Summary
Recently, we demonstrated that kinesin-1 can slide microtubules against each other providing the mechanical force required for initial neurite extension in Drosophila neurons. This sliding is only observed in young neurons actively forming neurites and is dramatically downregulated in older neurons. The downregulation is not caused by the global shut-down of kinesin-1, as the ability of kinesin-1 to transport membrane organelles is not diminished in mature neurons, suggesting that microtubule sliding is regulated by a dedicated mechanism [1]. Here, we have identified the “mitotic” kinesin Pavarotti (Pav-KLP) as an inhibitor of kinesin-1-driven microtubule sliding. Depletion of Pav-KLP in neurons strongly stimulated the sliding of long microtubules and neurite outgrowth, while its ectopic overexpression in the cytoplasm blocked both of these processes. Furthermore, postmitotic depletion of Pav-KLP in Drosophila neurons in vivo reduced embryonic/larval viability, with only a few animals surviving to the third instar larval stage. A detailed examination of motor neurons in the surviving larvae revealed the overextension of axons and mistargeting of neuromuscular junctions, resulting in uncoordinated locomotion. Taken together, our results identify a new role for Pav-KLP as a negative regulator of kinesin-1 driven neurite formation. These data suggest an important parallel between long microtubule-microtubule sliding in anaphase B and sliding of interphase microtubules during neurite formation.
Results
Pav-KLP negatively regulates microtubule sliding and process formation in Drosophila S2 cells
We performed a targeted RNAi screen in Drosophila S2 cells to determine whether microtubule motors besides kinesin-1 also participate in microtubule sliding. We previously demonstrated that microtubule-filled processes formed in S2 cells after actin depolymerization (Figure S1A) are generated by kinesin-1-dependent microtubule-microtubule sliding [2]. The length of these processes may reflect the extent of microtubule sliding. Thus, we initially screened for motors that affect the length of processes in interphase S2 cells treated with Cytochalasin D (CytoD). To our surprise, we found that depletion of the mitotic kinesin Pav-KLP, a Drosophila member of the kinesin-6 family, significantly increased the total length of processes (Figures 1A and 1C). This effect was independent of the increase in polyploidy caused by Pav-KLP knockdown (Figures S1B and S1C). To directly test whether this change in process length was caused by the effect of Pav-KLP on kinesin-1-dependent microtubule sliding, we directly measured sliding using the photoconvertible probe, Drosophila α-tubulin84B tagged with a tandem dimer of protein EOS2 (tdEOS-αtub)[3]. We restricted photoconversion of microtubules to a small area of the cytoplasm by placing a slit in the epifluorescent light path. This produced fiduciary marks on microtubules that allowed us to track and quantify their movement (see Supplemental Experimental Procedures for details) (Figure 1B). To verify the role of kinesin-1 in microtubule sliding, we depleted the endogenous motor with dsRNA treatment against kinesin heavy chain (KHC) 3′UTR. Knockdown of kinesin-1 significantly inhibited microtubule sliding (Figure 1D) [2]. The inhibition could be rescued by expressing full length KHC but not by KHC(MBD)*, a variant with mutations in the C-terminal microtubule binding domain (MBD) (Figure 1D) [4, 5]. Next, we characterized the function of Pav-KLP in the regulation of microtubule sliding. We found that Pav-KLP depletion dramatically stimulated microtubule sliding (Figures 1B and 1D; Movie S1). Process formation and microtubule sliding stimulated by Pav-KLP-depletion were dependent on kinesin-1 as shown by dual knockdown (Figures 1C and 1D, respectively), demonstrating that Pav-KLP is a negative regulator of kinesin-1-powered microtubule sliding and process formation.
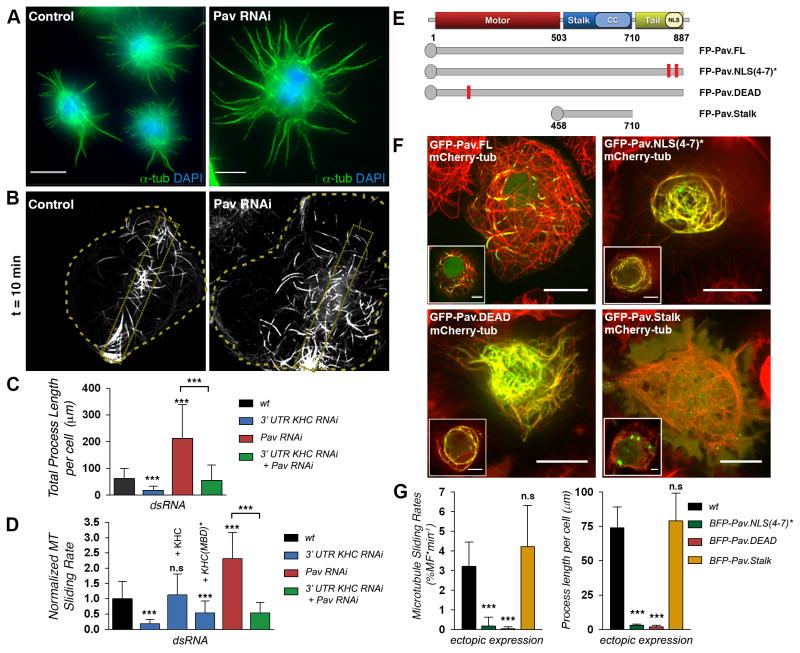
Figure 1. Pav-KLP controls kinesin-1 dependent microtubule sliding and process formation in Drosophila S2 cells.
(A) Pav-KLP knockdown increased the length of processes in S2 cells treated with CytoD. S2 cells were stained for microtubules (green) and nuclei (blue). See also Figure S1. Scale bars, 10 μm.
(B) Pav-KLP depletion increased microtubule sliding. Frames from time-lapse sequences of photoconverted S2 cells stably expressing tdEOS-αtub untreated (control) or treated with Pav-KLP dsRNA. Doted boxes inside the cell body represent the original photoconverted area. Scale bars, 10 μm. See Movie S1.
(C) Pav-KLP blocked process formation in a kinesin 1-dependent pathway. Quantification of process length of S2 cells untreated (wt) or treated with 3′UTR KHC, Pav-KLP, or KHC and Pav-KLP dsRNA (n>100 cells per condition; error bar, SD). *** p < 0.0005, unpaired t test.
(D) Pav-KLP blocked microtubule sliding in a kinesin 1-dependent pathway. Quantification of microtubule sliding from cells treated as in (C). In KHC 3′UTR knockdown cells, microtubule sliding rates could only be rescued by ectopic expression of KHC-BFP but not with KHC(MBD)*-BFP (n>15 cells; error bar, SD). *** p < 0.0005, unpaired t test.
