Abstract
Androgen receptor (AR) is widely expressed in breast cancer; however, there is limited information on the key molecular functions and gene targets of AR in this disease. In this study, gene expression data from a cohort of 52 breast cancer cell lines was analyzed to identify a network of AR co-expressed genes. A total of three hundred genes, which were significantly enriched for cell cycle and metabolic functions, showed absolute correlation coefficients (| CC |) of more than 0.5 with AR expression across the dataset. In this network, a subset of 35 “AR-signature” genes were highly co-expressed with AR (| CC| > 0.6) that included transcriptional regulators PATZ1, NFATC4, and SPDEF. Furthermore, gene encoding coagulation factor VII (F7) demonstrated the closest expression pattern with AR (CC= 0.716) in the dataset and factor VII protein expression was significantly associated to that of AR in a cohort of 209 breast tumors. Moreover, functional studies demonstrated that AR activation results in the induction of factor VII expression at both transcript and protein levels and AR directly binds to a proximal region of F7 promoter in breast cancer cells. Importantly, AR activation in breast cancer cells induced endogenous factor VII activity to convert factor X to Xa in conjunction with tissue factor. In summary, F7 is a novel AR target gene and AR activation regulates the ectopic expression and activity of factor VII in breast cancer cells. These findings have functional implications in the pathobiology of thromboembolic events and regulation of factor VII/tissue factor signaling in breast cancer.
Keywords: androgen receptor, breast cancer, factor VII, gene transcription
Introduction
Breast cancer is a heterogenous disease and therefore advancement in the molecular classification of breast cancer is a key step for the discovery of novel therapeutic targets and biomarkers in this disease. Traditionally breast cancer has been classified based on the expression of estrogen receptor (ER) and histopathological features. However, in the past decade gene expression profiling of primary breast tumors has resulted in the identification of several reproducible molecular subtypes [1–7]. Notably, these studies have led to a robust classification of ER-negative (ER-) breast cancer and revealed that basal and molecular apocrine subtypes represent two prominent molecular subgroups in ER- disease [3, 5, 7]. Molecular apocrine subtype is characterized by a steroid-response gene signature that includes androgen receptor (AR), FOXA1, Prolactin-Induced Protein (PIP), and a high frequency of ErbB2 overexpression [3–5]. It is notable that AR expression is present in 40 to 50% of ER-tumors and 60–70% of these cases also have ErbB2 overexpression [8–10]. Furthermore, AR is expressed in 88% of ER-positive (ER+) breast tumors [9].
Although the expression of AR in breast cancer has long been established [11, 12]; recent genomic findings have resulted in the re-emergence of an interest to study the functional and therapeutic implications of AR in breast cancer. In this respect, studies by our group and others have revealed that AR has a transcriptional and functional role in the regulation of key signaling pathways in breast cancer [13–18]. This includes a positive feedback loop between AR and ErbB2-extracellular signal-regulated kinase (ERK) signaling in molecular apocrine cells [13, 15, 18]. In this feedback loop, AR regulates ERK phosphorylation through the transcriptional activation of ErbB2 and, in turn, the ERK-CREB1 signaling regulates AR transcription [15]. Furthermore, AR acts as a transcriptional activator of PIP, a characteristic biomarker in breast cancer, which is required for cell cycle progression in both ER+ and ER- breast cancer cells [14, 17, 19–21]. Moreover, chromatin immunoprecipitation sequencing (ChIP-seq) for AR has been carried out in molecular apocrine cell line MDA-MB-453 and demonstrated that AR induces the WNT7B-ErbB3 signaling and there is a transcriptional interaction between AR and FOXA1 in MDA-MB-453 cells [16, 22]. Importantly, multiple preclinical studies have suggested a potential role for AR as a therapeutic target in breast cancer and currently this topic is under active investigation in clinical trials [4, 13, 18, 23–25].
Despite the emerging data on the importance of AR function in breast cancer, the available studies have only included a limited number of cell lines and the broader picture of AR transcriptional role and key targets of this gene in breast cancer have remained largely unclear. In this study, a comprehensive investigation of AR-transcriptional network was carried out using a large cohort of breast cancer cell lines and novel AR co-expressed genes and AR-associated pathways were identified. Furthermore, this study suggests that AR is a key regulator of coagulation factor VII transcription and function in breast cancer.
Materials and methods
Bioinformatics
a) Gene expression analysis
Gene expression data for a cohort of 52 breast cancer cell lines were extracted from a published microarray study by Neve et al. [26]. This cohort constituted of both ER+ and ER- cell lines and included a total of 22,216 gene expression data points for each line [26]. Extracted gene expression matrix was analysed to identify genes that were highly co-expressed with AR across the dataset. In this respect, Pearson correlation coefficients (CC) between AR expression and that of every gene in the dataset were calculated. Next, AR co-expressed genes were identified using two cutoffs of absolute CC values (|CC|) more than 0.5, p< 0.001 (AR co-expressed genes) and more than 0.6, p< 0.001 (AR gene-signature).
Furthermore, molecular functions of AR co-expressed genes (|CC|> 0.5, p<0.001) were assessed using Gene Ontology (GO) analysis. In this process, gene symbols of this gene set were first updated to the official symbols provide by Hugo Gene Nomenclature Committee [27]. Subsequently, functional annotation of the AR co-expressed genes was performed based on GO using The Database for Annotation, Visualization and Integrated Discovery (DAVID) Bioinformatics Resources v6.7 (National Institute of Allergy and Infectious Diseases, Bethesda, MD, USA), [28, 29]. In addition, a proximity matrix was obtained for AR gene-signature (|CC| > 0.6, p<0.001) followed by the hierarchical clustering of this signature using centroid linkage method with intervals measured by CC values. Data analysis to obtain CC values, proximity matrix, and clustering algorithms were performed using IBM SPSS Statistics 22 (Armonk, NY, USA).
b) Promoter analysis
Identification of putative AR binding sites in the 2 kb region of F7 promoter was carried out using PROMO software, which employs TRANSFAC version 8.3 [30, 31]. The sequence of the 2 kb region of F7 promoter was obtained using Ensembl Genome Browser (http://www.ensembl.org/index.html). Binding sites were predicted within a dissimilarity margin less or equal than 15%.
Tissue microarray cohort and immunohistochemistry
Three sets of breast cancer tissue microarray (TMA) slides were obtained from Pantomics (Richmond, CA, USA) that constituted of duplicate cores for a total of 209 malignant breast tumors (BRC1501-3). Immunohistochemistry (IHC) staining was performed as described before [32, 33]. Primary antibody incubation was carried out with rabbit polyclonal coagulation factor VII (FVII) antibody at 1:200 dilution (Novus Biologicals, Littleton, CO, USA) and mouse monoclonal AR antibody at 1:75 dilution (Dako, Carpinteria, CA, USA). Tumors with ≥10% nuclear-stained cells were considered positive for AR as previously published [8]. Since FVII has a cytoplasmic staining pattern, a semi-quantitaive scoring system that has been previously used for other cytoplasmic proteins was adapted to score FVII [34]. In this respect, FVII staining was scored based on the intensity of cytoplasmic staining as follows: no staining (score 0), weak staining (score 1), moderate staining (score 2), and intense staining (score 3).
Cell culture
Breast cancer cell lines T-47D and MFM-223 were obtained from American Type Culture Collection (Manassas, VA, USA). Culture media were obtained from Life Technologies (Grand Island, NY, USA). T-47D and MFM-223 cell lines were cultured in DMEM/F12 and DMEM media, respectively supplemented with 10% fetal bovine serum (FBS), (Fisher Scientific, Waltham, MA, USA). Cell culture treatment with 5α-Androstan-17β-ol-3-one (Dihydrotestosterone (DHT)), (Fisher Scientific) was carried out at 100 nM concentration to obtain an optimal AR response as described before [14, 15, 35–38]. DHT treatment was performed in phenol red-free media (Life Technologies) supplemented with 10% Charcoal/Dextran treated serum (Fisher Scientific) and cell lines were cultured in the media for 24 hours (h) or 48h prior to DHT treatment. AR inhibition was carried out using AR antagonist flutamide (Sigma-Aldrich, St. Louis, MO, USA) at 20 µM concentration for 48h in medium containing 10% FBS as described before [15, 23].
