Abstract
Perturbed biomechanical stimuli are thought to be critical for the pathogenesis of a number of congenital heart defects, including Hypoplastic Left Heart Syndrome (HLHS). While embryonic cardiomyocytes experience biomechanical stretch every heart beat, their molecular responses to biomechanical stimuli during heart development are poorly understood. We hypothesized that biomechanical stimuli activate specific signaling pathways that impact proliferation, gene expression and myocyte contraction. The objective of this study was to expose embryonic mouse cardiomyocytes (EMCM) to cyclic stretch and examine key molecular and phenotypic responses. Analysis of RNA-Sequencing data demonstrated that gene ontology groups associated with myofibril and cardiac development were significantly modulated. Stretch increased EMCM proliferation, size, cardiac gene expression, and myofibril protein levels. Stretch also repressed several components belonging to the Transforming Growth Factor-β (Tgf-β) signaling pathway. EMCMs undergoing cyclic stretch had decreased Tgf-β expression, protein levels, and signaling. Furthermore, treatment of EMCMs with a Tgf-β inhibitor resulted in increased EMCM size. Functionally, Tgf-β signaling repressed EMCM proliferation and contractile function, as assayed via dynamic monolayer force microscopy (DMFM). Taken together, these data support the hypothesis that biomechanical stimuli play a vital role in normal cardiac development and for cardiac pathology, including HLHS.
Keywords: Hypoplastic Left Heart Syndrome, Mechanical Stretch, Cardiomyocytes, Gene regulation, Contractility, Cardiac development
1. Introduction
Understanding the biomechanical and biochemical mechanisms of congenital heart disease is critical to reducing patient morbidity and mortality. Hypoplastic Left Heart Syndrome (HLHS), which occurs ~4 out of 10,000 live births, is one of the most severe and costly congenital defects[1]. HLHS presents with an insufficiently sized left ventricle that is unable to provide proper blood flow to the systemic circulation. While some genetic mutations have been correlated with HLHS[2–5], the majority of HLHS cases are idiopathic. This lack of understanding regarding the pathogenic drivers involved in this syndrome has impaired both the development of treatments and prognostic tools for HLHS. Interestingly, the proposed involvement of abrogated mechanical stimuli in the development of HLHS has led to the intriguing possibility that novel biomechanically modulated molecular pathways are causative for this disease.
Observations of fetal echocardiograms in HLHS patients have led to the hypothesis that disruption of biomechanical stimuli results in perturbed growth of the left ventricle[6–8]. Narrowing of the foramen ovale or mitral valve in utero decreases the diastolic filling of the left ventricle, reducing mechanical stretch stimuli on developing cardiomyocytes, and impairing left ventricular growth. This hypothesis is supported by data from model organisms (e.g. embryonic sheep and chicken), in which a reduction of left atrial size resulted in decreased diastolic filling of the left ventricle and development of a HLHS phenotype[9–13]. In addition to physiological changes, examination of postnatal cardiomyocytes from HLHS patients revealed a decrease in proliferation-related genes[14]. At the cellular level, animal models for HLHS were also shown to have decreased embryonic cardiomyocyte proliferation and increased apoptosis, recapitulating key features of the disease[10, 12]. Despite the progress in modeling HLHS, there is little information about the specific molecular signals that are impacted by biomechanical stimuli at the cellular level. Given this lack of knowledge about the molecular pathways involved in the pathogenesis of HLHS, understanding the response of embryonic cardiomyocytes under biomechanical stimuli is critical.
In this effort, we hypothesized that biomechanical stimuli promote embryonic cardiomyocyte growth via stretch-activated signaling pathways. To test this hypothesis, we utilized an in vitro model in which embryonic mouse cardiomyocytes (EMCMs) were exposed to biomechanical stretch. Our results demonstrated that in vitro stretch increased both proliferation and size, indicating a direct link of stretch loading to EMCM growth and proliferation. Additionally, stretch modulated the levels of key myofibrillar factors such as myosin heavy chain and Titin. Bioinformatic analyses of mRNA-sequencing (RNA-Seq) data from stretched and static cells demonstrated significant enrichment of gene ontology groups (GO) involved in myofibrillogenesis and heart development. In addition, previously identified stretch-responsive pathways (e.g. focal adhesion, GTPase, integrin, cytoskeletal, calcium ion binding, oxidoreductase activity) were modulated under biomechanical stretch. Together, these data demonstrated that cyclic stretch is sufficient to promote phenotypic and gene expression changes in EMCMs.
One molecular pathway that is suggested to be involved in HLHS pathology is the Tgf-β/SMAD signaling pathway[15, 16]. Tgf-β signaling has long been known to play crucial roles in development and disease. Indeed, activation of Tgf-β receptors controls the expression of Tgf-β-dependent genes by way of the SMAD proteins, which shuttle from the membrane-bound receptor to the nucleus to modulate gene-expression in a phosphorylation-dependent mechanism. During embryonic development, signaling through Tgf-β receptors is thought to play important roles in the selection of cell-lineage and cell-fate, as well as in the migration and homing of cells. Characterization of the Tgf-β/SMAD signaling pathway has provided insights into the plasticity of cell differentiation. Indeed, cells may undergo Tgf-β-dependent lineage transitions, for example epithelial-mesenchymal transdifferentiation (EMT), which is integral for normal embryo development and organogenesis[17]. In the heart, EMT is known to contribute to valve development[18]. Tgf-β2-knockout mice display perinatal lethality and congenital heart defects, with a hypercellular myocardium and an enlarged right ventricle[19]. Abnormal EMT caused by pathological Tgf-β signaling was shown to cause fibrosis and to play a role in tumor metastasis[17] During cardiomyopathy, Tgf-β signaling is thought to activate resident cardiac fibroblasts, leading to excessive fibroblast proliferation, cardiac fibrosis, and stiffening of the heart through excessive deposition of extracellular matrix. There is ongoing discussion that physiologic growth and pathologic hypertrophy of cardiomyocytes represent different pathways[20, 21]. Moreover, there may be a developmental stage specific (embryonic vs. neonatal/adult) difference in the cardiomyocyte response to Tgf-β signaling.
Intriguingly, pathological alterations to Tgf-β signaling have been implicated in the development of HLHS in humans. Specifically, the expression levels of Tgf-β pathway genes were lower in the right ventricles of HLHS patients compared to tissue samples from left and right ventricles of patients without heart disease[15]. Right ventricular tissue of HLHS patients experiences more stretch since these right ventricles have to pump the blood volume that is normally handled by both ventricles. Conversely, a recent report shows that left ventricular tissue from fetal HLHS patients, which experiences decreased stretch, has increased expression and activation of Tgf-β1[16]. Indeed, data from stem cell experiment provide evidence that activated Tgf-β signaling inhibits cardiomyogenesis[22–24]. Furthermore, data from cultured neonatal rat cardiomyocytes suggest that Tgf-β signaling inhibits cardiomyocyte proliferation in vitro[25, 26]. Together, these data support the concept that downregulation of Tgf-β signaling could play a role in proper cardiac development, and that pathological deregulation of this signaling pathway during development may drive the pathogenesis of HLHS.
Given these data, we chose to examine the Tgf-β pathway in detail to better understand the mechanism of biomechanical stretch. Herein, we observed that stretch stimulation was sufficient to decrease Tgf-β family member expression, protein levels, and signaling in vitro. EMCM size was increased when Tgf-β signaling was inhibited. In contrast, activation of Tgf-β signaling led to decreased proliferation, contractile force, and beating frequency of EMCMs. Bioinformatic analyses predicted SMAD3 binding sites within 5kb the transcriptional start sites of the majority of stretch responsive genes that are responsible for myofibrillogenesis and normal heart development. Taken together, our findings presented in this study demonstrate that the in vitro stretch of EMCMs can serve as a model to study the contribution that biomechanical stimuli play in normal cardiac development and in the development HLHS.
2. Methods
2.1 Embryonic mouse cardiomyocyte (EMCM) culture
EMCMs were harvested from E16.5 wild-type Swiss Webster mouse embryos (Charles River) using standard methods[27] under an IACUC-approved protocol.
2.2 Biomechanical EMCM stretch analyses
EMCMs were grown on Collagen-I-coated Bioflex plates (BF-3001C Flexcell International). EMCMs were concurrently exposed to cyclic stretch of 16% at 1 Hz for 24h using a Flexcell FX-5000 Tension system (Flexcell International), and to static condition (control) on Bioflex plates.
2.3 RNA Extraction
RNA was extracted from the EMCMs using the RNeasy Mini kit (Qiagen). The concentration of RNA was determined at 260 nm using ND-1000 (Nanodrop) and all RNA was assessed for integrity using an Agilent 2100 Bioanalyzer. Only samples with a RNA Integrity Number of >8 were used for mRNA sequencing experiments.
2.4 Library Construction
Libraries were generated from three biologic replicates for each condition. Purified RNA was sheared and used to prepare an Illumina sequencing library using the Illumina TruSeq RNA Sample lit (Illumina, San Diego, CA). Briefly, 1 μg of the total RNA was fragmented and converted to double stranded cDNA. Illumina-platform-specific adaptors were then ligated to the DNA, and library molecules were amplified using PCR in accordance with Illumina standard protocols. The first step involved purifying the polyA+ mRNA using polyT oligo-attached magnetic beads. Following purification, the mRNA was fragmented. The cleaved RNA fragments were copied into first strand cDNA using reverse transcriptase and random primers followed by second strand cDNA synthesis using DNA Polymerase I and RNase H. The resulting cDNA fragments were end-repaired via the addition of a single ‘A’ base, and Illumina sequencing adapters were ligated. The DNA products were purified and enriched via PCR to generate the final cDNA library. The sequencing library was quantified, and its insert sizes were validated by Agilent 2100 Bioanalyzer analysis.
