Abstract
Simian Immunodeficiency Viruses (SIVs) have been discovered in over 45 primate species; however, the pathogenic potential of most SIV strains remains unknown due to difficulties inherent in observing wild populations. Because those SIV infections that are pathogenic have been shown to induce changes in the host's gut microbiome, monitoring the microbiota present in fecal samples can provide a noninvasive means for studying the effects of SIV infection on the health of wild-living primates. Here, we examine the effects of SIVgor, a close relative of SIVcpz of chimpanzees and HIV-1 of humans, on the gut bacterial communities residing within wild gorillas, revealing that gorilla gut microbiomes are exceptionally robust to SIV infection. In contrast to the microbiomes of HIV-1 infected humans and SIVcpz-infected chimpanzees, SIVgor-infected gorilla microbiomes exhibit neither rises in the frequencies of opportunistic pathogens nor elevated rates of microbial turnover within individual hosts. Regardless of SIV infection status, gorilla microbiomes assort into enterotypes, one of which is compositionally analogous to those identified in humans and chimpanzees. The other gorilla enterotype appears specialized for a leaf-based diet and is enriched in environmentally derived bacterial genera. We hypothesize that the acquisition of this gorilla-specific enterotype was enabled by lowered immune-system control over the composition of the microbiome. Our results indicate differences between the pathology of SIVgor and SIVcpz/HIV-1 infections, demonstrating the utility of investigating host microbial ecology as a means for studying disease in wild primates of high conservation priority.
Introduction
The evolutionary history of Simian Immunodeficiency Virus (SIV) is characterized by numerous cases of host switching among the more than 45 primate species in which this lentivirus has been detected (Klatt et al., 2012). Infections can be either pathogenic or non-pathogenic depending on the host species and viral strain, and cross-species transmission of non-pathogenic SIVs can give rise to pathogenic strains in recipient hosts (Hahn and Sharp, 2010). However, in most cases, the health effects of naturally occurring SIV infections remain unknown due to the difficulties inherent in monitoring wild primate populations.
Within the great apes, SIV infections first appeared when SIVrcm from red-capped mangabeys (Cercocebus torquatus) and SIVgsb/mus/mon from greater spot-nosed, moustached or mona monkeys (Cercopithecus spp.) recombined to form SIVcpz (Bailes et al., 2003). That chimpanzee strain was subsequently transferred to humans on at least two separate occasions, developing into HIV-1 group N and group M (Gao et al., 1999), the latter being the etiological agent of the global AIDS epidemic (Keele et al., 2006). In a separate cross-species jump, SIVcpz was transferred to gorillas, generating SIVgor, which subsequently moved to humans, producing the HIV-1 group P (Takehisa et al., 2009), and possibly group O. Although closely related to both HIV-1 and SIVcpz, it is not known whether SIVgor is pathogenic and produces AIDS-like symptoms in its natural gorilla host.
A primary site of SIV/HIV replication is the gut, which contains nearly 70% of the body's T-cell-producing lymphoid tissue (Marchetti, 2013). In healthy individuals, this immune system regulates the gut microbiome (Kau et al., 2011), the largest reservoir of bacteria in the body (Turnbaugh et al., 2007). When hosts become immunocompromised, as in pathogenic SIV/HIV infections, they experience substantial alterations in the compositions of their gut microbial communities (Gori, 2008; Handley et al., 2012; Moeller et al., 2013b; Lozupone et al., 2013; Vujkovic-Cvijin et al., 2013; Mutlu, 2014; McHardy, 2013; Dillon, 2014) and often display gastrointestinal disorders, including colitis and microbial translocation from the gut to the bloodstream (Brenchley, 2006; Mentec et al., 1994). Chimpanzees infected with SIVcpz display increases in the prevalence of bacterial genera associated with opportunistic infections as well as augmented rates of change in gut microbiota composition over time (Moeller et al., 2013b). Similarly, HIV-positive humans display changes in the bacterial constituents of their gut microbiomes (Vujkovic-Cvijin et al., 2013; Lozupone et al., 2013; Mutlu, 2014; McHardy, 2013; Gori, 2008; Dillon, 2014), and rhesus macaques experimentally infected with a pathogenic variant of SIV display expanded diversity within their enteric viromes (Handley et al., 2012).