Quantitative real time-polymerase chain reaction
Quantitative real time-polymerase chain reaction (qRT-PCR) to assess the expression levels of gene encoding FVII (F7), (assay ID: Hs01551992_m) and PATZ1 (assay ID: Hs00204880_m1) was carried out using Taqman Gene Expression Assays (Life Technologies) as instructed by the manufacturer. Housekeeping genes RPLP0 and HPRT1 (Life Technologies) were applied as controls. Relative gene expression = gene expression in DHT-treated or flutamide-treated group/ average gene expression in control group.
Western blot analysis
Western blot analysis was carried out with rabbit polyclonal FVII antibody (Novus Biologicals) at 1:1000 dilution using 50 µg of T-47D cell lysate from each DHT-treated and control experiments. Membrane was stripped and immunoblotting with rabbit α-tubulin antibody (Abcam, Cambridge, UK) was applied to assess loading. To extract protein from the conditioned media, cell lines were cultured for 48 hours in serum-free media containing either DHT at 100 nM or control vehicle followed by concentration using Amicon Ultra-15 (3K) centrifugal filters (Millipore, Billerica, MA, USA). A total of 100 µg from each conditioned medium was precipitated and used for immunoblotting. Protein concentrations were measured using the BCA Protein Assay Kit (Fisher scientific). Immunoblot imaging and analysis of band densities were performed using ChemiDoc XRS system and Image Lab software, respectively (Bio-Rad, Hercules, CA, USA). Western blots were performed in three replicates and the average fold change was shown.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assay was performed in T-47D cell line using QuickChIP kit (Novus Biologicals) as instructed by the manufacturer. T-47D cells were treated with 100 nM of DHT for 48h before ChIP assays. DNA shearing was carried out by sonication at 100% output with ten pulses of 20 seconds each and a 2 minute rest on ice between each pulse and ChIP-grade rabbit polyclonal AR (Millipore) antibody was applied at 1:100 dilution for the assays. To quantify ChIP results, two primer sets for F7 promoter were used for qRT-PCR amplification by SYBR green method (Applied Biosystems). F7-promoter primers were forward primer 1: 5’ATCCCTCTGTCACCCTTGGA (start: −24), reverse primer 1: 5’CTGCCTGTTGACATTCCCCA (start: −90); and forward primer 2: 5’CCTCACACCTGTGTCCTCAAG (start: −1779), reverse primer 2: 5’GCAGGAACCGGGCTATCT (start: −1896). Amplification of 1% of input chromatin prior to immunoprecipitation was applied as the input control. ChIP assays using non-specific antibody (rabbit IgG) served as a negative control. The assays were carried out in six replicates and ChIP-signal was calculated as fold enrichment of F7-amplicon relative to the negative control for each experimental set.
Tissue factor-FVII activity assay
The activity of tissue factor (TF)-FVII was assessed by measuring the conversion of Factor X (FX) to activated FX (FXa) using the Tissue Factor Human Activity Assay Kit (AbCam). A total of 1.2 × 106 of T-47D cells was seeded in a 10 cm dish for each experiment. The next day, media were changed and cells were grown for 48h in phenol-free media supplemented with 10% Charcoal/Dextran treated serum followed by an additional 48h of culture in media containing either DHT at 100 nM (DHT group) or vehicle (control group). Subsequently, each sample was lysed with 75 µl of 15 mM octyl-β-D-glucopyranoside (Sigma-Aldrich) at 37°C for 15 minutes.
To assess the contribution of endogenous FVII to the overall TF activity, assays were carried out for each experiment in the absence (lysate) or presence of exogenous FVII (lysate + FVII). In this respect, TF-endogenous FVII activity was measured by adding 20 µl of each cell lysate to 10 µl of FX and 50 µl of assay diluent followed by an incubation at 37 °C for 30 min. Next, 20 µl of FXa substrate was added and absorbance was measured at 405 nm. In addition, total TF activity was assessed for each experiment by the addition of 10 µl of exogenous FVII to each lysate followed by the assay procedure as outlined above. Finally, the ratio of TF-FVII activity in each cell lysate was calculated relative to that of lysate + exogenous FVII to assess the contribution of endogenous FVII in the overall TF activity. All experiments were carried out in three replicates.
Statistical analysis
Biostatistics was carried out using IBM SPSS Statistics 22. Student t-test and paired sample t-test were applied for calculating the statistical significance. All error bars depict ± 2SEM. Receiver operating characteristic (ROC) analysis was applied to predict FVII-IHC scores based on the AR staining status in primary breast tumors. In this analysis, FVII scores ranged from 0–3 and AR status was 0 (< 10% AR staining) or 1 (≥ 10% AR staining).
Results
AR transcriptional network is enriched with cell cycle and metabolic genes
To identify the AR transcriptional network and key cellular functions associated with AR in breast cancer, expression data from a cohort of 52 breast cancer cell lines encompassing both ER+ and ER- subtypes was extracted from a study published by Neve et al. as explained in methods [26]. Correlation of AR expression with that of every gene in the dataset was calculated using the Pearson’s method to identify AR co-expressed genes. This analysis resulted in the identification of a total of three hundred AR co-expressed genes that had a |CC| of > 0.5 with AR expression (p< 0.001, Supplementary File 1). It is notable that among the AR co-expressed genes about 2/3 (a total of 195) had a positive correlation and about 1/3 (a total of 105) showed a negative correlation with AR expression (Supplementary File 1).
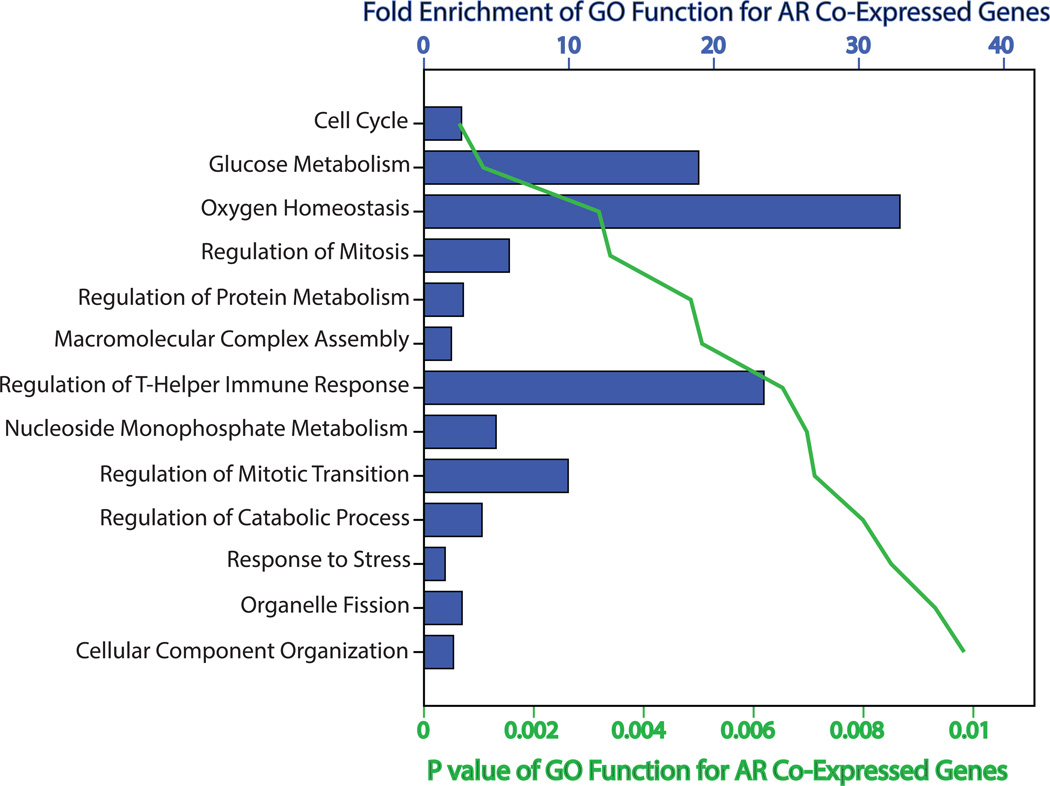
Moreover, functional annotation of the AR co-expressed genes was carried out based on GO using DAVID Bioinformatics Resources. This analysis identified a total of thirteen GO functions that significantly associated with the AR-transcriptional network (p< 0.01) and fold enrichment (FE) for these groups ranged from 1.5 to 33 (Figure 1 and Supplementary File 2). It is notable that the majority of these GO groups were related to cell cycle or cellular metabolism including the following functions: cell cycle, regulation of mitosis, regulation of mitotic metaphase/anaphase transition, glucose metabolism, oxygen homeostasis, positive regulation of protein metabolic process, macromolecular complex assembly, nucleoside monophosphate metabolic process, and regulation of catabolic process (Figure 1 and Supplementary File 2). Overall, these results suggest that there is a distinct network of AR co-expressed genes in breast cancer that is significantly enriched with cell cycle and metabolic genes.
