Abstract
To maintain daily cycles, the circadian clock must tightly regulate the rhythms of thousands of mRNAs and proteins with the correct period, phase, and amplitude to ultimately drive the wide range of rhythmic biological processes. Recent genomic approaches have revolutionized our view of circadian gene expression and highlighted the importance of post-transcriptional regulation in driving mRNA rhythmicity. Even after transcripts are made from DNA, subsequent processing and regulatory steps determine when, where, and how much protein will be generated. These post-transcriptional regulatory mechanisms can add flexibility to overall gene expression and alter protein levels rapidly without requiring transcript synthesis and are therefore beneficial for cells; however, the extent to which circadian post-transcriptional mechanisms contribute to rhythmic profiles throughout the genome and the mechanisms involved have not been fully elucidated. In this review, we will summarize how circadian genomics have revealed new insights into rhythmic post-transcriptional regulation in mammals and discuss potential implications of such regulation in controlling many circadian-driven physiologies.
In mammals, circadian rhythmicity is a fundamental aspect of temporal organization in essentially every cell in the body and modulates much of physiology, biochemistry, and behavior; therefore, it is not surprising that disruption of the clock can lead to many pathological states. Multiple signals originating in the suprachiasmatic nucleus (SCN) of the hypothalamus, the so-called “master circadian oscillator”, synchronize independent oscillators in each cell and peripheral tissue as well as other brain areas (reviewed in ref (1)). Within these cells and tissues, a set of clock genes and their protein products, which are highly conserved among animals, form transcription–translation feedback loops to generate cell-autonomous rhythms. This molecular clock, in turn, drives rhythmic gene expression, which will ultimately exert control over almost every biological, physiological, and behavioral process.1 Because the circadian clock is situated in such a pivotal position, disruption of the circadian clock by genetics and environmental conditions can result in dramatic changes in both mental and physical health, and therefore, understanding how circadian clocks maintain circadian rhythmicity is of central importance.
Although it has long been known that the circadian clock drives rhythmic transcription of clock-controlled genes (ccgs) that control rhythmic downstream processes, recent findings have challenged the current transcription-centric model. For example, proteome analyses have shown that as many as 50% of rhythmically expressed proteins do not exhibit rhythmicity in their mRNA levels.2,3 This inconsistency in rhythmic mRNA versus protein perhaps should not be too surprising, given that the correlation between mRNA and protein expression in general can be as low as 40%.4 In addition, mathematical modeling predicted more than 20 years ago that regulation of mRNA stability is essential for rhythmic mRNA expression.5 Therefore, in this review, we will examine how circadian genomics has enhanced our understanding of the relative roles of transcription and post-transcriptional control in generating the output rhythms of the cell in mammals. We will also discuss potential new applications of next-generation sequencing (NGS) technology in circadian genomics to explore novel post-transcriptional regulatory mechanisms. Regrettably, we will not cover epigenetic studies, such as circadian regulation of DNA methylation and histone modifications, although these have been extensively studied recently. Readers interested in this area are encouraged to refer to other studies.6−8
Clock Genes
The late 1990s to early 2000s was a time of great advancement in circadian biology, with the discovery of many core clock genes and the unraveling of the core circadian mechanism as an interlocking transcription–translation feedback loop.9−19 The core negative feedback loop consists of the transcription factor CLOCK interacting with BMAL1 to activate transcription of the Period (Per) and Cryptochrome (Cry) genes, resulting in high levels of these transcripts. The PER and CRY proteins then heterodimerize, translocate back to the nucleus, and interact with the CLOCK–BMAL1 complex to inhibit their own transcription. Subsequently, the PER–CRY repressor complex is degraded, and CLOCK–BMAL1 can now activate a new cycle of transcription. In addition to the primary negative feedback loop, there is a second feedback loop involving the nuclear hormone receptors Rev-erbs and RORs that negatively and positively regulate Bmal1 transcription, respectively. This secondary loop is thought to add robustness to the molecular clock.1,20 The entire cycle takes approximately 24 h to complete, and this is considered to be the driving force of the cell-autonomous clock.
Microarray-Based Circadian Genomics
By the beginning of the 21st Century, we had considerable knowledge about how the core oscillator generates circadian rhythms; however, less was known about how the oscillator regulates downstream biological, physiological, and behavioral processes that are under circadian control. On the basis of the core molecular clock mechanism, which consists of transcription–translation feedback loops, it was hypothesized that rhythmic ccg expression, driven by the core clock, would function as output molecules to control downstream processes.
On the other hand, in the 1990s, it was becoming clear that there was a need to develop a technique that could effectively measure the expression of tens of thousands of genes simultaneously. Microarray technology was thus developed to fulfill this goal and became commercially available with a reasonable cost. Therefore, many circadian biologists took advantage of this technique and identified hundreds of rhythmically expressed ccgs in different organisms and tissues. ccgs in the SCN were undoubtedly of prime importance in mammals; however, other tissues such as liver, kidney, heart, pineal gland, distal colon, and Rat-1 and NIH3T3 cells were also examined.21−29 Initial major findings from these microarrays were that each tissue expressed at least several hundred ccgs, while only a few dozen ccgs were identified in cell lines such as Rat-1 and NIH3T3 cells. Moreover, there was significant tissue specificity in ccgs with only a small proportion in common between each tissue.30 In fact, Per2 was the only mRNA that was commonly identified as being rhythmic among all microarray studies from both SCN and liver. Although this tissue specificity could simply be due to the absence of expression of particular genes in one tissue compared to another, and/or the differences in analytic parameters and assay stringency (see Limitations: How To Define “Rhythmicity”?), identification of ccgs and their specificity in each tissue/cell highlighted the fact that although the fundamental mechanisms driving clocks in each cell are the same, they drive rhythmic expression of specific subsets of genes that are relevant to that cell’s function.
Even though microarrays served as important tools in globally identifying ccgs in various different tissues, they also presented some limitations. For example, microarrays tend to underrepresent differences in gene expression; the amplitude of mRNA expression rhythms in most ccgs is <3-fold on a microarray, even though many have greater magnitude changes when measured by nonarray approaches such as quantitative polymerase chain reaction or Northern blotting.30 In addition, the choice of probe sets included on each array and errors in gene annotations for some of the probes (optimistically estimated to be approximately 1–5%31) were also concerns. Furthermore, the genes and specific probe sets on each microarray platform are not identical, thus making it difficult to compare data sets obtained from different platforms.
Next-Generation Sequencing-Based Circadian Genomics
While microarray technology flourished and was widely used in the early to mid 2000s, there was also a strong demand for low-cost DNA sequencing technologies that would potentially replace standard dye-terminator methods. In fact, the development of the prototypic NGS technology was already underway in the 1990s, although NGS sequencers did not come onto the market until 2004. As expected, these NGS technologies successfully reduced sequencing cost; for example, the cost to read the entire human genome dropped from almost US $100000 in 2002 to US $5000 in 2013.32 Furthermore, NGS has a broad range of possible applications for many different genomic studies, such as species classification and/or gene discovery, de novo assemblies of genomes that have not been sequenced in the past, SNP identification, epigenetic analysis, DNA methylation analysis, and, most notably, transcriptome analyses (RNA-seq) of cells, tissues, and organisms. In fact, microarrays are now being replaced by RNA-seq, because unlike a microarray, which requires that genes have representative sequences embedded on a platform, NGS allows one to quantify all known transcripts and to identify and quantify unknown transcripts anywhere in a given genome.