(E) Schematic cartoon of endogenous Pav-KLP and GFP- or BFP- tagged Pavarotti constructs used in this work. The red rectangles represent areas where the mutations were introduced.
(F) Ectopic expression pattern of Pav-KLP variants. S2 cells stably expressing mCherry-tubulin were transfected with GFP-Pav.FL or variants. Insets show the apical side of the same cells to highlight nuclear localization. Scale bars, 10 μm and 5 μm for main and inset images, respectively. Note that interaction of Pav-KLP with cytoplasmic microtubules blocks microtubule motility. See corresponding Movie S2.
(G) Ectopic expression of Pav-KLP variants blocked microtubule sliding and process formation. Quantification of microtubule sliding (left panel) and process length (right panel) in cells ectopically expressing BFP-tagged Pav-KLP variants (n>10 or >20 cells for sliding or process length quantification, respectively; error bars, SD). *** p < 0.0001, unpaired t test.
Pav-KLP together with Tumbleweed (RacGAP50C) forms the Centralspindlin complex [6], which both stabilizes the mitotic spindle by bundling antiparallel microtubules and recruits regulators of abscission like Pebble (Rho-GEF) [7]. Depletion of either Tumbleweed or Pebble promoted microtubule sliding and process formation (Figures S1D–G). However, depletion of small Rho GTPases, well-characterized downstream effectors of Centralspindlin in cytokinesis, had no or little effect on microtubule sliding and process formation in S2 cells (Figures S1D–F). Western blot analysis showed that knockdown of any of these three proteins (Pav-KLP, Tumbleweed, or Pebble) resulted in significant depletion of Pav-KLP (Figures S1H–I). Alltogether these data demonstrate that Pav-KLP/Centralspindlin regulate sliding of cytoplasmic microtubules independently of its effects on small GTPases.
Ectopic expression of Pav-KLP blocks microtubule sliding
To corroborate the Pav-KLP knockdown data, we overexpressed GFP-tagged Pav-KLP (GFP-Pav.FL) in S2 cells (Figure 1E). Time-lapse imaging of S2 cells expressing mCherry-Tubulin and GFP-Pav.FL showed that movement of interphase microtubules decorated with GFP-Pav.FL was completely blocked (Movie S2). To quantify the effect of Pav-KLP on microtubule sliding, we tagged Pav-KLP with the blue fluorescence protein mTagBFP2 [8] (creating BFP-Pav.FL) and coexpressed it with tdEOS-αtub. Figure 1F shows that BFP-Pav.FL bound to cytoplasmic microtubules in a cooperative fashion; some microtubules were heavily decorated, while others did not contain the label. Photoconversion experiments demonstrated that BFP-Pav.FL-decoration of microtubules blocked sliding while Pav.FL-free-microtubules in the same cell remained motile (Figures S1K–N and Movie S2). In addition, the expression of BFP-Pav.FL rescued sliding and process formation in Pav-KLP RNAi background (Figures S1O–P). To better characterize the inhibition of sliding by Pav-KLP, we increased the concentration of cytoplasmic Pav-KLP by generating Pav.NLS(4–7)*, a variant with an inactive nuclear localization sequences (NLS) (Figure 1E, 1F, and Movie S2) [9]. Expression of BFP-Pav.NLS(4–7)* suppressed microtubule sliding and process formation (Figure 1G). This was also observed in cells expressing a variant of Pav-KLP with a mutation that abolishes the ATPase activity of the motor domain (Pav.DEAD) (Figures 1E–G, and Movie S2) [9]. However, ectopic expression of the Pav.Stalk, a deletion mutant that cannot bind microtubules [9], did not affect microtubule sliding or process formation (Figures 1E–G, and Movie S2). Thus, Pav-KLP knockdown and ectopic expression data strongly suggest that Pav-KLP is the primary regulator of kinesin-1-driven microtubule sliding, and enhancement of microtubule sliding and process formation found in S2 cells after Tumbleweed or Pebble depletion can likely be explained by the simultaneous depletion of Pav-KLP. Analysis of peroxisome transport (Fig. S1J) shows that depletion of Pav-KLP had no effect on organelle transport by kinesin-1, and therefore, Pav-KLP specifically regulates microtubule sliding without affecting the conventional cargo-transporting function of kinesin-1.
Pav-KLP regulates microtubule sliding in Drosophila neurons
We have recently demonstrated that microtubule sliding by kinesin-1 provides the driving force required for neurite outgrowth (Figure S2A and Movie S3) [1] in cultured neurons obtained from dissociated Drosophila embryos. To test whether microtubules are transported as long polymers in neurons as we observed in S2 cells, we cultured neurons expressing tdEOS-αtub under the maternal αtub-Gal4 promoter (mat αtub>tdEOS-αtub). Time-lapse imaging of photoconverted microtubules in neurites clearly showed transport of long microtubules in neurons (Figure S2B and Movie S3). In order to confirm that kinesin-1-driven microtubule sliding is required for neurite outgrowth we cultured overnight primary neurons obtained from Khc27/Khc27 (a KHC protein null allele) embryos and quantified axon length. The maternal load of KHC mRNA in KHC null embryos was depleted by microinjection of dsRNA against KHC 3′UTR into embryos at the blastoderm stage before cellularization [1]. The axons in KHC-null neurons were significantly shorter than in control. The wild type phenotype was fully rescued by co-injecting the cDNA encoding KHC but not by KHC(MBD)* with KHC 3′UTR dsRNA (Figure S2C), further demonstrating that microtubule sliding is required for axon extension.