Figure 1.
Functional annotation of AR co-expressed genes based on Gene Ontology. Functional annotation of AR co-expressed genes was performed based on Gene Ontology (GO) using DAVID Bioinformatics Resources. Listed GO functions are significantly associated with the network of AR co-expressed genes (p< 0.01). Fold enrichment and p values are depicted for each GO group.
F7 and AR are highly co-expressed in breast cancer
The microarray dataset was further analyzed to identify genes that have a highly correlated expression pattern with AR across a cohort of 52 breast cancer cell lines. For this purpose a cutoff |CC| of > 0.6 (p< 0.001) with AR expression was adopted to identify this gene set; termed “AR gene-signature”. Notably, there were a total of 35 genes in the dataset, excluding the AR gene itself, which correlated with AR expression at this cutoff (Table 1). In addition, the direction of correlation with AR expression was positive in 60% and negative in 40% of these AR-signature genes (Table 1). Importantly, there were only three genes in the dataset, accounting for 1% of AR co-expressed genes that demonstrated a |CC|> 0.7 with AR expression (Table 1). Among these, gene encoding coagulation factor VII (F7) had the strongest positive correlation with AR across the cohort with a CC= 0.716 followed by PATZ1 (CC= 0.709). In addition, PRNP showed the strongest negative correlation with AR with a CC= −0.75 (Table 1). Therefore, genes that are highly co-expressed with AR represent only a small fraction of genome and among these F7 is the most positively correlated gene across the cohort.
Table 1.
List of genes that are highly co-expressed with AR in breast cancer
| Positively-correlated genes | CC | Negatively-correlated genes | CC |
|---|---|---|---|
| F7 | 0.716 | PRNP | −0.75 |
| PATZ1 | 0.709 | DONSON | −0.681 |
| ZNF205-AS1 | 0.699 | TTK | −0.662 |
| NFATC4 | 0.681 | EHBP1 | −0.654 |
| SPDEF | 0.67 | PICALM | −0.649 |
| MXD4 | 0.661 | NAB1 | −0.644 |
| CTNND2 | 0.651 | USP1 | −0.642 |
| SLCO2A1 | 0.645 | GALNT2 | −0.63 |
| RHOH | 0.643 | PGM1 | −0.628 |
| SGSM3 | 0.638 | GLS | −0.613 |
| TP53TG1 | 0.636 | STIL | −0.612 |
| FGFR4 | 0.634 | TOP2A | −0.612 |
| PYGO1 | 0.633 | BUB1 | −0.609 |
| MGAT5 | 0.632 | GART | −0.609 |
| PCDHA5 | 0.621 | ||
| DALRD3 | 0.616 | ||
| SLC9A1 | 0.614 | ||
| IGHM | 0.612 | ||
| AMBP | 0.603 | ||
| CRAT | 0.601 | ||
| MVK | 0.601 |
Each gene has an absolute Pearson correlation coefficient (|CC|) of > 0.6 with AR expression (p< 0.001) across a cohort of 52 breast cancer cell lines.
To further delineate the pattern of gene-expression in this AR-signature, a proximity matrix was generated based on the CC values for every pair of genes within the signature (Supplementary File 3). Next, hierarchical clustering of AR-signature genes was carried out using a centroid linkage method with intervals measured by CC values (Figure 2). Notably, there were two main clusters in AR gene-signature based on the direction of CC values with AR expression and these were further subdivided into four and two sub-clusters for the positively and negatively AR co-expressed arms of dendrogram, respectively (Figure 2). Importantly, F7 clustered at the closest proximity to AR in this dendrogram, which provides further evidence for a highly correlated expression pattern between these two genes across the cohort (Figure 2).
Figure 2.
Dendrogram for hierarchical clustering of AR gene-signature based on centroid linkage analysis. Hierarchical clustering of AR gene-signature was performed using centroid linkage method and intervals were measured by Pearson correlation coefficients. Arrow indicates the location of AR within the cluster. F7 has the closest proximity to AR in this dendrogram.
FVII protein expression is associated with AR in primary breast tumors
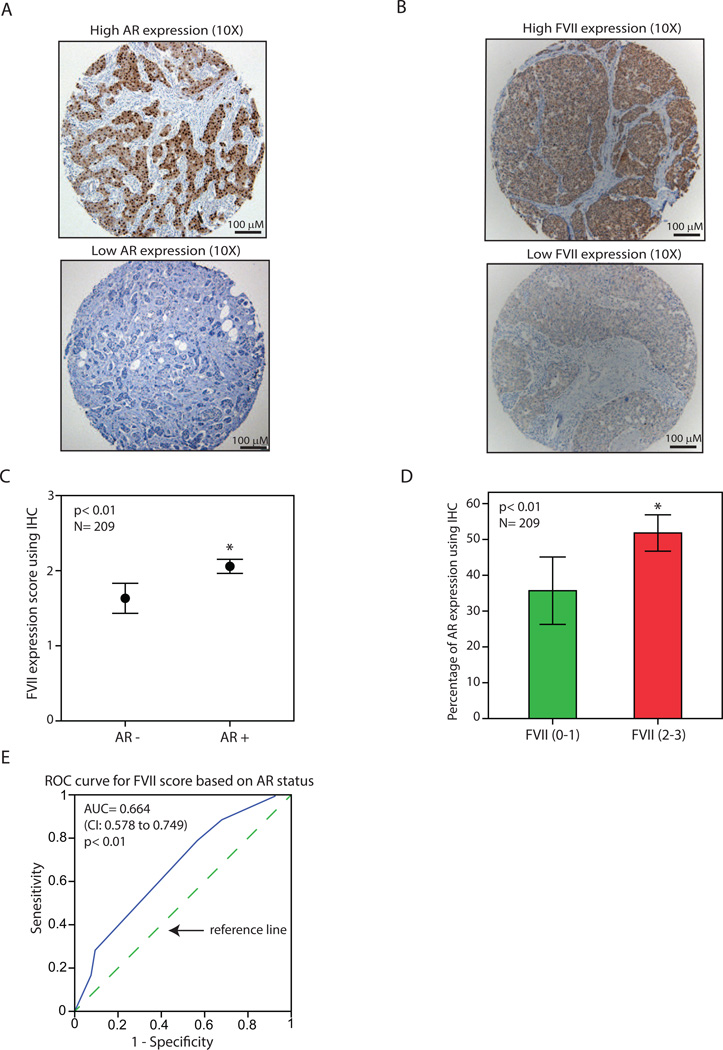
The association between FVII and AR expression in breast cancer was further investigated using IHC staining in a TMA cohort of 209 malignant breast tumors. In this respect, the pattern of AR expression was first characterized in the TMA cohort using IHC as described before [8, 19]. For each tumor sample the percentage of AR nuclear staining was measured as the average percentage of staining for duplicate cores and tumors with ≥10% nuclear-stained cells were considered positive for AR (Figure 3A and Supplementary File 4). In addition, FVII expression was assessed using an IHC scoring system ranging from 0 to 3 based on the intensity of cytoplasmic staining in each core as described in methods (Figure 3B and Supplementary File 4). Next, the average IHC score for duplicate cores of each sample was calculated to measure FVII staining in each tumor.
