Abstract
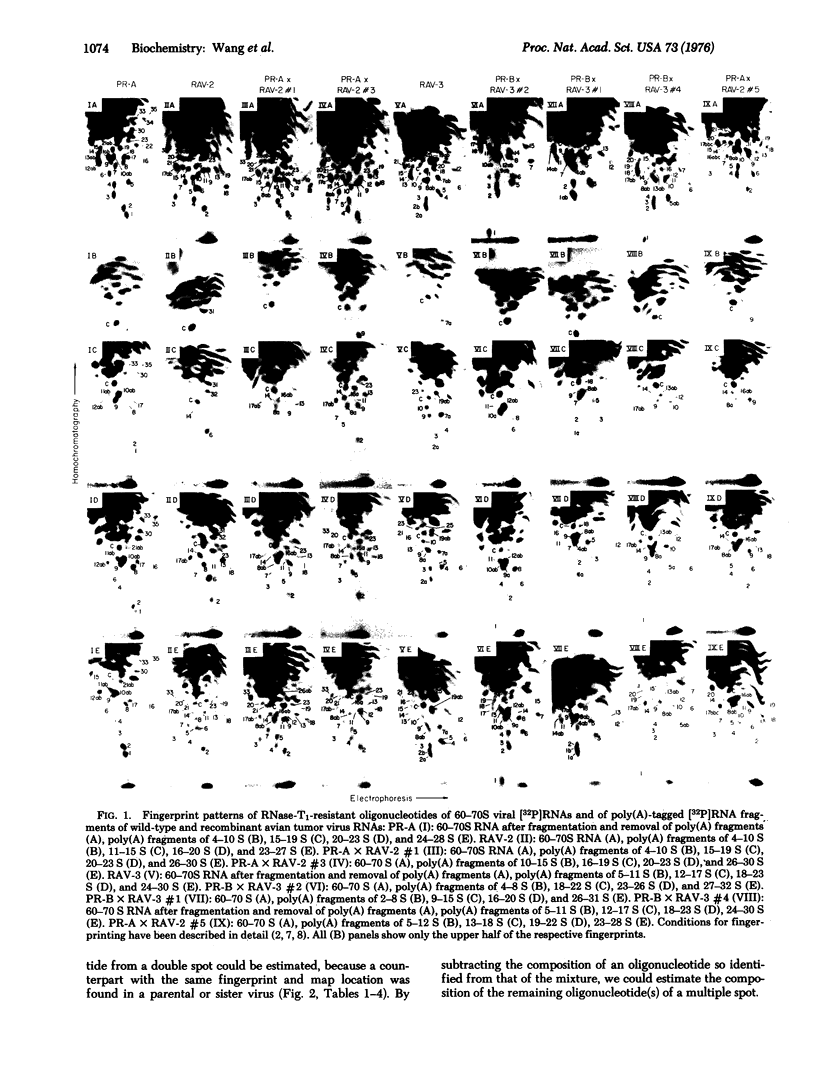
The distribution of leukosis-virus- and sarcoma-virus-specific oligonucleotide sequences was investigated in the RNAs of viral recombinants selected for an envelope gene (env) from a leukosis parent and a sarcoma gene (src) from a sarcoma parent. For this purpose 20 to 30 RNase-T1-resistant oligonucleotides were chemically analyzed and mapped within the 10,000 nucleotides of each viral RNA relative to the 3'-poly(A) end. The resulting oligonucleotide maps were compared. Proceeding from the 3' to the 5' end, the maps of four recombinants contained: (i) in a segment of 2000 nucleotides, three to four src-specific oligonucleotides, so identified because they were shared only with the sarcoma parent; and (ii) in a segment of 8000 nucleotides, 20 oligonucleotides shared with the leukosis parent, of which six to seven were also shared with the sarcoma parent. Two other recombinants contained: (1) in a segment of 2000 (one) or 3000 (the other) nucleotides, three src-specific oligonucleotides; (ii) in a segment of 3000 (one) or 2000 (the other) nucleotides, five (one) or four (the other) oligonucleotides, all or some of which are env-specific, because they were shared with the leukosis parent; (iii) in a segment of 5000 nucleotides (both), 11 functionally unidentified sarcoma-virus-derived oligonucleotides, of which seven were also shared with the leukosis parent. The map locations of parental oligonucleotides were not changed in recombinants and all viral strains tested shared six to eight highly conserved oligonucleotides at equivalent map locations. The partial map -env-src-poly(A) emerged from the analyses of these recombinants.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. Tumor viruses: 1974. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):1187–1200. doi: 10.1101/sqb.1974.039.01.137. [DOI] [PubMed] [Google Scholar]
- Beemon K., Duesberg P., Vogt P. Evidence for crossing-over between avian tumor viruses based on analysis of viral RNAs. Proc Natl Acad Sci U S A. 1974 Oct;71(10):4254–4258. doi: 10.1073/pnas.71.10.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billeter M. A., Parsons J. T., Coffin J. M. The nucleotide sequence complexity of avian tumor virus RNA. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3560–3564. doi: 10.1073/pnas.71.9.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P., Vogt P. K., Beemon K., Lai M. Avian RNA tumor viruses: mechanism of recombination and complexity of the genome. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):847–857. doi: 10.1101/sqb.1974.039.01.099. [DOI] [PubMed] [Google Scholar]
