11 | 1 | 0 | 1 | |||||||||||||||||
| HK639 | Temperate12 | 1 | 1* | 0 | 1 | |||||||||||||||||
| øES15 | Temperate13 | 1 | 1* | 0 | 1 | |||||||||||||||||
| HS2 | Temperate14 | 1 | 1* | 0 | 1 | |||||||||||||||||
| ENT47670 | Temperate15 | 1 | 1* | 0 | 1 | |||||||||||||||||
| ZF40 | Temperate16 | 1 | 1* | 0 | 1 | |||||||||||||||||
| øEt88 | Temperate17 | 1 | 1* | 0 | 1 | |||||||||||||||||
| epsilon15 | Temperate18 | 2 | 5 | 4 | 0 | 4 | ||||||||||||||||
| P1 | Temperate19 | 3 | 1 | 0 | 1 | |||||||||||||||||
| P2 | Temperate20 | 4 | 5 | 1 | 1 | 2 | 2 | 4 | ||||||||||||||
| ESSI-2 | Temperate21 | 1 | 1* | 0 | 1 | |||||||||||||||||
| Mu | Temperate22 | 2 | 1 | 0 | 1 | |||||||||||||||||
| SSU5 | Temperate23 | 2 | 2 | 1 | 1 | 1 | 1 | 2 | ||||||||||||||
| pEP-14 | Temperate24 | 1 | 1* | 0 | 1 | |||||||||||||||||
| Total phages | 163 | 77 | 22 | 17 | 13 | 12 | 8 | 5 | 4 | 3 | 3 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 132 |
Asterisks (*) indicate singleton clusters.
The host genera with the next most sequenced tailed phages are Yersinia (22 phages in 9 clusters), Erwinia (17 phages in 11 clusters), Klebsiella (13 phages in 9 clusters) and Cronobacter (12 phages in 9 clusters). Clusters are not limited to phages from one or several very closely related genera; for example, the 33 phages in the T7-like cluster infect nine genera, the 16 T1-like phages infect five genera, the seven T5-like phages infect four genera. Overall, the average number of host genera for the 38 non-singleton clusters is 3.2, indicating substantial variety of hosts within clusters. Although 29 of the 56 clusters have a single host genus, nearly all of these are low occupancy clusters with ≤4 phage members. Only 29% of the non-singleton clusters have a single host, and with only one exception, all 21 clusters with more than four members have more than one host genus. The single populous cluster with only one host species is the 933W-like cluster for which all eleven members infect E. coli. These phages all encode the Shiga-like toxin that is important in E. coli food poisoning and were isolated by induction from bacteria that cause the disease, so this could be the result of a sampling abnormality due to the human medical importance of this phage type. Thus, although phage clusters restricted to one or a few closely related hosts may well exist (particularly among the temperate phages, see below), the vast majority of the currently well-populated Enterobacteriaceae clusters are not limited to phages that infect a single host genus, and we expect the clusters that currently have low occupancy to gain additional host genera as phages of more hosts are studied.
Although no strong overall correlation between cluster and host is seen, our analysis reveals a strong correlation of subcluster membership with the host species, with a few notable exceptions. Overall, 83% of the 132 subclusters (including singleton clusters and subclusters) are populated by phages from a single host. If the 67 singleton subclusters are removed, 78% of the 65 multiple phage-containing subclusters are populated by phages from a single host (Table 5). Clearly there is a substantial correlation between subcluster and host genus among these tailed phages. The subcluster/host relationship is not equal between the lytic and temperate phages. The percentage of non-singleton subclusters with a single host is 83% among the temperate phages, while it is only 50% if only lytic phages are considered. The T7 and lambda superclusters afford clear examples of this lytic verses temperate difference in host/subcluster association. Among the 17 non-singleton subclusters in the lambda supercluster, 16 obey a one host genus per subcluster rule (the single exception is SPC-P1 in P22-like subcluster B), while only 2 of the 8 non-singleton subclusters in the T7 supercluster obey this rule. The T7-like cluster also contains an extreme exception to this rule. The Stenotrophomonas phage IME15, whose host belongs to a different bacterial family, has remarkable similarity to the E. coli phage T7-like subcluster A phages (as discussed above).
Table 5.
Host – subcluster relationships
| Clusters | Subclusters | Lytic subclusters | Temperate subclusters | T7 supercluster | Lambda supercluster | |
|---|---|---|---|---|---|---|
| Single Host Genus | 29 | 110 (43)1 | 68 (23) | 42 (20) | 13 (2) | 31 (16) |
| Multiple Host Genera | 27 | 22 (22) | 18 (18) | 4 (4) | 6 (6) | 1 (1) |
| Total | 56 | 132 (65) | 86 (41) | 46 (24) | 19 (8) | 32 (17) |
| Percentage Single Host | 52% | 83% (78%) | 79% (50%) | 91% (83%) | 68% (25%) | 97% (94%) |
Values in parentheses ignore singleton subclusters.
We also note that this analysis does not account for differences in host range. Many phages are highly specific for their host species or serovar, but the few phages with broader host range add another level of complexity to such analyses. A large majority of the phages in our panel have not been tested systematically for infectivity on Enterobacteriaceae species other than the host upon which they were isolated (for example, although it is not always known if it can replicate in such distant hosts, phage P1 is reported to be able to inject transducing DNA into other Enterobacteriaceae hosts such as Pectobacterium carotovorum (Burova and Tovkach, 2006) and even hosts from other families or classes such as Vibrio harveyi (Belas et al., 1984) and Myxococcus xanthus (Kuner and Kaiser, 1981). A recent analysis of mycobacteriophages affords an approximation of the fraction of phages to expect to have a broad or narrow host range. Jacobs-Sera et al. (2012) reported that most of 221 phages tested that infect M. smegmatis mc2155 are highly specific for that host; only 23 phages (10.4%) are capable of efficient Mycobacterium tuberculosis infection.