When this new technology was applied to mammalian circadian biology, ∼1000–2000 ccgs were identified in mouse liver,33−37 which is 2–10-fold more the number identified by microarrays. This is probably due to the more sensitive and comprehensive nature of RNA-seq and the increased sensitivity in algorithms to extract rhythmicity, although microarray technology also identified thousands of ccgs with an increased time resolution in the sampling interval (every one vs 4 h).29 In addition to increased power in identifying ccgs, these series of new RNA-seq studies also shed light on rhythmically expressed nonprotein coding transcripts, such as long noncoding RNAs (lncRNAs) or microRNAs (miRNAs). Recent findings suggest more than 90% of the entire genome is transcribed in mammals,38 and the majority of these transcripts account for nonprotein coding transcripts. Some of these noncoding transcripts such as miRNAs and lncRNAs have regulatory functions and likely contribute to the complexity of the organism by exerting additional control over protein expression. Contributions of circadian clock function to miRNA expression or vice versa have been widely accepted and well established. In particular, miR-132 and miR-219 seem to be directly involved in the clock system and regulate the circadian period or response to light.39 The expression of many miRNAs is found to be rhythmic, not only in liver but also in retina and SCN,39−43 and bioinformatics, as well as experimental evidence, suggests that many core clock genes are under the control of miRNA.44−49 A recent study has also shown that up to 30% of ccgs undergo miRNA-mediated regulation to perhaps control the phase and amplitude of rhythmic mRNA expression patterns post-transcriptionally.50 In addition to miRNAs, the expression of lncRNAs is rhythmic in liver and pineal gland.35,51 Although the precise roles for most lncRNAs remain largely elusive, studies of some lncRNAs have revealed that they exert a diverse spectrum of regulatory mechanisms across a variety of cellular pathways, ranging from embryonic stem cell differentiation, imprinting, X-chromosome inactivation, cell cycle regulation, and neuronal development, as well as diseases such as cancer or neurological disorders (reviewed in ref (52)). Therefore, it would not be surprising that these rhythmically expressed lncRNAs take part in regulating rhythmic processes. One notable lncRNA is an antisense transcript of Per2,34,35,37 whose expression is antiphasic to the sense Per2 mRNA. Antisense transcripts make up a class of lncRNAs that are transcribed from the opposite DNA strand of the sense RNA transcripts with which they share sequence complementarity,53−56 and the existence of a long antisense transcript for core clock genes has also been reported in Neurospora and Antheraea pernyi (silkmoth).57,58 The conservation of antisense transcripts to core clock genes across kingdoms seems to imply that this is an important mechanism for circadian clock regulation.
Another unexpected observation from circadian NGS analyses was that rhythmic mRNA expression relies to a great extent upon post-transcriptional regulation. By analyzing circadian NGS data using reads mapped not only to exons but also to introns (as an indicator of pre-mRNA expression), Koike et al.37 found that approximately 80% of ccg mRNAs did not undergo rhythmic de novo transcription. Similar findings were shown by Menet et al.34 using a method called Nascent-seq that directly assesses rhythmic de novo transcription by measuring nascent RNA levels. Furthermore, 50–70% of transcripts that are rhythmic in de novo transcription do not exhibit rhythmic mRNA expression.34 This type of observation is unique to NGS and cannot be typically made from microarray studies, because standard microarrays measure only the amount of transcript that hybridizes to the pre-embedded specific probes, making it difficult to distinguish pre-RNA from mRNA unless samples are prepared to represent only nascent RNAs (i.e., from nuclear run-ons) or custom arrays are specifically designed to contain probe sets to introns. All these data strongly suggest that post-transcriptional regulation plays a major role in driving mRNA oscillation rhythms.
Post-transcriptional Circadian Genomics
Even before genomic studies became popular in the circadian biology field, several nongenomic approaches had already revealed that the mammalian circadian clock system utilizes various post-transcriptional regulatory mechanisms such as splicing,59 alternative polyadenylation,60 and poly(A) tail length regulation61−63 to control rhythmic gene expression. Theoretical models also predicted that mRNA stability regulation was important for cycling mRNAs, as mRNA half-life impacts their amplitude, and the more stable the transcript, the lower the amplitude of its cycling.5 The key components of post-transcriptional regulation are typically trans-acting factors (i.e., miRNAs and RNA-binding proteins) acting on cis elements residing in target mRNAs, which leads to the consequent regulation.
The first evidence of post-transcriptional regulation in circadian biology came from the study of the Drosophila Period (Per) gene, showing that the stability of Per mRNA is under circadian control and that its mRNA stability changes around the clock.64 Similarly, the mRNA stability of one of the mammalian Per homologue genes, Period1, was also found to be regulated post-transcriptionally.65 The noncoding 3′UTR portion of Per1 mRNA plays a role in its stability control, and the post-transcriptional regulators, LARK RNA-binding proteins, activate translation of PER1 protein expression.66,67 Subsequently, other cycling mRNAs, such as Per2, Per3, and Cry1, were also shown to be under mRNA stability control in a clock-dependent manner, and their mRNAs are more stable during the rising phase of mRNA cycling and less stable during the declining phase. Three heterogeneous ribonucleoprotein particles (hnRNPs), hnRNP I/PTB, hnRNP Q, and hnRNP D/AUF1, appear to take part in the regulation of mRNA stability of the clock genes mentioned above, as well as regulating the temporal translation of CRY1, PER1, REV-ERBα, and AANAT, a rate-limiting enzyme in the melatonin-producing pathway.68−75
In addition, the circadian clock may regulate translation more broadly, because the activity of the translation initiation complex, including the eukaryotic translational initiation factor 4E-binding protein 1 (4E-BP1) as well as the mTORC1 pathway (both of which are indispensable for protein synthesis), is under circadian control in both SCN and liver.76−78 4E-BP1 also appears to desynchronize core clock function and/or attenuate its light responsiveness by repressing the translation of vasoactive intestinal peptide (VIP). Because VIP is a key neuropeptide that synchronizes SCN cells to the environmental light–dark cycle, as well as transmitting environmental cues (i.e., light) between individual cells within the SCN,76 4E-BP1-mediated translation perhaps controls entrainment and synchrony of the master clock. However, because 4E-BP1 acts generally in translation, it is still unclear why circadian regulation of 4E-BP1 seems to cause specific effects on VIP translation.
The emergence of genomic approaches has improved our knowledge of post-transcriptional regulation from a few specific examples to a more global view. For example, a study with Affymetrix mouse exon arrays discovered that 0.4% of genes were under the control of rhythmic alternative splicing,79 although this is a surprisingly small effect considering that alternative splicing is a widespread and highly regulated event in mammals, affecting ∼80% of mouse genes.79 Interestingly, circadian alternative splicing correlated with rhythmic mRNA expression, supporting the idea that circadian alternative splicing occurs cotranscriptionally.80 This could also explain why only a low percentage of genes undergo circadian alternative splicing, as cotranscriptional splicing is approximately 2-fold less efficient in mouse liver.81 Another microarray-based study, “poly(A)denylome analysis”, which measured poly(A) tail lengths of individual mRNAs, showed that approximately 2.5% of mRNAs have rhythmic poly(A) tail lengths in mouse liver, and importantly, the fluctuation in the poly(A) tail length, not the mRNA levels, correlated with the rhythmicity of the protein levels.82 Furthermore, ribosome profiling analyses, a technique used to isolate actively translated RNAs from polyribosome fractions, discovered that approximately 2% of expressed genes are translated rhythmically, independent of the rhythmicity of steady-state mRNA.78 Corresponding protein expression was validated as being rhythmic, indicating that these mRNAs undergo post-transcriptional regulation to be translated rhythmically.