Because pathogenic SIV infections induce alterations in gut microbial communities, assaying gut microbial communities can provide a means for monitoring SIV pathogenicity in wild host populations. In particular, characterization of the microbiota present in fecal samples can allow detection of pathology even in the absence of direct or continual contact with hosts. Here, we examine the gut microbiota of SIVgor-infected and uninfected gorillas and find, in contrast to studies of chimpanzees and humans, no changes in the gut microbiota associated with infection status. Thus, the pathological effects of SIVcpz and HIV-1 on gut microbiomes appear to be absent in gorillas infected with the closely related SIVgor.
Methods
Sample preparation
Fecal samples (n=186) were collected from field sites in southern Cameroon and preserved at -80°C in RNAlater (Ambion) (Neel et al., 2010; Etienne et al., 2012). Samples were considered as infected with SIVgor when cross-reactive antibodies were detected after dialysis during the INNO-LIA HIV score confirmation test (Innogenetics, Ghent, Belgium) and/or when SIVgor fragments were detected after RT-PCR reactions on fecal RNA using SIVgor and SIVgor/SIVcpz/HIV-1 consensus primers in env (gp41 ectodomain, 315 or 440 bp) and/or pol (245 or 330 bp) as previously reported (Neel et al., 2010; Etienne et al., 2012). To identify individuals, samples were genotyped at seven loci in two multiplex PCRs (D18s536, D4s243, D10s676, D9s922 and D2s1326, D2s1333, D4s1627). Homozygous loci were amplified from three to seven times to minimize allelic dropout, and when multiplex PCR reactions yielded poor results, fecal DNA was extracted again and a new set of PCRs was performed as previously reported (Navidi et al., 1992; Taberlet et al., 1996; Bonin, et al., 2004; Etienne et al., 2012). Total DNA from 50 μl aliquots of thawed fecal material was extracted by a bead-beating procedure described in Goodman et al. (2011). PCR amplifications of the 16S rDNA V4 region bounded by primers 515F and 806R were performed as in Caporaso et al. (2011). Amplicons were sequenced on an Illumina MiSeq at the GSAF, UT Austin following methods outlined by Degnan and Ochman (2012).
Sequence filtering
Reads were processed in QIIME (Caporaso et al., 2010). FASTQ files were filtered for quality with split_libraries.py using default settings. OTUs at 97% identity were chosen through the uclust algorithm as implemented in pick_otus.py and taxonomically assigned with the RDP classifier using a confidence threshold of 0.80. To eliminate the effect of sequencing errors and to focus our analyses on established residents of the gut microbiome, any OTU represented by fewer than 50 reads was removed, following the methods of Moeller et al. (2013a). The resulting dataset was rarefied to an even depth of 10,000 sequences per sample. Two samples were excluded from subsequent analyses due to insufficient sampling depth.
Statistical analyses
Beta-diversity distances and dissimilarities (weighted and unweighted UniFrac, Bray Curtis, and Euclidean) were calculated for all pairwise comparisons of samples, and alpha diversity (Chao1 and total species counts) was calculated for all samples in QIIME. For analyses assessing compositional differences between SIV-positive and SIV-negative samples, only the 163 samples for which SIV status was determined and for which greater than 10,000 reads were acquired were included. Two-tailed t-tests were performed to identify OTUs and genera over- or under-represented within SIV-positive microbiomes relative to SIV-negative microbiomes, and a Bonferroni-corrected p-value threshold of 0.01 was applied. Two-tailed t-tests of beta-diversity measures were used to assess whether SIV-positive samples were more divergent from SIV-negative samples than SIV-negative samples were from one another and to test whether SIV-positive samples were more divergent from SIV-positive samples than SIV-negative samples were from one another. Two-tailed t-tests of beta-diversity measures were also used to evaluate whether the microbiomes of gorillas from the same field site within Cameroon were more similar than those of gorillas from different field sites.