Conclusions
The Enterobacteriaceae tailed phage analysis provided here forms a robust framework for understanding their diversity and relationships, for relating newly discovered phages to the extant Enterobacteriaceae phages, for examining host/phage evolutionary relationships, and for comparing phages that infect this bacterial family with phages that infect other families. In addition, this analysis illustrates the need for utilizing several different methods for determining phage relatedness, including nucleotide and encoded AA sequence similarity, as well as conservation of gene content and order. Nucleotide sequence is most useful for studying closely related phages, while encoded protein sequence as well as gene content and order can show more distant relationships. We have grouped phage clusters with such more distant (but syntenic) relationships into superclusters of phages with very similar molecular lifestyles; such related clusters can have little remaining sequence similarity. We agree with Hatfull and colleagues (2010; 2013) that nucleotide sequence dot plot analysis is extremely useful for first level cluster assignment, since it naturally and robustly groups phages operationally into clusters that contain only phages with similar lifestyles, and it does not depend on detailed knowledge of a phage’s molecular lifestyle. In addition, dot plot analysis confirms genome synteny and shows both sequence similarity and mosaic differences between related phages, which are important in understanding the more detailed relationships among phages. Genome mosaicism appears to be present in all clusters found here when a sufficient number of phages have been studied. Methods that simply build trees from whole genome or encoded protein sequences or which calculate the fraction of shared nucleotide sequence or genes lose sight of these important aspects of phage diversity. In addition, annotation differences can drastically alter gene product analysis as discussed in the øKT comparison above.
Clearly, in spite of speculations to the contrary less than a decade ago (Hendrix, 2002; Casjens, 2005), the Enterobacteriaceae tailed phages can be, for the most part, unambiguously parsed into clusters of related phages (i. e., they are not uniformly distributed in ‘genome sequence space’), and ambiguous cases can be understood in terms of relatively infrequent recent horizontal transfer of genetic information between clusters. Not surprisingly, the horizontal transfer among these phages that is responsible for the mosaic nature of their genomes appears to occur much more frequently within clusters than between them, and between related clusters within superclusters more frequently than between other clusters. Nonetheless, several striking examples of sequence similarities between clusters outside of superclusters show that such transfers do occasionally happen. Yet, in spite of such exchanges that are exemplified by the long tail fiber exchange between phage lambda- and T4-like clusters (George et al., 1983) and S. enterica serovar Typhimurium receptor-specific tailspikes among members of the P22-, SP6, SETP3-, Vi01- and 9NA-like clusters (Walter et al., 2008; De Lappe et al., 2009; Casjens and Thuman-Commike, 2011; Andres et al., 2012), overall we find very little evidence for wholesale and rapid exchange between clusters, and horizontal exchange is not so rapid that it obscures the existence of clusters or subclusters.
We identified 56 Enterobacteriaceae tailed phage clusters of which 18 are singletons. Among these are 34 and 23 clusters with members that infect Escherichia/Shigella or Salmonella hosts, and of these 9 and 5 are singletons, respectively. Although the existence of such a large fraction of singletons indicates that there are certainly undiscovered clusters that infect these genera, it is nonetheless likely that most of the common phage types have been identified for these two genera. Thus, if we optimistically assume that half of the clusters that infect these genera have been identified, there would be 50–70 different phage clusters whose members infect each Enterobacteriaceae genus (there is no reason to suspect that the above two genera are particularly rich in phages). The number of tailed phage clusters that might infect members of the whole Enterobacteriaceae family is much harder to predict at this juncture. Sixteen of the 56 clusters have no members that infect Escherichia or Salmonella, and 15 clusters infect only these two genera. Most of these two sets of clusters are low occupancy, so no strong statistical argument can be made for the existence of clusters that infect one or a small number of genera, but the possibility of genus-specific clusters certainly exists. Thus, given that no phages have been characterized from at least three-quarters of the Enterobacteriaceae genera, the possibility of hundreds of Enterobacteriaceae tailed phage clusters seems quite reasonable at this point.
A comparison of our 56 cluster study with the analysis of the sequences of 471 tailed phages that infect the Gram positive host Mycobacteria smegmatis reveals a similar diversity of 30 clusters (Hatfull et al., 2013; Hatfull, 2014). This diversity, analyzed with rules essentially identical to those used here, gave a comparable number of clusters to those delineated here for E. coli or S. enterica. The mosaic nature of the mycobacteriophage genomes and the presence of subcluster structure within clusters is also similar to the Enterobacteriaceae phages. Thus, the extent and nature of tailed phage diversity appears to be relatively constant for these two very distantly related, free-living bacterial hosts. These studies agree that there are limits to phage diversity, perhaps 30–50 phage clusters for each host genus. In spite of these predicted limits, this still implies an very large extant overall phage diversity.
The number of gene types present in these phage clusters is also huge, with over 50,000 different gene products reported for ~500 mycobacteriophages (Hatfull, 2014). Phages are perhaps the largest reservoir for understanding the possibilities of protein diversity within a particular function. There have been several reports of phage proteins that have diverged beyond the point of either nucleotide or amino acid sequence recognition, yet maintain a similar function and/or fold. Several such examples are provided above in the lambda and T7 supercluster discussions as well as the MCP discussion, where phages have chosen from ‘menus’ of protein options to fulfill particular functions; however, without detailed protein structural information it is impossible to distinguish between nonhomologous genes and very divergent homologous genes.
In spite of host range uncertainties and sampling insufficiencies discussed above, our results show a rather strong correlation between closely-related phages and their hosts (78% of the Enterobacteriaceae phage non-singleton subclusters are populated by phages from a single host), but this correlation breaks down when comparisons are made at the cluster level. Thus, host switching by phages, even within a bacterial family, is not so rapid that it completely obscures the relationship between subclusters and hosts. We also found that the temperate phage subclusters correlate significantly better with their host than do the lytic phages (83% versus 50%, respectively, of non-singleton subclusters infect a single genus). This lower host diversity within subclusters in the temperate phages is likely due to the large number of delicate host interactions temperate phages must utilize during the lysogenic life cycle. Lytic phages can perform their necessary functions without regard to host survival and so are expected to have fewer host-specific interactions. This difference is apparent in the comparison of the T7 and lambda superclusters above and suggests that different types of phages vary greatly in the evolutionary restrictions that are based on the host they affect. Since 18 of the clusters we identify contain only one phage, the continued isolation and characterization of many more Enterobacteriaceae phages will be required for a full understanding of the relationship of phage type to bacterial host differences. We also note that a large majority of the temperate phages in our panel were discovered after induction of a prophage from a bacterial isolate and not as virions in environmental samples, so the under-representation of temperate phages in genera other than Escherichia or Salmonella (only 15 of 112 temperate phages) may be due to the fact that most phages for those other hosts were isolated from environmental virion samples.