2.5 RNA-Seq
The RNA sequencing libraries were loaded onto the Illumina cBot Cluster Station (Illumina, San Diego, CA) where they bound to complementary adapter oligos grafted onto a proprietary flow cell substrate. Libraries were generated from three static and three stretched samples. Isothermal amplification of the cDNA construct was carried out creating clonal template clusters of approximately 1,000 copies each. The Illumina HiSeq2500 directly sequenced the resulting high-density array of template clusters on the flow cell using sequencing by synthesis. Four proprietary fluorescently labeled reversible terminator nucleotides were utilized to sequence the millions of clusters base by base, in parallel. Single-end sequencing was performed which generated 30 million short reads, 50bp in length per sample. The data generated by the sequencer was formatted to FASTQ format using the Illumina data analysis pipeline with sequences and associated Phred quality scores. An Illumina default quality score of Q30 was used equating with 99.9% base call accuracy. Low quality reads were filtered out to exclude those most likely representing sequencing errors.
Analysis of sequencing data involved the following steps: mapping the sequencing tags to the reference mouse transcriptome using the Bowtie aligner[28], assembling read counts via transcript-level expression summaries, normalizing the data based on total counts to adjust the counts for variance in sequencing depths, significance analysis of sequencing data and false discovery rate (FDR) analysis using the samR package, generation of significant transcripts lists, and Gene Ontology (GO) and Pathway Analysis.
2.6 samR analysis
samR[29] was used to detect differential expression in the RNA-Seq data. In order to calculate the FDR, the repeated permutation of the data was done to determine if the expression of any genes is significantly related to the response. The output file that SAM generated consisted of all the significant genes with the SAM score or the statistic value and also the associated q value. The significant genes were sorted based on their q value and the fold change, and a significant gene list with the most significant genes at the top was generated.
2.7 Statistical analysis of pathways and gene ontology terms
Each gene ontology term or pathway was treated simply as a set of genes. The transcript list, sorted by q-value in ascending order, was translated into Entrez gene ID’s and parsed so several different probes represent the same gene. Only the highest-ranking transcript was kept for further analysis. The sorted list of genes was subjected to a non-parametric variant of the Gene Set Enrichment Analysis, in which the p-value of a gene set of size n was defined as follows: let us denote the kth highest rank in gene set as rk, and define pk as the probability that out of n randomly chosen ranks (without replacement) the k-th highest is not smaller than rk. The p-value of the gene set was defined as min k [Pk] and was designed to detect the overrepresented gene sets at the top of the list.
These data were then subjected to a Bonferroni adjustment of gene set p-values for the number of gene sets tested. There were often several gene sets with overlapping gene content (and therefore statistically dependent). This overlap was partly due to the design of the gene ontology database and partly because genes tend to be involved in multiple processes. Thus, significant terms should not be reported as independent gene sets. Instead, we clustered the significant gene sets using variation of information as the distance metric[30] and presented them graphically in this context. Gene sets that clustered together generally shared a number of member genes.
Heatmaps of expression levels were created using in-house hierarchical clustering software. The colors qualitatively corresponded to fold changes with respect to a reference, which was calculated as the mid-point between compared groups.
2.8 Proliferation quantification
5–ethynyl–2–deoxyuridine (EdU) staining was performed using the Click-it EDU kit (Invitrogen). The n cited in the figures represented biological replicates. Data was presented as previously described [31].
2.9 Flow cytometry
Single cell suspensions of EMCMs were fixed with 4% PFA, filtered, and the acquisition of cells was obtained using the Accuri flow cytometer (BD Biosciences). Sided scatter on live cells and mean fluorescent intensity were quantified using Flowjo software (Tree Star). The n cited in the figures represented biological replicates.
2.10 Real-Time PCR
cDNA was prepared using Superscript III (Invitrogen). Quantitative Real Time PCR (qPCR) was performed using TaqMan primers (Applied Biosystems) and SYBR Green primers for a panel of cardiac and Tgf-β genes. ΔΔCT values were calculated; Gapdh was used as the endogenous control. The n represented biologic replicates. Nkx2.5, Notch1, Srf, and Tgfbr1 primers were Taqman primers. SYBR primer sequences are listed below and were validated using MIQE protocols as previously described[32].
| Myocd | Forward-GGGTGCGCACAGATGACTGGT Reverse-TCTTCCCTGGCATCGGCTGG |
| Myh6 | Forward-CTGCTGGAGAGGTTATTCCTCG Reverse-GGAAGAGTGAGCGGCGCATCAAGG |
| Myh7 | Forward-CTACCCTCAGGTGGCTCC Reverse-TGTGCCTCCAGCCTCTCCTT |
| Tgfb1 | Forward-CTCCCGTGGCTTCTAGTGC Reverse-GCCTTAGTTTGGACAGGATCTG |
| Tgfb2 | Forward-CTTCGACGTGACAGACGCT Reverse-GCAGGGGCAGTGTAAACTTATT |
| Tgfb3 | Forward-CCTGGCCCTGCTGAACTTG Reverse-TTGATGTGGCCGAAGTCCAAC |
| Tgfbr2 | Forward-CCGCTGCATATCGTCCTGTG Reverse-AGTGGATGGATGGTCCTATTACA |
| Ttn2ba | Forward-GGCGGTGCCTGCAGGTTAGA Reverse-GGTGGTTCTTGCACAATCAGGGC |
| Ttn2b | Forward-GCCCCCGAGTCGATTCTGCAT Reverse-AGGCTAGCAATTTCTTTCATCTCCA |
| Map4k4 | Forward-GCTAAGCAAACGGGCAGAGGA Reverse-CAGGTGGGACCACTGTACCTGT |
2.11 Western blotting
Western Blotting was performed as previously described[33]. Briefly, samples were boiled and loaded onto 4–20% polyacrylamide gels (BioRad). Protein samples were run at ~120V for one hour and transferred onto nitrocellulose membranes via wet transfer. Protein loading levels were checked using Ponceau staining and blocked in either 5% milk/TBST or 3.5% BSA/TBST. Membranes were then probed using primary antibodies for Rabbit anti-Tgf-β1, Tgf-β2, Tgf-β3, Tgf-βr1, Tgf-βr2, SMAD-3, Phospho-SMAD-3, total JNK (Cell Signaling), mouse anti-sarcomeric myosin heavy chain (clone 1025, Developmental Studies Hybridoma Bank) and mouse anti-GAPDH (Santa Cruz). For analysis of Titin isoforms and protein levels, vertical agarose gel electrophoresis (VAGE) was used as previously described[34, 35]. Briefly, cells were homogenized into VAGE sample buffer (8M urea, 2M thiourea, 3% SDS, 75mM DTT, 50mM Tris-HCl, pH 6.8). Separation of protein samples was done using 1% SDS-agarose gels (384mM glycine, 30% glycerol (v/v), 0.1% SDS (w/v), 50mM Tris-HCl, pH8.8), followed by western blotting onto nitrocellulose membranes. Protein loading was determined by Ponceau staining, and membranes were incubated with Titin-T11 antibody (Sigma Aldrich) in 5% BSA/TBST over night at 4°C. After washing off unbound primary antibody using TBST, membranes were incubated with HRP-linked anti mouse antibodies (Jackson) for 2 hours at room temperature. After washing in TBST, membranes were developed and exposed to X-ray films. Densitometry to determine protein amount was performed as previously described[33]. Sample size n represents biologic replicates.
2.12 Prediction of stretch-responsive transcription factors
The 5kb gene promoters of stretch responsive genes from specific GO-terms were scanned for statistical enrichment of evolutionarily conserved (human to mouse) transcription-factor-binding sites in the TRANSFAC professional database using Whole Genome RVista[36].
2.13 TGF-β2 treatment
EMCMs were treated with 1ng/ml of TGF-β2 (R&D Systems) and then exposed to static or stretch conditions for 24h as described above. Untreated EMCMs were used as controls.
2.14 ITD-1 treatment
EMCMs were treated with 5μM ITD-1, aTGFβR2 inhibitor[23], for 24h. Given, ITD-1 is dissolved in DMSO, DMSO treatment was used as the control condition.
2.15 Dynamic Monolayer Force Microscopy (DMFM)
We developed Dynamic Monolayer Force Microscopy to quantify the traction forces applied by a confluent cell sheet on their substratum and the cumulative mechanical stresses transmitted through the cell sheet. EMCMs were plated on polyacrylamide gel pads with Young modulus E = 10 KPa. The gel pads were seeded with 0.2-μm green fluorescent beads and their surface was coated with collagen (Fig 3A and supplemental Video). A spinning-disk confocal microscope at 20X was used to acquire time-lapse sequences of the periodic cell contraction at a time resolution of 20 Hz. As the cells contracted, they generated deformations in their substratum, which were measured by tracking the motion of the fluorescent beads using standard image cross-correlation techniques (supplemental Video). The traction stresses were determined from the measured deformations by solving the partial differential equations of equilibrium for the polyacrylamide substratum, which behaved like a linearly elastic medium, as previously described[37, 38]. Dynamic analysis of the deformation[39] allowed for the traction stresses to be determined during several contraction cycles without removing the cells from the gel pads. The cumulative intracellular stresses transmitted through the cell sheet were determined by solving the Kirchhoff-Love equations of mechanical equilibrium for a thin elastic plate subject to the forcing created by the measured traction stresses, using previously described methods[40, 41]. Overall intracellular stress was quantified for each sample by spatially averaging the maximum principal value of the intracellular stress at the instant of peak contraction.