Other genome-wide analyses also provided evidence that post-transcriptional events are critical for determining the pace and amplitude of the circadian clock. The m6A RNA methylation-dependent RNA processing pathway contributes to the period length,83 and cold-inducible RNA-binding protein (CIRBP) regulates the amplitude.84 Genetic or pharmacological disturbance of the m6A RNA methylation pathway resulted in the elongation of the circadian period via the retention of methylated RNAs in the nucleus. Notably, the m6A RIP (RNA immunoprecipitation)-seq analysis revealed that core clock mRNAs such as Per1-3, Dbp, and Nr1d1,2 (Rev-erbα and Rev-erbβ), among other nonclock mRNAs, have methylated adenines in their mRNAs, suggesting that these mRNAs are substrates of the RNA methylation-dependent RNA processing pathway. Given that m6A modification is ubiquitous, affecting >7000 genes in humans,85 global regulation of mRNAs with m6A modification, not just core clock mRNAs, might contribute to the lengthening of the circadian period. In contrast, depletion of CIRBP led to dampening of the rhythms, and this was accompanied by the reduction in the level of expression of several clock proteins, such as CLOCK, DBP, and PER2, with an only minor effect on their mRNA levels; however, it is still unclear whether CIRBP directly binds to pre-RNA/mRNA of these clock genes and controls their protein expression post-transcriptionally. Given the broad impact of CIRBP on circadian amplitude regulation, it is plausible that the effect of CIRBP is indirect.
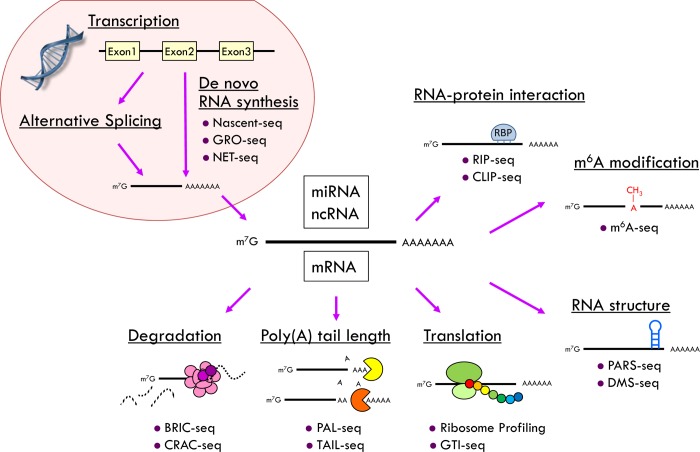
There are likely other post-transcriptional mechanisms yet to be discovered that control circadian clock function and/or circadian gene expression. A number of different sequencing methods have been developed that can be used to demonstrate circadian control of post-transcriptional events, including Nascent-seq, GRO (global run-on)-seq, and NET (native elongation transcript)-seq, to assess de novo RNA synthesis;34,86,87 RIP (RNA immunoprecipitation)-seq and CLIP (cross-linking immunoprecipitation)-seq to globally identify target RNA recognition sequences of an RNA-binding protein;88,89 BRIC (5′-bromouridine immunoprecipitation chase)-seq90 for mRNA stability; CRAC (in vivo RNA cross-linking)-seq91 to identify target mRNAs of exosome-mediated mRNA decay; PAL [poly(A) tail length]-seq or TAIL-seq92,93 for poly(A) tail length regulation; GTI (global translation initiation)-seq for translation initiation sites; PARS (parallel analysis of RNA structure)-seq and DMS (dimethyl sulfate)-seq for RNA secondary structure;94,95 and m6A-seq for m6A modifications.96 Future studies using these and other techniques will likely uncover a plethora of regulatory mechanisms that will provide insight into how the circadian clock controls rhythmic gene expression post-transcriptionally without relying on rhythmic de novo transcription (Figure 1).
Figure 1.
Potential new applications of NGS technology in circadian genomics to explore novel post-transcriptional regulatory mechanisms. Nascent-seq,34 GRO-seq,87 NET-seq,86 RIP-seq,89 CLIP-seq,88 BRIC-seq,90 CRAC-seq,91 PAL-seq or TAIL-seq,92,93 GTI-seq,107 PARS-seq or DMS-seq,94,95 and m6A-seq.96 RBP denotes the RNA-binding protein.
Circadian Proteomics
Considering the fact that proteins, not mRNAs, are the functional entities that ultimately drive biological processes, identification of rhythmically expressed proteins should be a priority; however, this task has been difficult to achieve, mainly because of the difficulties and expense of quantitative proteomic approaches. A recently developed technique, SILAC (stable isotope labeling by amino acids in cell culture), is beginning to overcome this issue. SILAC compares two groups of samples, one of which is labeled with a “light” or normal amino acid while the other is labeled with a “heavy” amino acid (i.e., 2H vs 1H, 13C vs 12C, or 15N vs 14N). The samples are mixed, and then mass spectrometry analysis is performed. The ratio of peak intensities is then measured. Each peptide should appear as a pair, differing only in the mass shift between the two samples with light versus heavy amino acids, and this ratio of peak intensities represents the ratio of the amount of each protein.97
When this technique was applied to circadian proteomics, 5–10% of all expressed proteins were determined to be rhythmic in mouse liver,3,98 a percentage that is very similar to the percentage of ccgs identified in genomic analyses. Among these proteins, however, only 50–80% had rhythmically expressed corresponding mRNAs, further indicating that rhythmicity in mRNA and rhythmicity in protein do not necessarily correlate. Perplexingly, neither SILAC study identified any core clock proteins among the rhythmic proteins, even though most circadian genomic studies, as well as Western blot analyses, have found them to be rhythmically expressed. This was probably due to the low expression levels of clock proteins that are below the limit of detection of current technologies.
Limitations: How To Define “Rhythmicity”?
Although genomic approaches have been widely used and have identified many new ccgs, they also have limitations. The results from genomic and proteomic studies are somewhat inconsistent, and it is thus often unclear which genes and/or proteins are actually playing important roles in rhythmic processes. Even in cases when the exact same tissue was examined, only a handful of genes were found to be in common between different experiments. For example, of 97 or 408 genes identified as ccgs in mouse SCN from two individual studies, only 27 genes were shared between the data sets.21,23 In addition, of 395, 338, 524, 892, 1126, 1204, 1262, 2037, and 2741 ccgs identified in various studies from mouse liver (four microarray and five RNA-seq studies), only 30 genes were conserved in all four independent microarray studies, 60 genes were shared in all five independent RNA-seq studies, and just five ccgs (Hsd3b5, Lgals9, Per2, Por, and Usp2) were common in all nine studies.21−23,29,33−37 Obviously, a lowered threshold of commonality increases the number of common ccgs; 19, 32, 80, and 126 genes were commonly detected as being rhythmic from eight, seven, six, and five of all nine studies, respectively. The majority of common ccgs consist of clock genes [Arntl (Bmal1), Clock, Cry1, Nr1d1 (Rev-erbα), Nr1d2 (Rev-erbβ), and Rorc] and other relatively high-amplitude ccgs [Ccrn4l (Nocturnin) and Avpr1a (arginine vasopressin receptor 1A)]; however, Per1 and Bmal were missing in some of these data sets, and Dbp was not extracted as a ccg in the three microarray studies from early days,21−23 in spite of their widely recognized robust hepatic rhythmic expression.99 This inconsistency issue was not restricted to transcriptome analyses; other global circadian approaches revealed a similar problem. The overlapping genes between Nascent-seq and intron counts are still small,34,37 although it is possible that differences in criteria (intron cycling genes vs nascent transcripts) affected the outcomes, even though both should indicate the status of active de novo transcription. Moreover, two independent SILAC proteome studies identified only 54 proteins shared between the data sets, of 186 and 476 rhythmic proteins identified in each study. This could be due to several factors: differences in the experimental design, such as light versus dark condition (i.e., LD vs DD), sampling intervals (i.e., every one vs four vs 6 h), or sampling duration [i.e., one vs two circadian cycle(s)], experimental conditions to lyse and/or homogenize cells (i.e., solubility of proteins), sensitivity of mass spectrometry/microarray/RNA-seq, methods used for circadian rhythmicity analyses, or a combination of any of these. Biological variability may also explain some discrepant results, but perhaps the most critical factor is the analytical method used to define whether the expression of a particular transcript is rhythmic or not; this further emphasizes the difficulty of extracting ccgs from large data sets.