MATERIALS AND METHODS
Phage sequences
For ease of interpretation of matrix dot plot comparisons the phage genome sequences were manually curated into a database in which genomes are oriented so that within each cluster all members have the same orientation. Only three reported Enterobacteriaceae phage genomes were not included in our database. The reported phage ES2 genome (Accession No. JF314845) is considerably smaller than other tailed phages and is reported to have a contractile tail; however, its reported sequence does not appear to contain enough genes to build a typical contractile tail. In addition, its reported coat protein amino acid (AA) sequence fragment does not appear to be encoded by the reported ES2 sequence (Lee et al., 2011). We also note that sequence of the putative Staphylococcus ‘phage SA1’ genome (Accession No. GU169904) appears to be an inadvertent fusion of two complete genomes that are very similar to two different Salmonella phages that are Chi- and Felix-O1-like. Since nearly identical tailed phages are not known to infect hosts as distantly related as the Firmicute Staphylococcus and Proteobacteria Salmonella genera, and since such a natural fusion of two complete phage genomes has never been documented, it seems likely that the SA1 GenBank entry is the result of inadvertent co-assembly of two different Enterobacteriaceae phage genomes rather than a single Staphylococcus phage genome. Finally, the reported “complete” genome sequence of the SETP3-like Salmonella phage L13 (accession No. KC832325) is missing nearly half of its sequence compared to its close relatives. We believe this is likely due to a sequencing error. We recommend these three sequences not be considered legitimate until further characterization of these phages is reported. Database details and the manipulated phage genome sequences are available from the corresponding authors or can be downloaded at the following world-wide web site (http://enterbacteriophage.byu.edu). There are often multiple GenBank (Benson et al., 2013) entries for a single phage genome, with somewhat different annotations. GenBank accession numbers used in this analysis is provided in Table S1. When a revision to an original GenBank file is used (NC_… accession numbers), GenBank allows easy manipulation back to the original annotation.
Comparison of phage genome sequences
Comparisons of phage genomes were made using matrix dot plot generating computer programs Gepard (Krumsiek et al., 2007) and DNA Strider (Douglas, 1995). The definition of such cluster was ≥50% of the nucleotide sequence of the genome is recognizably similar and syntenic (50% homology span length by dot plot analysis at word length 10) to other members of the cluster. We include phages in a cluster whose genome is cumulatively >50% similar by dot plot analysis. Note that Gepard compares both strands, so parallel orientation of nucleotide sequences is not essential for detection of similarity, and that “word length” is a measure of sequence match stringency in Gepard; longer word lengths are more stringent and show less background in the plots. The word length is variable in the figures of this manuscript (10–12 for nucleotide plots) due to the high variance in the number of phages belonging to a cluster (longer word lengths were used to decrease background in large clusters). Whole genome (gene product) AA Gepard dot plots were performed on tandem arrays of GenBank (Benson et al., 2013) annotated gene product (encoded protein) AA sequences that were generated by Phamerator (Cresawn et al., 2011). Average nucleotide identity was determined by ANI (Lassmann and Sonnhammer, 2005), genomic maps were produced by DNAmaster (http://cobamide2.bio.pitt.edu) or Phamerator (Cresawn et al., 2011), and conserved gene product content was determined using CoreGenes 3.5 at the default BLASTP threshold of 75 (Zafar et al., 2002; Mahadevan et al., 2009). Neighbor-joining trees were created with Clustal X (Larkin et al., 2007).
Supplementary Material
Acknowledgments
This work was supported by NIH grant RO1-AI074825 (SRC), U. of Utah Pathology Department funding (SRC), and the College of Life Sciences and the Department of Microbiology and Molecular Biology of Brigham Young University (JHG). We thank Roger Hendrix, Graham Hatfull, Justin Leavitt, Adam Clayton, Colin Dale, Derek Pickard and Andrey Letarov for access to unpublished information. We also thank Garrett Jensen for assistance in collecting genomes for Phamerator analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REEFERENCES
- Adriaenssens EM, Ceyssens PJ, Dunon V, Ackermann HW, Van Vaerenbergh J, Maes M, De Proft M, Lavigne R. Bacteriophages LIMElight and LIMEzero of Pantoea agglomerans, belonging to the “phiKMV-like viruses”. Appl Environ Microbiol. 2011;77:3443–50. doi: 10.1128/AEM.00128-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anderson TF. The activity of a bacteriostatic substance in the reaction between bacterial virus and host. Science. 1945;101:565–6. doi: 10.1126/science.101.2631.565. [DOI] [PubMed] [Google Scholar]
- Andres D, Roske Y, Doering C, Heinemann U, Seckler R, Barbirz S. Tail morphology controls DNA release in two Salmonella phages with one lipopolysaccharide receptor recognition system. Mol Microbiol. 2012;83:1244–53. doi: 10.1111/j.1365-2958.2012.08006.x. [DOI] [PubMed] [Google Scholar]
- Baker J, Limberger R, Schneider SJ, Campbell A. Recombination and modular exchange in the genesis of new lambdoid phages. New Biol. 1991;3:297–308. [PubMed] [Google Scholar]