Since the cell proliferation and size changed between the test groups, it is important to demonstrate that the calculated internal cell sheet stresses were independent of both cell size and the height of the cell sheet. First, let us prove that the Kirchoff-Love hypothesis was still valid even if the cell culture under study was not a monolayer sensu stricto. In order for this hypothesis to hold, it is sufficient that the lateral dimension L of the cell sheet is much larger than its thickness, h. In our experiments, the lateral dimension of the field of view was L>300 microns both in x and y. Now, it is quite safe to expect that h did not exceed two or three cell heights, as existing data indicates that cell proliferation increases by no more than 30% after 24 hours[42]. Thus, in the worst-case scenario and assuming a conservative cell height of 10 microns, we would have sheet thickness of h = 30 microns. Even in that worst-case scenario, we would have L > 10 × h. Thus, Kirchhoff–Love theory still held even if the cell sheet was not a pure monolayer. Second, let us demonstrate that the measured cell sheet stresses in our experiments were independent of 1) cell area 2) cell height and the thickness of the sheet. This independence was a direct consequence of the form of the equilibrium equations employed to determine the internal cell sheet stresses[41], where the forcing term was the measured traction stress normalized by the thickness of the cell sheet, T/h. Because traction stresses were already normalized for cell area and changes in sheet thickness were accounted for by the denominator of the forcing term, the measured cell sheet stresses were not affected by cell area, cell height or the thickness of the sheet.
Statistical comparison of populations was performed using the non-parametric Mann-Whitney U test (also known as Wilcoxon rank-sum test) to test the null hypothesis that meadian intracellular stresses in two populations were the same, against the alternative hypothesis that one particular population had higher values than the other.
2.16 Statistics
Unless otherwise stated (e.g. the RNA-Seq and DMFM), p-values were calculated using Student t-test. p<0.05 was considered statistically significant. The error bars in all of the graphs represents standard error of the mean expect for the DMFM data which has a box and whisker plot.
3. Results
3.1 RNA-Seq GO-terms
Perturbed biomechanical stimuli are thought to be critical for the pathogenesis of Hypoplastic Left Heart Syndrome (HLHS). Understanding the effects of biomechanical stimuli can therefore yield key insight into cardiac development and pathogenesis. To investigate the effect of these stimuli on EMCMs and to identify key stretch-responsive pathways, we performed unbiased examination of the cardiac transcriptome of EMCMs exposed to stretch via mRNA-Sequencing (RNA-Seq). Bioinformatic analyses of the RNA-Seq data demonstrated a significant enrichment of gene ontology (GO) groups important for cardiac development and function, e.g myofibril (p=3.75×10−25) and heart development (p=8.37×10−20) (Table 1). Other novel GO-terms in the RNA-Seq data that were modulated by stretch include terms associated with transcription (9.72×10−4) and translation regulation (7.97×10−18) (Supplemental Figures 2–4).
Table 1. Table of Gene Ontology (GO) terms most modulated by cyclic stretch.
Bioinformatics analysis of the RNA-Seq data from EMCMs exposed to 16% stretch at 1 Hz or static conditions for 24 h. The GO-terms were selected from the biologic process, cellular component, and molecular function based upon having the lowest p-value.
| Gene Ontology (GO) Group | Bonferroni corrected p-value |
|---|---|
| Contractile fiber part | 2.72E-25 |
| Myofibril | 3.75E-25 |
| Contractile fiber | 4.37E-25 |
| Regulation of cellular component organization | 1.08E-24 |
| Sarcomere | 3.94E-24 |
| Cardiovascular system development | 6.33E-22 |
| Circulatory system development | 6.33E-22 |
| Regulation of cellular protein metabolic process | 1.03E-21 |
| Heart development | 8.37E-20 |
| Organelle envelope | 2.06E-19 |
In addition to our observation that stretch modulates GO-terms associated with cardiac and myocyte development, bioinformatics analysis also identified GO-terms that have components known to be stretch-responsive[43–48]. Specifically, GO-terms associated with focal adhesion (3.69×10−4), GTPase regulator activity (1.04×10−5), integrin binding (4.95×10−6), cytoskeletal (1.21×10−18), calcium ion binding (1.43×10−6), oxidoreductase activity (3.65×10−10), and intracellular protein kinase cascade (3.63 x10−6), were modulated by cyclic stretch. We further explored the role of stretch-modulated kinases. Particularly, we examined if stretch was able to modulate the c-Jun N-terminal kinase (JNK) pathway, as components of this pathway had been previously associated with the stretch response of cardiomyocytes[48]. Expression of mitogenic-activated protein kinase kinase kinase kinase isoform 4 (Map4k4), an activator of JNK, was increased under stretch in both RNA-Seq analysis (17%, p=6.60 × 10−6) and qPCR (67%) confirmation. Additionally, we found that the protein levels of JNK were increased by cyclic stretch (Supplemental figure 5), corroborating our data that cyclic stretch increased Map4k4 expression and activation of the JNK pathway.
Our RNA-Seq data demonstrated that cyclic stretch modulates a number of known and novel stretch-mediated pathways at the transcriptional and protein level. The modulation of GO-terms associated with myofibrillar and heart development, transcription and translation regulation provides further evidence that biomechanical stretch is an important stimulus for EMCMs. Taken together, these findings support the idea that cyclic stretch is important for cardiomyocyte development and maturation, and that our experimental system modulated canonical stretch response pathways, like the JNK signaling pathway.
3.2 Cyclic stretch increases expression of cardiac genes, proliferation, EMCM size, and myofibril protein levels
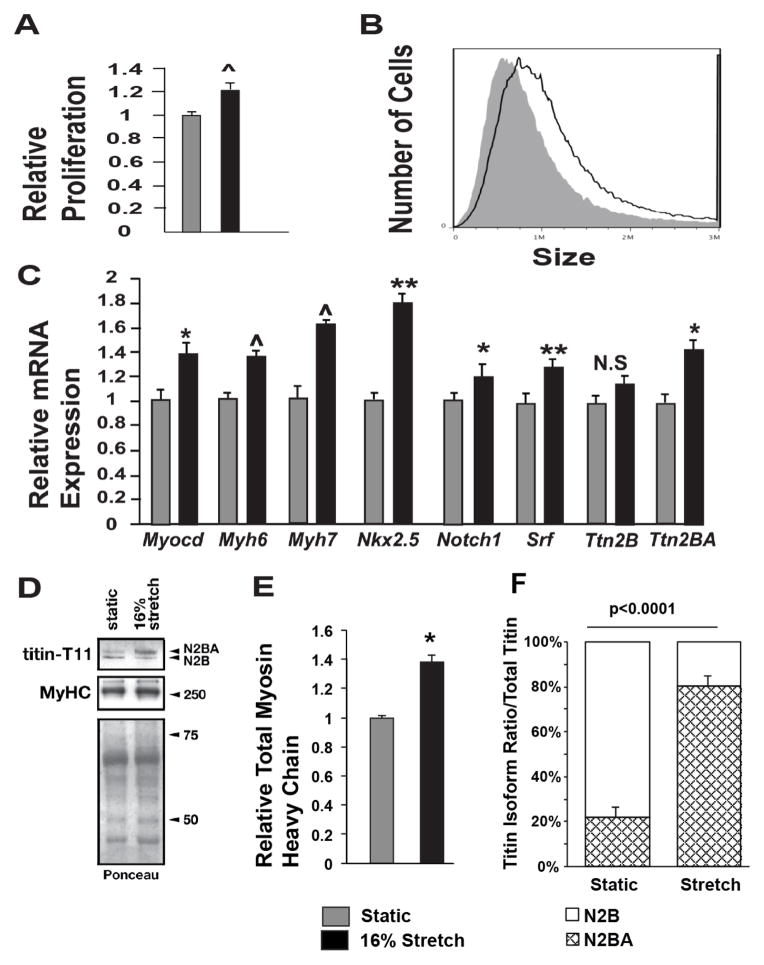
Given that stretch modulated GO-terms associated with cardiac growth and development, we performed phenotypic analyses along with qPCR examination of key cardiac genes. We observed that EMCMs exposed to cyclic stretch exhibited a 21% increase in proliferation (p<0.001) (Figure 1A). Flow cytometry side scatter analyses (SSC)[49] were used to quantify EMCM size. Stretched EMCMs were 33% larger (p<0.001) compared to static controls (Figure 1B). We performed qPCR for candidate key cardiac genes that have important roles in EMCM development and/or function. We concentrated on Myocardin (Myocd), Nkx2.5, Notch1, and Serum Response Factor (Srf) due to their important roles in EMCM gene development and function, as well as myosin heavy chains (Myh6, Myh7) and Titin (Ttn) for their role in myofibrillogenesis and cardiac differentiation. These key genes displayed significantly elevated mRNA levels in stretched EMCMs versus static controls (Figure 1C). Intriguingly, Titin mRNA levels were only increased for the longer N2BA isoform, while increases in the mRNA levels for the shorter N2A isoform were not statistically significant (Figure 1C, right). The increase in myosin heavy-chain and Titin was further validated by immunoblot analysis. Expression levels of sarcomeric myosin heavy-chains increased 1.4 fold in stretched versus static EMCMs (Figure 1E), corroborating the increase in expression levels of Myh genes. While the overall Titin protein level remained unchanged (relative Titin expression levels of 1.0±0.2 static vs. 1.2±0.6 stretch, p=0.39), cyclic stretch modulated Titin isoform expression (Figure 1D). Isoform specific analysis of Titin immunoblots revealed a remarkable shift towards the longer, more compliant N2BA isoform in stretched samples, compared to samples under static condition, where the majority of Titin was in the shorter less compliant N2A isoform (Figure 1D, lower left panel). Together, these data demonstrated that cyclic stretch is sufficient to increase the proliferation, growth, and cardiac gene expression in EMCMs.