To determine whether the time since infection was associated with the composition of the gut microbiota, we applied a two-tailed t-test to evaluate whether SIV-positive samples collected at least one year after the acquisition of SIV infection were more divergent from one another than SIV-negative samples were from one another. A one-tailed t-test was used to test whether consecutive SIV-positive samples from longitudinally sampled individuals were more divergent than consecutive SIV-negative samples. To determine the influence of time of sampling on the composition of the gut microbiome, two-tailed t-tests of beta-diversity measures were applied to assess whether samples collected on the same day or in the same year were more similar to one another than were samples collected on different days or in different years, respectively.
Enterotype clustering was performed in R using previously described methodologies (Arumugam et al., 2011). The dataset was partitioned into a specified number of clusters (1-15) through PAM clustering, and the Calinski-Harabasz index was calculated for each clustering scheme to determine which scheme best fit the data. Fisher's exact tests were used to evaluate whether any particular enterotype was overrepresented within SIV-positive or SIV-negative samples and to identify OTUs overrepresented within certain clusters. Only significant differences based on Bonferroni-corrected p-value threshold of 0.01 are reported.
Results
SIVgor-positive and SIVgor-negative gut microbiota are indistinguishable
We sequenced the V4 region of the 16S rDNAs present in 69 fecal samples from SIV-positive and 96 fecal samples from SIV-negative western lowland gorillas (Gorilla gorilla gorilla) residing at four locations in southern Cameroon. We recovered 3,666,475 high-quality reads, which were subsampled to 10,000 reads per sample, yielding a dataset spanning 67 SIV-positive and 96 SIV-negative samples. The mapping file containing metadata used in the analyses is presented in Table S1.
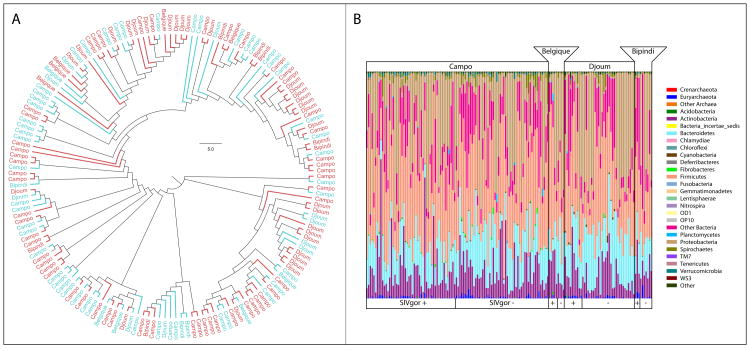
We calculated beta-diversity metrics and similarity indices to test whether the microbiota in SIVgor-positive and SIVgor-negative samples differed compositionally. Taken as a whole, SIV-positive and SIV-negative microbiomes were indistinguishable from one another: SIV-positive microbiomes were as compositionally similar to SIV-negative microbiomes as SIV-negative microbiomes were to other SIV-negative microbiomes (t-test, p > 0.6). Time since SIV infection did not appear to influence the composition of the gut microbiome, as SIV-positive samples collected a year or more after initial infection remained compositionally indistinguishable from SIV-negative samples. A plot of all samples against the first two principal coordinates of pairwise Bray-Curtis dissimilarities shows no clear differentiation between the compositions of SIV-positive and SIV-negative samples, nor among the compositions of samples from different collection sites within Cameroon, although there is substantial variation in the gut communities in the population at large (Figure 1, Figure 2, Figure S2).
Figure 1. Variation in the gut microbiome among gorillas is not associated with SIVgor-infection status or geography.
A. Cladogram based on Euclidean distances among the OTU-level compositional profiles of gorilla microbiomes. Tips of the cladogram are labeled to indicate the field site from which each sample was collected, with SIVgor-negative and SIVgor-positive samples shown in blue and red, respectively. B. Stacked colored bars showing the relative abundances of bacterial phyla within samples. Colors correspond to bacterial phyla, and each bar represents the microbiome recovered from a single sample. Samples are grouped according to field site and SIVgor-infection status.