For the detection of circadian rhythmicity, several algorithms have been used historically, including JTK_CYCLE, ARSER, COSOPT, Fisher’s G test, and CircWave (http://www.euclock.org/results/item/circ-wave.html).21,100,101 Each algorithm has unique characteristics for detecting ccgs from noisy data sets, and multiple factors such as tolerance to noise (i.e., outliers) and fit to sinusoid can significantly affect the results. These algorithms run using “periodic regression analysis”, which utilizes the principles of regression analysis and tests the goodness of fit between experimental data and a sinusoid, and/or “spectrum analysis”, which calculates a periodic function that would best fit the experimental data and analyzes periodic component(s) within the data. More sensitive algorithms return larger numbers of ccgs because they can detect low-amplitude rhythms. Moreover, the numbers of ccgs can also be significantly affected by the normalization process, especially for weakly rhythmic genes. In contrast to microarray studies, in which quantile normalization has been the default methodology, NGS technology still seeks an optimal normalization method,102 and this issue can also introduce variability into the detection of rhythmicity. This raises the following serious question: are these weakly rhythmic genes biologically meaningful? There is no clear answer to the question yet, but the existence of ccgs with low amplitudes may indicate that the circadian clock modulates output physiology in a subtle manner.
As such, it is clear that we have to be careful in interpreting the data from genomic analyses. Just like any other biological experiments, it is extremely difficult to exclude false positives without creating false negatives and to set an arbitrary but significant threshold. Less stringent parameters will most likely result in an increased number of rhythmic genes and thus more common ccgs across the data sets, although it is still unclear what would be the appropriate boundary to distinguish rhythmic from nonrhythmic and biologically meaningful rhythms from noise.
Concluding Remarks and Future Perspectives
There is no doubt that these various global approaches are powerful and that it is important to identify rhythmically expressed genes and, furthermore, to understand the level at which the clock exerts its control to generate a rhythmic output; however, the identification of the true ccgs, which are ultimately important for controlling circadian physiology and behavior, can be challenging. What are “bona fide” ccgs with functional importance? How can these be defined? Within the 54 proteins commonly identified as being rhythmic in SILAC studies, 11 proteins exhibited rhythmicity at the mRNA level in transcriptome analyses (Table 1). These genes appear to be good candidates for “bona fide” ccgs; however, it is dangerous to rely solely on gene expression data based on whether a given mRNA or protein is rhythmic, because rhythmicity is determined with an arbitrary significance threshold and it is virtually impossible to eliminate all the false positives and/or negatives, as mentioned above. Even if the amplitude of the expression rhythm of a gene is too low to detect, output rhythms can be amplified by having multiple weak ccgs functioning within the same pathway, and the rhythmic expression of a key (i.e., rate-limiting) enzyme might be able to confer rhythmicity to the entire pathway, in spite of a lack of rhythmicity in other genes involved in the pathway.
Table 1. Rhythmically Expressed Genes at Protein and mRNA Levels in Mouse Liver.
| gene | no. of positivesa | molecular function |
|---|---|---|
| POR | 9 | cytochrome P450 reductase |
| FKBP4 | 8 | peptidyl-prolyl cis–trans isomerase |
| TARS | 8 | threonyl-tRNA synthetase |
| ALAS1 | 7 | 5-aminolevulinate synthase |
| ABCC2 | 6 | canalicular multispecific organic anion transporter |
| CLPX | 6 | ATP-dependent Clp protease ATP-binding subunit |
| CROT | 6 | peroxisomal carnitine O-octanoyltransferase |
| SLC7A2 | 6 | low-affinity cationic amino acid transporter |
| FMO5 | 5 | dimethylaniline monooxygenase |
| GNE | 5 | bifunctional UDP-N-acetylglucosamine |
| MAN2A1 | 5 | α-mannosidase 2 |
Numbers of studies that identified a given gene as being rhythmic.
Indeed, gene ontology analyses support the idea that there are particular pathways that are common between experiments even though specific ccgs are not. For instance, between SCN and liver, genes involved in “Ubl conjugation (i.e., proteins that are post-translationally modified by the attachment of at least one ubiquitin-like modifier protein, such as ubiquitin and SUMO)” are enriched in data sets from both tissues (Tables 2 and 3). Given the involvement of ubiquitin ligases, FBXL3 and FBXL21, in the core clock mechanism, this result from ontology analysis seems quite reasonable.103−105 Additionally, keywords describing “Nucleotide-Binding” and “Apoptosis” appear to be under circadian control (Tables 2 and 3), although the mechanistic analyses that link these pathways to the circadian clock are still lacking. It is possible that the circadian clock regulates the same pathways in different tissues by utilizing different genes, even though there are only a few overlapping ccgs between different tissues. It would be interesting to pursue the possible mechanistic links between the circadian clock and these processes.
Table 2. Pathway/Domain Terms Shared in Two Independent SCN ccg Data Sets21,23.
| pathwaya | description |
|---|---|
| circadian rhythm | GO:007623, Mmu04710 |
| acetylation | SP_PIR_KEYWORDS |
| apoptosis | SP_PIR_KEYWORDS, GO:006915 |
| programmed cell death | GO:0012501 |
| methylation | SP_PIR_KEYWORDS |
| nucleus | SP_PIR_KEYWORDS |
| phosphoprotein | SP_PIR_KEYWORDS |
| Ubl conjugation | SP_PIR_KEYWORDS |
| nucleotide binding | GO:0000166, IPR012677, SP_PIR_KEYWORDS |
| membrane-enclosed lumen | GO:0031974 |
| organelle lumen | GO:0043233 |
| intracellular organelle lumen | GO:0070013 |
| RNA recognition motif (RRM) | IPR000504, SM00360 |
| cell death | GO:0008219 |
| death | GO:0016265 |
| nuclear receptor ROR | IPR003079 |
| PAS | IPR000014, IPR013655, SM00086, UP_SEQ_FEATURE |
| PAC motif | IPR001610, SM00091 |
DAVID returns pathways that are enriched in a given gene data set with each P value being up to 0.1.