Figure 3.
Association of factor VII and AR protein expression in breast tumors using immunohistochemistry. (A) Immunohistochemistry (IHC) to demonstrate breast tumors with a high level (top panel) and a low level (bottom panel) of AR expression. Magnifications are at 10X. (B) IHC to demonstrate breast tumors with a high level (top panel) and a low level (bottom panel) of coagulation factor VII (FVII) expression. Magnifications are at 10X. (C) FVII expression scores using IHC for AR+ and AR- breast tumors. *, p< 0.01 is for AR+ vs. AR- groups. (D) Percentage of AR expression using IHC for breast tumors with a low FVII staining (scores: 0–1) and those with a high level of FVII (scores: 2–3). *, p< 0.01 is for FVII (2–3) vs. FVII (0–1). (E) Receiver operating characteristic (ROC) analysis to predict FVII-IHC scores based on the AR status in primary breast tumors. FVII scores ranged from 0–3 and AR status was 0 (< 10% AR staining) or 1 (≥ 10% AR staining). CI: confidence interval. Dashed line is a diagonal reference line. All Error Bars: ± 2SEM.
IHC results were subsequently analyzed to examine an association between FVII and AR expression in breast tumors. In this process, IHC scores for FVII were compared between AR+ and AR- breast tumors across the TMA cohort and AR+ tumors showed a significantly higher level of FVII expression compared to that of AR- cancers with average scores of 2.1 and 1.6, respectively (p< 0.01, Figure 3C). In addition, the percentage of AR staining was compared between breast tumors that had a relatively low FVII staining (scores: 0–1) and those with a relatively high level of FVII (scores: 2–3). Notably, tumors with higher levels of FVII also demonstrated a significantly higher percentage of AR expression across the cohort with AR staining at 52% (± 2.5) and 36% (± 4.7) for FVII (2–3) and FVII (0–1) groups, respectively (p< 0.01, Figure 3D). Finally, ROC analysis was applied to predict FVII expression scores based on the positive or negative status of AR staining in breast tumors. Importantly, AR status could reliably predict FVII expression with an area under the curve (AUC) of 0.664 (p< 0.01, confidence interval: 0.578–0.749, Figure 3E). Altogether, these findings suggest that there is a positive association between the protein levels of FVII and AR in primary breast tumors.
F7 is a target gene of AR in breast cancer
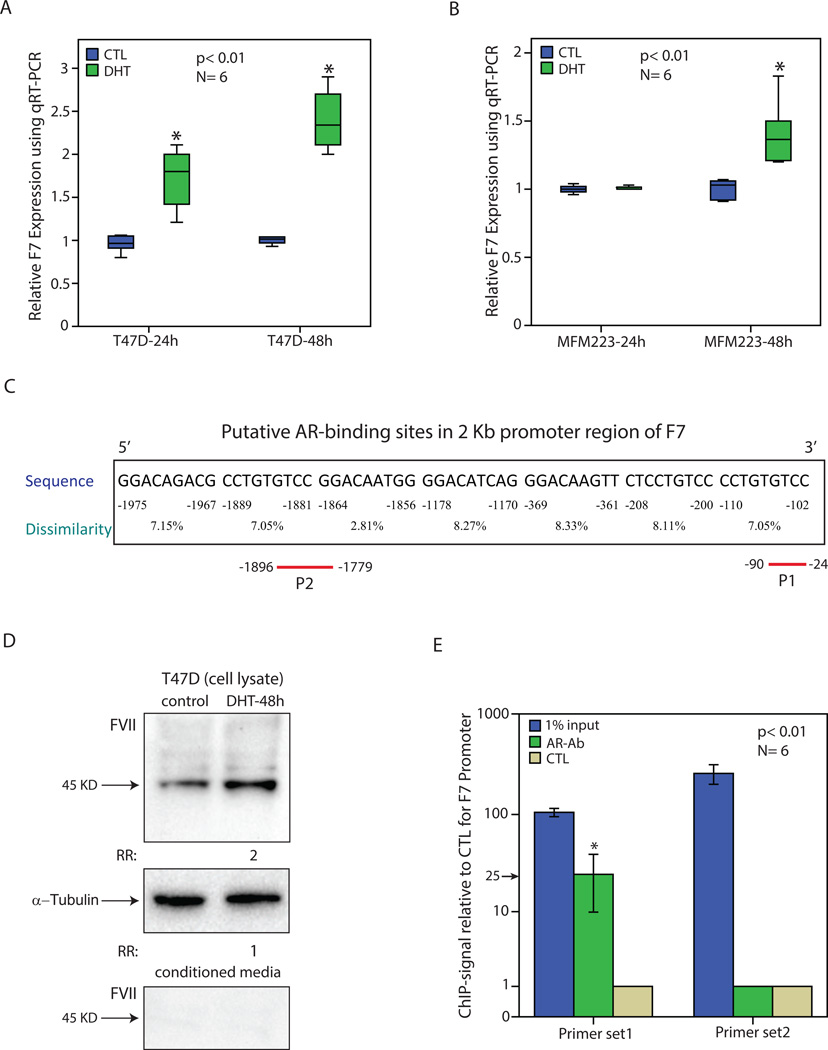
In view of the highly correlated expression pattern between AR and factor VII at transcript and protein levels, the possibility of F7 transcriptional regulation by AR was investigated in breast cancer cells. To examine this hypothesis, AR+/ER+ cell line T-47D and AR+/ER- line MFM-223 [19, 39], were tested to assess the effect of AR activation by DHT on F7 transcription. Incubation with DHT at 100 nM was carried out at two time points of 24h and 48h in phenol red-free media supplemented with 10% Charcoal/Dextran treated serum. Next, total RNA was extracted at each time-point from DHT-treated and control experiments followed by qRT-PCR assessment of F7 expression in T-47D and MFM-223 cell lines. Notably, AR activation by DHT significantly increased F7 expression in T-47D cells by approximately 2 and 2.5-fold at 24h and 48h time points, respectively (p< 0.01, Figure 4A). In addition, in MFM-223 cell line, F7 expression was significantly increased by approximately 1.5-fold (p< 0.01) following DHT treatment for 48h without a change in the expression of this gene at the 24h time-point (Figure 4B). These findings indicate that AR activation results in an induction of F7 transcription in breast cancer cells.
Figure 4.
Regulation of factor VII transcript and protein levels by AR in breast cancer. (A) Relative expression of F7 using qRT-PCR in T-47D cell line following treatment with DHT for 24 and 48 hours. Expression values are presented relative to the control (CTL) experiments at each time-point. Experiments were carried out in 6 replicates and *, p< 0.01 is for DHT-treated vs. control groups. (B) Relative expression of F7 in MFM-223 cell line following treatment with DHT as outlined in (A). (C) Putative AR-binding sites in the 2 Kb promoter region of F7 gene using PROMO software. Binding sites were predicted within a dissimilarity margin ≤ 15%. Location with respect to the ATG start codon, sequence, and dissimilarity margin is depicted for each putative binding site. P1 and P2 represent the amplicons used for ChIP assays. (D) Western blot analysis to show FVII level following DHT treatment for 48h in T-47D cell line using cell lysates or conditioned media. Fold change (RR) in band density was measured relative to the control in three replicate experiments. Rabbit α-tubulin antibody was applied to assess loading for cell lysates. (E) Chromatin immunoprecipitation (ChIP) with two primer sets for F7 promoter and AR antibody (AR-Ab). Amplification of 1% of input chromatin was used as the input control and non-specific antibody (CTL) served as a negative control. Each assay was carried out in six replicates and ChIP-signal was calculated as fold enrichment of F7-amplicon relative to the negative control. *, p< 0.01 is for AR-Ab vs. CTL. Error Bars: ± 2SEM.
A similar approach was followed to assess the effect of AR activation with DHT on PATZ1 expression, which is another gene that is highly co-expressed with AR (Table 1). However, as opposed to F7, DHT treatment did not stimulate PATZ1 expression in breast cancer cell lines (Supplementary File 5).
