Abstract
In this study, we examined colonization dynamics of segmented filamentous bacteria (SFB) in intestine of Swiss Webster (SW) mice infected with Helicobacter hepaticus (Hh). At 8 weeks post-inoculation with Hh (WPI), cecal and colonic SFB levels in the control males were significantly lower compared to those at 16 WPI. Hh infection in both genders did not alter SFB levels in the jejunum and ileum, but increased SFB levels in the cecum and colon of males compared to the controls (P<0.05) at 8 WPI. At 16 WPI, the Hh-infected females contained lower levels of SFB in the jejunum, cecum and colon compared to the female controls. Irrespective of gender, aging and Hh infection, the Il-17A mRNA levels decreased from the small intestine to the cecum and then to the colon, whereas the Foxp3 mRNA levels were comparable in these intestinal regions. There were significant differences in Il-17A mRNA levels in the ileum (P<0.05, R2 = 0.31), with females having greater Il-17A mRNA levels than males, and higher SFB colonization levels related to more Il-17A mRNA. These results indicate that aging and gender play an important role in colonization dynamics of intestinal SFB and ileal SFB-associated Th17 response.
Keywords: Segmented filamentous bacteria, Colonization dynamics, H. hepaticus, Il-17A expression, Swiss Webster mice
1. Introduction
The intestinal mucosal surfaces of humans and other animals are colonized by complex microbiota which confer multiple benefits to the host such as providing essential nutrients, energy harvest, the maturation and regulation of the mucosal immune system and resistance to colonization by invading microorganisms [1, 2]. However, perturbation of the commensal microbiota is also associated with human diseases such as obesity, IBD and colon cancer [1, 2]. A well characterized example of commensal bacteria contributing to development of host immunity is segmented filamentous bacteria (SFB) that recently have been shown to promote a robust Th17 cell response [3, 4]. SFB, a group of spore-forming gram-positive bacteria, were previously named Candidatus Arthromitus because of their morphological resemblance to bacterial filaments and are currently given a provisional name, Candidatus Savagella, in honor of the American gut microbiologist Dwayne C. Savage [5]. These bacteria are members of the gut commensal microbiota in a wide spectrum of hosts including mice, rats, dogs, rabbits, fish and humans [6–9]. They are phylogenetically most closely related to the genus Clostridium and to date are uncultivable. The inability to culture SFB is likely in part due to the lack of pathways for synthesizing essential amino acids as suggested by their genomic sequences [10–13].
SFB predominantly colonize the terminal ileum where they act as a strong inducer of proinflammatory T helper 17 (Th17) cell responses [3, 4, 14, 15]. Crucial roles of Th17 cells have been implicated in bacterial clearance, autoimmune diseases and cancer [16]. SFB also play an important role in inducing mucosal IgA response [15, 17, 18]. Thus, potential effects of SFB on biomedical research in animal models require further investigations [9]. We previously reported that colonization dynamics of select species of 8 altered Schaedler flora (ASF) in Swiss Webster (SW) mice is changed by aging, gender and infection with a murine pathogen Helicobacter hepaticus. In this study, we tested a hypothesis that these factors could also influence intestinal colonization dynamics of SFB in the same set of SW mice.
2. Materials and Methods
2.1. Mice and infection
This is a retrospective cohort study of Swiss Webster mice focusing on colonization dynamics of SFB not included in a previous study [19]. The experimental protocols for housing and infecting mice with H. hepaticus as well as collection of the intestinal samples were previously described [19]. Briefly, 4-to-6-week-old male and female mice free of known murine viruses, Helicobacter spp. and parasites, were obtained from Taconic Farms (Germantown, NY). The mice were maintained in static microisolater cages in an Association for Accreditation and Assessment of Laboratory Animal Care, International-accredited facility, and fed a diet (ProlabRMH3000) from PMI Nutrition International (Richmond, IN). After being in quarantine for a week, the mice were inoculated with H. hepaticus 3B1 (3B1) or sham-dosed with Brucella broth as a control, respectively. Mice received 0.2 ml of fresh inoculum (~2 × 108 organisms) by gastric gavage every other day for a total of three inoculations.