Figure 2. Gorillas maintain enterotypes that are robust SIVgor infection.
A. Principal coordinates plot of the Bray-Curtis dissimilarities among samples reveal almost complete overlap and no significant compositional differences between SIV-positive (red circles) and SIV-negative (blue triangles) gut microbiomes. X-axis explains 28% of the variation in the dataset and reflects a tradeoff between the abundances of environmentally derived bacteria, such as Empedobacter, and common great-ape gut symbionts, such as Prevotella. Note that both SIV-positive and SIV-negative samples populate all quadrants of the plot. B. Histogram of the distribution of samples across the first principal coordinate. The bimodal distribution reflects enterotype clustering.
We also tested if particular OTUs were differentially represented in SIVgor-positive and SIVgor-negative cohorts and found that none was significantly associated with either infection status. Moreover, we observed no differences in alpha diversity between SIVgor-positive and SIVgor-negative samples, contrasting the situation reported for HIV-infected humans (Lozupone et al., 2013; Mutlu, 2014; McHardy, 2013; Gori, 2008; Dillon, 2014).
Gut microbiota of individual hosts do not display instability after SIVgor infection
In chimpanzees, SIV-infected hosts show accelerated rates of change in the composition of their gut microbiomes (Moeller et al., 2013a), so we searched for similar changes in gorillas by calculating pairwise beta-diversity distances and dissimilarities between consecutive samples from 19 individuals sampled either before or after SIVgor infection and 2 individuals sampled both before and after SIVgor infection. The analysis revealed no differences in the rate of divergence between consecutive SIVgor-positive and consecutive SIVgor-negative samples (Figure 3), showing that, unlike the situation in chimpanzees, SIV infections in gorillas do not influence the rate at which the composition of the gut microbiome changes.
Figure 3. Rates of change in the composition of gorilla gut microbiota are unaffected by SIVgor infection.
Shown are average pairwise Bray-Curtis dissimilarities between samples collected consecutively from individual SIV-negative and SIV-positive hosts. The dataset included 17 pairs of consecutive SIV-negative samples and 18 pairs of consecutive SIV-positive samples. Error bars represent 95% confidence intervals.YRD
Gorillas maintain characteristic gut enterotypes, even when infected by SIV
The gut microbiota of humans and chimpanzees assort into enterotypes, communities that differ in the relative abundances of specific bacterial genera (Arumugam et al., 2011; Moeller et al., 2012). We tested for the presence of gut enterotypes in gorillas of known SIVgor-infection status using methods originally described (Arumugam et al., 2011). We observed the strongest support for two clusters of gut communities based on the relative abundances of bacterial genera. (Relative abundances of the bacterial genera that differentiate enterotypes are presented in Table 1.) There was no association between enterotype and the presence of SIVgor in these samples (t-test, p > 0.5), nor between enterotype and sample site (t-test, p > 0.5). Compositionally analogous enterotypes were recovered when only SIV-negative samples or only SIV-positive samples were included in the analysis, again showing that enterotypes occur independently of SIV-infection status. Longitudinal analyses of individuals' enterotype assignments revealed that hosts can change enterotype over time: 13 of 21 hosts assayed over time changed enterotype assignment at least once during the sampling period.
Table 1.
Frequencies of bacterial taxa overrepresented within gorilla enterotypes:
| Taxa Overrepresented in Frequency in Gorilla Enterotype 1 | Frequency in Enterotype 1 | Frequency in Enterotype 2 |
|---|---|---|
| Acinetobacter | 0.1924 | 0.0383 |
| unclassified Flavobacteriaceae | 0.0400 | 0.0010 |
| Stenotrophomonas | 0.0364 | 0.0008 |
| Empedobacter | 0.0333 | 0.0008 |
| Spingobacterium | 0.0256 | 0.0002 |
| Leadbetterella | 0.0053 | 0.0001 |
| Taxa Overrepresented in Frequency in Gorilla Enterotype 2 | Frequency in Enterotype 1 | Frequency in Enterotype 2 |
|---|---|---|
| unclassified Ruminococcaceae* | 0.0049 | 0.0346 |
| Dialister* | 0.0066 | 0.0237 |
| Prevotella* | 0.0026 | 0.0218 |
Listed are bacterial genera that distinguish the two gorilla enterotypes. Taxa listed in order of relative abundance within the enterotype in which they are overrepresented.