The induction of F7 by AR activation raises the question whether F7 is a direct AR target gene in breast cancer. This possibility was first investigated by the bioinformatics analysis of the 2 kb promoter region of F7 gene using PROMO software, which demonstrated a total of seven putative AR binding sites in this promoter region (Figure 4C). Moreover, the effect of AR activation on FVII protein level was assessed following DHT treatment of T-47D cell line for 48h and cell lysates and conditioned media were examined separately for fold-changes in FVII level relative to the controls using western blot analysis. Importantly, there was a 2-fold increase in FVII protein level following DHT treatment in T-47D cell lysates compared to the control samples (Figure 4D). In addition, it is notable that FVII protein was not detectable in the condition media of T-47D cells (Figure 4D). Therefore, in agreement with the F7 transcription data, FVII is induced at the protein level following AR activation and there are several putative AR binding sites in F7 promoter region.
To investigate whether F7 is a direct AR target gene, ChIP assays were carried out using a ChIP-grade AR antibody and two primer-sets in the 2 kb region of F7 promoter. These primer sets were designed to amplify regions that contained putative AR bindings in F7 promoter (Figure 4C). In this respect, primer set 1 and primer set 2 were applied to generate amplicons for −24 to −90 bp (P1) and −1779 to −1896 bp (P2) regions of F7 promoter, respectively (Figure 4C). Amplification of 1% of input chromatin was applied as the input control and ChIP assays using non-specific IgG served as a negative control. Following qRT-PCR, ChIP-signal was calculated as fold enrichment of F7-amplicon relative to the negative control for each experimental set. Importantly, ChIP with AR antibody for the P1 amplicon region demonstrated a significant enrichment by approximately 25-fold compared to the negative control antibody (p< 0.01, Figure 4E). In contrast, P2 amplicon did not have an enrichment using ChIP assays (Figure 4E). In addition, P1 and P2 regions both showed amplification using the input chromatin (Figure 4E). These data suggest that AR binds to F7 promoter in a region close to the ATG start codon. Therefore, we can conclude that AR is a direct transcriptional activator of F7 in breast cancer.
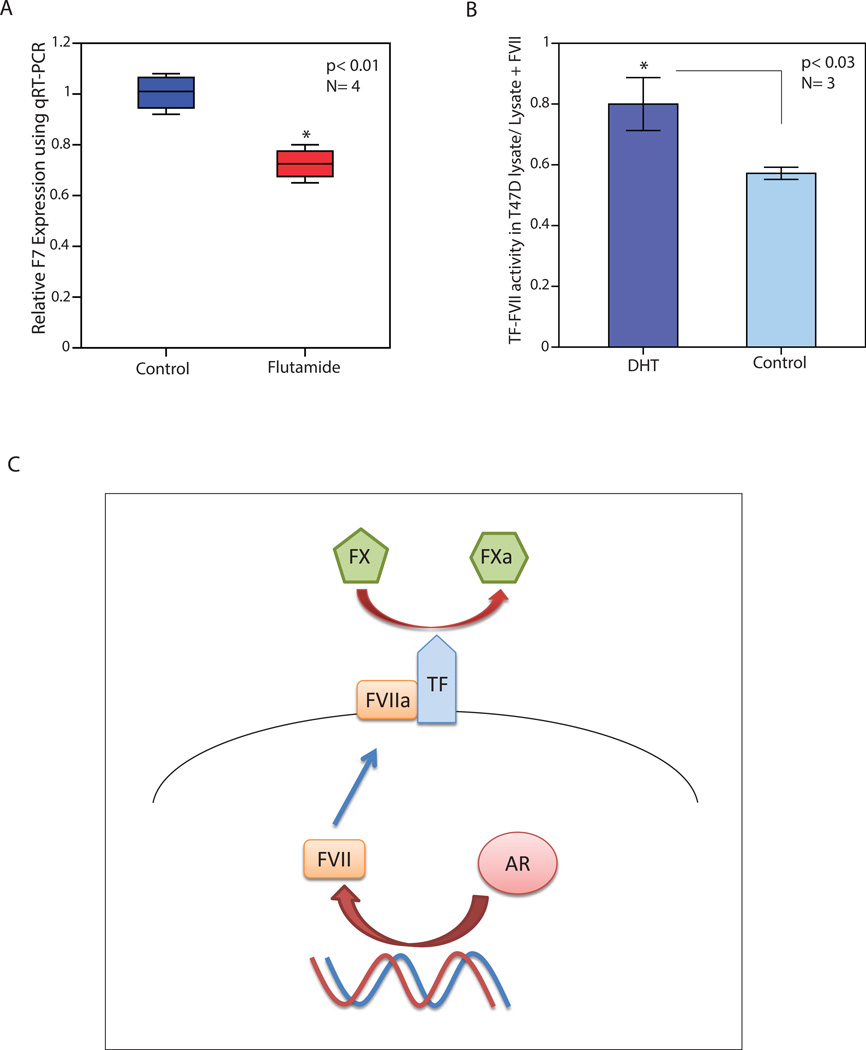
Moreover, T-47D cell line was treated with AR antagonist flutamide at 20 µM concentration for 48h and expression of F7 was assessed relative to the control using qRT-PCR. Notably, there was a reduction in F7 transcription by 30% following AR inhibition (p< 0.01, Figure 5A). This finding indicates that AR activity is necessary for the baseline expression of F7 in T-47D cells and further supports a key role for AR in the transcriptional regulation of F7.
Figure 5.
The effect of AR inhibition on F7 expression and AR activation on factor VII activity, and a schematic model for AR induction of factor VII. (A) Relative expression of F7 using qRT-PCR in T-47D cell line following treatment with flutamide (FLU) for 48 hours. Expression values are presented relative to the control (CTL) experiments. *, p< 0.01 is for FLU-treated vs. control groups. (B) The effect of AR activation by DHT on FVII in T-47D cell line using Tissue Factor (TF)-FVII activity assay. T-47D cells were treated either with DHT or vehicle control for 48h followed by cell lysate extraction and measurement of TF-FVII activity. The ratio of TF-FVII activity in each cell lysate was calculated relative to that of lysate + exogenous FVII and compared between the DHT-treated and control groups. Error Bars: ± 2SEM. (C) A schematic model for the AR-mediated induction of factor VII in breast cancer. In this model, AR induces FVII expression and leads to an increased activity of FVIIa/TF complex in breast cancer cells, which in turn activates FX to FXa conversion. TF: Tissue Factor, FVII: factor VII, FX: factor X, FXa: activated factor X, red arrows indicate a stimulatory effect.
FVII activity is induced by AR in T-47D breast cancer cell line
The effect of AR-mediated induction of FVII on the activity of this protein was assessed by measuring the conversion of FX to FXa in breast cancer cells. T-47D cell line was treated with either DHT at 100 nM or vehicle control for 48h followed by cell lysate extraction and measurement of TF-FVII activity. To assess the contribution of endogenous FVII to the overall TF activity, assays were carried out for each experiment in the absence (lysate) or presence of exogenous FVII (lysate + FVII) as described in methods. Next, the ratio of TF-FVII activity in each cell lysate was calculated relative to that of lysate + exogenous FVII and compared between the DHT-treated and control groups. Importantly, AR activation significantly increased the ratio of endogenous TF-FVII activity/total TF activity from 0.57 (± 0.01) in the control group to 0.80 (± 0.04) in the DHT-treated cells (p< 0.03, Figure 5B). This finding indicates that AR activation induces FVII activity in breast cancer cells.
Discussion
Although the functional significance of AR in prostate cancer has long been established, the importance of this nuclear receptor in breast cancer biology has been the subject of more recent studies [4, 13, 15, 16, 23, 40, 41]. In this respect, emerging data suggest that there are functional cross-talks between AR and some of the key signaling pathways in breast cancer including ErbB2, ERK-CREB1, PTEN, and WNT7B-ErbB3 [13, 15, 16, 41]. In addition, there is evidence for a close transcriptional cooperation between AR and FOXA1 in molecular apocrine cells [22]. Despite these findings, due to the fact that breast cancer is highly heterogenous, a comprehensive assessment of various molecular subtypes of this disease is required to elucidate the overall function of AR in breast cancer. In addition, AR target genes that have a key role in the AR signaling pathway have remained largely unknown. In this study, a transcriptional network of AR co-expressed genes was identified using a cohort of 52 breast cancer cell lines. It is notable that we have previously carried out a similar approach to discover a gene-signature for PIP expression that highly correlated with the cellular functions of this gene [19]. Importantly, the identification of this AR-network provides the opportunity to investigate AR-associated cellular functions and novel targets of this gene in breast cancer.