Five male and 5 female mice from each group were euthanized at 8 WPI (15 weeks of age) and 16 WPI (23 weeks of age), respectively. Immediately after euthanasia with CO2, contents in the intestine were removed by rinsing with sterile saline. One-cm segments of jejunum, ileum, cecum, and colon for RNA/DNA isolation were collected and frozen in liquid nitrogen immediately after sampling and stored at −70°C prior to use. Total DNA and RNA from jejunum, ileum, cecum and colon were isolated using Trizol Reagents following the supplier’s procedure (Invitrogen, Carlsbad, CA).
2.2. Fecal and cecal DNA extraction
Three fecal pellets from each of 10 female C57BL/6 mice, 5 from the Jackson Laboratory (Bar Harbor, ME) and 5 from Taconic Farm, were collected and stored at −20°C prior to use. Fecal DNA was prepared using the QIAamp Fast DNA Stool Mini Kit according to the supplier’s protocol (Qiagen Inc., Valencia, CA). Additionally, cecal DNA from 2 female C57BL/6 mice of the Jackson Laboratory was extracted using Trizol Reagents.
2.3. PCR, molecular cloning and qPCR
Two pairs of the SFB 16S rRNA gene-based qPCR primers were described previously [18, 20]: SFB225F (5′-AGGAGGAGTCTGCGGCACATTAGC-3′)/SFB558R (5′-CGCATCCTTTACGCCCAGTTATTC-3′) and SFB736F (5′-GACGCTGAGGCATGAGAGCAT-3′)/SFB844R (5′-GACGGCACGGATTGTTATTCA-3′). PCR with primers SFB225F and SFB844R (Figure 1A) gave rise to approximately 620 bp fragment from fecal DNA of the Taconic C57BL/6 mice using PCR conditions as follows: in a 50 μl-volume containing ~100 ng of DNA, 500 nM of each primer, 1 X reaction buffer #2 (Roche Diagnostics GmbH, Mannheim, Germany), high-fidelity Taq DNA polymerase. A thermocycling program consisted of: at 94°C for 5 min, 35 cycles of 94°C for 1 min/58°C for 1 min/72°C for 1 min, followed by 72 °C for 5 min. The PCR DNA fragment was cloned into a TOPO vector following the supplier’s instruction (Invitrogen, Carlsbad, CA) and plasmid DNA was prepared using the Aquick Mini Kit (Qiagen Inc.). The identity of recombinant plasmids containing the SFB 16S-rRNA gene fragment was confirmed by DNA sequencing in the ABI 310 sequencer (Life Technologies, Grand Island, NY). Concentrations of the plasmid DNA were determined using a spectrophotometer GeneQuan Pro (Amersham Biosciences, Piscataway, NJ).
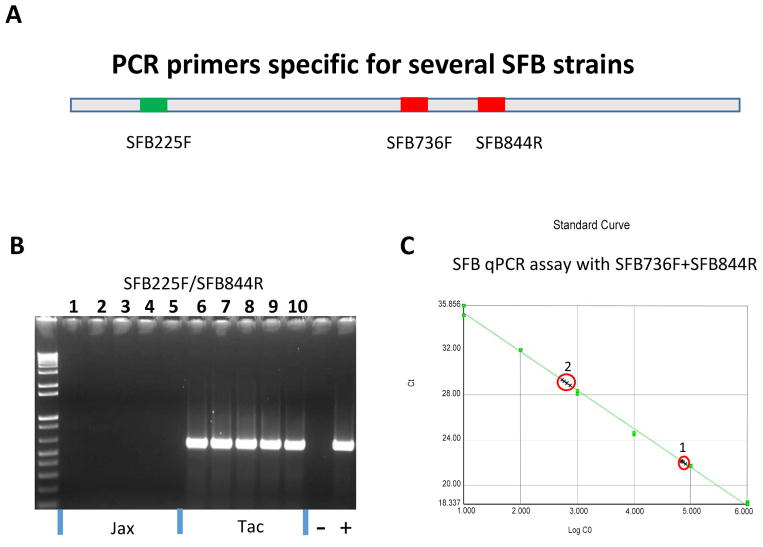
Figure 1.