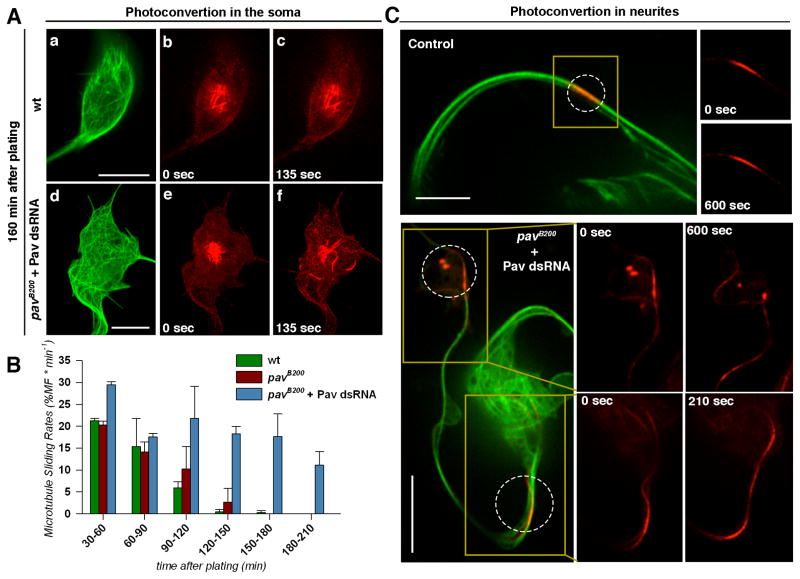
We hypothesized that in neurons, like in S2 cells, Pav-KLP negatively regulates microtubule sliding, thereby controlling neurite outgrowth. To test this hypothesis, we photoconverted microtubules in the soma or neurites of control and Pav-KLP-depleted Drosophila neurons. We depleted Pav-KLP by combining the strongest loss-of-function pav allele, pavB200 with injection of Pav-KLP dsRNA (see details in Supplemental Experimental Procedures). In control neurons, we observed that microtubules were actively sliding during the first hour (Figure 2B and Movie S3), but their sliding became undetectable 120–150 min after plating (Figures 2A and 2B; Movie S4). These data are consistent with the kinetics of neurite outgrowth where the most active axon extension takes place in the first three hours after plating, as we previously described [1]. We found that Pav-KLP depletion by the combination of pavB200 mutation and Pav-KLP dsRNA injection dramatically delayed the downregulation of microtubule sliding (Figures 2A–C; Movie S4). Thus, in neurons, like in cultured S2 cells, Pav-KLP is a negative regulator of microtubule sliding.
Figure 2. Pav-KLP regulates microtubule sliding both in the soma and neurites of cultured Drosophila neurons.
(A) Time-lapse images of cultured neurons expressing photoconvertible tdEOS-αtub show that Pav-KLP depletion stimulated microtubule sliding in the soma. (a–c) a control neuron and (d–f) a pav-RNAi neuron. The first column shows the green channel before photoconversion, and the second and third show the red channel after photoconversion. Scales bars, 5 μm. See corresponding Movie S3.
(B) Quantification of microtubule sliding in cultured neurons of different genotypes as a function of time after plating. Quantification averaged from two independent experiments (n>9 neurons per category; error bars, SD).
(C) Time-lapse images of cultured neurons expressing photoconvertible tdEOS-αtub shows that Pav-KLP depletion stimulated microtubule sliding in the neurites. Note that in neurons plated after 160min, microtubule sliding was not detectable in control neurons, however microtubules slid actively in Pav-KLP RNAi neurons. Scales bars, 10 μm. See corresponding Movie S3.
Pav-KLP regulates initial neurite outgrowth in cultured Drosophila neurons
Next, we explored the role of Pav-KLP in neurite outgrowth. We hypothesized that Pav-KLP knockdown in neurons, which prolonged microtubule sliding, would generate longer neurites. To test this prediction, we cultured neurons for 16 hours and quantified the length of their axons. Measurement of axon length revealed that pavB200 homozygous neurons generated significantly longer neurites than controls, (Figures 3A and 3B). Further reduction of Pav-KLP levels by dsRNA microinjection into pavB200 homozygous embryos generated even longer axons (Figures 3A and 3B). These results are in good agreement with the effects of Pav-KLP depletion on microtubule sliding (Figure 2).
Figure 3. Pav-KLP controls axon outgrowth in Drosophila neurons.
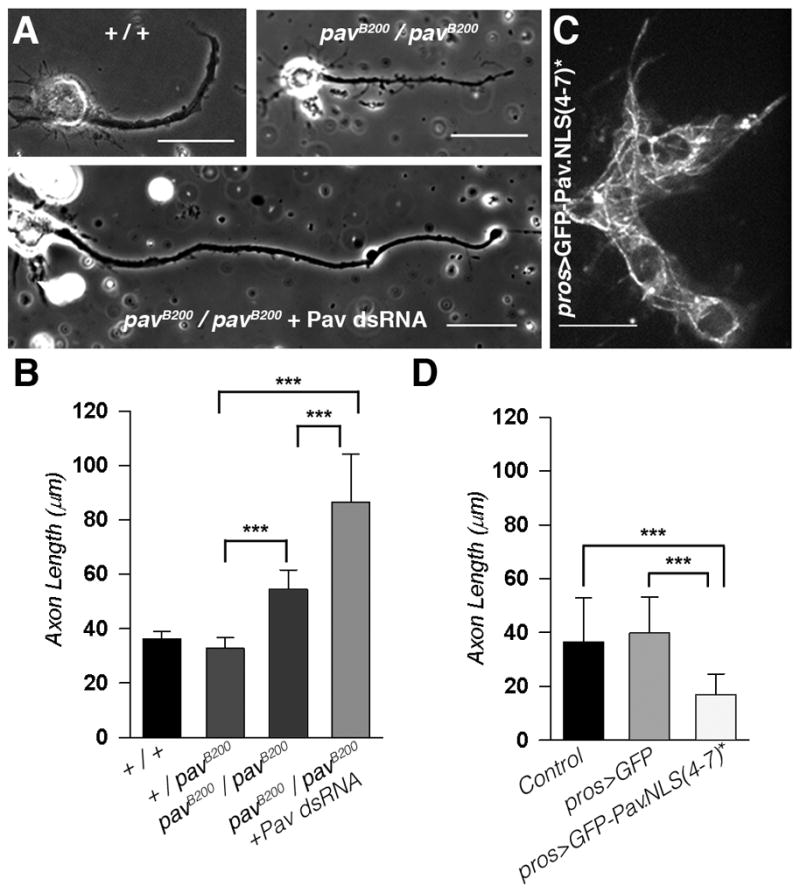
(A) Pav-KLP depletion induced formation of longer axons. Representative phase contrast images of neurons (after 16h in culture with 5 μM CytoD) from three different genotypes (+/+, control; pavB200/pavB200; and pavB200/pavB200 +Pav-KLP dsRNA neurons). Scale bars, 20 μm.
(B) Pav-KLP knockdown induced the formation of longer axons. Axon length measurements of neurons from the three genotypes (+/+;pavB200/+; pavB200/pavB200; and pavB200/pavB200 + Pav-KLP dsRNA neurons). The mean and SD are shown from two independent experiments (n>29 neurons per category; ***p < 0.0001, unpaired t test).
(C) Mutations in the NLS of Pav-KLP induced its accumulation on cytoplasmic microtubules. Confocal image of neurons expressing GFP-Pav.NLS(4–7)* under control of pros-Gal4 after 16h in culture with 5 μM CytoD. Scale bar, 10 μm.
(D) Accumulation of Pav-KLP in the cytoplasm blocked neurite outgrowth. Measurement of axon length of neurons ectopically expressing pros>GFP or pros>GFP-Pav.NLS(4–7)*. (n>30 neurons per category; error bars, SD ***p < 0.0001, unpaired t test.