Figure 1. Cyclic stretch increases EMCM proliferation, size, cardiac gene expression, and myofibrillar protein levels.
A. EDU staining comparing the proliferation rates of static and stretched EMCMs. Stretched EMCMs have a 21.3% increase in relative proliferation compared to static controls (1.21 +/− 6% vs. 1 +/− 3.2%; p<0.001; n=8–9). B. EMCMs exposed to stretch are 33% larger compared to static controls as quantified by flow cytometry (1.42×106 +/− 0.01 vs 1.07×106 +/− 0.04; p<0.001; n=3). C. qPCR for key cardiac genes demonstrates increased expression in stretched EMCMs compared to static controls. Gapdh was used as control. n=4–6. D. Immunoblotting showing representative total sarcomeric myosin heavy chain expression and Titin isoform levels. Total myosin heavy chain levels are increased in stretched EMCMs. Titin N2BA is the predominate isoform in stretched EMCMs, as compared to Titin N2B as is seen in static controls. E. Stretched EMCMs have increased Myosin heavy chain levels (1.38 +/− 0.10 vs 1 +/− 0.03; p=0.014; n=4). F. Quantification of Titin isoforms. 80.8% +/− 8.6% of Titin in stretched EMCMs is of the N2BA isoform. In contrast, static cardiomyocytes contain 22.8% +/− 9.4% of the Titin N2BA isoform. (the rest is Titin N2B; n=4; *p<0.05, **p<0.005, ^p<0.001).
3.3 Stretch decreases expression, protein levels, and activity of the Tgf-β pathway
Published reports examining samples from HLHS patients indicated that Tgf-β signaling is altered in vivo. Specifically, right ventricles of HLHS patients were found to display changes in the Tgf-β/SMAD signaling pathway[15]. Additionally, left ventricles of fetal HLHS patients displayed increased expression of Tgf-β1[16]. These data obtained from patient samples suggest that Tgf-β and its pathway components may be critically involved in HLHS pathology. Interestingly, examination of our RNA-Seq data also identified the Tgf-β signaling GO-group as significantly modulated by stretch (p=3.95×10−5). We therefore examined the role that Tgf-β activation plays for cardiac development and function using our stretch-activated EMCMs.
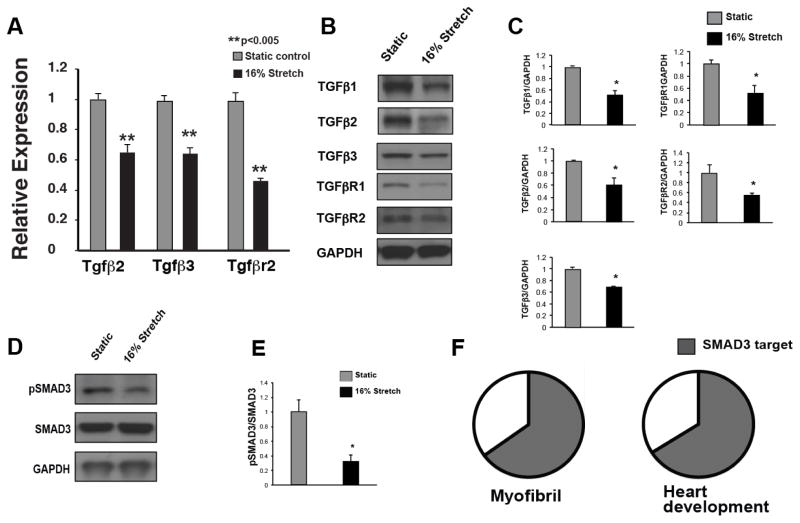
Stretch significantly decreased mRNA and protein levels of Tgf-β family members as well as their receptors in EMCMs (Figure 2A–C). Canonical activation of Tgf-β leads to subsequent phosphorylation of SMAD2/3, which shuttles into the nucleus to modulate gene expression. To assess the activity of Tgf-β signaling in stretched EMCMs, we examined SMAD3 phosphorylation levels. Stretched EMCMs had significantly decreased phospho-SMAD3 levels compared to controls (Figure 2D and E). Given that activated SMAD2/3 can modulate the transcription of target genes, we examined if SMAD3 binding sites were enriched in the proximal promoters of stretch responsive genes. To identify and examine stretch-responsive targets of Tgf-β signaling, we utilized Global regulatory VISTA (rVISTA), a bioinformatic tool that identifies conserved transcription factor binding sites in promoter regions of genes that are located within 5kb of the transcriptional start site[36]. Global rVISTA analyses predicted conserved SMAD3 binding sites in 65% of stretch-responsive myofibrillar genes (p=3.03×10−8) and in 66% of genes involved in heart development (p=4.96×10−36; Figure 2F). In addition, SMAD3 binding sites were also enriched in the proximal promoter regions of stretch responsive genes in GO-terms associated with focal adhesion, integrin binding, cytoskeletal, calcium ion binding, transcription, and intracellular protein kinase cascade (Supplemental figure 6).
Figure 2. Cyclic stretch decreased Tgf-β expression and signaling.
Since the GO-term associated with Tgf-β was modulated by stretch (p=3.95×10−5), the role of Tgf-β signaling in stretched EMCMs was examined. A. qPCR for Tgf-β expression demonstrates that cyclic stretch inhibits expression of Tgf-β2, Tgf-β3, and Tgfβr2. B. and C. Protein levels of Tgf-β ligands and receptors are decreased in stretched EMCMs. D and E. Western blotting for phospho-SMAD3 demonstrates decreased Tgf-β/SMAD signaling in stretched EMCMs. F. Global rVISTA analysis (which examines up to 5kb proximal to the transcriptional start site) predicts that 65% of stretch responsive myofibrillar (p=3.03×10−8) and 66% of cardiac development genes (p=4.96×10−36) have SMAD binding sites within 5 kb. (n=3 panels A–E; *p<0.05, **p<0.005).
These data demonstrated that cyclic stretch downregulates mRNA and protein levels of Tgf-β family members along with repressed Tgf-β signaling, as shown by decreased phospho-SMAD3 levels. Additionally, rVISTA analysis predicted that the majority of stretch-responsive genes in GO-terms associated myofibrillar and cardiac development are targets of Tgf-β signaling.
3.4 Tgf-β2 inhibits stretch-mediated proliferation
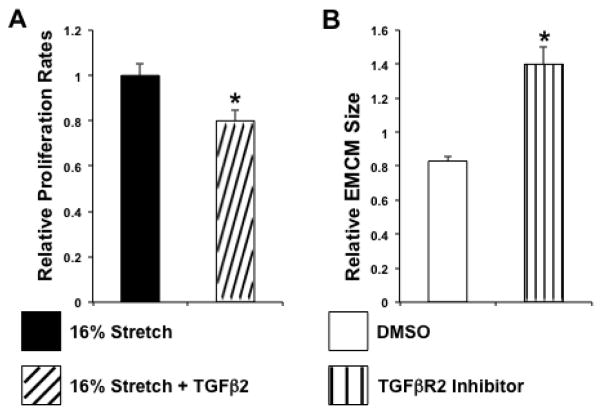
Tgf-β signaling influences a great number of target pathways that alter cell function. We therefore examined key functions in EMCM cellular physiology (proliferation, growth, and contractility). Cell-biological studies that disrupted the Tgf-β pathway in cardiomyocytes (embryonic to neonatal) and cardiac stem cells suggested that Tgf-β signaling represses EMCM proliferation under static conditions[23–26]. To investigate if Tgf-β repression is necessary for the stretch-mediated increase in EMCM proliferation, we treated EMCMs with Tgf-β2 and exposed these cells to cyclic stretch. Our results showed that Tgf-β2-treated stretched EMCMs had a 20% reduction in proliferation compared to untreated stretched controls (p<0.05; Figure 3A). Given that Tgf-β is repressed during stretch-mediated proliferation of EMCMs, and now that the addition of Tgf-β2 during stretch abrogates the proliferative response, we concluded that Tgf-β signaling negatively modulates the stretch-mediated proliferation of EMCMs. Furthermore, our results demonstrated that the repression of Tgf-β signaling, via stretch, positively modulates stretch-mediated proliferation.
Figure 3. TGFβ2 modulates proliferation and growth of EMCMs.
A. The addition of 1 ng/mg Tgf-β2 during stretch results in 20% reduction in proliferation (1 +/− 0.05 vs 0.80 +/− 0.05; p<0.05; n=3). B. Embryonic cardiomyocytes treated with ITD-1, a Tgfβr2 inhibitor, are 69% larger (1.40×106 +/−0.10 vs 0.83×106+/− 0.03; p<0.001; n=3).