- Battaglioli EJ, Baisa GA, Weeks AE, Schroll RA, Hryckowian AJ, Welch RA. Isolation of generalized transducing bacteriophages for uropathogenic strains of Escherichia coli. Appl Environ Microbiol. 2011;77:6630–5. doi: 10.1128/AEM.05307-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson BL, Allen HK, Brunelle BW, Lee IS, Casjens SR, Stanton TB. The agricultural antibiotic carbadox induces phage-mediated gene transfer in Salmonella. Front Microbiol. 2014;5:52. doi: 10.3389/fmicb.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belas R, Mileham A, Simon M, Silverman M. Transposon mutagenesis of marine Vibrio spp. J Bacteriol. 1984;158:890–6. doi: 10.1128/jb.158.3.890-896.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41:D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh O, Borsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–8. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- Bielke L, Higgins S, Donoghue A, Donoghue D, Hargis BM. Salmonella host range of bacteriophages that infect multiple genera. Poult Sci. 2007;86:2536–40. doi: 10.3382/ps.2007-00250. [DOI] [PubMed] [Google Scholar]
- Born Y, Fieseler L, Marazzi J, Lurz R, Duffy B, Loessner MJ. Novel virulent and broad-host-range Erwinia amylovora bacteriophages reveal a high degree of mosaicism and a relationship to Enterobacteriaceae phages. Appl Environ Microbiol. 2011;77:5945–54. doi: 10.1128/AEM.03022-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D. A theory of modular evolution in bacteriophages. Ann N Y Acad Sci. 1980;354:484–491. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- Boyd EF. Bacteriophage-encoded bacterial virulence factors and phage-pathogenicity island interactions. Adv Virus Res. 2012;82:91–118. doi: 10.1016/B978-0-12-394621-8.00014-5. [DOI] [PubMed] [Google Scholar]
- Brussaard CP, Wilhelm SW, Thingstad F, Weinbauer MG, Bratbak G, Heldal M, Kimmance SA, Middelboe M, Nagasaki K, Paul JH, Schroeder DC, Suttle CA, Vaque D, Wommack KE. Global-scale processes with a nanoscale drive: the role of marine viruses. ISME J. 2008;2:575–8. doi: 10.1038/ismej.2008.31. [DOI] [PubMed] [Google Scholar]
- Brussow H, Hendrix RW. Phage genomics: small is beautiful. Cell. 2002;108:13–6. doi: 10.1016/s0092-8674(01)00637-7. [DOI] [PubMed] [Google Scholar]
- Burova LM, Tovkach FI. Expression of genes of prophage P1 Escherichia coli in cells of phytopathogenic Erwinia. Mikrobiol Z. 2006;68:39–47. [PubMed] [Google Scholar]
- Cairns J, Stent G, Watson J, editors. Phage and the origins of molecular biology. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory of Quantitative Biology; 1966. [Google Scholar]
- Canchaya C, Fournous G, Brussow H. The impact of prophages on bacterial chromosomes. Mol Microbiol. 2004;53:9–18. doi: 10.1111/j.1365-2958.2004.04113.x. [DOI] [PubMed] [Google Scholar]
- Cardinale D, Carette N, Michon T. Virus scaffolds as enzyme nano-carriers. Trends Biotechnol. 2012;30:369–76. doi: 10.1016/j.tibtech.2012.04.001. [DOI] [PubMed] [Google Scholar]
- Casas V, Maloy S. Role of bacteriophage-encoded exotoxins in the evolution of bacterial pathogens. Future Microbiol. 2011;6:1461–73. doi: 10.2217/fmb.11.124. [DOI] [PubMed] [Google Scholar]
- Casjens S. Prophages and bacterial genomics: what have we learned so far? Mol Microbiol. 2003;49:277–300. doi: 10.1046/j.1365-2958.2003.03580.x. [DOI] [PubMed] [Google Scholar]
- Casjens S, Gilcrease D. Determining DNA packaging strategy by analysis of the termini of the chromosomes in tailed-bacteriophage virions. In: Clokie M, Kropinski A, editors. Bacteriophages: methods and protocols. Humana Press; Totowa, NJ: 2009. pp. 91–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens S, Hatfull G, Hendrix R. Evolution of dsDNA tailed-bacteriophage genomes. Sem Virol. 1992;3:383–397. [Google Scholar]
- Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The Bacteriophages. Vol. 1. Plenum Press; New York City: 1988. pp. 15–91. [Google Scholar]
- Casjens S, Hendrix R. Bacteriophages and the bacterial genome. In: Higgins NP, editor. The bacterial chromosome. ASM Press; Washingtion, D. C: 2005. pp. 39–52. [Google Scholar]
- Casjens SR. Comparative genomics and evolution of the tailed-bacteriophages. Curr Opin Microbiol. 2005;8:451–8. doi: 10.1016/j.mib.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Casjens SR, Thuman-Commike PA. Evolution of mosaically related tailed bacteriophage genomes seen through the lens of phage P22 virion assembly. Virology. 2011;411:393–415. doi: 10.1016/j.virol.2010.12.046. [DOI] [PubMed] [Google Scholar]
- Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323:139–41. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- Chen Z, Schneider TD. Comparative analysis of tandem T7-like promoter containing regions in enterobacterial genomes reveals a novel group of genetic islands. Nucleic Acids Res. 2006;34:1133–47. doi: 10.1093/nar/gkj511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibeu A, Lingohr EJ, Masson L, Manges A, Harel J, Ackermann HW, Kropinski AM, Boerlin P. Bacteriophages with the ability to degrade uropathogenic Escherichia coli biofilms. Viruses. 2012;4:471–87. doi: 10.3390/v4040471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau AM, Bertrand C, Letarov A, Tetart F, Krisch HM. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology. 2007;362:384–96. doi: 10.1016/j.virol.2006.12.031. [DOI] [PubMed] [Google Scholar]