Development of PCR assays for detecting SFB. A, Schematic representation of PCR primer locations on the SFB 16S rRNA gene. B, Primers SFB225F and SFB844R specifically PCR-amplified a product from SFB. Lanes 1–5, Fecal DNA templates from SFB-negative C57BL/6 mice of the Jackson Laboratory; Lanes 6–10, fecal DNA from SFB-positive C57BL/6 mice of Taconic Farm. C, SFB qPCR assay with optimal amplification efficiency and linear regression (slope, −3.4; r2=0.996). 1 and 2 represent 105 and 103 copeis of the SFB 16S RNA gene spiked in 50 ng of fecal and cecal DNA from two SFB-negative C57BL mice from the Jackson Laboratory, respectively. No fluorescent signal was detected in non-spiked controls (50 ng).
For generating a standard curve of SFB qPCR, a serial 10-fold dilution of 10, 100, 103, 104, 105, 106 copies of the recombinant plasmid DNA containing the SFB 16S rRNA gene fragment was used. SFB levels in each of the respective intestinal segments were quantified with the primer pair of SFB736F and SFB844R based on standard curves by qPCR in the 7500 Fast Real-Time PCR System (Life Technologies). Briefly, a 20-μl mixture contained 5 μl of each of DNA templates of 50 to 100 ng (in duplicate), 1μl of the primer mixture containing 200 nM of each of the forward and reverse primers, 10 μl of 2 x SYGR green Fast master mix (Life Technologies), and 4 μl of ddH2O; the qPCR conditions followed the manufacturer’s default setting added with the dissociation curve function. The copy numbers of the SFB 16S-rRNA gene were then normalized to μg of mouse chromosomal DNA whose quantities in the samples were measured by qPCR using the 18S rRNA gene-based primers and probe mixture (Life Technologies).
2.4. Expression of select cytokines
For relative mRNA quantitation of selected genes, total RNA from jejunum, ileum, cecum and colon of SW mice was prepared using Trizol reagent according to manufacturer’s recommendations (Invitrogen). Five μg of total RNA from each sample was converted into cDNA using the High Capacity cDNA Archive kit (Life Technologies). Levels of Il-17A and Foxp3 mRNA were measured by qPCR using commercial primers and probes (TaqMan Gene Expression Assays) in the 7500 FAST Real-time PCR System. Transcript levels were normalized to the endogenous control glyceraldehyde-3-phosphate dehydrogenase mRNA levels (Gapdh). Since the values of ΔCts are inversely related to the quantity of target genes, the arbitral values of 20 minus ΔCt were used to represent positive correlations between the values and relative quantities of each target.
2.5. Statistical analysis
Multiple factor analysis of variance (ANOVA) tests using STATA/IC 13.0 for Mac (StataCorp, College Station, TX) were performed to determine if SFB colonization levels (continuous variable) varied by H. hepaticus infection status (binary variable) in the jejunum, ileum, cecum and colon, considering sex (binary variable), and H. hepaticus duration (binary variable; 8 or 16 WPI) with subsequent stratification. ANOVA testing was also employed to determine if Il-17A mRNA levels (continuous variable) varied by H. hepaticus infection status or by H. hepaticus colonization levels (continuous variable) and SFB colonization levels, also considering sex and H. hepaticus duration. Additionally, data on the levels of SFB and cytokine expression were also analyzed between two groups using a student t test for normally distributed data or Mann-Whitney U test for abnormally distributed data. Normality of the data sets was analyzed using the Kolmogorov-Smirnov test. Values of P <0.05 were considered significant.
3. Results
3.1. Development of conventional PCR and qPCR assays for detecting SFB in clinical and environmental samples
Given that SFB as a potent inducer of proinflammatory Th17 responses potentially influences the immune responses and pathology of a SFB-positive host to pathogenic infection, it will be very useful to develop conventional and quantitative PCR assays for monitoring SFB status in experimental animals used in biomedical research. Several previous studies described SFB-specific qPCR assays but the relevant assay conditions were not given in detail [11, 18, 20]. Based on two sets of the published primers SFB225F/SFB558 and SFB736F/844R, a pair of primers SFB225F and SFB844R gave rise to a ~620-pb PCR amplicon from SFB-positive C57BL/6 mice (Taconic Farms, Germantown, NY) but not from SFB-negative mice (The Jackson Laboratory, Bar Harbor, ME), which can be visualized on an 1.2 % agarose gel with ethidium bromide staining (Figure 1A and B). Our results demonstrate that this primer pair are SFB-specific under the PCR conditions described in this study, which can be readily applied to detect SFB in clinical and environmental samples.