Table 3. Pathway/Domain Terms Shared in Nine Independent Liver ccg Data Sets21−23,29,33−37.
| criteria | pathway | description |
|---|---|---|
| P < 0.05 in all | biological rhythms | SP_PIR_KEYWORDS |
| circadian rhythms | mmu04710 | |
| cytoplasm | SP_PIR_KEYWORDS | |
| endoplasmic reticuluma | SP_PIR_KEYWORDS, GO:0005783 | |
| lyase | SP_PIR_KEYWORDS | |
| phosphoproteina | SP_PIR_KEYWORDS | |
| steroid metabolic processa | GO:0008202 | |
| steroid hormone receptor | IPR001723 | |
| nuclear hormone receptor | IPR000536, IPR008946 | |
| zinc finger, nuclear hormone receptor type | IPR001628 | |
| basic leucine zipper | IPR011700 | |
| ZnF_C4 | SM00399 | |
| ligand-binding domain of hormone receptors | SM00430 | |
| binding site: substrate | UP_SEQ_FEATURE | |
| nucleotide bindinga | SP_PIR_KEYWORDS | |
| NADPa | SP_PIR_KEYWORDS | |
| nucleotide bindinga | GO:0000166 | |
| purine nucleotide binding | GO:0017076 | |
| ribonucleotide binding | GO:0032553 | |
| purine ribonucleotide binding | GO:0032555 | |
| P < 0.05 in eight | rhythmic process | GO:0048511 |
| circadian rhythm | GO:0007623 | |
| cofactor metabolic process | GO:0051186 | |
| ligand-dependent nuclear receptor activity | GO:004879 | |
| cytosol | GO:0005829 | |
| lipid biosynthetic process | GO:0008610 | |
| amine biosynthetic process | GO:0009309 | |
| regulation of cell death | GO:0010941 | |
| regulation of apoptosis | GO:0042981 | |
| regulation of programmed cell death | GO:0043067 | |
| endomembrane systema | GO:0012505 | |
| organelle membrane | GO:0031090 | |
| oxidation reductiona | GO:0055114 | |
| oxidoreducatsea | SP_PIR_KEYWORDS | |
| transferase | SP_PIR_KEYWORDS | |
| pyridoxal phosphate-dependent transferase | IPR015421 | |
| pyridoxal phosphate | SP_PIR_KEYWORDS | |
| Ubl conjucation | SP_PIR_KEYWORDS | |
| mutagenesis site | UP_SEQ_FEATURE | |
| BRLZ (basic leucine-zipper motif) | SM00338 | |
| NAD(P)-binding domain | IPR016040 |
Pathways/domains that were also enriched in proteome analyses.
In addition, integrative approaches involving many different types of analysis will provide additional insight and power and will likely be necessary to elucidate how the circadian clock regulates specific biological pathways. For example, it is now widely recognized that the circadian clock regulates metabolism, and disruption of the clock leads to metabolic disorders, including obesity and diabetes, although the detailed mechanisms are still being uncovered. To fill this gap, an interesting approach, CircadiOmics, was performed that combined gene ontology (pathway) analysis, transcriptome (microarray), protein–protein interaction, Bmal1 ChIP (chromatin immunoprecipitation), transcription factor motif analysis, and metabolomics assays. This approach predicted the uracil salvage pathway should be under circadian control, and this prediction was experimentally demonstrated.106
Recent developments in NGS technology in the past five years have completely transformed our view of circadian genomics and unveiled that the gene regulatory networks are far more complicated than we have ever anticipated. A growing body of evidence suggests that transcriptional mechanisms are not sufficient to sustain all rhythmic mRNA expression, and many layers of regulation, particularly post-transcriptional regulation, play significant roles in driving rhythmic gene expression. Despite its significance, understanding of post-transcriptional mechanisms still lags far behind that of transcriptional processes, and genome-wide regulation of the post-transcriptional networks has yet to be examined. Furthermore, this new concept also raises a series of critical questions: What are the real entities that control circadian output pathways? Are they ccgs or rhythmically expressed proteins? If so, what defines the period, phase, and amplitude of rhythmic gene expression, especially when global translation and de novo transcription (nascent transcription) peak at early night?37,78 If not, why do mRNAs/proteins need to be rhythmically generated? It is possible that what has been considered “rhythmic transcription” is a default mechanism that induces transcription once a day, as evidenced by the fact that global transcription occurs predominantly at early night.37 Perhaps post-transcriptional mechanisms serve to determine the phase and amplitude of each cycling mRNA. The circadian system ensures that the overt output rhythms are sustained continuously and periodically by employing multiple different regulatory mechanisms. Further studies will be needed to answer these questions to fully understand the global impact that the circadian clock imposes on downstream physiologies.
Acknowledgments
We thank all the Green lab members for helpful discussions and comments, especially C. Rosensweig and Dr. M. de Groot for a critical reading of the manuscript.
Glossary
Abbreviations
- Ccg
clock-controlled gene
- NGS
next-generation sequencing.
This work was supported by the National Institutes of Health (Grants R01GM090247, R21NS079986, and R01AG045795 to C.B.G.) and the Tomizawa Jun-ichi & Keiko Fund of Molecular Biology Society of Japan for Young Scientist and the Brain and Behavior Research Foundation (NARSAD) (to S.K.).
The authors declare no competing financial interest.
Funding Statement
National Institutes of Health, United States
References
- Lowrey P. L.; Takahashi J. S. (2004) Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet. 5, 407–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A. B.; Karp N. A.; Maywood E. S.; Sage E. A.; Deery M.; O’Neill J. S.; Wong G. K.; Chesham J.; Odell M.; Lilley K. S.; Kyriacou C. P.; Hastings M. H. (2006) Circadian orchestration of the hepatic proteome. Curr. Biol. 16, 1107–1115. [DOI] [PubMed] [Google Scholar]
- Mauvoisin D.; Wang J.; Jouffe C.; Martin E.; Atger F.; Waridel P.; Quadroni M.; Gachon F.; Naef F. (2014) Circadian clock-dependent and -independent rhythmic proteomes implement distinct diurnal functions in mouse liver. Proc. Natl. Acad. Sci. U.S.A. 111, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel C.; Marcotte E. M. (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat. Rev. Genet. 13, 227–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J.; Falvey E.; Lavery D.; Talbot D.; Schmidt E.; Ossipow V.; Fonjallaz P.; Schibler U. (1992) The role of the transcriptional activator protein DBP in circadian liver gene expression. J. Cell Sci., Suppl. 16, 123–127. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K.; Sassone-Corsi P. (2013) Epigenetic regulation of the molecular clockwork. Prog. Mol. Biol. Transl. Sci. 119, 29–50. [DOI] [PubMed] [Google Scholar]
- Aguilar-Arnal L.; Sassone-Corsi P. (2013) The circadian epigenome: How metabolism talks to chromatin remodeling. Curr. Opin. Cell Biol. 25, 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D.; Lazar M. A. (2012) Clocks, metabolism, and the epigenome. Mol. Cell 47, 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tei H.; Okamura H.; Shigeyoshi Y.; Fukuhara C.; Ozawa R.; Hirose M.; Sakaki Y. (1997) Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 389, 512–516. [DOI] [PubMed] [Google Scholar]
- Sun Z. S.; Albrecht U.; Zhuchenko O.; Bailey J.; Eichele G.; Lee C. C. (1997) RIGUI, a putative mammalian ortholog of the Drosophila period gene. Cell 90, 1003–1011. [DOI] [PubMed] [Google Scholar]