3.5 ITD-1 promotes EMCM growth
Reduced Tgf-β signaling had been previously linked to increased proliferation of cardiomyocytes (embryonic to neonatal) and cardiac stem cells [23–26]. However, the role of Tgf-β signaling on cardiomyocyte growth had not been previously investigated. To determine if Tgf-β inhibition was sufficient to increase cardiomyocyte size, we treated EMCMs with the Tgf-βr2 inhibitor ITD-1[23]. We targeted Tgf-βr2 since it is the non-promiscuous receptor required for canonical Tgf-β signaling. Moreover, Tgf-βr2 was previously reported to be decreased in myocardial samples from HLHS patients exposed to increased stretch[15]. In our experiments, ITD-1 treated cells were 69% larger than DMSO treated control EMCMs (p<0.001; Figure 3B). Thus, our data demonstrated that inhibition of Tgf-β signaling by ITD-1 is sufficient to increase EMCM size.
3.6 Tgf-β2 decreased cardiomyocyte contractility
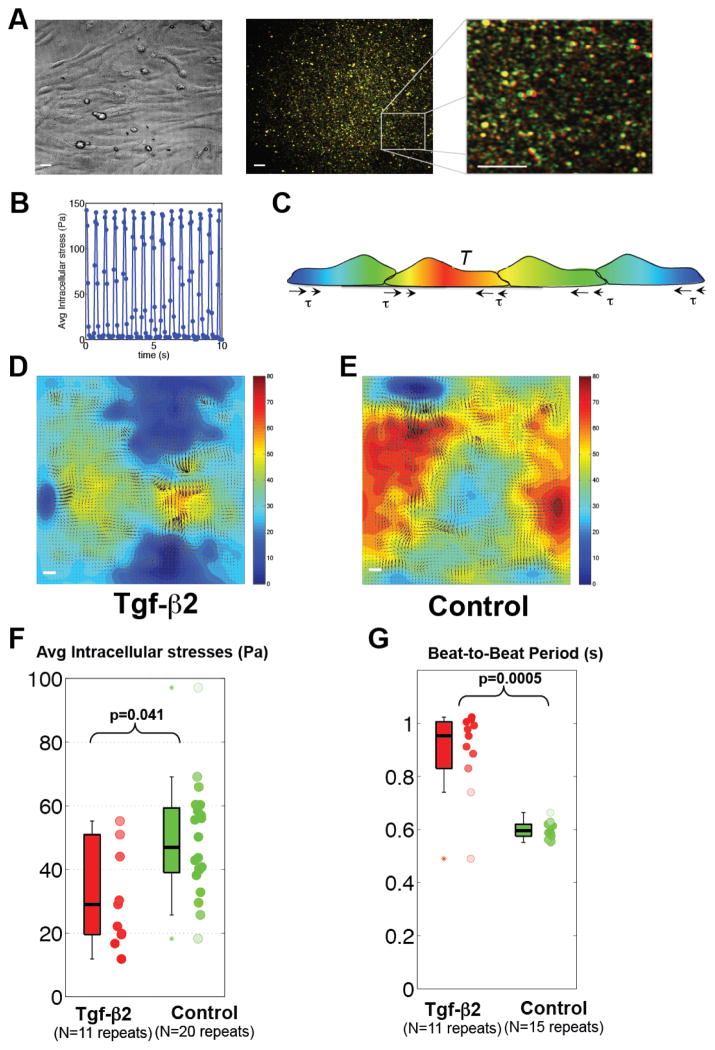
Systolic function is critical to maintain cardiac output. We examined if Tgf-β is able to modulate EMCM contractility in vitro using a novel dynamic monolayer force microscopy (DMFM) analysis (Figs 4A–C and Supplemental Video 1). In order to quantify how strongly EMCMs contracted against their substrate, we quantified the mechanical stresses generated by a confluent cell sheet of spontaneously beating EMCMs on the substratum. We used these data to deduce the spatio-temporal distribution of intracellular stress generated by and transmitted through the cell sheet by invoking mechanical equilibrium of the sheet (heat map in Figs. 4D, E and Supplemental Videos 2 and 3). Using this DMFM analysis, we observed that Tgf-β2 treated EMCMs had a 40% reduction in intracellular stresses as compared to untreated controls (median 29 Pa vs. 47 Pa, p<0.05; Figure 4F), and a 60% increase in beating period (median 0.95 s vs. 0.60 s, p<0.001; Figure 4G). Based upon these data, we concluded that Tgf-β2 is sufficient to repress EMCM contractile force and beating frequency.
Figure 4. Tgf-β2 is sufficient to reduce EMCM contractility.
A. Bright field and fluorescent images of beating EMCMs obtained for DMFM. Fluorescence images at maximum contraction (green) and relaxation (red) are overlaid, leading to red/green patterns in regions of large deformation and yellow patterns in regions of small deformation. B. Mean squared deformations in the whole fluorescence field determined from tracking bead motion. Peaks correspond to maximum contraction while valleys correspond to relaxation. C. Model of the equilibrium principle used to determine intracellular stresses[40, 41]. D and E. Comparison of traction stresses (arrows) and intracellular stresses (heat map, in Pa) in EMCMs treated with 1ng/ml of Tgf-β2 compared to untreated controls, as measured by DMFM. Red represents higher intracellular stress. F. Boxplot showing that EMCMs treated with Tgf-β2 display decreased intracellular stress compared to controls (p<0.05, WT n=20, Tgf-β2 N=11). G. Boxplot showing that EMCMs treated with Tgf-β2 exhibit slower beating period compared to controls (p<0.001, WT n=15, Tgf-β2 n=11). Scale bars are 20 μm long.
4. Discussion
Uncovering biomechanically modulated molecular pathways can offer key insight into the role of mechanical stimuli in normal cardiac development and cardiac pathology (e.g. HLHS). Examinations of patients with HLHS indicate that their hearts have significant changes in biomechanical loading and signaling in utero when compared to normally developing hearts. These alterations in biomechanical load lead to the hypothesis that abrogated mechanical stimuli directly result in aberrant cellular signaling. In this study, we utilized EMCMs exposed to cyclic stretch to unveil and characterize mechano-sensitive signaling pathways through RNA-Seq. Our results showed that stretch stimulates cardiac gene expression, proliferation, cell-growth, and myofibril proteins. We also found that Tgf-β/SMAD pathway is repressed by cyclic stretch, and that inhibition of Tgf-β signaling results in increased EMCM growth. Furthermore, Tgf-β2 inhibited both stretch-mediated EMCM proliferation and decreased collective EMCM contractility, as measured by DMFM.
Mechanical stretch regulates Tgf-β/SMAD signaling in EMCMs
Pathological deregulation of the Tgf-β pathway had been previously identified in HLHS patients [15, 16]. In these studies, right ventricles of HLHS patients experienced increased hemodynamic loading, leading to lower expression levels of Tgf-β family members[15]. In contrast, the left ventricles of HLHS patients were observed to experience decreased stretch and subsequently increased expression of Tgf-β1[16]. These data indicated that the Tgf-β pathway is modulated by mechanical stretch in a physiological or pathological way.
These findings led to our hypothesis that mechanical strain is sufficient to regulate Tgf-β signaling and Tgf-β dependent gene expression. In agreement with this hypothesis, we observed that cyclic stretch of EMCMs was sufficient to repress the levels of Tgf-β ligands and their receptors on the mRNA and protein levels. Moreover, cyclic stretch inhibited the downstream activation of SMAD3 in vitro. Also consistent with our hypothesis, we found that SMAD3 binding sites were enriched in stretch-modulated genes associated with cardiomyocyte differentiation, myofibrillogenesis and contractility. These data corroborated studies on neonatal rat cardiomyocytes, which found that inhibition of Tgf-β signaling resulted in increased cardiomyocyte proliferation and myofibrillogenesis[25, 26]. Taken together, our results from stretched EMCMs reflect the in vivo situation wherein ventricular embryonic/neonatal cardiomyocytes experience physiologic stretch during normal cardiac development.
Tgf-β signaling modulates EMCM proliferation and size
Given that we observed direct changes to Tgf-β/SMAD signaling under stretch, we subsequently sought to understand the effects of this pathway on cellular physiology. To investigate the role of Tgf-β signaling on stretched EMCMs, we administered Tgf-β2, thereby activating Tgf-β/SMAD signaling. Intriguingly, Tgf-β2-treated EMCMs experienced decreased proliferation despite being stimulated by cyclic stretch. Moreover, treatment with the Tgf-βr2 inhibitor ITD-1 was sufficient to increase the size of EMCMs. We did not observe a change in size between stretched EMCMs and stretch/TGF-β2 treated EMCMs. Given that stretch reduces the levels of TGF-β ligands and receptors, it might be necessary to increase the levels of the TGF-β receptors during stretch to block the stretch-mediated increase in size. Concurrent with these findings, recent studies on left ventricles of HLHS patients[16] and experiments on stem cells[22–24] show that activation of Tgf-β signaling was sufficient to inhibit cardiomyogenesis.