The functional annotation of AR co-expressed genes showed a significant enrichment for the process of cell cycle including mitosis (Figure 1, Supplementary File 2). This is in agreement with the studies that demonstrated AR modulation using either ligand-mediated activation or blockage of this receptor with specific inhibitors would significantly alter proliferation of breast cancer cells [4, 13, 16, 18, 23]. In addition, these findings are compatible with a suggested role for AR in cell cycle progression of prostate cancer cells [42–44]. Interestingly, functional annotations revealed an overlap between some of the cell cycle functions associated with the AR co-expressed genes and those of PIP-signature. In particular, both AR and PIP co-expressed genes had significant enrichments for some of the key mitotic transition genes including BUB1, CDC20, and TTK (Supplementary File 2), [19]. Moreover, PIP itself is co-expressed with AR and is an established AR target gene with a significant function in cell cycle progression (Supplementary File 1, [17, 19, 21]). Therefore, we can conclude that AR and PIP may be involved in similar cell cycle pathways in breast cancer.
Another group of GO functions that showed a significant level of enrichment among the AR co-expressed genes belonged to cellular metabolism of glucose, protein, and nucleoside as well as oxygen homeostasis (Figure 1). These findings are intriguing since there is a growing appreciation for the fact that metabolic signals are integrated and coupled to cell cycle progression [45]. Therefore, the combined enrichments for cell cycle and metabolic functions would suggest that AR may play a role in the metabolic regulation of cell cycle in breast cancer cells. Moreover, the association of AR-network with the metabolic pathways in breast cancer has a similar pattern to the findings of a genomic study on LNCaP cell line that showed AR is involved in the regulation of metabolism and biosynthesis in prostate cancer cells [44], indicating that this may represent a preserved AR function across different types of cancer. Altogether, functional annotations of AR co-expressed genes suggest that cell cycle and metabolism are the main cellular functions associated with AR expression in breast cancer.
In the AR-transcriptional network, a subset of 35 genes showed a highly correlated expression pattern with AR across the dataset; “AR gene-signature” (Table 1). The set of genes with the strongest positive correlation with AR included F7 as well as a group of transcriptional regulators namely PATZ1, NFATC4, and SPDEF that have been associated with AR but their function in breast cancer remains largely unknown. Among these PATZ is a coregulator of AR that attenuates the RNF4-mediated enhancement of AR-dependent transcription in prostate cancer cells [46]. Moreover, PATZ, RNF4 and AR belong to the same protein complex in prostate cancer cell line LNCaP [46]. In view of PATZ co-regulatory function with AR and the fact that our data does not show a direct induction of PATZ1 expression by AR activation, it is likely that the observed co-expression between AR and PATZ1 is due to a common transcriptional regulatory mechanism for these genes in breast cancer.
Another AR-signature gene with transcriptional activity is NFATC4, which is a member of nuclear factors of activated T cells DNA-binding transcription complex (NFAT) [47]. NFAT is a transcription factor family that has been associated with the expression of constitutively active AR variants in prostate cancer and links these variants to cell proliferation [48]. Furthermore, SPDEF encodes a protein that belongs to the ETS family of transcription factors, which directly interacts with the DNA binding domain of AR and enhances androgen-mediated activation of the prostate-specific antigen promoter in prostate cancer cells [49]. Moreover, the most negatively correlated AR-signature gene is PRNP that encodes a prion protein, which is involved in the pathogenesis of several neurodegenerative disorders as well as regulation of apoptosis [50, 51]. Therefore, the AR gene-signature contains several transcriptional regulators that have been associated with AR in prostate cancer and their role in the biology of breast cancer deserves further investigation.
It is notable that F7 had the strongest positive correlation with AR expression in the dataset and clustered at the closest proximity to AR in hierarchical clustering of the gene-signature (Figure 2). F7 encodes coagulation FVII protein that is constitutively produced in various cancer cells and after binding to TF on the cell surface forms TF/FVIIa complex, which in turn activates the extrinsic coagulation cascade mediated through FX [52, 53]. Published data suggest that the ectopic expression of FVII by cancer cells is involved in the pathobiology of thromboembolic events in malignancies and promotes cancer cell proliferation, invasion, and migration through the activation of protease-activated receptor-2 (PAR2) signaling [52–56]. Importantly, this ectopic production of FVII would circumvent the requirement for FVII from the blood circulation for TF/VIIa/PAR2 signaling [54], and therefore has a key function in the biology of cancer cells.
Mechanisms involved in the ectopic expression of FVII by malignancies including breast cancer are poorly understood. Although, it is known that binding of transcription factor HNF-4 is crucial for F7 gene activation in hepatocytes, HNF-4 is not responsible for F7 expression in breast cancer cells [57]. On the other hand, Sp1 binding to the F7 promoter region is essential for the constitutive activation of this gene in breast cancer and epigenetics regulation by histone acetyltransferases p300 and CBP may also be involved in the process of ectopic FVII expression [57]. In this study, gene expression and IHC data demonstrated that FVII and AR are highly co-expressed at both transcript and protein levels in breast cancer. Importantly, these findings are explained by the fact that F7 is a novel AR target gene as evident from the induction of FVII transcription and protein expression by AR activation in breast cancer cells in addition to the direct binding of AR to a proximal region of F7 promoter (Figure 4). Furthermore, the absence of secreted FVII in conditioned media of T-47D cells (Figure 4D) is in agreement with a previous study that showed unlike hepatocytes, FVII is not secreted into culture media of cancer cells [57], suggesting that ectopically synthesized FVII is involved in TF/FVIIa complex formation on the surface of cancer cells in an autocrine manner.
Moreover, AR activation induces endogenous FVII activity in breast cancer cells indicating that the AR-mediated transcriptional activation of F7 has functional implications by increasing the conversion of FX to FXa. Therefore, we can propose a model for the regulation of ectopic FVII expression by AR in breast cancer (Figure 5C). In this process, AR induces FVII expression and leads to an increased activity of FVIIa/TF complex in breast cancer cells, which in turn activates FX to FXa conversion (Figure 5C). Importantly, this activation of coagulation FVII by AR provides a novel mechanism for the transcriptional regulation of ectopic FVII expression in cancer cells. In addition, this model implicates a potential role for AR signaling in the pathobiology of thromboembolic events and the regulation of FVII/TF signaling pathway in breast cancer.
Conclusions
This study has identified a transcriptional network for AR co-expressed genes in breast cancer that is significantly enriched for cell cycle and metabolic functions. In this network, a set of 35 “AR-signature” genes were highly co-expressed with AR that included several transcriptional regulators. Furthermore, F7 demonstrated the strongest positive correlation with AR expression in the genomic dataset and FVII protein level was significantly associated with that of AR in a cohort of breast tumors. These findings were explained by demonstrating that F7 is a novel AR target gene in breast cancer. Moreover, this study suggests that AR activation leads to the induction of FVII activity in FVII/TF complex. Therefore, we can conclude that AR regulates the ectopic expression and activity of FVII in breast cancer.
Supplementary Material
A network of AR co-expressed genes was identified in breast cancer.
AR co-expressed genes are enriched for cell cycle and metabolic functions.
Factor VII is strongly co-expressed with AR at transcriptional and protein levels.
Gene encoding factor VII is a novel AR target gene.
AR induces factor VII activity to convert factor X to Xa.
Acknowledgements
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA086862.
Abbreviations
- FVII
coagulation factor VII protein
- F7
gene encoding coagulation factor VII
- AR
androgen receptor
- ER
estrogen receptor
- PIP
Prolactin-Induced Protein
- TF
Tissue Factor
- FX
factor X
- FXa
activated factor X
- DHT
dihydrotestosterone
- ERK
extracellular signal-regulated kinase
- PAR2
protease-activated receptor-2
- qRT-PCR
quantitative real time-polymerase chain reaction
- ChIP
chromatin immunoprecipitation
- IHC
immunohistochemistry
- TMA
tissue microarray
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- GO
Gene Ontology
- CC
correlation coefficient
- ROC
receiver operating characteristic
- AUC
area under the curve
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author has no conflicts of interest to disclose.