- Comeau AM, Arbiol C, Krisch HM. Gene network visualization and quantitative synteny analysis of more than 300 marine T4-like phage scaffolds from the GOS metagenome. Mol Biol Evol. 2010;27:1935–1944. doi: 10.1093/molbev/msq076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresawn SG, Bogel M, Day N, Jacobs-Sera D, Hendrix RW, Hatfull GF. Phamerator: a bioinformatic tool for comparative bacteriophage genomics. BMC Bioinformatics. 2011;12:395. doi: 10.1186/1471-2105-12-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick FH, Barnett L, Brenner S, Watts-Tobin RJ. General nature of the genetic code for proteins. Nature. 1961;192:1227–32. doi: 10.1038/1921227a0. [DOI] [PubMed] [Google Scholar]
- Culpepper BK, Morris DS, Prevelige PE, Bellis SL. Engineering nanocages with polyglutamate domains for coupling to hydroxyapatite biomaterials and allograft bone. Biomaterials. 2013;34:2455–62. doi: 10.1016/j.biomaterials.2012.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin C. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- d’Herelle F. The Bacteriophage and Its Behavior. Williams & Wilkins; Baltimore, MD: 1926. [Google Scholar]
- De Lappe N, Doran G, O’Connor J, O’Hare C, Cormican M. Characterization of bacteriophages used in the Salmonella enterica serovar Enteritidis phage-typing scheme. J Med Microbiol. 2009;58:86–93. doi: 10.1099/jmm.0.000034-0. [DOI] [PubMed] [Google Scholar]
- De Paepe MM, Hutinet G, Son O, Amarir-Bouhram J, Schbath S, Petit M. Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: the role of Rad52-like recombinase. PLoS Genet. 2014;10:e1004181. doi: 10.1371/journal.pgen.1004181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbruck M. Bacterial viruses or bacteriophages. Biol Rev Camb Philos Soc. 1946;21:30–40. [PubMed] [Google Scholar]
- Demerec M, Fano U. Bacteriophage-Resistant Mutants in Escherichia coli. Genetics. 1945;30:119–36. doi: 10.1093/genetics/30.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbins AT, George M, Jr, Basham DA, Ford ME, Houtz JM, Pedulla ML, Lawrence JG, Hatfull GF, Hendrix RW. Complete genomic sequence of the virulent Salmonella bacteriophage SP6. J Bacteriol. 2004;186:1933–44. doi: 10.1128/JB.186.7.1933-1944.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas SE. DNA Strider. An inexpensive sequence analysis package for the Macintosh. Mol Biotechnol. 1995;3:37–45. doi: 10.1007/BF02821333. [DOI] [PubMed] [Google Scholar]
- Dykhuizen D. Species Numbers in Bacteria. Proc Calif Acad Sci. 2005;56:62–71. [PMC free article] [PubMed] [Google Scholar]
- Dykhuizen DE. Santa Rosalia revisited: why are there so many species of bacteria? Antonie Van Leeuwenhoek. 1998;73:25–33. doi: 10.1023/a:1000665216662. [DOI] [PubMed] [Google Scholar]
- Falgenhauer L, Yao Y, Fritzenwanker M, Schmiedel J, Imirzalioglu C, Chakraborty T. Complete genome sequence of phage-like plasmid pECOH89, encoding CTX-M-15. Genome Announc. 2014;2:e00356–14. doi: 10.1128/genomeA.00356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas ME, Aanei IL, Behrens CR, Tong GJ, Murphy ST, O’Neil JP, Francis MB. PET Imaging and biodistribution of chemically modified bacteriophage MS2. Mol Pharm. 2013;10:69–76. doi: 10.1021/mp3003754. [DOI] [PubMed] [Google Scholar]
- Farr R, Choi DS, Lee SW. Phage-based nanomaterials for biomedical applications. Acta Biomater. 2014;10:1641–1750. doi: 10.1016/j.actbio.2013.06.037. [DOI] [PubMed] [Google Scholar]
- Federhen F. The NCBI Taxonomy database. Nucleic Acids Res. 2012;40:D136–143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix A, Callow BR. Typing of paratyphoid B bacilli by Vi bacteriophage. Br Med J. 1943;2:127–30. doi: 10.1136/bmj.2.4308.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filee J, Tetart F, Suttle CA, Krisch HM. Marine T4-type bacteriophages, a ubiquitous component of the dark matter of the biosphere. Proc Natl Acad Sci U S A. 2005;102:12471–12476. doi: 10.1073/pnas.0503404102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filee J, Bapteste E, Susko E, Krisch HM. A selective barrier to horizontal gene transfer in the T4-type bacteriophages that has preserved a core genome with the viral replication and structural genes. Mol Biol Evol. 2006;23:1688–96. doi: 10.1093/molbev/msl036. [DOI] [PubMed] [Google Scholar]
- Fortier LC, Sekulovic O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence. 2013;4:354–65. doi: 10.4161/viru.24498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Mohan R, Gupta RS. Phylogenomics and protein signatures elucidating the evolutionary relationships among the Gammaproteobacteria. Int J Syst Evol Microbiol. 2009;59:234–47. doi: 10.1099/ijs.0.002741-0. [DOI] [PubMed] [Google Scholar]
- George DG, Yeh LS, Barker WC. Unexpected relationships between bacteriophage lambda hypothetical proteins and bacteriophage T4 tail-fiber proteins. Biochem Biophys Res Commun. 1983;115:1061–8. doi: 10.1016/s0006-291x(83)80043-6. [DOI] [PubMed] [Google Scholar]
- Grose JH, Belnap DM, Jensen JD, Mathis AD, Prince JT, Merrill BD, Burnett SH, Breakwell DP. The genomes, proteomes and structure of three novel phages that infect the Bacillus cereus group and carry putative virulence factors. J Virol. 2014;88 doi: 10.1128/JVI.01364-14. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggard-Ljungquist E, Halling C, Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol. 1992;174:1462–77. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hambly E, Suttle CA. The viriosphere, diversity, and genetic exchange within phage communities. Curr Opin Microbiol. 2005;8:444–50. doi: 10.1016/j.mib.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Hambly E, Tetart F, Desplats C, Wilson WH, Krisch HM, Mann NH. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc Natl Acad Sci U S A. 2001;98:11411–6. doi: 10.1073/pnas.191174498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF. Mycobacteriophages: windows into tuberculosis. PLoS Pathog. 