Those taxa marked with an asterisk were previously shown to be associated with enterotype clustering in chimpanzees and/or humans (Aruguman et al., 2011, Moeller et al., 2012)
Association of microbiota with geographic and temporal factors
Previous surveys of apes living throughout equatorial Africa have shown that chimpanzees and gorilla living on opposite sides of the continent harbor microbiota that are more compositionally divergent than those of chimpanzees and gorillas residing within the same range (Moeller et al., 2013a). To examine whether small-scale geographic separation promotes compositional divergence between gorilla gut microbiomes, we tested whether the microbiomes of gorillas from the same field site in Cameroon were more similar than those of gorillas from different field sites in Cameroon (Figure 1). Based on the tested distance and dissimilarity measures, samples collected from the same field-site were not more similar to one another than were samples from different field sites (p > 0.3). Similarly, samples collected during the same year but on different days were not more similar than one another than were samples collected during different years (p > 0.1). Conversely, samples collected on the same day were more similar to one another than were samples collected on different days (p <0.01).
Discussion
Monitoring the microbial ecology of wild primate populations can provide a non-invasive means for studying the effects of diseases in these hosts. In the present study, we analyzed gorilla fecal microbiomes to remotely investigate the consequences and pathology of SIVgor infection. Unlike infections of SIVcpz in chimpanzees (Moeller et al., 2013b) and of HIV-1 in humans (Lozupone et al., 2013; Vujkovic-Cvijin et al., 2013; Mutlu, 2014; McHardy, 2013; Gori, 2008; Dillon, 2014), SIVgor did not induce changes in gut microbial communities observed in other great apes (Figure 1, Figure 2). Neither the pathogen-associated bacterial genera within SIVcpz-infected chimpanzees (Moeller et al., 2013b) nor those bacterial taxa associated with inflammatory bowel disease in HIV-1-infected humans (Png et al., 2010) bloomed to high abundances in SIVgor-infected gorillas. Moreover, in contrast to SIVcpz-infected chimpanzees, SIVgor-infected gorillas did not exhibit elevated rates of change in gut-microbiota composition within individuals (Figure 3). Thus, gorillas do not display symptoms of gastrointestinal dysfunction common to pathogenic SIV and HIV infections that have been observed in humans and chimpanzees.
It has been proposed that the changes in the gut microbiota in response to SIVcpz/HIV-1 infection contribute to the progression of AIDS by exacerbating systemic immune activation through the translocation of microbial products from the gut to the bloodstream (Brenchley, 2006). Recent work in HIV-1 infected humans has revealed associations between gut microbiome dysbiosis and markers of mucosal immune disruption, T cell activation, and chronic inflammation (Vujkovic-Cvijin et al., 2013). Thus, the absence of gut-microbiota changes in SIVgor-infected gorillas is surprising. While the possibility that gorillas do not experience the immune-system deterioration characteristic of SIVcpz and HIV-1 infection cannot be formally excluded, the close evolutionary relatedness of SIVgor and SIVcpz/HIV-1, and the physiological similarities and close evolutionary relationship among the great apes, would suggest that a decline of immune functions due to CD4 T-cell killing also occurs in SIVgor-infected gorillas. SIVmac causes AIDS in experimentally infected macaques, but several studies have shown that the bacterial component of the macaque gut microbiome is surprisingly stable even in end-stage SIVmac infection (McKenna, 2008; Handley, 2012; Brenchley, 2013). The SIVmac associated disease course is greatly accelerated compared to SIVcpz and HIV-1 infection, with animals progressing to AIDS in a significantly shorter time frame. The lack of microbial changes in the fecal samples of SIVgor infected gorillas may thus point to a similarly accelerated disease course. However, post-mortem studies of wild-living gorillas will ultimately be required to determine whether SIVgor infection causes CD4+ T-cell depletion and immunodeficiency similar to SIVcpz and HIV-1 infections.