DMFM was used to measure Tgf-β’s effect on collective EMCM contractility
Many of the GO-terms identified in our stretch experiments were linked to myofibrillogenesis and cardiac differentiation. This finding led to the intriguing possibility that Tgf-β signaling directly influences contractility. To examine this possibility, we developed a novel technique that allowed us to directly observe changes in the contraction of EMCMs. When cells contract, they tug on their substratum as well as on their neighboring cells. Dynamic monolayer force microscopy (DMFM) allows for the quantification of the forces applied by a confluent cell sheet on their substratum and, more important, the cumulative mechanical forces collectively transmitted through the cell sheet. The latter are relevant to understanding how cellular forces build-up to achieve contraction at the tissue and organ level. Cells were seeded on a flexible substratum of known elastic properties and the resulting substratum deformation was measured to calculate these forces. While force microscopy has been used in other settings, especially in cell migration[37, 38, 41, 50–57], it has rarely been used with cardiomyocytes or has been limited to single-cell assays[39, 58–60]. Published measurements of force build-up in confluent cardiomyocyte sheets are non-existent. Force microscopy usually requires removing cells from the substratum to obtain undeformed substratum images to use as reference to determine deformations. Nevertheless, the development of DMFM has allowed us to measure dynamic intracellular stresses in pulsating cell sheets for the first time based on dynamic analysis of substratum deformation[39]. This procedure allows for the mechanical stresses to be determined during several contraction cycles without removing the cells from the substratum (Figure 3). The broader impact of DMFM is as a powerful, multifaceted tool in cardiac biology since it is a novel way to quantify cardiomyocyte contractility in vitro. Using DMFM, we observed that Tgf-β2 represses both EMCM contractile force and beating frequency and further characterize the role of this biomechanically repressed factor on cardiomyocyte function.
Tgf-β/SMAD signaling provide a mechanism by which stretch modulates cardiac gene expression
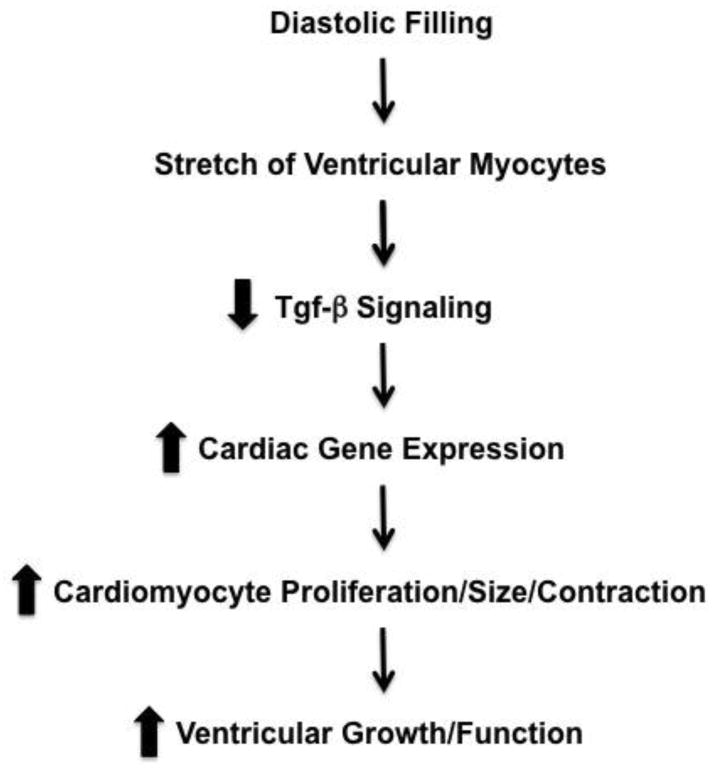
Based upon our data, it is clear that Tgf-β signaling plays a key role in cardiomyocyte development and physiology. However, little is known about the direct regulation of genes in developing cardiomyocytes by Tgf-β/SMAD. It was previously demonstrated that Tgf-β/SMAD signaling results in transcriptional regulatory complexes that can activate or repress cardiac gene expression[61]. As a result of our bioinformatics analyses of RNA-Seq data and global rVISTA analyses, we propose the model that cyclic stretch repression of Tgf-β/SMAD signaling results in activation of key cardiac genes (Figure 5). These key genes are involved in myofibrillar and cardiac development, focal adhesion, integrin binding, cytoskeleton, calcium ion binding, and transcriptional regulation. Our model is based on the finding that the majority of these key stretch-responsive genes associated are predicted to have conserved SMAD3 sites within the genes’ proximal promoters. Indeed, we found that many key cardiac transcription factors and myofibril genes were predicted to be under the control of Tgf-β/SMAD-dependent signaling (including Gata4, Notch1, Srf and Myocardin, the myosin heavy chain genes Myh6 and Myh7, and Titin). In addition to our data, previous findings that Tgf-β/SMAD is perturbed in HLHS patients[15, 16] support our proposed model.
Figure 5. Model of how stretch modulates Tgf-β signaling and cardiac development.
Based upon our data, we propose that physiologic stretch of developing cardiomyocytes represses Tgf-β signaling, which in turn results in increased cardiomyocyte proliferation, size, gene expression, and contractile function.
An additional mechanism by which stretch modulates biologic processes would be via isoform switching. In fact, one of the biological processes most impacted in hearts of HLHS patients was recently shown to be mRNA isoform switches[62]. Our RNA-Seq data, qPCR and immunoblot assays revealed changes in the stretch-induced alternative splicing of Titin, which switched from the Titin-N2A to the Titin-N2BA isoform in biomechanically stimulated cardiomyocytes. This is result is potentially interesting because Titin-N2A is shorter than Titin-N2BA, and Titin isoform length is known to regulate myofibrillar compliance[63]. Longer Titin isoforms, which incorporate additional exons encoding for unstructured regions result in enhanced muscle compliance, whereas shorter isoforms were associated with a decrease in myofibrillar passive compliance. Titin isoform switches are developmentally regulated[64–66] and may be influenced in cardiac pathology by growth factors, hormones and activation of PI3K/AKT signaling[67, 68]. Generally it was found that fetal and neonatal myocardium express the more compliant N2BA isoforms of Titin, whereas healthy adult myocardium typically contains the less compliant N2B isoform. Our finding that the major Titin isoform in stretched EMCMs is the more compliant N2BA-isoform recapitulates to a greater extent the in vivo situation in healthily developing fetal myocardium. The increase in N2BA observed in stretched ECMCs contrasts with the static control case, where the shorter Titin N2A isoform was more expressed. Previous work[67] found no stretch-induced changes in alternative Titin isoform expression. However, species differences (rat versus mouse) and differences in the stretch protocol (10% stretch at 0.02Hz or 1Hz intervals for 96h, versus 16% stretch at 1Hz intervals for 24h) may account for these divergent results.
Recent findings demonstrated that alternative splicing of several key cardiac genes, including Titin, is under control of the muscle specific splice factor Rbm20[69]. Using a spontaneously mutated animal model[70], these data showed that relatively modest changes in the Rbm20 expression level, such as in heterozygous animals, result in dramatic changes to Titin isoform distribution. Lower Rbm20 protein levels shifted the Titin isoform distribution to the more compliant N2BA variant, whereas the complete loss of the splice factor resulted in a giant N2BA variant. Intrigued by these data, we examined if Rbm20 was altered in stretched EMCMs versus static controls. Indeed, biomechanical strain significantly reduced Rbm20 mRNA levels in our RNA-seq data compared to controls (6% downregulation; p=4.55×10−2). More intriguingly, Rbm20 is among the Tgf-β target genes identified by our rVISTA analysis. These data demonstrate that biomechanical strain and Tgf-β signaling control the regulation of cardiac transcription, splice factors and myofibrillar genes.
In summary, we showed that stretch is able to regulate cardiac gene-expression, proliferation and cell-growth in a Tgf-β/SMAD dependent manner, which supports our hypothesis that biomechanical stimuli play a vital role for normal cardiac development. Clinical data along with our in vitro data further demonstrate that cyclic stretch is sufficient to repress Tgf-β signaling both in vitro and in vivo. The spatio-temporal deregulation of this signaling pathway likely has important clinical implications for HLHS patients.
5. Conclusions
Taken together, these data indicate that repression of Tgf-β signaling is key to the stretch response in EMCMs (Figure 5). While we focused on Tgf-β/SMAD, we anticipate that additional stretch responsive pathways also play roles in cardiac development and HLHS. Analyses of our RNA-Seq data identified additional candidates for stretch responsive pathways, which will be examined in future studies and will lend further insight into cardiac development and disease. Indeed, modulation of these key pathways could have a significant impact on HLHS and other conditions that operate on the biomechanical-molecular axis.
Supplementary Material
Highlights.
Biomechanical stimuli are altered in Hypoplastic Left Heart Syndrome.
Mechanical stimuli activate signaling pathways and regulate embryonic cardiomyocytes function.
Tgf-β signaling altered embryonic cardiomyocytes function (proliferation and size).
Dynamic Force Microscopy found decreased contractile function under Tgf-β stimulation.
Data supports the hypothesis that mechanical stimuli are key for cardiac development.
Acknowledgments
The authors would like to acknowledge the Biomedical Genomics Microarray Core Facility University of California San Diego for their assistance with the RNA-Seq.
Sources of Funding:
V.N. was partially funded by NIH K08 HL086775. RM and JCA were partially supported by NIH grants 1R01GM084227 and 1R21 HL108268-01. IB was funded by AHA Postdoctoral Fellowship (12POST12030256). Funding for SL was provided by an NIH grant (HL107744). Additionally, the UCSD Department of Pediatrics and Rady’s Children’s Heart Institute provided funding.