To further confirm that Pav-KLP regulates neurite outgrowth, we ectopically expressed GFP-Pav.NLS(4–7)* in neurons under a neuroblast-specific promoter, pros-Gal4. GFP-Pav.NLS(4–7)* strongly decorated microtubules in neurons (Figure 3C). Neurites of these neurons were dramatically shorter than those in controls (Figure 3D), consistent with a role of Pav-KLP in inhibiting neurite outgrowth. Thus, the data obtained in cultured embryonic neurons demonstrate that Pav-KLP is a negative regulator of interphase microtubule sliding and neurite outgrowth in Drosophila neurons.
Pav-KLP regulates axon length in vivo
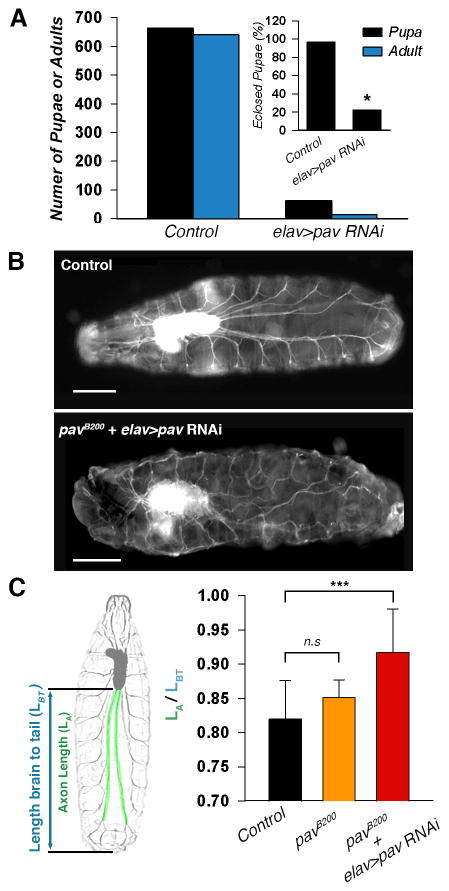
Next, we tested whether Pav-KLP has a role in Drosophila neurodevelopment in vivo. A number of Pav-KLP mutations have been described in the literature that have obvious effects on neurodevelopment [10, 11], but these results are difficult to interpret as they could be explained by problems with cytokinesis rather than effects of Pav-KLP on microtubule sliding. To avoid the effects of Pav-KLP depletion on cell division, we drove pav-RNAi (targeting pav coding sequence 907–927) by the pan-neuronal driver elav-Gal4. elav-Gal4 is expressed in all neurons post-mitotically starting from stage 11 [12], therefore elav>pav-RNAi would not affect cell division. We noticed that the survival rate of elav>pav-RNAi pupae was much lower (20%) than in control (95%) (Figure 4A), suggesting that Pav-KLP has an important role in neurodevelopment in vivo.
Figure 4. Depletion of Pav-KLP in postmitotic neurons results in overextension of axons outgrowth in Drosophila larvae.
(A) Postmitotic depletion of Pav-KLP in neurons reduced Drosophila viability. *Note that elav>pav RNAi adults died at the few hours after eclosing.
(B) Ventral view of early second instar larvae expressing GFP-Nvr2. Representative wide-field images of second instar larvae; control (upper panel) or elav>pav RNAi from pavB200/+ mothers (bottom panel). Scale bars, 200 μm
(C) Quantification of the motor neuron axon length in Drosophila early second instar larvae. Left panel schemically represents the strategy used to quantify axon length in the larvae. The length of the longest motor neuron axons of second instar larvae (in green) were normalized by the distance between the ventral ganglion and the posterior end (in blue). Right panel shows the axon length quantification of control, pavB200, and elav>pav RNAi with maternal pavB200/+. The mean and SD are shown (n>10 axons per category; ***p < 0.0005, unpaired t test).
We visualized motor neurons in intact early second instar larvae using the GFP gene trap inserted within the endogenous Nervana2 locus (GFP-Nrv2) [13]. Nervana2, a Sodium/potassium-transporting ATPase channel subunit, specifically labels larval brain, ventral ganglions, and segmental nerves. A ventral view of an early second instar control larva showed a uniform nerve system pattern with stretched segmental nerves innervating each hemisegment (Figure 4B, upper panel). However, depletion of Pav-KLP by elav>pav-RNAi in flies with halved pav maternal load (pavB200/+ mothers) caused major defects in nervous system development. elav>pav-RNAi early second instar larvae had curved segmental nerves, most likely due to axon overgrowth (Figure 4B, lower panel). Quantification of the length of the longest nerves in Drosophila larvae revealed that depletion of Pav-KLP in neurons induced significant neurite overgrowth (Figure 4C).
The few larvae expressing elav>pav-RNAi that were able to develop to the third instar stage probably had a lower efficiency of Pav-KLP depletion resulting in a milder phenotype. A lateral view of those larvae showed a characteristic curling of the posterior end (the tail-flipping phenotype, Figure S3A). This phenotype is caused by paralysis of dorsal musculature and is typically observed in animals with mutations that cause degeneration of motor neurons [14, 15]. In addition, locomotion of the surviving elav>pav-RNAi third instar larvae was dramatically affected; these larvae were very sluggish and their crawling trajectories were typically circular (Figure S3B and Movie S5). Examination of neuromuscular junctions (NMJs) using the transgenic reporter CD8-GFP-Shaker showed that NMJs were missing in one side of elav>pav-RNAi third instar larvae (Figures S3C–E). These data clearly show that Pav-KLP is necessary to control axon outgrowth and proper NMJ formation in Drosophila.
DISCUSSION
In our previous work, we showed that microtubule sliding by kinesin-1 drives initial neurite outgrowth in Drosophila neurons and that sliding is downregulated as neurons mature [1]. In this paper, we demonstrate that the “mitotic” kinesin Pav-KLP functions as a negative regulator of interphase microtubule sliding both in Drosophila S2 cells and in Drosophila neurons. Knockdown of Pav-KLP stimulated microtubule sliding producing longer axons, while overexpression of Pav-KLP inhibited both sliding and neurite outgrowth. Increased length of axons after Pav-KLP depletion was also observed in vivo in Drosophila. Therefore, Pav-KLP attenuates neurite outgrowth by downregulation of kinesin-1-powered microtubule-microtubule sliding.