2014;10:e1003953. doi: 10.1371/journal.ppat.1003953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Jacobs-Sera D, Lawrence JG, Pope WH, Russell DA, Ko CC, Weber RJ, Patel MC, Germane KL, Edgar RH, Hoyte NN, Bowman CA, Tantoco AT, Paladin EC, Myers MS, Smith AL, Grace MS, Pham TT, O’Brien MB, Vogelsberger AM, Hryckowian AJ, Wynalek JL, Donis-Keller H, Bogel MW, Peebles CL, Cresawn SG, Hendrix RW. Comparative genomic analysis of 60 Mycobacteriophage genomes: genome clustering, gene acquisition, and gene size. J Mol Biol. 2010;397:119–43. doi: 10.1016/j.jmb.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF, Pedulla ML, Jacobs-Sera D, Cichon PM, Foley A, Ford ME, Gonda RM, Houtz JM, Hryckowian AJ, Kelchner VA, Namburi S, Pajcini KV, Popovich MG, Schleicher DT, Simanek BZ, Smith AL, Zdanowicz GM, Kumar V, Peebles CL, Jacobs WR, Jr, Lawrence JG, Hendrix RW. Exploring the mycobacteriophage metaproteome: phage genomics as an educational platform. PLoS Genet. 2006;2:e92. doi: 10.1371/journal.pgen.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull, G. F., Science Education Alliance Phage Hunters Advancing, G., Evolutionary Science, P., KwaZulu-Natal Research Institute for, T., Course, H. I. V. M. G., University of California-Los Angeles Research Immersion Laboratory in, V., Phage Hunters Integrating, R., and Education, P. Complete genome sequences of 63 mycobacteriophages. Genome Announc. 2013;1 doi: 10.1128/genomeA.00847-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser R, Blasche S, Dokland T, Haggard-Ljungquist E, von Brunn A, Salas M, Casjens S, Molineux I, Uetz P. Bacteriophage protein-protein interactions. Adv Virus Res. 2012;83:219–98. doi: 10.1016/B978-0-12-394438-2.00006-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix R, Casjens S. Bacteriophage λ and its genetic neighborhood. In: Calendar R, editor. The Bacteriophages. 2. Oxford Press; New York City, N.Y: 2006. pp. 409–447. [Google Scholar]
- Hendrix RW. Bacteriophages: evolution of the majority. Theor Popul Biol. 2002;61:471–80. doi: 10.1006/tpbi.2002.1590. [DOI] [PubMed] [Google Scholar]
- Hendrix RW. Bacteriophage genomics. Curr Opin Microbiol. 2003;6:506–11. doi: 10.1016/j.mib.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Hershey AD, Chase M. Independent functions of viral protein and nucleic acid in growth of bacteriophage. J Gen Physiol. 1952;36:39–56. doi: 10.1085/jgp.36.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Fan H, Pei G, Fan H, Zhang Z, An X, Mi Z, Shi T, Tong Y. Complete genome sequence of IME15, the first T7-like bacteriophage lytic to pan-antibiotic-resistant Stenotrophomonas maltophilia. J Virol. 2012;86:13839–40. doi: 10.1128/JVI.02661-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman P. Bacteriophages and nanostructured materials. Adv Appl Microbiol. 2012;78:55–73. doi: 10.1016/B978-0-12-394805-2.00003-8. [DOI] [PubMed] [Google Scholar]
- Jacobs-Sera D, Marinelli LJ, Bowman C, Broussard GW, Guerrero Bustamante C, Boyle MM, Petrova ZO, Dedrick RM, Pope WH, Modlin RL, Hendrix RW, Hatfull GF Science Education Alliance Phage Hunters Advancing Genomics, Evolutionary Science Sea-Phages Program . On the nature of mycobacteriophage diversity and host preference. Virology. 2012;434:187–201. doi: 10.1016/j.virol.2012.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen EC, Schrader HS, Rieland B, Thompson TL, Lee KW, Nickerson KW, Kokjohn TA. Prevalence of broad-host-range lytic bacteriophages of Sphaerotilus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl Environ Microbiol. 1998;64:575–80. doi: 10.1128/aem.64.2.575-580.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhala RJ, Ford ME, Duda RL, Youlton A, Hatfull GF, Hendrix RW. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J Mol Biol. 2000;299:27–51. doi: 10.1006/jmbi.2000.3729. [DOI] [PubMed] [Google Scholar]
- Kale A, Bao Y, Zhou Z, Prevelige PE, Gupta A. Directed self-assembly of CdS quantum dots on bacteriophage P22 coat protein templates. Nanotechnology. 2013;24:045603. doi: 10.1088/0957-4484/24/4/045603. [DOI] [PubMed] [Google Scholar]
- Kim M, Kim S, Ryu S. Complete genome sequence of bacteriophage SSU5 specific for Salmonella enterica serovar Typhimurium rough strains. J Virol. 2012;86:10894. doi: 10.1128/JVI.01796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, Adams M, Carstens E, Lefkowitz E, editors. In Virus Taxonomy: classification and nomenclature of viruses: IXth report of the International Committee on Taxonomy of Viruses. San Diego. CA: Elsevier Academic Press; 2012. [Google Scholar]
- Kropinski AM. Sequence of the genome of the temperate, serotype-converting, Pseudomonas aeruginosa bacteriophage D3. J Bacteriol. 2000;182:6066–74. doi: 10.1128/jb.182.21.6066-6074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski AM, Kovalyova IV, Billington SJ, Patrick AN, Butts BD, Guichard JA, Pitcher TJ, Guthrie CC, Sydlaske AD, Barnhill LM, Havens KA, Day KR, Falk DR, McConnell MR. The genome of epsilon15, a serotype-converting, Group E1 Salmonella enterica-specific bacteriophage. Virology. 2007;369:234–44. doi: 10.1016/j.virol.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumsiek J, Arnold R, Rattei T. Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–8. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- Kuhn J, Suissa M, Chiswell D, Azriel A, Berman B, Shahar D, Reznick S, Sharf R, Wyse J, Bar-On T, Cohen I, Giles R, Weiser I, Lubinsky-Mink S, Ulitzur S. A bacteriophage reagent for Salmonella: molecular studies on Felix O1. Int J Food Microbiol. 2002;74:217–27. doi: 10.1016/s0168-1605(01)00682-1. [DOI] [PubMed] [Google Scholar]