Abbreviations
- HLHS
Hypoplastic Left Heart Syndrome
- GO
Gene ontology
- DMFM
Dynamic monolayer force microscopy
- RNA-Seq
mRNA-sequencing
- FDR
False discovery rate
- EdU
5–ethynyl–2′–deoxyuridine
- qPCR
Quantitative Real Time PCR
- SSC
Flow cytometry side scatter
Footnotes
Subject Codes: Pediatric and congenital heart disease, Physiologic and pathologic control of gene expression, Contractile Function, Other myocardial biology, Cell biology/structural biology
Disclosures:
The authors have no disclosures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Menon SC, Keenan HT, Weng HY, Lambert LM, Burch PT, Edwards R, et al. Outcome and resource utilization of infants born with hypoplastic left heart syndrome in the Intermountain West. The American journal of cardiology. 2012;110:720–7. doi: 10.1016/j.amjcard.2012.04.050. [DOI] [PubMed] [Google Scholar]
- 2.Dasgupta C, Martinez AM, Zuppan CW, Shah MM, Bailey LL, Fletcher WH. Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE) Mutat Res. 2001;479:173–86. doi: 10.1016/s0027-5107(01)00160-9. [DOI] [PubMed] [Google Scholar]
- 3.Elliott DA, Kirk EP, Yeoh T, Chandar S, McKenzie F, Taylor P, et al. Cardiac homeobox gene NKX2–5 mutations and congenital heart disease: associations with atrial septal defect and hypoplastic left heart syndrome. J Am Coll Cardiol. 2003;41:2072–6. doi: 10.1016/s0735-1097(03)00420-0. [DOI] [PubMed] [Google Scholar]
- 4.Reamon-Buettner SM, Hecker H, Spanel-Borowski K, Craatz S, Kuenzel E, Borlak J. Novel NKX2–5 mutations in diseased heart tissues of patients with cardiac malformations. Am J Pathol. 2004;164:2117–25. doi: 10.1016/S0002-9440(10)63770-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McBride KL, Riley MF, Zender GA, Fitzgerald-Butt SM, Towbin JA, Belmont JW, et al. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum Mol Genet. 2008;17:2886–93. doi: 10.1093/hmg/ddn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trines J, Hornberger LK. Evolution of Congenital Heart Disease in Utero. Pediatr Cardiol. 2004 doi: 10.1007/s00246-003-0592-2. [DOI] [PubMed] [Google Scholar]
- 7.Simpson JM, Sharland GK. Natural history and outcome of aortic stenosis diagnosed prenatally. Heart. 1997;77:205–10. doi: 10.1136/hrt.77.3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hornberger LK, Sanders SP, Rein AJ, Spevak PJ, Parness IA, Colan SD. Left heart obstructive lesions and left ventricular growth in the midtrimester fetus. A longitudinal study. Circulation. 1995;92:1531–8. doi: 10.1161/01.cir.92.6.1531. [DOI] [PubMed] [Google Scholar]
- 9.deAlmeida A, McQuinn T, Sedmera D. Increased ventricular preload is compensated by myocyte proliferation in normal and hypoplastic fetal chick left ventricle. Circulation research. 2007;100:1363–70. doi: 10.1161/01.RES.0000266606.88463.cb. [DOI] [PubMed] [Google Scholar]
- 10.deAlmeida A, Sedmera D. Fibroblast Growth Factor-2 regulates proliferation of cardiac myocytes in normal and hypoplastic left ventricles in the developing chick. Cardiol Young. 2009;19:159–69. doi: 10.1017/S1047951109003552. [DOI] [PubMed] [Google Scholar]
- 11.Fishman NH, Hof RB, Rudolph AM, Heymann MA. Models of congenital heart disease in fetal lambs. Circulation. 1978;58:354–64. doi: 10.1161/01.cir.58.2.354. [DOI] [PubMed] [Google Scholar]
- 12.Sedmera D, Hu N, Weiss KM, Keller BB, Denslow S, Thompson RP. Cellular changes in experimental left heart hypoplasia. Anat Rec. 2002;267:137–45. doi: 10.1002/ar.10098. [DOI] [PubMed] [Google Scholar]
- 13.Sedmera D, Pexieder T, Rychterova V, Hu N, Clark EB. Remodeling of chick embryonic ventricular myoarchitecture under experimentally changed loading conditions. Anat Rec. 1999;254:238–52. doi: 10.1002/(SICI)1097-0185(19990201)254:2<238::AID-AR10>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 14.Gambetta K, Al-Ahdab MK, Ilbawi MN, Hassaniya N, Gupta M. Transcription repression and blocks in cell cycle progression in hypoplastic left heart syndrome. Am J Physiol Heart Circ Physiol. 2008;294:H2268–75. doi: 10.1152/ajpheart.91494.2007. [DOI] [PubMed] [Google Scholar]
- 15.Ricci M, Mohapatra B, Urbiztondo A, Birusingh RJ, Morgado M, Rodriguez MM, et al. Differential changes in TGF-beta/BMP signaling pathway in the right ventricular myocardium of newborns with hypoplastic left heart syndrome. J Card Fail. 2010;16:628–34. doi: 10.1016/j.cardfail.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 16.Gaber N, Gagliardi M, Patel P, Kinnear C, Zhang C, Chitayat D, et al. Fetal reprogramming and senescence in hypoplastic left heart syndrome and in human pluripotent stem cells during cardiac differentiation. The American journal of pathology. 2013;183:720–34. doi: 10.1016/j.ajpath.2013.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Derynck R, Akhurst RJ. Differentiation plasticity regulated by TGF-beta family proteins in development and disease. Nature cell biology. 2007;9:1000–4. doi: 10.1038/ncb434. [DOI] [PubMed] [Google Scholar]
- 18.Desgrosellier JS, Mundell NA, McDonnell MA, Moses HL, Barnett JV. Activin receptor-like kinase 2 and Smad6 regulate epithelial-mesenchymal transformation during cardiac valve formation. Developmental biology. 2005;280:201–10. doi: 10.1016/j.ydbio.2004.12.037. [DOI] [PubMed] [Google Scholar]
- 19.Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, et al. TGFbeta2 knockout mice have multiple developmental defects that are non-overlapping with other TGFbeta knockout phenotypes. Development. 1997;124:2659–70. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorn GW., 2nd The fuzzy logic of physiological cardiac hypertrophy. Hypertension. 2007;49:962–70. doi: 10.1161/HYPERTENSIONAHA.106.079426. [DOI] [PubMed] [Google Scholar]
- 21.Kehat I, Molkentin JD. Molecular pathways underlying cardiac remodeling during pathophysiological stimulation. Circulation. 2010;122:2727–35. doi: 10.1161/CIRCULATIONAHA.110.942268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kitamura R, Takahashi T, Nakajima N, Isodono K, Asada S, Ueno H, et al. Stage-specific role of endogenous Smad2 activation in cardiomyogenesis of embryonic stem cells. Circulation research. 2007;101:78–87. doi: 10.1161/CIRCRESAHA.106.147264. [DOI] [PubMed] [Google Scholar]
- 23.Willems E, Cabral-Teixeira J, Schade D, Cai W, Reeves P, Bushway PJ, et al. Small molecule-mediated TGF-beta type II receptor degradation promotes cardiomyogenesis in embryonic stem cells. Cell stem cell. 2012;11:242–52. doi: 10.1016/j.stem.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen WP, Wu SM. Small molecule regulators of postnatal Nkx2.5 cardiomyoblast proliferation and differentiation. J Cell Mol Med. 2012;16:961–5. doi: 10.1111/j.1582-4934.2011.01513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kardami E. Stimulation and inhibition of cardiac myocyte proliferation in vitro. Mol Cell Biochem. 1990;92:129–35. doi: 10.1007/BF00218130. [DOI] [PubMed] [Google Scholar]
- 26.Sheikh F, Hirst CJ, Jin Y, Bock ME, Fandrich RR, Nickel BE, et al. Inhibition of TGFbeta signaling potentiates the FGF-2-induced stimulation of cardiomyocyte DNA synthesis. Cardiovascular research. 2004;64:516–25. doi: 10.1016/j.cardiores.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 27.Lim BK, Peter AK, Xiong D, Narezkina A, Yung A, Dalton ND, et al. Inhibition of Coxsackievirus-associated dystrophin cleavage prevents cardiomyopathy. The Journal of clinical investigation. 2013;123:5146–51. doi: 10.1172/JCI66271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Tibshirani R. Finding consistent patterns: a nonparametric approach for identifying differential expression in RNA-Seq data. Stat Methods Med Res. 2013;22:519–36. doi: 10.1177/0962280211428386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kozak I, Sasik R, Freeman WR, Sprague LJ, Gomez ML, Cheng L, et al. A degenerative retinal process in HIV-associated non-infectious retinopathy. PloS one. 2013;8:e74712. doi: 10.1371/journal.pone.0074712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hara M, Matsumori A, Ono K, Kido H, Hwang MW, Miyamoto T, et al. Mast cells cause apoptosis of cardiomyocytes and proliferation of other intramyocardial cells in vitro. Circulation. 1999;100:1443–9. doi: 10.1161/01.cir.100.13.1443. [DOI] [PubMed] [Google Scholar]
- 32.Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–22. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 33.Banerjee I, Fuseler JW, Intwala AR, Baudino TA. IL-6 loss causes ventricular dysfunction, fibrosis, reduced capillary density, and dramatically alters the cell populations of the developing and adult heart. American journal of physiology Heart and circulatory physiology. 2009;296:H1694–704. doi: 10.1152/ajpheart.00908.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lange S, Ouyang K, Meyer G, Cui L, Cheng H, Lieber RL, et al. Obscurin determines the architecture of the longitudinal sarcoplasmic reticulum. Journal of cell science. 2009;122:2640–50. doi: 10.1242/jcs.046193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Warren CM, Krzesinski PR, Greaser ML. Vertical agarose gel electrophoresis and electroblotting of high-molecular-weight proteins. Electrophoresis. 2003;24:1695–702. doi: 10.1002/elps.200305392. [DOI] [PubMed] [Google Scholar]