Pav-KLP and its orthologs (members of the kinesin 6 family) were originally identified as essential components for central spindle assembly and cleavage furrow formation. Pav-KLP depletion induces defects in morphology of the mitotic spindle at telophase and failure to recruit contractile ring components [9, 10]. However, the work of Baas and colleagues has demonstrated that CHO1/MKLP1, the mammalian ortholog of Pav-KLP, has an additional function in neurodevelopment. Their data show that CHO1/MKLP1 plays a role in establishing dendrite identity in differentiated neurons. Depletion of CHO1/MKLP1 induces progressive loss of dendrites. The authors concluded that CHO1/MKLP1 organizes microtubules in dendrites by transporting short minus-end-distal microtubule fragments into the dendrites [16]. Their more recent work revisited the role of CHO1/MKLP1 in developing neurons and suggested that CHO1/MKLP1 can regulate neurite outgrowth. They showed that depletion of CHO1/MKLP1 increased transport of short microtubule fragments [17]. Our data are in agreement with the idea that Pav-KLP regulates formation of neurites. However, the mechanisms reported here are clearly different from the results obtained by the Baas group in two significant aspects: First, we have shown that the reorganization of microtubules required for neurite formation is driven by kinesin-1 [1]. Second, our visualization technique clearly demonstrates that microtubules in developing Drosophila neurons are moved as long polymers. It is possible that the differences between our results and the work by the Baas lab are explained by different models (Drosophila versus mammalian neurons). A more attractive idea is that similar mechanisms work in both systems, but further work is required to understand the role of kinesin-1 in neurite outgrowth in mammalian neurons [18].
Interestingly, work by several groups has shown that proteins that function together with kinesin-6 in the cytokinesis pathway could also regulate neuronal morphogenesis. For example, Tumbleweed [19] or Ect2/Pebble/RhoGEF [20] depletion increase the extent of neurite outgrowth suggesting that Tumbleweed and RhoGEF control neurite outgrowth through actin reorganization. However, our results demonstrate that the primary regulator of neurite outgrowth is the essential partner of these two proteins, the kinesin-6, Pav-KLP. Furthermore, the effect of Pav-KLP on neurite formation is independent of actin or small GTPases (although we cannot completely exclude more subtle effects of Tumbleweed or Ect2 on the actin cytoskeleton in developing neurons). Indeed, a recent work concluded that an actin-signaling pathway regulated by the Centralspindlin complex controls protrusive activity required for directional neuronal migration [21].
The original idea that mitotic motors regulate cytoplasmic microtubules in neurons was proposed by Baas and colleagues, who suggested that microtubule arrays in neurons are established by mechanisms that are analogous to those that organize the mitotic spindle [22]. Supporting this idea, they demonstrated that inhibition of other mitotic motors, i.e. kinesin-5, affected the length axons [23, 24]. Advancing this concept, we propose that Pav-KLP/kinesin-6 directly regulates cytoplasmic microtubule arrangement by cross-linking them. It has been shown that loss of function mutations on ZEN-4/MKLP1, the C. elegans form of Pav-KLP, produce longer spindles [25], suggesting that kinesin-6 motors inhibit sliding of microtubules against each other during anaphase B. If this is indeed the case, our results suggest an important functional similarity between the molecular mechanisms of cell division and process formation in neurons. While Anaphase B is driven in part by microtubule-microtubule sliding powered by bipolar kinesin-5 and negatively regulated by kinesin-6 (mammalian MKLP1/C. elegans Zen-4/Drosophila Pav-KLP), the initial formation of neurites requires microtubule-microtubule sliding by kinesin-1 and, similarly to Anaphase B, is negatively regulated by kinesin-6. Thus, kinesin-6 motors together with other components of the Centralspindlin complex can function as general brakes of microtubule-microtubule sliding during both cell division and postmitotic neurite formation.
The fact that microtubule sliding is inhibited by Pav-KLP in mature but not in young neurons suggests that Pav-KLP itself is temporally regulated. One potential mechanism that could affect the ability of Pav-KLP (MNLP-1) to regulate microtubule sliding is Pav-KLP phosphorylation. It has been shown by Mishima and colleagues that phosphorylation of Ser710 in MKLP-1 (Ser743 in Drosophila Pav-KLP) promotes its binding to protein 14-3-3, preventing MKLP-1 from clustering on microtubules [6]. Future studies using phosphomimetic variants of Pav-KLP may help to test this mechanism.
Supplementary Material
Related to Figure 1. Time-lapse images of photoconverted microtubules in control (left panel) or Pav-KLP RNAi (right panel) of S2 cells expressing tdEOS-αtub. Scale bars, 10 μm.
Example 1 is a time-lapse movie of S2 cells expressing mCherry-tubulin and GFP-tagged Pav-KLP variants shown in Figure 1F. Note that motility is inhibited only in microtubules decorated with GFP-tagged Pav-KLP. Example 2 is a time-lapse movie of photoconverted microtubules in an S2 cell expressing tdEOS-αtub and BFP-Pav.FL. Note that microtubules decorated with BFP-Pav.FL did not slide. Scale bars, 10 μm.
Related to Figures S2. Example 1 is a time-lapse movie of the young neurons shown in Figure S2A. These neurons expressed Jupiter-GFP and their plasma membrane is labeled with the membrane dye, Deep Red. Note that microtubules pushed and advanced the plasma membrane. Examples 2 and 3 (shown in Figure S2B) are time-lapse movies of photoconverted microtubules (red channel) in young embryonic neurons. Note that long photoconverted microtubules slided from the photoconverted area either to the nascent neurites (Example 2) or to the cell body (Example 3). Scale bars, 10 μm.
Related with Figure 2. Example 1 (shown in Figure 2A) is a time-lapse movie of microtubules in the soma of a mature cultured neuron. Examples 2 and 3 are time-lapse movies of microtubules photoconverted in the neurites of control or Pav-KLP RNAi mature neurons (shown in Figure 2C). Note that while microtubule sliding was inhibited in control, microtubules were still actively sliding in neurons obtained from dissociated pavB200 mutant embryos microinjected with Pav-KLP dsRNA. Scale bars, 5 μm in Example 1, and 10 μm in Examples 2 and 3.
Related with Figure S3. Pav-KLP depletion in neurons induced locomotion defects in third instar larvae. Time-lapses of larvae crawling on black agarose plates showed that the locomotion of elav>pav-RNAi larvae (right panel) is impaired in comparison with control larvae (left panel).