An alternative explanation for the lack of effects of SIVgor infection on the composition of gorilla gut microbiomes is that the stability of the gorilla gut microbiome is less dependent on host immune function than is the stability of the chimpanzee or the human gut microbiome. Gorillas, like other herbivores, cultivate gut microbiota dominated by environmentally derived bacterial taxa that aid in the digestion of cellulose (Muegge et al., 2011). In contrast, human and chimpanzee microbiota are dominated by co-adapted bacteria that have evolved alongside, and are actively regulated by, their hosts immune systems (Ley et al., 2008; Ochman et al., 2010). The acquisition by gorillas and other herbivores of environmental bacteria may have been facilitated by a relaxation of immune system control over the composition of the microbiota. If the dominant gorilla gut bacteria have transitioned to a host-associated lifestyle in recent evolutionary time, their persistence within gorillas may not depend on interactions with the host immune system to the degree to which does the persistence of the dominant constituents of chimpanzee and human gut microbiomes. Under this view, SIVgor infections may cause deterioration of the immune system as do SIVcpz and HIV infections, but unlike the gut microbiomes of chimpanzees and humans, gorilla microbiomes remain compositionally stable despite the absence of normal immune-system regulation.
Each gorilla microbiome in our dataset could be assigned to one of two genus-level enterotypes, which were well-supported statistically, and not associated with SIVgor-infection or sample site (Figure 2). Although both enterotypes contained high abundances of environmentally derived bacteria, one gorilla enterotype contained relatively high abundances of Prevotella, paralleling enterotypes observed in human, chimpanzee, and mouse populations (Table 1; Arumugam et al., 2011; Moeller et al., 2012; Wang et al., 2014), and suggesting that community structures dominated by this genus arose within host populations before the diversification of the great apes. However, the second gorilla enterotype was enriched in environmentally derived bacterial genera absent from other great-ape gut microbiomes. The presence of these taxa, many of which appear to be capable of digesting complex plant polysaccharides (Muegge et al., 2011), may reflect adaptations to a folivorous lifestyle. Because the maintenance of enterotypes within host populations has been linked to dietary variation (Wang et al., 2014), our results therefore suggest dietary variation within the sampled gorilla population, in which some individuals rely more strongly on a folivorous diet. Indeed, observational studies have indicated that the diets of gorillas in Cameroon can vary alongside fluctuating food abundances (Deblauwe, 2009), and longitudinal analyses of the gut microbiomes of individual gorillas revealed that a host's enterotype can change over time, as has been previously observed in chimpanzees (Moeller et al., 2012) and mice (Wang et al., 2014).
Previous analyses of great ape microbiota have shown that hosts residing in the same range share gut microbes at higher rates than hosts residing in different ranges (Moeller et al., 2013a). Moreover, migration of chimpanzees between separated social groups induces compositional shifts in the gut microbiome away from the microbiota of the group of origin and toward the microbiota of the receiving group (Moeller and Ochman, 2013). In contrast, the compositional variation among gorilla microbiomes examined in the present study was not significantly associated with sample site (Figure 1). Our results indicate that this effect of geography on gut microbiomes is not evident when hosts share a range that enables occasional contact. Thus, small scale and temporary geographic separation among gorillas in Cameroon does not promote the compositional divergence of gut microbial communities.
Previous surveys of chimpanzee populations have observed that the gut microbiomes of co-occurring hosts experience parallel compositional shifts, such that time of sampling is strongly associated with the composition of the gut microbiome (Degnan et al., 2012a). Similarly, we observed that sampling date was strongly associated with the composition of gorilla gut microbiomes. Thus, the gut microbiomes of gorillas in Cameroon also appear to experience parallel compositional shifts, potentially driven by shared environmental factors such as dietary variation or microbial dispersal events.