- HUEBNER R. J., ARMSTRONG D., OKUYAN M., SARMA P. S., TURNER H. C. SPECIFIC COMPLEMENT-FIXING VIRAL ANTIGENS IN HAMSTER AND GUINEA PIG TUMORS INDUCED BY THE SCHMIDT-RUPPIN STRAIN OF AVIAN SARCOMA. Proc Natl Acad Sci U S A. 1964 May;51:742–750. doi: 10.1073/pnas.51.5.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman A. C., Green R. W., Bolognesi D. P., Vanaman T. C. Comparative chemical properties of avian oncornavirus polypeptides. Virology. 1975 Apr;64(2):339–348. doi: 10.1016/0042-6822(75)90110-5. [DOI] [PubMed] [Google Scholar]
- Joho R. H., Billeter M. A., Weissmann C. Mapping of biological functions on RNA of avian tumor viruses: location of regions required for transformation and determination of host range. Proc Natl Acad Sci U S A. 1975 Dec;72(12):4772–4776. doi: 10.1073/pnas.72.12.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangel W. F., Delius H., Duesberg P. H. Structure and molecular weight of the 60-70S RNA and the 30-40S RNA of the Rous sarcoma virus. Proc Natl Acad Sci U S A. 1974 Nov;71(11):4541–4545. doi: 10.1073/pnas.71.11.4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowinski R. C., Watson K. F., Yaniv A., Spiegelman S. Serological analysis of the deoxyribonucleic acid polymerase of avian oncornaviruses. II. Comparison of avian deoxyribonucleic acid polymerases. J Virol. 1972 Nov;10(5):959–964. doi: 10.1128/jvi.10.5.959-964.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks W. P., Scolnick E. M., Ross J., Todaro G. J., Aaronson S. A. Immunological relationships of reverse transcriptases from ribonucleic acid tumor viruses. J Virol. 1972 Jan;9(1):110–115. doi: 10.1128/jvi.9.1.110-115.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P. H., Kawai S., Hanafusa H. Location of envelope-specific and sarcoma-specific oligonucleotides on RNA of Schmidt-Ruppin Rous sarcoma virus. Proc Natl Acad Sci U S A. 1976 Feb;73(2):447–451. doi: 10.1073/pnas.73.2.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P., Beemon K., Vogt P. K. Mapping RNase T1-resistant oligonucleotides of avian tumor virus RNAs: sarcoma-specific oligonucleotides are near the poly(A) end and oligonucleotides common to sarcoma and transformation-defective viruses are at the poly(A) end. J Virol. 1975 Oct;16(4):1051–1070. doi: 10.1128/jvi.16.4.1051-1070.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Duesberg P. Properties and location of poly(A) in Rous sarcoma virus RNA. J Virol. 1974 Dec;14(6):1515–1529. doi: 10.1128/jvi.14.6.1515-1529.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]