Figure S1. Pav-KLP regulates microtubule sliding and process formation independently of small Rho GTPases in S2 cells. Related to Figure 1. (A) CytoD treatment induced microtubule-filled processes in Drosophila S2. Scale bar, 5 μm.<1br>(B and C) Polyploid cells induced by long treatment of CytoD did not develop longer processes than control cells. (B) Treatment of Pav-KLP dsRNA induced formation of multinuclear cells due to cytokinesis impairment. Alternatively, the number of S2 polynuclear cells could be increased by long treatment of the actin-fragmentation drug CytoD. (n>100 cells for each category.) (C) Quantification of process length in mononuclear (control) and multinuclear cells treated with either Pav-KLP dsRNA or CytoD. (n>80 cells for each catergory; error bars, SD).<1br>(D–G) Depletion of Centralspindlin complex components activated microtubule sliding and process formation independently of small Rho GTPases in S2 cells. Process length (D) and number of processes (E) were quantified per cell after knockdown of Centralspindlin components or small Rho GTPases family proteins. (n>70 cells for each catergory; error bars, SD) ***, p < 0.0005, unpaired t test. (F) Quantification of microtubule sliding in S2 cells treated with dsRNA. (n>10 cells for each catergory; error bars, SD) ***, p < 0.0005; *, p= 0.045. (G) Simultaneous treatment of Pav-KLP, Tumbleweed and Pebble dsRNA does not have an additive effect on microtubule sliding activation. (n>15 cells for each catergory; error bars, SD).<1br>(H–I) Pav-KLP stability was diminished when Tumbleweed or Pebble are depleted by dsRNA. (H) Protein levels were quantified after western blot in two independent experiments.(I) Representative western blots of the S2 lysates described in (H). Dilutions of untreated cells provided to estimate degree of knockdown.<1br>(J) Quantification of peroxisome transport in S2 cells. Peroxisomes located in the S2 processes were tracked for 1 minute, and retrograde and anterograde run-trajectories for each peroxisome were determined by DiaTracker (Semasopht). Peroxisome displacement was normalized to the untreated control (n>663 peroxisomes from 25 cells for each condition).<1br>(K–N) Ectopic expression of BFP-Pav.FL blocked microtubule sliding. An S2 cell coexpressing tdEOS-αtub and BFP-Pav.FL was photoconverted in two different areas and microtubule sliding was quantified. (K) Confocal images of green and red channel before photoconversion. Scale bar, 10 μm. (L) The first and last frame (20 minutes) of a time-lapse in the red channel are depicted. Note that photoconverted microtubules in zone 2 were static. See movie S2. (M) After the time-lapse acquisition, BFP-Pav.FL localization was detected with blue light. Note that Pav-KLP decorated Microtubules were static while microtubules in the other zone without Pav-KLP moved freely. Scale bar, 10 μm. (N) Quantification of the relative red signal remaining in the photoconverted zone.<1br>(O–P) Expression of the Pav-KLP full length protein rescues sliding rates and process formation. Quantification of microtubule sliding (O) and process formation (P) in S2 cells treated with dual dsRNAs against the Pav-KLP 5′ and 3′ UTR sequences. BFP-Pav.FL rescued both sliding and process formation. ***, p < 0.001; **, p> 0.0025 (n>10 cells for each category; error bars, SD).
Figure S2. Microtubule sliding drives neurite outgrowth in young neurons. Related to Figure 2.
(A) A confocal image of young cultured neurons expressing endogenous GFP-tagged Jupiter with the plasma membrane stained with Deep Red (upper panel). Time-lapse acquisition showed that neurite outgrowth is coupled to microtubule motility (bottom panels). See movie S3. Scale bar, 5 μm.
(B) Microtubules slide as long fragments in young Drosophila neurons. The arrows track the movement of a long microtubule after photoconversion in the neurite of a young neuron expressing mat αtub>tdEOS-αtub. See movie S3. Scale bar, 5 μm.
(C) Kinesin-1 drives axon outgrowth in Drosophila neurons. Depletion of KHC by zygotic protein null (khc27) and injection of dsRNA against the 3′UTR of KHC resulted in neurons with very short neurites. Coinjection of pMT-KHC-BFP, but not of pMT-KHC(MBD)*-BFP, rescued the length of the axons. The mean and SD are shown (n>10 neurons per category; **p= 0.0019, and *p>0.0425, unpaired t test).
Figure S3. Pav-KLP regulates neuromuscular junction formation. Related to Figure 4.
(A) When crawling, wild type third instar larvae maintained a flat body posture. In contrast, most elav>pav RNAi larvae exhibited a posterior paralysis and a classical tail-flipping phenotype indicating a defect in proper motor neuron development. Scale bar, 1 mm.
(B) Third instar larvae expressing elav>pav RNAi have severe locomotion defects. Crawling trajectories of surviving larvae were represented as multicolor merge images using the Temporal-Color Code plugin in Fiji (n=7 larvae per condition). See corresponding Movie S5.
(C) Cartoon representing a ventral view section of the third instar larval musculature. The segment used to analyse NMJs is highlighted with the red rectangle.<1br>(D) Schematic diagrams of NMJs on ventral muscles 6/7 of a wild-type larva. The muscles and NMJs are highlighted with dark and light green, respectively.
(E and F) Some neuromuscular terminals are missing in elav>pav RNAi knockdown third instar larvae. Confocal images (Z-projections, maximal intensity) of NMJs labeled by MHC-CD8-GFP-SH in segment A6 of control (E) and elav>pav RNAi (F) intact third instar larvae. Arrowheads indicate the NMJs that innervate muscles 6 and 7. Note that this NMJ is missing in the right hemisegment of the elav>pav RNAi larva. Scale bars, 100 μm.
Highlights.
Pav-KLP downregulates kinesin-1-powered sliding of long cytoplasmic microtubules.
Pav-KLP is a negative regulator of neurite outgrowth in cultured Drosophila neurons.
Pav-KLP is required for proper neurite extension of Drosophila neurons in vivo.