In sum, SIVgor-infected gorillas do not display the gastrointestinal dysregulation characteristic of AIDS progression seen in SIVcpz-infected chimpanzees or HIV-1-infected humans. The robustness of gut microbial communities to SIVgor infection may reflect an accelerated disease course in gorillas and/or alterations in the immune-system control over the gorilla gut microbiome, which consists largely of environmentally derived bacteria that have only come to live within hosts in recent evolutionary time. These results illustrate the viability of characterizing gut microbiomes in order to remotely assess gastrointestinal symptoms of SIV in wild primate populations.
Supplementary Material
Acknowledgments
We thank the staff of project PRESICA for fieldwork in Cameroon and the Cameroonian Ministries of Health, Forestry and Wildlife, and Scientific Research and Innovation for permission to collect samples in Cameroon. We thank Kim Hammond for assistance with the preparation of figures. This work was supported by grants from the National Institutes of Health (R01 AI 091595, R37 AI 050529, R01 AI 058715 to B.H.H.; R01 GM101209 to H.O.), a National Science Foundation Graduate Research Fellowship to A.H.M., and grants 12182 and 12255 from Agence National de Recherche sur le SIDA (ANRS) to M.P.
Footnotes
Conflict of Interest. The authors declare no conflict of interest.
Supporting Information. Supporting information is available at the Molecular Ecology website.
Author Contributions. AHM performed analyses and wrote the manuscript. MP, AA, EMN, AE and BHH provided samples and reagents. HO wrote the manuscript.
Data Accessibility. All microbiome sequence data and metadata produced for this study have been deposited to NCBI SRA under accession number SRX720662 and to Dryad; DOI: 10.5061/dryad.2n6p6. Data are also available upon request from Howard Ochman at howard.ochman@austin.utexas.edu
References
- Arumugam M, Raes J, Pelletier E, et al. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailes E, Gao F, Bibollet-Ruche F, et al. Hybrid origin of SIV in chimpanzees. Science. 2003;300:1713–1713. doi: 10.1126/science.1080657. [DOI] [PubMed] [Google Scholar]
- Bonin A, Bellemain E, Bronken Eidesen P, et al. How to track and assess genotyping errors in population genetics studies. Mol Ecol. 2004;13:3261–73. doi: 10.1111/j.1365-294X.2004.02346.x. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Schacker TW, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Brenchley JM. Mucosal immunity in human and simian immunodeficiency lentivirus infections. Mucosal Immunol. 2013;6:657–665. doi: 10.1038/mi.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courgnaud V, Lauré F, Fultz PN, et al. Genetic differences accounting for evolution and pathogenicity of simian immunodeficiency virus from a sooty mangabey monkey after cross-species transmission to a pig-tailed macaque. J Virol. 1992;66:414–419. doi: 10.1128/jvi.66.1.414-419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deblauwe I. Temporal variation in insect-eating by chimpanzees and gorillas in southeast Cameroon: extension of niche differentiation. Int J Primat. 2009;30:229–252. [Google Scholar]
- Degnan PH, Pusey AE, Losdorf E, Goodall J, Wroblewski EE, Wilson ML, Rudicell RS, Hahn BH, Ochman H. Factors associated with the diversification of the gut microbial communities within chimpanzees from Gombe National Park. Proc Natl Acad Sci USA. 2012a;109:13034–13039. doi: 10.1073/pnas.1110994109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degnan PH, Ochman H. Illumina-based analysis of microbial community diversity. ISME J. 2012b;6:183–194. doi: 10.1038/ismej.2011.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SM, Lee EJ, Kotter CV, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne L, Locatelli S, Ayouba A, et al. Noninvasive follow-up of simian immunodeficiency virus infection in wild-living nonhabituated western lowland gorillas in Cameroon. J Virol. 2012;86:9760–9772. doi: 10.1128/JVI.01186-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald F, Harris K, Doyle R, et al. Short Communication: Evidence that microbial translocation occurs in HIV-infected children in the United Kingdom. AIDS Res Human Retroviruses. 2013;29:1589–1593. doi: 10.1089/aid.2013.0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Bailes E, Robertson DL, et al. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- Goodman AL, Wu M, Gordon JI. Identifying microbial fitness determinants by insertion sequencing using genome-wide transposon mutant libraries. Nat Prot. 2011;6:1969–1980. doi: 10.1038/nprot.2011.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori A, Tincati C, Rizzardini G, et al. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J Clin Microbiol. 2008;46:757–758. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadalupe M, Reay E, Sankaran S, et al. Severe CD4+ T-cell depletion in gut lymphoid tissue during primary human immunodeficiency virus type 1 infection and substantial delay in restoration following highly active antiretroviral therapy. J Virol. 2003;77:11708–11717. doi: 10.1128/JVI.77.21.11708-11717.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley SA, Thackray LB, Zhao G, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kau AL, Ahern PP, Griffin NW, et al. Human nutrition, the gut microbiome and the immune system. Nature. 2011;474:327–336. doi: 10.1038/nature10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Van Heuverswyn F, Li Y, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Silvestri G, Hirsch V. Nonpathogenic simian immunodeficiency virus infections. Cold Spring Harbor Perspectives in Medicine. 2012;2 doi: 10.1101/cshperspect.a007153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Li M, Campbell TB, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26:2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy IH, Li X, Tong M, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna P, Hoffmann C, Minkah N, et al. The macaque gut microbiome in health, lentiviral infection, and chronic enterocolitis. PLoS Pathog. 2008;4:e20. doi: 10.1371/journal.ppat.0040020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentec H, Leport C, Leport J, et al. Cytomegalovirus colitis in HIV-1-infected patients: a prospective research in 55 patients. AIDS. 1994;84:61–468. doi: 10.1097/00002030-199404000-00007. [DOI] [PubMed] [Google Scholar]
- Moeller AH, Degnan PH, Pusey AE, et al. Chimpanzees and humans harbour compositionally similar gut enterotypes. Nat Comm. 2012;3:1179. doi: 10.1038/ncomms2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Ochman H. Factors that drive variation among gut microbial communities. Gut Microbes. 2013;4:403–408. doi: 10.4161/gmic.26039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Peeters M, Ndjango JB, et al. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 2013a;23:1715–1720. doi: 10.1101/gr.154773.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller AH, Shilts M, Li Y, et al. SIV-induced instability of the chimpanzee gut microbiome. Cell Host Microbe. 2013b;14:340–345. doi: 10.1016/j.chom.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu EA, Keshavarzian A, Losurdo J, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV-infected subjects. PLoS Pathogens. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navidi W, Arnheim N, Waterman MS. A multiple-tubes approach for accurate genotyping of very small DNA samples by using PCR: statistical considerations. Am J Hum Genet. 1992;50:347–359. [PMC free article] [PubMed] [Google Scholar]
- Neel C, Etienne L, Li Y, et al. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. J Virol. 2010;84:1464–1476. doi: 10.1128/JVI.02129-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Png CW, Lindén SK, Gilshenan KS, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Amer J Gastroent. 2010;105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Hahn BH. The evolution of HIV-1 and the origin of AIDS. Philos Trans R Soc Lond B Biol Sci. 2010;365:2487–94. doi: 10.1098/rstb.2010.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberlet P, Griffin S, Goossens B, et al. Reliable genotyping of samples with very low DNA quantities using PCR. Nuc Acids Res. 1996;24:3189–3194. doi: 10.1093/nar/24.16.3189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takehisa J, Kraus MH, Ayouba A, et al. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J Virol. 2009;83:1635–1648. doi: 10.1128/JVI.02311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Hamady M, et al. The human microbiome project: exploring the microbial part of ourselves in a changing world. Nature. 2007;449:804. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandeWoude S, Apetrei C. Going wild: lessons from naturally occurring T-lymphotropic lentiviruses. Clin Microbiol Rev. 2006;19:728–762. doi: 10.1128/CMR.00009-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I, Dunham RM, Iwai S, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Science Trans Med. 2013;5:193ra91. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Linnenbrink M, Kunzel S, et al. Dietary history contributes to enterotype-like clustering and functional metagenomic content in the intestinal microbiome of wild mice. Proc Natl Acad Sci USA. 2014;111:E2703–E2710. doi: 10.1073/pnas.1402342111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.