Reference
- 1.Sorlie T, Perou CM, Tibshirani R, Asas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffery SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Eystein Lonning P, Borresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sorlie T, Tibshirani R, Parker J, Hastie T, Marron JS, Nobel A, Deng S, Johnsen H, Pesich R, Geisler S, Demeter J, Perou CM, Lonning PE, Brown PO, Borresen-Dale AL, Botstein D. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farmer P, Bonnefoi H, Becette V, Tubiana-Hulin M, Fumoleau P, Larsimont D, Macgrogan G, Bergh J, Cameron D, Goldstein D, Duss S, Nicoulaz AL, Brisken C, Ficke M, Delorenzi M, Iggo R. Identification of molecular apocrine breast tumours by microarray analysis. Oncogene. 2005;24:4660–4671. doi: 10.1038/sj.onc.1208561. [DOI] [PubMed] [Google Scholar]
- 4.Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, Gerald WL. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25:3994–4008. doi: 10.1038/sj.onc.1209415. [DOI] [PubMed] [Google Scholar]
- 5.Teschendorff AE, Naderi A, Barbosa-Morais NL, Pinder SE, Ellis IO, Aparicio S, Brenton JD, Caldas C. A consensus prognostic gene expression classifier for ER positive breast cancer. Genome Biol. 2006;7:R101. doi: 10.1186/gb-2006-7-10-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guedj M, Marisa L, de Reynies A, Orsetti B, Schiappa R, Bibeau F, MacGrogan G, Lerebours F, Finetti P, Longy M, Bertheau P, Bertrand F, Bonnet F, Martin AL, Feugeas JP, Bieche I, Lehmann-Che J, Lidereau R, Birnbaum D, Bertucci F, de The H, Theillet C. A refined molecular taxonomy of breast cancer. Oncogene. 2012;31:1196–1206. doi: 10.1038/onc.2011.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S, Koo J, Park HS, Kim JH, Choi SY, Lee JH, Park BW, Lee KS. Expression of androgen receptors in primary breast cancer. Ann Oncol. 2010;21:488–492. doi: 10.1093/annonc/mdp510. [DOI] [PubMed] [Google Scholar]
- 9.Collins LC, Cole KS, Marotti JD, Hu R, Schnitt SJ, Tamimi RM. Androgen receptor expression in breast cancer in relation to molecular phenotype: results from the Nurses' Health Study. Mod Pathol. 2011;24:924–931. doi: 10.1038/modpathol.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niemeier LA, Dabbas DJ, Beriwal S, Striebel JM, Bhargava R. Androgen receptor in breast cancer: expression in estrogen receptor-positive tumors and in estrogen receptor-negative tumors with apocrine differentiation. Mod Pathol. 2009;23:205–212. doi: 10.1038/modpathol.2009.159. [DOI] [PubMed] [Google Scholar]
- 11.Wagner RK, Gorlich L, Jungblut PW. Dihydrotestosterone receptor in human mammary cancer. Acta endocrinologica. Supplementum. 1973;173:65. [PubMed] [Google Scholar]
- 12.Allegra JC, Lippman ME, Thompson EB, Simon R, Barlock A, Green L, Huff KK, Do HM, Aitken SC. Distribution, frequency, and quantitative analysis of estrogen, progesterone, androgen, and glucocorticoid receptors in human breast cancer. Cancer Res. 1979;39:1447–1454. [PubMed] [Google Scholar]
- 13.Naderi A, Hughes-Davies L. A functionally significant cross-talk between androgen receptor and ErbB2 pathways in estrogen receptor negative breast cancer. Neoplasia. 2008;10:542–548. doi: 10.1593/neo.08274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naderi A, Meyer M. Prolactin-induced protein mediates cell invasion and regulates integrin signaling in estrogen receptor-negative breast cancer. Breast Cancer Res. 2012;14:R111. doi: 10.1186/bcr3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chia KM, Liu J, Francis GD, Naderi A. A feedback loop between androgen receptor and ERK signaling in estrogen receptor-negative breast cancer. Neoplasia. 2011;13:154–166. doi: 10.1593/neo.101324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni M, Chen Y, Lim E, Wimberly H, Bailey ST, Imai Y, Rimm DL, Liu XS, Brown M. Targeting androgen receptor in estrogen receptor-negative breast cancer. Cancer Cell. 2011;20:119–131. doi: 10.1016/j.ccr.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baniwal SK, Little GH, Chimge NO, Frenkel B. Runx2 controls a feed-forward loop between androgen and prolactin-induced protein (PIP) in stimulating T47D cell proliferation. J Cell Physiol. 2012;227:2276–2282. doi: 10.1002/jcp.22966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naderi A, Liu J. Inhibition of androgen receptor and Cdc25A phosphatase as a combination targeted therapy in molecular apocrine breast cancer. Cancer Lett. 2010;298:74–87. doi: 10.1016/j.canlet.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Naderi A, Vanneste M. Prolactin-induced protein is required for cell cycle progression in breast cancer. Neoplasia. 2014;16:329–342. doi: 10.1016/j.neo.2014.04.001. e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baniwal SK, Chimge NO, Jordan VC, Tripathy D, Frenkel B. Prolactin-induced protein (PIP) regulates proliferation of luminal A type breast cancer cells in an estrogen-independent manner. PLoS One. 2013;8:e62361. doi: 10.1371/journal.pone.0062361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carsol JL, Gingras S, Simard J. Synergistic action of prolactin (PRL) and androgen on PRL-inducible protein gene expression in human breast cancer cells: a unique model for functional cooperation between signal transducer and activator of transcription-5 and androgen receptor. Mol Endocrinol. 2002;16:1696–1710. doi: 10.1210/mend.16.7.0875. [DOI] [PubMed] [Google Scholar]
- 22.Robinson JL, Macarthur S, Ross-Innes CS, Tilley WD, Neal DE, Mills IG, Carroll JS. Androgen receptor driven transcription in molecular apocrine breast cancer is mediated by FoxA1. Embo J. 2011;30:3019–3027. doi: 10.1038/emboj.2011.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naderi A, Chia KM, Liu J. Synergy between inhibitors of androgen receptor and MEK has therapeutic implications in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13:R36. doi: 10.1186/bcr2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cochrane DR, Bernales S, Jacobsen BM, Cittelly DM, Howe EN, NC DA, Spoelstra NS, Edgerton SM, Jean A, Guerrero J, Gomez F, Medicherla S, Alfaro IE, McCullagh E, Jedlicka P, Torkko KC, Thor AD, Elias AD, Protter AA, Richer JK. Role of the Androgen Receptor in Breast Cancer and Preclinical Analysis of Enzalutamide. Breast Cancer Res. 2014;16:R7. doi: 10.1186/bcr3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gucalp A, Tolaney S, Isakoff SJ, Ingle JN, Liu MC, Carey LA, Blackwell K, Rugo H, Nabell L, Forero A, Stearns V, Doane AS, Danso M, Moynahan ME, Momen LF, Gonzalez JM, Akhtar A, Giri DD, Patil S, Feigin KN, Hudis CA, Traina, C TA. Translational Breast Cancer Research, Phase II trial of bicalutamide in patients with androgen receptor-positive, estrogen receptor-negative metastatic Breast Cancer. Clin Cancer Res. 2013;19:5505–5512. doi: 10.1158/1078-0432.CCR-12-3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray KA, Daugherty LC, Gordon SM, Seal RL, Wright MW, Bruford EA. Genenames.org: the HGNC resources in 2013. Nucleic Acids Res. 2013;41:D545–D552. doi: 10.1093/nar/gks1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Messeguer X, Escudero R, Farre D, Nunez O, Martinez J, Alba MM. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. 2002;18:333–334. doi: 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 31.Farre D, Roset R, Huerta M, Adsuara JE, Rosello L, Alba MM, Messeguer X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003;31:3651–3653. doi: 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naderi A, Liu J, Francis GD. A feedback loop between BEX2 and ErbB2 mediated by c-Jun signaling in breast cancer. Int J Cancer. 2012;130:71–82. doi: 10.1002/ijc.25977. [DOI] [PubMed] [Google Scholar]