Acknowledgments
We would like to acknowledge D. Glover, C. Doe, E. Ferguson, B. McCabe, the Bloomington Stock Center (NIH P40OD018537), and Yale GFP Flytrap Database for fly stocks and E. Griffis, J. Scholey, V. Verkhusha and M. Murray’s labs for antibodies and plasmids. Our special thanks are to Stephen Rogers, Gary Banker, Peter Hollenbeck, Michael Glozter, Kari Barlan, Caroline Hookway and Masha Gelfand for their fruitful comments on earlier versions of this manuscript and our work on microtubule sliding in general. Research reported in this publication was supported by the National Institute of General Medical Science of the National Institutes of Health under award number R01GM052111 to V.I.G. and by Basque Government Department of Education, Universities and Research under award number BFI-2011-295 to U.d.C.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lu W, Fox P, Lakonishok M, Davidson MW, Gelfand VI. Initial neurite outgrowth in Drosophila neurons is driven by Kinesin-powered microtubule sliding. Curr Biol. 2013;23:1018–1023. doi: 10.1016/j.cub.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jolly AL, Kim H, Srinivasan D, Lakonishok M, Larson AG, Gelfand VI. Kinesin-1 heavy chain mediates microtubule sliding to drive changes in cell shape. Proc Natl Acad Sci U S A. 2010;107:12151–12156. doi: 10.1073/pnas.1004736107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barlan K, Lu W, Gelfand VI. The microtubule-binding protein ensconsin is an essential cofactor of Kinesin-1. Curr Biol. 2013;23:317–322. doi: 10.1016/j.cub.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seeger MA, Rice SE. Microtubule-associated protein-like binding of the kinesin-1 tail to microtubules. J Biol Chem. 2010;285:8155–8162. doi: 10.1074/jbc.M109.068247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan J, Chao DL, Toba S, Koyasako K, Yasunaga T, Hirotsune S, Shen K. Kinesin-1 regulates dendrite microtubule polarity in Caenorhabditis elegans. Elife. 2013;2:e00133. doi: 10.7554/eLife.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- 7.Somers WG, Saint R. A RhoGEF and Rho family GTPase-activating protein complex links the contractile ring to cortical microtubules at the onset of cytokinesis. Dev Cell. 2003;4:29–39. doi: 10.1016/s1534-5807(02)00402-1. [DOI] [PubMed] [Google Scholar]
- 8.Subach OM, Cranfill PJ, Davidson MW, Verkhusha VV. An enhanced monomeric blue fluorescent protein with the high chemical stability of the chromophore. PLoS One. 2011;6:e28674. doi: 10.1371/journal.pone.0028674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minestrini G, Mathe E, Glover DM. Domains of the Pavarotti kinesin-like protein that direct its subcellular distribution: effects of mislocalisation on the tubulin and actin cytoskeleton during Drosophila oogenesis. J Cell Sci. 2002;115:725–736. doi: 10.1242/jcs.115.4.725. [DOI] [PubMed] [Google Scholar]
- 10.Adams RR, Tavares AA, Salzberg A, Bellen HJ, Glover DM. pavarotti encodes a kinesin-like protein required to organize the central spindle and contractile ring for cytokinesis. Genes Dev. 1998;12:1483–1494. doi: 10.1101/gad.12.10.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Salzberg A, D’Evelyn D, Schulze KL, Lee JK, Strumpf D, Tsai L, Bellen HJ. Mutations affecting the pattern of the PNS in Drosophila reveal novel aspects of neuronal development. Neuron. 1994;13:269–287. doi: 10.1016/0896-6273(94)90346-8. [DOI] [PubMed] [Google Scholar]
- 12.Campos AR, Rosen DR, Robinow SN, White K. Molecular analysis of the locus elav in Drosophila melanogaster: a gene whose embryonic expression is neural specific. EMBO J. 1987;6:425–431. doi: 10.1002/j.1460-2075.1987.tb04772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun B, Xu P, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proc Natl Acad Sci U S A. 1999;96:10438–10443. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurd DD, Saxton WM. Kinesin mutations cause motor neuron disease phenotypes by disrupting fast axonal transport in Drosophila. Genetics. 1996;144:1075–1085. doi: 10.1093/genetics/144.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Djagaeva I, Rose DJ, Lim A, Venter CE, Brendza KM, Moua P, Saxton WM. Three routes to suppression of the neurodegenerative phenotypes caused by kinesin heavy chain mutations. Genetics. 2012;192:173–183. doi: 10.1534/genetics.112.140798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu W, Cook C, Sauter C, Kuriyama R, Kaplan PL, Baas PW. Depletion of a microtubule-associated motor protein induces the loss of dendritic identity. J Neurosci. 2000;20:5782–5791. doi: 10.1523/JNEUROSCI.20-15-05782.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin S, Liu M, Mozgova OI, Yu W, Baas PW. Mitotic motors coregulate microtubule patterns in axons and dendrites. J Neurosci. 2012;32:14033–14049. doi: 10.1523/JNEUROSCI.3070-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira A, Niclas J, Vale RD, Banker G, Kosik KS. Suppression of kinesin expression in cultured hippocampal neurons using antisense oligonucleotides. J Cell Biol. 1992;117:595–606. doi: 10.1083/jcb.117.3.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein AY, Jan YN, Luo L. Function and regulation of Tumbleweed (RacGAP50C) in neuroblast proliferation and neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102:3834–3839. doi: 10.1073/pnas.0500748102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuji T, Higashida C, Aoki Y, Islam MS, Dohmoto M, Higashida H. Ect2, an ortholog of Drosophila pebble, regulates formation of growth cones in primary cortical neurons. Neurochem Int. 2012 doi: 10.1016/j.neuint.2012.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falnikar A, Tole S, Liu M, Liu JS, Baas PW. Polarity in migrating neurons is related to a mechanism analogous to cytokinesis. Curr Biol. 2013;23:1215–1220. doi: 10.1016/j.cub.2013.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baas PW. Microtubules and neuronal polarity: lessons from mitosis. Neuron. 1999;22:23–31. doi: 10.1016/s0896-6273(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 23.Myers KA, Baas PW. Kinesin-5 regulates the growth of the axon by acting as a brake on its microtubule array. J Cell Biol. 2007;178:1081–1091. doi: 10.1083/jcb.200702074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haque SA, Hasaka TP, Brooks AD, Lobanov PV, Baas PW. Monastrol, a prototype anti-cancer drug that inhibits a mitotic kinesin, induces rapid bursts of axonal outgrowth from cultured postmitotic neurons. Cell Motil Cytoskeleton. 2004;58:10–16. doi: 10.1002/cm.10176. [DOI] [PubMed] [Google Scholar]
- 25.Dechant R, Glotzer M. Centrosome separation and central spindle assembly act in redundant pathways that regulate microtubule density and trigger cleavage furrow formation. Dev Cell. 2003;4:333–344. doi: 10.1016/s1534-5807(03)00057-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Related to Figure 1. Time-lapse images of photoconverted microtubules in control (left panel) or Pav-KLP RNAi (right panel) of S2 cells expressing tdEOS-αtub. Scale bars, 10 μm.
Example 1 is a time-lapse movie of S2 cells expressing mCherry-tubulin and GFP-tagged Pav-KLP variants shown in Figure 1F. Note that motility is inhibited only in microtubules decorated with GFP-tagged Pav-KLP. Example 2 is a time-lapse movie of photoconverted microtubules in an S2 cell expressing tdEOS-αtub and BFP-Pav.FL. Note that microtubules decorated with BFP-Pav.FL did not slide. Scale bars, 10 μm.
Related to Figures S2. Example 1 is a time-lapse movie of the young neurons shown in Figure S2A. These neurons expressed Jupiter-GFP and their plasma membrane is labeled with the membrane dye, Deep Red. Note that microtubules pushed and advanced the plasma membrane. Examples 2 and 3 (shown in Figure S2B) are time-lapse movies of photoconverted microtubules (red channel) in young embryonic neurons. Note that long photoconverted microtubules slided from the photoconverted area either to the nascent neurites (Example 2) or to the cell body (Example 3). Scale bars, 10 μm.