- King D. P.; Zhao Y.; Sangoram A. M.; Wilsbacher L. D.; Tanaka M.; Antoch M. P.; Steeves T. D.; Vitaterna M. H.; Kornhauser J. M.; Lowrey P. L.; Turek F. W.; Takahashi J. S. (1997) Positional cloning of the mouse circadian clock gene. Cell 89, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearman L. P.; Zylka M. J.; Weaver D. R.; Kolakowski L. F. Jr.; Reppert S. M. (1997) Two period homologs: Circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron 19, 1261–1269. [DOI] [PubMed] [Google Scholar]
- Takumi T.; Taguchi K.; Miyake S.; Sakakida Y.; Takashima N.; Matsubara C.; Maebayashi Y.; Okumura K.; Takekida S.; Yamamoto S.; Yagita K.; Yan L.; Young M. W.; Okamura H. (1998) A light-independent oscillatory gene mPer3 in mouse SCN and OVLT. EMBO J. 17, 4753–4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangoram A. M.; Saez L.; Antoch M. P.; Gekakis N.; Staknis D.; Whiteley A.; Fruechte E. M.; Vitaterna M. H.; Shimomura K.; King D. P.; Young M. W.; Weitz C. J.; Takahashi J. S. (1998) Mammalian circadian autoregulatory loop: A timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron 21, 1101–1113. [DOI] [PubMed] [Google Scholar]
- Zylka M. J.; Shearman L. P.; Levine J. D.; Jin X.; Weaver D. R.; Reppert S. M. (1998) Molecular analysis of mammalian timeless. Neuron 21, 1115–1122. [DOI] [PubMed] [Google Scholar]
- Koike N.; Hida A.; Numano R.; Hirose M.; Sakaki Y.; Tei H. (1998) Identification of the mammalian homologues of the Drosophila timeless gene, Timeless1. FEBS Lett. 441, 427–431. [DOI] [PubMed] [Google Scholar]
- Takumi T.; Nagamine Y.; Miyake S.; Matsubara C.; Taguchi K.; Takekida S.; Sakakida Y.; Nishikawa K.; Kishimoto T.; Niwa S.; Okumura K.; Okamura H. (1999) A mammalian ortholog of Drosophila timeless, highly expressed in SCN and retina, forms a complex with mPER1. Genes Cells 4, 67–75. [DOI] [PubMed] [Google Scholar]
- Lowrey P. L.; Shimomura K.; Antoch M. P.; Yamazaki S.; Zemenides P. D.; Ralph M. R.; Menaker M.; Takahashi J. S. (2000) Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 288, 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph M. R.; Menaker M. (1988) A mutation of the circadian system in golden hamsters. Science 241, 1225–1227. [DOI] [PubMed] [Google Scholar]
- Takahashi J. S.; Hong H. K.; Ko C. H.; McDearmon E. L. (2008) The genetics of mammalian circadian order and disorder: Implications for physiology and disease. Nat. Rev. Genet. 9, 764–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S.; Antoch M. P.; Miller B. H.; Su A. I.; Schook A. B.; Straume M.; Schultz P. G.; Kay S. A.; Takahashi J. S.; Hogenesch J. B. (2002) Coordinated transcription of key pathways in the mouse by the circadian clock. Cell 109, 307–320. [DOI] [PubMed] [Google Scholar]
- Storch K. F.; Lipan O.; Leykin I.; Viswanathan N.; Davis F. C.; Wong W. H.; Weitz C. J. (2002) Extensive and divergent circadian gene expression in liver and heart. Nature 417, 78–83. [DOI] [PubMed] [Google Scholar]
- Ueda H. R.; Chen W.; Adachi A.; Wakamatsu H.; Hayashi S.; Takasugi T.; Nagano M.; Nakahama K.; Suzuki Y.; Sugano S.; Iino M.; Shigeyoshi Y.; Hashimoto S. (2002) A transcription factor response element for gene expression during circadian night. Nature 418, 534–539. [DOI] [PubMed] [Google Scholar]
- Hoogerwerf W. A.; Sinha M.; Conesa A.; Luxon B. A.; Shahinian V. B.; Cornelissen G.; Halberg F.; Bostwick J.; Timm J.; Cassone V. M. (2008) Transcriptional profiling of mRNA expression in the mouse distal colon. Gastroenterology 135, 2019–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kita Y.; Shiozawa M.; Jin W.; Majewski R. R.; Besharse J. C.; Greene A. S.; Jacob H. J. (2002) Implications of circadian gene expression in kidney, liver and the effects of fasting on pharmacogenomic studies. Pharmacogenetics 12, 55–65. [DOI] [PubMed] [Google Scholar]
- Humphries A.; Klein D.; Baler R.; Carter D. A. (2002) cDNA array analysis of pineal gene expression reveals circadian rhythmicity of the dominant negative helix-loop-helix protein-encoding gene, Id-1. J. Neuroendocrinol. 14, 101–108. [DOI] [PubMed] [Google Scholar]
- Duffield G. E.; Best J. D.; Meurers B. H.; Bittner A.; Loros J. J.; Dunlap J. C. (2002) Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr. Biol. 12, 551–557. [DOI] [PubMed] [Google Scholar]
- Grundschober C.; Delaunay F.; Puhlhofer A.; Triqueneaux G.; Laudet V.; Bartfai T.; Nef P. (2001) Circadian regulation of diverse gene products revealed by mRNA expression profiling of synchronized fibroblasts. J. Biol. Chem. 276, 46751–46758. [DOI] [PubMed] [Google Scholar]
- Hughes M. E.; DiTacchio L.; Hayes K. R.; Vollmers C.; Pulivarthy S.; Baggs J. E.; Panda S.; Hogenesch J. B. (2009) Harmonics of circadian gene transcription in mammals. PLoS Genet. 5, e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffield G. E. (2003) DNA microarray analyses of circadian timing: The genomic basis of biological time. J. Neuroendocrinol. 15, 991–1002. [DOI] [PubMed] [Google Scholar]
- Knight J. (2001) When the chips are down. Nature 410, 860–861. [DOI] [PubMed] [Google Scholar]
- Hayden E. C. (2014) Technology: The $1,000 genome. Nature 507, 294–295. [DOI] [PubMed] [Google Scholar]
- Yoshitane H.; Ozaki H.; Terajima H.; Du N. H.; Suzuki Y.; Fujimori T.; Kosaka N.; Shimba S.; Sugano S.; Takagi T.; Iwasaki W.; Fukada Y. (2014) CLOCK-controlled polyphonic regulations of circadian rhythms through canonical and non-canonical E-boxes. Mol. Cell. Biol. 34, 1776–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet J. S.; Rodriguez J.; Abruzzi K. C.; Rosbash M. (2012) Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. eLife 1, e00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C.; Schmitz R. J.; Nathanson J.; Yeo G.; Ecker J. R.; Panda S. (2012) Circadian oscillations of protein-coding and regulatory RNAs in a highly dynamic mammalian liver epigenome. Cell Metab. 16, 833–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G.; Canella D.; Symul L.; Migliavacca E.; Gilardi F.; Liechti R.; Martin O.; Harshman K.; Delorenzi M.; Desvergne B.; Herr W.; Deplancke B.; Schibler U.; Rougemont J.; Guex N.; Hernandez N.; Naef F.; Cycli X. C. (2012) Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 10, e1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N.; Yoo S. H.; Huang H. C.; Kumar V.; Lee C.; Kim T. K.; Takahashi J. S. (2012) Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 338, 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derrien T.; Johnson R.; Bussotti G.; Tanzer A.; Djebali S.; Tilgner H.; Guernec G.; Martin D.; Merkel A.; Knowles D. G.; Lagarde J.; Veeravalli L.; Ruan X.; Ruan Y.; Lassmann T.; Carninci P.; Brown J. B.; Lipovich L.; Gonzalez J. M.; Thomas M.; Davis C. A.; Shiekhattar R.; Gingeras T. R.; Hubbard T. J.; Notredame C.; Harrow J.; Guigo R. (2012) The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H. Y.; Papp J. W.; Varlamova O.; Dziema H.; Russell B.; Curfman J. P.; Nakazawa T.; Shimizu K.; Okamura H.; Impey S.; Obrietan K. (2007) microRNA modulation of circadian-clock period and entrainment. Neuron 54, 813–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi A.; Vaz C.; Bhattacharya A.; Ramaswamy R. (2009) miRNA-regulated dynamics in circadian oscillator models. BMC Syst. Biol. 3, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K.; Wang R. (2012) MicroRNA-mediated regulation in the mammalian circadian rhythm. J. Theor. Biol. 304, 103–110. [DOI] [PubMed] [Google Scholar]