- Kuhn JC. Detection of Salmonella by bacteriophage Felix O1. Methods Mol Biol. 2007;394:21–37. doi: 10.1007/978-1-59745-512-1_2. [DOI] [PubMed] [Google Scholar]
- Kuhnert P, Korczak BM, Stephan R, Joosten H, Iversen C. Phylogeny and prediction of genetic similarity of Cronobacter and related taxa by multilocus sequence analysis (MLSA) Int J Food Microbiol. 2009;136:152–8. doi: 10.1016/j.ijfoodmicro.2009.02.022. [DOI] [PubMed] [Google Scholar]
- Kuner JM, Kaiser D. Introduction of transposon Tn5 into Myxococcus for analysis of developmental and other nonselectable mutants. Proc Natl Acad Sci U S A. 1981;78:425–9. doi: 10.1073/pnas.78.1.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Sonnhammer EL. Kalign--an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavigne R, Darius P, Summer EJ, Seto D, Mahadevan P, Nilsson AS, Ackermann HW, Kropinski AM. Classification of Myoviridae bacteriophages using protein sequence similarity. BMC Microbiol. 2009;9:224. doi: 10.1186/1471-2180-9-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Hatfull GF, Hendrix RW. Imbroglios of viral taxonomy: genetic exchange and failings of phenetic approaches. J Bacteriol. 2002;184:4891–905. doi: 10.1128/JB.184.17.4891-4905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg E. Lysogenicity in E. coli K-12. Genetics. 1951;36:560. [Google Scholar]
- Lee TJ, Schwartz C, Guo P. Construction of bacteriophage ø29 DNA packaging motor and its applications in nanotechnology and therapy. Ann Biomed Eng. 2009;37:2064–81. doi: 10.1007/s10439-009-9723-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YD, Kim JY, Park JH, Chang H. Genomic analysis of bacteriophage ESP2949-1, which is virulent for Cronobacter sakazakii. Arch Virol. 2012;157:199–202. doi: 10.1007/s00705-011-1147-0. [DOI] [PubMed] [Google Scholar]
- Lee YD, Park JH, Chang HI. Genomic sequence analysis of virulent Cronobacter sakazakii bacteriophage ES2. Arch Virol. 2011;156:2105–8. doi: 10.1007/s00705-011-1096-7. [DOI] [PubMed] [Google Scholar]
- Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevan P, King JF, Seto D. Data mining pathogen genomes using GeneOrder and CoreGenes and CGUG: gene order, synteny and in silico proteomes. Int J Comput Biol Drug Des. 2009;2:100–14. doi: 10.1504/ijcbdd.2009.027586. [DOI] [PubMed] [Google Scholar]
- McConnell MR, Schoelz JE. Evidence for shorter average O-polysaccharide chainlength in the lipopolysaccharide of a bacteriophage Felix 01-sensitive variant of Salmonella anatum A1. J Gen Microbiol. 1983;129:3177–84. doi: 10.1099/00221287-129-10-3177. [DOI] [PubMed] [Google Scholar]
- Modi SR, Lee HH, Spina CS, Collins JJ. Antibiotic treatment expands the resistance reservoir and ecological network of the phage metagenome. Nature. 2013;499:219–22. doi: 10.1038/nature12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molineux I. The T7 group. In: Calendar R, editor. The Bacteriophages. 2. Oxford University Press; Oxford: 2006. pp. 277–301. [Google Scholar]
- Moran NA, Russell JA, Koga R, Fukatsu T. Evolutionary relationships of three new species of Enterobacteriaceae living as symbionts of aphids and other insects. Appl Environ Microbiol. 2005;71:3302–10. doi: 10.1128/AEM.71.6.3302-3310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam KT, Kim DW, Yoo PJ, Chiang CY, Meethong N, Hammond PT, Chiang YM, Belcher AM. Virus-enabled synthesis and assembly of nanowires for lithium ion battery electrodes. Science. 2006;312:885–8. doi: 10.1126/science.1122716. [DOI] [PubMed] [Google Scholar]
- Ohmori H, Haynes LL, Rothman-Denes LB. Structure of the ends of the coliphage N4 genome. J Mol Biol. 1988;202:1–10. doi: 10.1016/0022-2836(88)90512-8. [DOI] [PubMed] [Google Scholar]
- Oliver KM, Degnan PH, Hunter MS, Moran NA. Bacteriophages encode factors required for protection in a symbiotic mutualism. Science. 2009;325:992–4. doi: 10.1126/science.1174463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent K, Tang J, Cardonne G, Gilcrease E, Janssen M, Olson N, Casjens S, Baker T. Three-dimensional reconstructions of the bacteriophage CUS-3 virion reveal a conserved coat protein I-domain but a distinct tailspike receptor-binding domain. Virology. 2014 doi: 10.1016/j.virol.2014.06.017. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent KN, Gilcrease EB, Casjens SR, Baker TS. Structural evolution of the P22-like phages: comparison of Sf6 and P22 procapsid and virion architectures. Virology. 2012;427:177–88. doi: 10.1016/j.virol.2012.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov VM, Ratnayaka S, Nolan JM, Miller ES, Karam JD. Genomes of the T4-related bacteriophages as windows on microbial genome evolution. Virol J. 2010;7:292. doi: 10.1186/1743-422X-7-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard D, Thomson NR, Baker S, Wain J, Pardo M, Goulding D, Hamlin N, Choudhary J, Threfall J, Dougan G. Molecular characterization of the Salmonella enterica serovar Typhi Vi-typing bacteriophage E1. J Bacteriol. 2008;190:2580–7. doi: 10.1128/JB.01654-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickard D, Toribio AL, Petty NK, van Tonder A, Yu L, Goulding D, Barrell B, Rance R, Harris D, Wetter M, Wain J, Choudhary J, Thomson N, Dougan G. A conserved acetyl esterase domain targets diverse bacteriophages to the Vi capsular receptor of Salmonella enterica serovar Typhi. J Bacteriol. 2010;192:5746–54. doi: 10.1128/JB.00659-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pupo GM, Lan R, Reeves PR. Multiple independent origins of Shigella clones of Escherichia coli and convergent evolution of many of their characteristics. Proc Natl Acad Sci USA. 2000;97:10567–72. doi: 10.1073/pnas.180094797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi S, Liepold LO, Abedin MJ, Johnson B, Prevelige P, Frank JA, Douglas T. P22 viral capsids as nanocomposite high-relaxivity MRI contrast agents. Mol Pharm. 2013;10:11–7. doi: 10.1021/mp300208g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravin V, Ravin N, Casjens S, Ford ME, Hatfull GF, Hendrix RW. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J Mol Biol. 2000;299:53–73. doi: 10.1006/jmbi.2000.3731. [DOI] [PubMed] [Google Scholar]