- 36.Dubchak I, Munoz M, Poliakov A, Salomonis N, Minovitsky S, Bodmer R, et al. Whole-Genome rVISTA: A Tool to Determine Enrichment of Transcription Factor Binding Sites in Gene Promoters from Transcriptomic Data. Bioinformatics. 2013 doi: 10.1093/bioinformatics/btt318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Del Alamo JC, Meili R, Alonso-Latorre B, Rodriguez-Rodriguez J, Aliseda A, Firtel RA, et al. Spatio-temporal analysis of eukaryotic cell motility by improved force cytometry. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:13343–8. doi: 10.1073/pnas.0705815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Alamo JC, Meili R, Alvarez-Gonzalez B, Alonso-Latorre B, Bastounis E, Firtel R, et al. Three-dimensional quantification of cellular traction forces and mechanosensing of thin substrata by fourier traction force microscopy. PloS one. 2013;8:e69850. doi: 10.1371/journal.pone.0069850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacot JG, McCulloch AD, Omens JH. Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys J. 2008;95:3479–87. doi: 10.1529/biophysj.107.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hur SS, del Alamo JC, Park JS, Li YS, Nguyen HA, Teng D, et al. Roles of cell confluency and fluid shear in 3-dimensional intracellular forces in endothelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11110–5. doi: 10.1073/pnas.1207326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tambe DT, Croutelle U, Trepat X, Park CY, Kim JH, Millet E, et al. Monolayer stress microscopy: limitations, artifacts, and accuracy of recovered intercellular stresses. PloS one. 2013;8:e55172. doi: 10.1371/journal.pone.0055172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soonpaa MH, Kim KK, Pajak L, Franklin M, Field LJ. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–9. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- 43.Torsoni AS, Constancio SS, Nadruz W, Jr, Hanks SK, Franchini KG. Focal adhesion kinase is activated and mediates the early hypertrophic response to stretch in cardiac myocytes. Circulation research. 2003;93:140–7. doi: 10.1161/01.RES.0000081595.25297.1B. [DOI] [PubMed] [Google Scholar]
- 44.Torsoni AS, Marin TM, Velloso LA, Franchini KG. RhoA/ROCK signaling is critical to FAK activation by cyclic stretch in cardiac myocytes. Am J Physiol Heart Circ Physiol. 2005;289:H1488–96. doi: 10.1152/ajpheart.00692.2004. [DOI] [PubMed] [Google Scholar]
- 45.Zhang SJ, Truskey GA, Kraus WE. Effect of cyclic stretch on beta1D-integrin expression and activation of FAK and RhoA. Am J Physiol Cell Physiol. 2007;292:C2057–69. doi: 10.1152/ajpcell.00493.2006. [DOI] [PubMed] [Google Scholar]
- 46.Cadre BM, Qi M, Eble DM, Shannon TR, Bers DM, Samarel AM. Cyclic stretch down-regulates calcium transporter gene expression in neonatal rat ventricular myocytes. Journal of molecular and cellular cardiology. 1998;30:2247–59. doi: 10.1006/jmcc.1998.0788. [DOI] [PubMed] [Google Scholar]
- 47.Pimentel DR, Amin JK, Xiao L, Miller T, Viereck J, Oliver-Krasinski J, et al. Reactive oxygen species mediate amplitude-dependent hypertrophic and apoptotic responses to mechanical stretch in cardiac myocytes. Circulation research. 2001;89:453–60. doi: 10.1161/hh1701.096615. [DOI] [PubMed] [Google Scholar]
- 48.Pan J, Singh US, Takahashi T, Oka Y, Palm-Leis A, Herbelin BS, et al. PKC mediates cyclic stretch-induced cardiac hypertrophy through Rho family GTPases and mitogen-activated protein kinases in cardiomyocytes. J Cell Physiol. 2005;202:536–53. doi: 10.1002/jcp.20151. [DOI] [PubMed] [Google Scholar]
- 49.Lopez JE, Myagmar BE, Swigart PM, Montgomery MD, Haynam S, Bigos M, et al. beta-myosin heavy chain is induced by pressure overload in a minor subpopulation of smaller mouse cardiac myocytes. Circulation research. 2011;109:629–38. doi: 10.1161/CIRCRESAHA.111.243410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bastounis E, Meili R, Alonso-Latorre B, del Alamo JC, Lasheras JC, Firtel RA. The SCAR/WAVE complex is necessary for proper regulation of traction stresses during amoeboid motility. Molecular biology of the cell. 2011;22:3995–4003. doi: 10.1091/mbc.E11-03-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Butler JP, Tolic-Norrelykke IM, Fabry B, Fredberg JJ. Traction fields, moments, and strain energy that cells exert on their surroundings. American journal of physiology Cell physiology. 2002;282:C595–605. doi: 10.1152/ajpcell.00270.2001. [DOI] [PubMed] [Google Scholar]
- 52.Dembo M, Oliver T, Ishihara A, Jacobson K. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophys J. 1996;70:2008–22. doi: 10.1016/S0006-3495(96)79767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dembo M, Wang YL. Stresses at the cell-to-substrate interface during locomotion of fibroblasts. Biophys J. 1999;76:2307–16. doi: 10.1016/S0006-3495(99)77386-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Iwadate Y, Yumura S. Actin-based propulsive forces and myosin-II-based contractile forces in migrating Dictyostelium cells. Journal of cell science. 2008;121:1314–24. doi: 10.1242/jcs.021576. [DOI] [PubMed] [Google Scholar]
- 55.Meili R, Alonso-Latorre B, del Alamo JC, Firtel RA, Lasheras JC. Myosin II is essential for the spatiotemporal organization of traction forces during cell motility. Molecular biology of the cell. 2010;21:405–17. doi: 10.1091/mbc.E09-08-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Serra-Picamal X, Conte V, Vincent R, Anon E, Tambe DT, Bazellieres E, et al. Mechanical waves during tissue expansion. Nature Physics. 2012;8:628–U66. [Google Scholar]
- 57.Tambe DT, Hardin CC, Angelini TE, Rajendran K, Park CY, Serra-Picamal X, et al. Collective cell guidance by cooperative intercellular forces. Nat Mater. 2011;10:469–75. doi: 10.1038/nmat3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hersch N, Wolters B, Dreissen G, Springer R, Kirchgessner N, Merkel R, et al. The constant beat: cardiomyocytes adapt their forces by equal contraction upon environmental stiffening. Biol Open. 2013;2:351–61. doi: 10.1242/bio.20133830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCain ML, Lee H, Aratyn-Schaus Y, Kleber AG, Parker KK. Cooperative coupling of cell-matrix and cell-cell adhesions in cardiac muscle. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:9881–6. doi: 10.1073/pnas.1203007109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rodriguez AG, Han SJ, Regnier M, Sniadecki NJ. Substrate Stiffness Increases Twitch Power of Neonatal Cardiomyocytes in Correlation with Changes in Myofibril Structure and Intracellular Calcium. Biophys J. 2011;101:2455–64. doi: 10.1016/j.bpj.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen CR, Kang Y, Siegel PM, Massague J. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 62.Ricci M, Xu Y, Hammond HL, Willoughby DA, Nathanson L, Rodriguez MM, et al. Myocardial alternative RNA splicing and gene expression profiling in early stage hypoplastic left heart syndrome. PloS one. 2012;7:e29784. doi: 10.1371/journal.pone.0029784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Linke WA, Hamdani N. Gigantic business: titin properties and function through thick and thin. Circulation research. 2014;114:1052–68. doi: 10.1161/CIRCRESAHA.114.301286. [DOI] [PubMed] [Google Scholar]
- 64.Lahmers S, Wu Y, Call DR, Labeit S, Granzier H. Developmental control of titin isoform expression and passive stiffness in fetal and neonatal myocardium. Circulation research. 2004;94:505–13. doi: 10.1161/01.RES.0000115522.52554.86. [DOI] [PubMed] [Google Scholar]
- 65.Opitz CA, Leake MC, Makarenko I, Benes V, Linke WA. Developmentally regulated switching of titin size alters myofibrillar stiffness in the perinatal heart. Circulation research. 2004;94:967–75. doi: 10.1161/01.RES.0000124301.48193.E1. [DOI] [PubMed] [Google Scholar]
- 66.Greaser ML, Krzesinski PR, Warren CM, Kirkpatrick B, Campbell KS, Moss RL. Developmental changes in rat cardiac titin/connectin: transitions in normal animals and in mutants with a delayed pattern of isoform transition. J Muscle Res Cell Motil. 2005;26:325–32. doi: 10.1007/s10974-005-9039-0. [DOI] [PubMed] [Google Scholar]
- 67.Kruger M, Sachse C, Zimmermann WH, Eschenhagen T, Klede S, Linke WA. Thyroid hormone regulates developmental titin isoform transitions via the phosphatidylinositol-3-kinase/AKT pathway. Circulation research. 2008;102:439–47. doi: 10.1161/CIRCRESAHA.107.162719. [DOI] [PubMed] [Google Scholar]
- 68.Kruger M, Babicz K, von Frieling-Salewsky M, Linke WA. Insulin signaling regulates cardiac titin properties in heart development and diabetic cardiomyopathy. Journal of molecular and cellular cardiology. 2010;48:910–6. doi: 10.1016/j.yjmcc.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 69.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nature medicine. 2012;18:766–73. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Greaser ML, Warren CM, Esbona K, Guo W, Duan Y, Parrish AM, et al. Mutation that dramatically alters rat titin isoform expression and cardiomyocyte passive tension. Journal of molecular and cellular cardiology. 2008;44:983–91. doi: 10.1016/j.yjmcc.2008.02.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.