The 620-pb PCR amplicon cloned into the TOPO plasmid vector was used to generate a standard curve consisting of a serial 10-fold dilution of 10, 100, 103, 104, 105, and 106 copies. The primer pair SFB225F and SFB558R failed to produce quantitative measurement of SFB (data not shown). In contrast, the assay efficacy and linear regression of the standard curve with SyBr green using primers SFB736F and SFB844R satisfied qPCR requirements as presented in Figure 1C. To test whether the DNA templates from murine feces or intestinal tissues have inhibitory effect on the established qPCR assay, 103 or 105 copies of the recombinant plasmid DNA used in generating the SFB standard curve were spiked into fecal and cecal DNA (~50 ng) from SFB-free C57BL/6 from the Jackson Laboratory. The result demonstrated that there was no significant inhibition on the copies of the spiked SFB 16S rRNA gene noted in the assay, whereas SFB was absent in the non-spiked controls (Figure 1C). The low detection limit is ≥10 copies of the SFB 16S rRNA gene.
3.2. Colonization levels of SFB were increased in the large intestine during aging
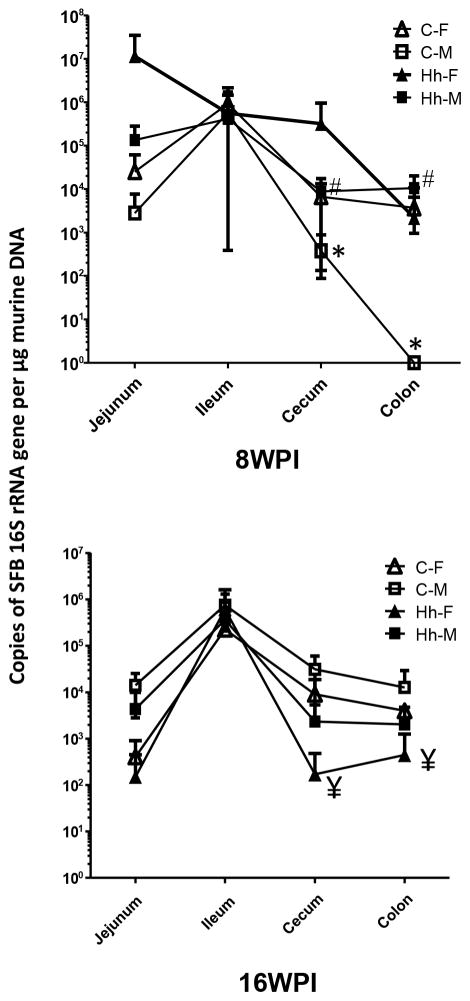
SFB levels in the jejunum, ileum, cecum and colon of each mouse were quantified using the qPCR assay established in this study (Figure 2). At 8 WPI, all 5 females were positive for SFB in the jejunum and ileum; all 5 male mice were SFB-positive in the ileum and 4 of 5 in the jejunum. In the cecum, 4 of the 5 females and 5 males, respectively, were positive for SFB, while 2 of the 5 females and no males were colonized by SFB in the colon. At 16 WPI, all 5 female mice were positive for SFB in ileum, cecum and colon, and 3 of the 5 females were colonized by SFB in the jejunum. All male mice were positive for SFB in the jejunum, ileum and cecum; 4 of 5 also were SFB-positive in the colon. Consistent with previous reports, our results indicated that the small intestine of control mice is a primary colonization niche of SFB. The colonization levels of SFB in the cecum and colon of males was significantly increased at 16 WPI compared to 8 WPI (P <0.05).
Figure 2.
Colonization dynamics of intestinal SFB is influenced by aging and H. hepaticus infection in a gender-dependent manner. Graphs are plotted based mean ± standard deviation of each group. All P values are <0.05, * 8 vs 16 WPI for sham males; # the infected vs sham males at 8WPI; ¥ the infected vs sham females at 16 WPI. All comparisons represent the corresponding intestinal regions.