- Rizzo AA, Suhanovsky MM, Baker ML, Fraser LCR, Jones LM, Rempel DL, Gross ML, Chiu W, Alexandrescu AT, Teschke CM. Multiple functional roles of the accessory I-domain of bacteriophage P22 coat protein revealed by NMR structure and cryoEM modeling. Structure. 2014;22:1–12. doi: 10.1016/j.str.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier H, Iida S, Arber W. DNA inversion regions Min of plasmid p15B and Cin of bacteriophage P1: evolution of bacteriophage tail fiber genes. J Bacteriol. 1992;174:3936–44. doi: 10.1128/jb.174.12.3936-3944.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Coulson AR, Hong GF, Hill DF, Petersen GB. Nucleotide sequence of bacteriophage lambda DNA. J Mol Biol. 1982;162:729–73. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- Scholl D, Adhya S, Merril CR. Bacteriophage SP6 is closely related to phages K1-5, K5, and K1E but encodes a tail protein very similar to that of the distantly related P22. J Bacteriol. 2002;184:2833–6. doi: 10.1128/JB.184.10.2833-2836.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholl D, Merril C. The genome of bacteriophage K1F, a T7-like phage that has acquired the ability to replicate on K1 strains of Escherichia coli. J Bacteriol. 2005;187:8499–503. doi: 10.1128/JB.187.24.8499-8503.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Bao N, Prevelige PE, Gupta A. Fabrication of ordered nanostructures of sulfide nanocrystal assemblies over self-assembled genetically engineered P22 coat protein. J Am Chem Soc. 2010;132:17354–7. doi: 10.1021/ja107080b. [DOI] [PubMed] [Google Scholar]
- Shen PS, Domek MJ, Sanz-Garcia E, Makaju A, Taylor RM, Hoggan R, Culumber MD, Oberg CJ, Breakwell DP, Prince JT, Belnap DM. Sequence and structural characterization of Great Salt Lake bacteriophage CW02, a member of the T7-like supergroup. J Virol. 2012;86:7907–17. doi: 10.1128/JVI.00407-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M, Davis RW, Davidson N. Heteroduplexs of DNA moleculaes of lambdoid pahges: physical mapping of their base sequence relationships by electron microscopy. In: Hershey AD, editor. The bacteriophage lambda. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1971. pp. 313–328. [Google Scholar]
- Smith KC, Castro-Nallar E, Fisher JN, Breakwell DP, Grose JH, Burnett SH. Phage cluster relationships identified through single gene analysis. BMC Genomics. 2013;14:410. doi: 10.1186/1471-2164-14-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267:865–80. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stummeyer K, Schwarzer D, Claus H, Vogel U, Gerardy-Schahn R, Muhlenhoff M. Evolution of bacteriophages infecting encapsulated bacteria: lessons from Escherichia coli K1-specific phages. Mol Microbiol. 2006;60:1123–35. doi: 10.1111/j.1365-2958.2006.05173.x. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Viruses in the sea. Nature. 2005;437:356–61. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- Suttle CA. Marine viruses--major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- Volkova VV, Lu Z, Besser T, Grohn YT. Modeling infection dynamics of bacteriophages in enteric Escherichia coli: estimating the contribution of transduction to antimicrobial gene spread. Appl Environ Microbiol. 2014 doi: 10.1128/AEM.00446-14. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Fiedler C, Grassl R, Biebl M, Rachel R, Hermo-Parrado XL, Llamas-Saiz AL, Seckler R, Miller S, van Raaij MJ. Structure of the receptor-binding protein of bacteriophage Det7: a podoviral tail spike in a myovirus. J Virol. 2008;82:2265–73. doi: 10.1128/JVI.01641-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland BC, Szybalski W, Ris H. Mapping of deletions and substitutions in heteroduplex DNA molecules of bacteriophage lambda by electron microscopy. Science. 1969;163:1343–8. doi: 10.1126/science.163.3873.1343. [DOI] [PubMed] [Google Scholar]
- Wilhelm SW, Jeffrey WH, Suttle CA, Mitchell DL. Estimation of biologically damaging UV levels in marine surface waters with DNA and viral dosimeters. Photochem Photobiol. 2002;76:268–73. doi: 10.1562/0031-8655(2002)076<0268:eobdul>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Wittmann J, Dreiseikelmann B, Rohde M, Meier-Kolthoff JP, Bunk B, Rohde C. First genome sequences of Achromobacter phages reveal new members of the N4 family. Virol J. 2014;11:14. doi: 10.1186/1743-422X-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–83. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafar N, Mazumder R, Seto D. CoreGenes: a computational tool for identifying and cataloging “core” genes in a set of small genomes. BMC Bioinformatics. 2002;3:12. doi: 10.1186/1471-2105-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Guo H, Jin L, Czornyj E, Hodes A, Hui WH, Nieh AW, Miller JF, Zhou ZH. A new topology of the HK97-like fold revealed in Bordetella bacteriophage by cryoEM at 3.5 A resolution. Elife. 2013;2:e01299. doi: 10.7554/eLife.01299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.