- Na Y. J.; Sung J. H.; Lee S. C.; Lee Y. J.; Choi Y. J.; Park W. Y.; Shin H. S.; Kim J. H. (2009) Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp. Mol. Med. 41, 638–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.; Witmer P. D.; Lumayag S.; Kovacs B.; Valle D. (2007) MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem. 282, 25053–25066. [DOI] [PubMed] [Google Scholar]
- Gatfield D.; Le Martelot G.; Vejnar C. E.; Gerlach D.; Schaad O.; Fleury-Olela F.; Ruskeepaa A. L.; Oresic M.; Esau C. C.; Zdobnov E. M.; Schibler U. (2009) Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev. 23, 1313–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S.; Gatfield D.; Esau C. C.; Green C. B. (2010) MicroRNA-122 modulates the rhythmic expression profile of the circadian deadenylase Nocturnin in mouse liver. PLoS One 5, e11264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shende V. R.; Neuendorff N.; Earnest D. J. (2013) Role of miR-142-3p in the post-transcriptional regulation of the clock gene Bmal1 in the mouse SCN. PLoS One 8, e65300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W.; Li Y.; Wang X.; Wu L.; Wang Y. (2011) MiR-206-mediated dynamic mechanism of the mammalian circadian clock. BMC Syst. Biol. 5, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H.; Kim S. H.; Lee H. R.; Kim W.; Kim D. Y.; Shin J. C.; Yoo S. H.; Kim K. T. (2013) MicroRNA-185 oscillation controls circadian amplitude of mouse Cryptochrome 1 via translational regulation. Mol. Biol. Cell 24, 2248–2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y. E.; Pan Y.; Park E.; Konstantinopoulos P.; De S.; D’Andrea A.; Chowdhury D. (2014) MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. eLife 3, e02445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du N. H.; Arpat A. B.; De Matos M.; Gatfield D. (2014) MicroRNAs shape circadian hepatic gene expression on a transcriptome-wide scale. eLife 3, e02510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon S. L.; Munson P. J.; Cherukuri P. F.; Sugden D.; Rath M. F.; Möller M.; Clokie S. J.; Fu C.; Olanich M. E.; Rangel Z.; Werner T.; Program N. C. S.; Mullikin J. C.; Klein D. C. (2012) Circadian changes in long noncoding RNAs in the pineal gland. Proc. Natl. Acad. Sci. U.S.A. 109, 13319–13324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. T.; Bartolomei M. S. (2013) X-inactivation, imprinting, and long noncoding RNAs in health and disease. Cell 152, 1308–1323. [DOI] [PubMed] [Google Scholar]
- Chen J.; Sun M.; Hurst L. D.; Carmichael G. G.; Rowley J. D. (2005) Human antisense genes have unusually short introns: Evidence for selection for rapid transcription. Trends Genet. 21, 203–207. [DOI] [PubMed] [Google Scholar]
- Chen J.; Sun M.; Hurst L. D.; Carmichael G. G.; Rowley J. D. (2005) Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 21, 326–329. [DOI] [PubMed] [Google Scholar]
- Katayama S.; Tomaru Y.; Kasukawa T.; Waki K.; Nakanishi M.; Nakamura M.; Nishida H.; Yap C. C.; Suzuki M.; Kawai J.; Suzuki H.; Carninci P.; Hayashizaki Y.; Wells C.; Frith M.; Ravasi T.; Pang K. C.; Hallinan J.; Mattick J.; Hume D. A.; Lipovich L.; Batalov S.; Engstrom P. G.; Mizuno Y.; Faghihi M. A.; Sandelin A.; Chalk A. M.; Mottagui-Tabar S.; Liang Z.; Lenhard B.; Wahlestedt C. (2005) Antisense transcription in the mammalian transcriptome. Science 309, 1564–1566. [DOI] [PubMed] [Google Scholar]
- Kiyosawa H.; Yamanaka I.; Osato N.; Kondo S.; Hayashizaki Y. (2003) Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 13, 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauman I.; Reppert S. M. (1996) Circadian clock neurons in the silkmoth Antheraea pernyi: Novel mechanisms of Period protein regulation. Neuron 17, 889–900. [DOI] [PubMed] [Google Scholar]
- Kramer C.; Loros J. J.; Dunlap J. C.; Crosthwaite S. K. (2003) Role for antisense RNA in regulating circadian clock function in Neurospora crassa. Nature 421, 948–952. [DOI] [PubMed] [Google Scholar]
- Belanger V.; Picard N.; Cermakian N. (2006) The circadian regulation of Presenilin-2 gene expression. Chronobiol. Int. 23, 747–766. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Hu W.; Murakawa Y.; Yin J.; Wang G.; Landthaler M.; Yan J. (2013) Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci. Rep. 3, 2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagampang F. R.; Yang J.; Nakayama Y.; Fukuhara C.; Inouye S. T. (1994) Circadian variation of arginine-vasopressin messenger RNA in the rat suprachiasmatic nucleus. Brain Res. 24, 179–184. [DOI] [PubMed] [Google Scholar]
- Robinson B. G.; Frim D. M.; Schwartz W. J.; Majzoub J. A. (1988) Vasopressin mRNA in the suprachiasmatic nuclei: Daily regulation of polyadenylate tail length. Science 241, 342–344. [DOI] [PubMed] [Google Scholar]
- Gerstner J. R.; Vanderheyden W. M.; Lavaute T.; Westmark C. J.; Rouhana L.; Pack A. I.; Wickens M.; Landry C. F. (2012) Time of day regulates subcellular trafficking, tripartite synaptic localization, and polyadenylation of the astrocytic fabp7 mRNA. J. Neurosci. 32, 1383–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So W. V.; Rosbash M. (1997) Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J. 16, 7146–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbacher L. D.; Yamazaki S.; Herzog E. D.; Song E. J.; Radcliffe L. A.; Abe M.; Block G.; Spitznagel E.; Menaker M.; Takahashi J. S. (2002) Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice invivo. Proc. Natl. Acad. Sci. U.S.A. 99, 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S.; Matsumoto K.; Hirose M.; Shimada M.; Nagano M.; Shigeyoshi Y.; Hoshino S.; Ui-Tei K.; Saigo K.; Green C. B.; Sakaki Y.; Tei H. (2007) LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. U.S.A. 104, 1859–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S.; Hirose M.; Tokunaga K.; Sakaki Y.; Tei H. (2003) Structural and functional analysis of 3′ untranslated region of mouse Period1 mRNA. Biochem. Biophys. Res. Commun. 301, 1–7. [DOI] [PubMed] [Google Scholar]
- Kim D. Y.; Woo K. C.; Lee K. H.; Kim T. D.; Kim K. T. (2010) hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb α via IRES-mediated translation. Nucleic Acids Res. 38, 7068–7078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H.; Kim S. H.; Kim H. J.; Kim W.; Lee H. R.; Jung Y.; Choi J. H.; Hong K. Y.; Jang S. K.; Kim K. T. (2014) AUF1 contributes to Cryptochrome1 mRNA degradation and rhythmic translation. Nucleic Acids Res. 42, 3590–3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H.; Woo K. C.; Kim D. Y.; Kim T. D.; Shin J.; Park S. M.; Jang S. K.; Kim K. T. (2012) Rhythmic interaction between Period1 mRNA and hnRNP Q leads to circadian time-dependent translation. Mol. Cell. Biol. 32, 717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T. D.; Woo K. C.; Cho S.; Ha D. C.; Jang S. K.; Kim K. T. (2007) Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev. 21, 797–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D. Y.; Kwak E.; Kim S. H.; Lee K. H.; Woo K. C.; Kim K. T. (2011) hnRNP Q mediates a phase-dependent translation-coupled mRNA decay of mouse Period3. Nucleic Acids Res. 39, 8901–8914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K. C.; Ha D. C.; Lee K. H.; Kim D. Y.; Kim T. D.; Kim K. T. (2010) Circadian amplitude of cryptochrome 1 is modulated by mRNA stability regulation via cytoplasmic hnRNP D oscillation. Mol. Cell. Biol. 30, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo K. C.; Kim T. D.; Lee K. H.; Kim D. Y.; Kim W.; Lee K. Y.; Kim K. T. (2009) Mouse period 2 mRNA circadian oscillation is modulated by PTB-mediated rhythmic mRNA degradation. Nucleic Acids Res. 37, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak E.; Kim T. D.; Kim K. T. (2006) Essential role of 3′-untranslated region-mediated mRNA decay in circadian oscillations of mouse Period3 mRNA. J. Biol. Chem. 281, 19100–19106. [DOI] [PubMed] [Google Scholar]