Related with Figure 2. Example 1 (shown in Figure 2A) is a time-lapse movie of microtubules in the soma of a mature cultured neuron. Examples 2 and 3 are time-lapse movies of microtubules photoconverted in the neurites of control or Pav-KLP RNAi mature neurons (shown in Figure 2C). Note that while microtubule sliding was inhibited in control, microtubules were still actively sliding in neurons obtained from dissociated pavB200 mutant embryos microinjected with Pav-KLP dsRNA. Scale bars, 5 μm in Example 1, and 10 μm in Examples 2 and 3.
Related with Figure S3. Pav-KLP depletion in neurons induced locomotion defects in third instar larvae. Time-lapses of larvae crawling on black agarose plates showed that the locomotion of elav>pav-RNAi larvae (right panel) is impaired in comparison with control larvae (left panel).
Figure S1. Pav-KLP regulates microtubule sliding and process formation independently of small Rho GTPases in S2 cells. Related to Figure 1. (A) CytoD treatment induced microtubule-filled processes in Drosophila S2. Scale bar, 5 μm.<1br>(B and C) Polyploid cells induced by long treatment of CytoD did not develop longer processes than control cells. (B) Treatment of Pav-KLP dsRNA induced formation of multinuclear cells due to cytokinesis impairment. Alternatively, the number of S2 polynuclear cells could be increased by long treatment of the actin-fragmentation drug CytoD. (n>100 cells for each category.) (C) Quantification of process length in mononuclear (control) and multinuclear cells treated with either Pav-KLP dsRNA or CytoD. (n>80 cells for each catergory; error bars, SD).<1br>(D–G) Depletion of Centralspindlin complex components activated microtubule sliding and process formation independently of small Rho GTPases in S2 cells. Process length (D) and number of processes (E) were quantified per cell after knockdown of Centralspindlin components or small Rho GTPases family proteins. (n>70 cells for each catergory; error bars, SD) ***, p < 0.0005, unpaired t test. (F) Quantification of microtubule sliding in S2 cells treated with dsRNA. (n>10 cells for each catergory; error bars, SD) ***, p < 0.0005; *, p= 0.045. (G) Simultaneous treatment of Pav-KLP, Tumbleweed and Pebble dsRNA does not have an additive effect on microtubule sliding activation. (n>15 cells for each catergory; error bars, SD).<1br>(H–I) Pav-KLP stability was diminished when Tumbleweed or Pebble are depleted by dsRNA. (H) Protein levels were quantified after western blot in two independent experiments.(I) Representative western blots of the S2 lysates described in (H). Dilutions of untreated cells provided to estimate degree of knockdown.<1br>(J) Quantification of peroxisome transport in S2 cells. Peroxisomes located in the S2 processes were tracked for 1 minute, and retrograde and anterograde run-trajectories for each peroxisome were determined by DiaTracker (Semasopht). Peroxisome displacement was normalized to the untreated control (n>663 peroxisomes from 25 cells for each condition).<1br>(K–N) Ectopic expression of BFP-Pav.FL blocked microtubule sliding. An S2 cell coexpressing tdEOS-αtub and BFP-Pav.FL was photoconverted in two different areas and microtubule sliding was quantified. (K) Confocal images of green and red channel before photoconversion. Scale bar, 10 μm. (L) The first and last frame (20 minutes) of a time-lapse in the red channel are depicted. Note that photoconverted microtubules in zone 2 were static. See movie S2. (M) After the time-lapse acquisition, BFP-Pav.FL localization was detected with blue light. Note that Pav-KLP decorated Microtubules were static while microtubules in the other zone without Pav-KLP moved freely. Scale bar, 10 μm. (N) Quantification of the relative red signal remaining in the photoconverted zone.<1br>(O–P) Expression of the Pav-KLP full length protein rescues sliding rates and process formation. Quantification of microtubule sliding (O) and process formation (P) in S2 cells treated with dual dsRNAs against the Pav-KLP 5′ and 3′ UTR sequences. BFP-Pav.FL rescued both sliding and process formation. ***, p < 0.001; **, p> 0.0025 (n>10 cells for each category; error bars, SD).
Figure S2. Microtubule sliding drives neurite outgrowth in young neurons. Related to Figure 2.
(A) A confocal image of young cultured neurons expressing endogenous GFP-tagged Jupiter with the plasma membrane stained with Deep Red (upper panel). Time-lapse acquisition showed that neurite outgrowth is coupled to microtubule motility (bottom panels). See movie S3. Scale bar, 5 μm.
(B) Microtubules slide as long fragments in young Drosophila neurons. The arrows track the movement of a long microtubule after photoconversion in the neurite of a young neuron expressing mat αtub>tdEOS-αtub. See movie S3. Scale bar, 5 μm.
(C) Kinesin-1 drives axon outgrowth in Drosophila neurons. Depletion of KHC by zygotic protein null (khc27) and injection of dsRNA against the 3′UTR of KHC resulted in neurons with very short neurites. Coinjection of pMT-KHC-BFP, but not of pMT-KHC(MBD)*-BFP, rescued the length of the axons. The mean and SD are shown (n>10 neurons per category; **p= 0.0019, and *p>0.0425, unpaired t test).
Figure S3. Pav-KLP regulates neuromuscular junction formation. Related to Figure 4.
(A) When crawling, wild type third instar larvae maintained a flat body posture. In contrast, most elav>pav RNAi larvae exhibited a posterior paralysis and a classical tail-flipping phenotype indicating a defect in proper motor neuron development. Scale bar, 1 mm.
(B) Third instar larvae expressing elav>pav RNAi have severe locomotion defects. Crawling trajectories of surviving larvae were represented as multicolor merge images using the Temporal-Color Code plugin in Fiji (n=7 larvae per condition). See corresponding Movie S5.
(C) Cartoon representing a ventral view section of the third instar larval musculature. The segment used to analyse NMJs is highlighted with the red rectangle.<1br>(D) Schematic diagrams of NMJs on ventral muscles 6/7 of a wild-type larva. The muscles and NMJs are highlighted with dark and light green, respectively.
(E and F) Some neuromuscular terminals are missing in elav>pav RNAi knockdown third instar larvae. Confocal images (Z-projections, maximal intensity) of NMJs labeled by MHC-CD8-GFP-SH in segment A6 of control (E) and elav>pav RNAi (F) intact third instar larvae. Arrowheads indicate the NMJs that innervate muscles 6 and 7. Note that this NMJ is missing in the right hemisegment of the elav>pav RNAi larva. Scale bars, 100 μm.