- 33.Naderi A, Meyer M, Dowhan DH. Cross-Regulation between FOXA1 and ErbB2 signaling in estrogen receptor-negative breast cancer. Neoplasia. 2012;14:283–296. doi: 10.1593/neo.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borre M, Nerstrom B, Overgaard J. Association between immunohistochemical expression of vascular endothelial growth factor (VEGF), VEGF-expressing neuroendocrine-differentiated tumor cells, and outcome in prostate cancer patients subjected to watchful waiting. Clin Cancer Res. 2000;6:1882–1890. [PubMed] [Google Scholar]
- 35.Leotoing L, Manin M, Monte D, Baron S, Communal Y, Lours C, Veyssiere G, Morel L, Beaudoin C. Crosstalk between androgen receptor and epidermal growth factor receptor-signalling pathways: a molecular switch for epithelial cell differentiation. J Mol Endocrinol. 2007;39:151–162. doi: 10.1677/JME-07-0021. [DOI] [PubMed] [Google Scholar]
- 36.Tan PY, Chang CW, Chng KR, Wansa KD, Sung WK, Cheung E. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol Cell Biol. 2012;32:399–414. doi: 10.1128/MCB.05958-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gamble SC, Odontiadis M, Waxman J, Westbrook JA, Dunn MJ, Wait R, Lam EW, Bevan CL. Androgens target prohibitin to regulate proliferation of prostate cancer cells. Oncogene. 2004;23:2996–3004. doi: 10.1038/sj.onc.1207444. [DOI] [PubMed] [Google Scholar]
- 38.Obinata D, Takayama K, Urano T, Murata T, Ikeda K, Horie-Inoue K, Ouchi Y, Takahashi S, Inoue S. ARFGAP3, an androgen target gene, promotes prostate cancer cell proliferation and migration. Int J Cancer. 2012;130:2240–2248. doi: 10.1002/ijc.26224. [DOI] [PubMed] [Google Scholar]
- 39.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y, Pietenpol JA. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Culig Z, Klocker H, Bartsch G, Hobisch A. Androgen receptors in prostate cancer. Endocr Relat Cancer. 2002;9:155–170. doi: 10.1677/erc.0.0090155. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y, Romigh T, He X, Tan MH, Orloff MS, Silverman RH, Heston WD, Eng C. Differential regulation of PTEN expression by androgen receptor in prostate and breast cancers. Oncogene. 2011;30:4327–4338. doi: 10.1038/onc.2011.144. [DOI] [PubMed] [Google Scholar]
- 42.Schiewer MJ, Augello MA, Knudsen KE. The AR dependent cell cycle: mechanisms and cancer relevance. Mol Cell Endocrinol. 2012;352:34–45. doi: 10.1016/j.mce.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarkar S, Brautigan DL, Parsons SJ, Larner JM. Androgen receptor degradation by the E3 ligase CHIP modulates mitotic arrest in prostate cancer cells. Oncogene. 2014;33:26–33. doi: 10.1038/onc.2012.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Massie CE, Lynch A, Ramos-Montoya A, Boren J, Stark R, Fazli L, Warren A, Scott H, Madhu B, Sharma N, Bon H, Zecchini V, Smith DM, Denicola GM, Mathews N, Osborne M, Hadfield J, Macarthur S, Adryan B, Lyons SK, Brindle KM, Griffiths J, Gleave ME, Rennie PS, Neal DE, Mills IG. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. Embo J. 2011;30:2719–2733. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee IH, Finkel T. Metabolic regulation of the cell cycle. Current opinion in cell biology. 2013;25:724–729. doi: 10.1016/j.ceb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pero R, Lembo F, Palmieri EA, Vitiello C, Fedele M, Fusco A, Bruni CB, Chiariotti L. PATZ attenuates the RNF4-mediated enhancement of androgen receptor-dependent transcription. J Biol Chem. 2002;277:3280–3285. doi: 10.1074/jbc.M109491200. [DOI] [PubMed] [Google Scholar]
- 47.Hoey T, Sun YL, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 48.Lapouge G, Marcias G, Erdmann E, Kessler P, Cruchant M, Serra S, Bergerat JP, Ceraline J. Specific properties of a C-terminal truncated androgen receptor detected in hormone refractory prostate cancer. Adv Exp Med Biol. 2008;617:529–534. doi: 10.1007/978-0-387-69080-3_53. [DOI] [PubMed] [Google Scholar]
- 49.Oettgen P, Finger E, Sun Z, Akbarali Y, Thamrongsak U, Boltax J, Grall F, Dube A, Weiss A, Brown L, Quinn G, Kas K, Endress G, Kunsch C, Libermann TA. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem. 2000;275:1216–1225. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]
- 50.Sander P, Hamann H, Drogemuller C, Kashkevich K, Schiebel K, Leeb T. Bovine prion protein gene (PRNP) promoter polymorphisms modulate PRNP expression and may be responsible for differences in bovine spongiform encephalopathy susceptibility. J Biol Chem. 2005;280:37408–37414. doi: 10.1074/jbc.M506361200. [DOI] [PubMed] [Google Scholar]
- 51.Paitel E, Alves da Costa C, Vilette D, Grassi J, Checler F. Overexpression of PrPc triggers caspase 3 activation: potentiation by proteasome inhibitors and blockade by anti-PrP antibodies. J Neurochem. 2002;83:1208–1214. doi: 10.1046/j.1471-4159.2002.01234.x. [DOI] [PubMed] [Google Scholar]
- 52.Koizume S, Jin MS, Miyagi E, Hirahara F, Nakamura Y, Piao JH, Asai A, Yoshida A, Tsuchiya E, Ruf W, Miyagi Y. Activation of cancer cell migration and invasion by ectopic synthesis of coagulation factor VII. Cancer Res. 2006;66:9453–9460. doi: 10.1158/0008-5472.CAN-06-1803. [DOI] [PubMed] [Google Scholar]
- 53.Yokota N, Koizume S, Miyagi E, Hirahara F, Nakamura Y, Kikuchi K, Ruf W, Sakuma Y, Tsuchiya E, Miyagi Y. Self-production of tissue factor-coagulation factor VII complex by ovarian cancer cells. Br J Cancer. 2009;101:2023–2029. doi: 10.1038/sj.bjc.6605406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van den Berg YW, Osanto S, Reitsma PH, Versteeg HH. The relationship between tissue factor and cancer progression: insights from bench and bedside. Blood. 2012;119:924–932. doi: 10.1182/blood-2011-06-317685. [DOI] [PubMed] [Google Scholar]
- 55.Hjortoe GM, Petersen LC, Albrektsen T, Sorensen BB, Norby PL, Mandal SK, Pendurthi UR, Rao LV. Tissue factor-factor VIIa-specific up-regulation of IL-8 expression in MDA-MB-231 cells is mediated by PAR-2 and results in increased cell migration. Blood. 2004;103:3029–3037. doi: 10.1182/blood-2003-10-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fan L, Yotov WV, Zhu T, Esmailzadeh L, Joyal JS, Sennlaub F, Heveker N, Chemtob S, Rivard GE. Tissue factor enhances protease-activated receptor-2-mediated factor VIIa cell proliferative properties. Journal of thrombosis and haemostasis : JTH. 2005;3:1056–1063. doi: 10.1111/j.1538-7836.2005.01250.x. [DOI] [PubMed] [Google Scholar]
- 57.Koizume S, Yokota N, Miyagi E, Hirahara F, Nakamura Y, Sakuma Y, Yoshida A, Kameda Y, Tsuchiya E, Ruf W, Miyagi Y. Hepatocyte nuclear factor-4-independent synthesis of coagulation factor VII in breast cancer cells and its inhibition by targeting selective histone acetyltransferases. Mol Cancer Res. 2009;7:1928–1936. doi: 10.1158/1541-7786.MCR-09-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.