- Cao R.; Robinson B.; Xu H.; Gkogkas C.; Khoutorsky A.; Alain T.; Yanagiya A.; Nevarko T.; Liu A. C.; Amir S.; Sonenberg N. (2013) Translational control of entrainment and synchrony of the suprachiasmatic circadian clock by mTOR/4E-BP1 signaling. Neuron 79, 712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R.; Anderson F. E.; Jung Y. J.; Dziema H.; Obrietan K. (2011) Circadian regulation of mammalian target of rapamycin signaling in the mouse suprachiasmatic nucleus. Neuroscience 181, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouffe C.; Cretenet G.; Symul L.; Martin E.; Atger F.; Naef F.; Gachon F. (2013) The circadian clock coordinates ribosome biogenesis. PLoS Biol. 11, e1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlincy N. J.; Valomon A.; Chesham J. E.; Maywood E. S.; Hastings M. H.; Ule J. (2012) Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol. 13, R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J. E. Jr. (2013) Reflections on the history of pre-mRNA processing and highlights of current knowledge: A unified picture. RNA 19, 443–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodor Y. L.; Menet J. S.; Tolan M.; Rosbash M. (2012) Cotranscriptional splicing efficiency differs dramatically between Drosophila and mouse. RNA 18, 2174–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S.; Sher-Chen E. L.; Green C. B. (2012) Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev. 26, 2724–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin J. M.; Doi M.; Yamaguchi Y.; Hida H.; Nishimura S.; Yoshida M.; Isagawa T.; Morioka M. S.; Kakeya H.; Manabe I.; Okamura H. (2013) RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. [DOI] [PubMed] [Google Scholar]
- Morf J.; Rey G.; Schneider K.; Stratmann M.; Fujita J.; Naef F.; Schibler U. (2012) Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science 338, 379–383. [DOI] [PubMed] [Google Scholar]
- Dominissini D.; Moshitch-Moshkovitz S.; Schwartz S.; Salmon-Divon M.; Ungar L.; Osenberg S.; Cesarkas K.; Jacob-Hirsch J.; Amariglio N.; Kupiec M.; Sorek R.; Rechavi G. (2012) Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- Churchman L. S., and Weissman J. S. (2012) Native elongating transcript sequencing (NET-seq). Current Protocols in Molecular Biology (Ausubel F. M., et al. , Eds.) Chapter 4, Unit 4, 14, pp 11–17, Wiley, New York. [DOI] [PubMed] [Google Scholar]
- Core L. J.; Waterfall J. J.; Lis J. T. (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen K. B.; Darnell R. B. (2008) CLIP: Crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol. Biol. 488, 85–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J.; Ohsumi T. K.; Kung J. T.; Ogawa Y.; Grau D. J.; Sarma K.; Song J. J.; Kingston R. E.; Borowsky M.; Lee J. T. (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 40, 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani H.; Akimitsu N. (2012) Genome-wide technology for determining RNA stability in mammalian cells: Historical perspective and recent advantages based on modified nucleotide labeling. RNA Biol. 9, 1233–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C.; Kudla G.; Wlotzka W.; Tuck A.; Tollervey D. (2012) Transcriptome-wide analysis of exosome targets. Mol. Cell 48, 422–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H.; Lim J.; Ha M.; Kim V. N. (2014) TAIL-seq: Genome-wide Determination of Poly(A) Tail Length and 3′ End Modifications. Mol. Cell 53, 1044–1052. [DOI] [PubMed] [Google Scholar]
- Subtelny A. O.; Eichhorn S. W.; Chen G. R.; Sive H.; Bartel D. P. (2014) Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508, 66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y.; Qu K.; Zhang Q. C.; Flynn R. A.; Manor O.; Ouyang Z.; Zhang J.; Spitale R. C.; Snyder M. P.; Segal E.; Chang H. Y. (2014) Landscape and variation of RNA secondary structure across the human transcriptome. Nature 505, 706–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouskin S.; Zubradt M.; Washietl S.; Kellis M.; Weissman J. S. (2014) Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Lu Z.; Gomez A.; Hon G. C.; Yue Y.; Han D.; Fu Y.; Parisien M.; Dai Q.; Jia G.; Ren B.; Pan T.; He C. (2014) N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. (2006) Functional and quantitative proteomics using SILAC. Nat. Rev. Mol. Cell Biol. 7, 952–958. [DOI] [PubMed] [Google Scholar]
- Robles M. S.; Cox J.; Mann M. (2014) In-vivo quantitative proteomics reveals a key contribution of post-transcriptional mechanisms to the circadian regulation of liver metabolism. PLoS Genet. 10, e1004047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J.; Schibler U. (1990) Expression of the liver-enriched transcriptional activator protein DBP follows a stringent circadian rhythm. Cell 63, 1257–1266. [DOI] [PubMed] [Google Scholar]
- Yang R.; Su Z. (2010) Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. Bioinformatics 26, i168–i174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M. E.; Hogenesch J. B.; Kornacker K. (2010) JTK_CYCLE: An efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J. Biol. Rhythms 25, 372–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M. D.; Oshlack A. (2010) A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka S. M.; Yoo S. H.; Park J.; Song W.; Kumar V.; Hu Y.; Lee C.; Takahashi J. S. (2007) Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell 129, 1011–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. H.; Mohawk J. A.; Siepka S. M.; Shan Y.; Huh S. K.; Hong H. K.; Kornblum I.; Kumar V.; Koike N.; Xu M.; Nussbaum J.; Liu X.; Chen Z.; Chen Z. J.; Green C. B.; Takahashi J. S. (2013) Competing E3 ubiquitin ligases govern circadian periodicity by degradation of CRY in nucleus and cytoplasm. Cell 152, 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A.; Yumimoto K.; Tsunematsu R.; Matsumoto M.; Oyama M.; Kozuka-Hata H.; Nakagawa T.; Lanjakornsiripan D.; Nakayama K. I.; Fukada Y. (2013) FBXL21 regulates oscillation of the circadian clock through ubiquitination and stabilization of cryptochromes. Cell 152, 1106–1118. [DOI] [PubMed] [Google Scholar]
- Eckel-Mahan K. L.; Patel V. R.; Mohney R. P.; Vignola K. S.; Baldi P.; Sassone-Corsi P. (2012) Coordination of the transcriptome and metabolome by the circadian clock. Proc. Natl. Acad. Sci. U.S.A. 109, 5541–5546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J.; Qian S. B. (2014) TISdb: A database for alternative translation initiation in mammalian cells. Nucleic Acids Res. 42, D845–D850. [DOI] [PMC free article] [PubMed] [Google Scholar]