3.3. H. hepaticus infection altered colonization dynamics of SFB in a gender-dependent manner
Compared to the sham controls, infection of both genders with H. hepaticus did not significantly alter SFB colonization levels in the jejunum and ileum of mice at both time points. Multiple factor ANOVA stratified by H. hepaticus duration revealed a significant difference in SFB colonization levels at 16 WPI in the jejunum [F(2, 13) = 4.15, P<0.05, R2 = 0.39], but this was related to sex, with male mice having higher SFB colonization levels than females. However, H. hepaticus infection did significantly increase SFB levels in the cecum and colon of males at 8 WPI (Figure 2, P <0.05). In contrast, infection in the females significantly decreased cecal and colonic SFB colonization levels compared to sham controls at 16 WPI (P<0.05). There was no correlation between the colonization levels of H. hepaticus and SFB (data not shown). Further, H. hepaticus infected mice were found by multiple factor ANOVA testing to have lower SFB colonization levels in the cecum at 16 WPI, compared to non-infected mice [F(2, 17) = 4.10, P<0.05, R2 = 0.33]; the sex of the mice was found non-significant, but when included in the model explained 9.5% more variance in cecal SFB colonization levels than when excluded, suggesting again that males had higher levels of SFB.
3.4. Analysis of intestinal Il-17A and Foxp3 expression levels
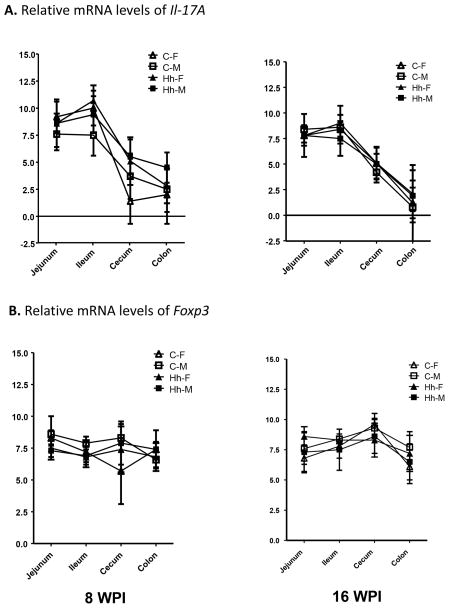
It has been reported that the predominant colonization of SFB in the ileum increases the robust accumulation of Th17 cells, but SFB colonization has little impact on the population of Foxp3-expressing Treg cells in the ileum of SW mice [3]. To ascertain whether SFB colonization levels or H. hepaticus infection affect intestinal Th17 cell response, we examined mRNA levels of Il-17A and Foxp3 mRNA levels in jejunum, ileum, cecum and colon. Expression of Il-17A in sham controls was significantly higher in the jejunum and ileum than in the cecum and colon at both time points (Figure 3A, P<0.05). In contrast, there were no significant changes in Foxp3 mRNA levels between the small and large intestine regions of the sham controls (Figure 3B). Statistically significant differences in Il-17A mRNA levels in the ileum [F(4, 30) = 3.37, P<0.05, R2 = 0.31] were found by multiple factor ANOVA testing, with females having greater Il-17A mRNA levels than males, and higher SFB colonization levels related to more Il-17A mRNA. Also, Il-17A mRNA levels in the colon were greater at 8 WPI, compared to 16 WPI [F(4, 34) = 2.79, P<0.05, R2 = 0.25]. Infection with H. hepaticus did not significantly alter the mRNA levels of both Foxp3 and Il-17A in any of the sampled regions except for in the cecum of the infected females where there was a significant increase in Il-17A mRNA expression compared to the sham controls (Figure 3A, P<0.05).
Figure 3.
Relative expression of intestinal Il-17A and Foxp3 mRNA. A. Higher levels of Il-17A mRNA in the small intestine compared to the large bowel. B. There were no significant differences in mRNA levels of Foxp3 irrespective of aging, intestinal regions, genders or H. hepaticus infection.
4. Discussion
By use of SFB-qPCR assay established in this study, we examined the colonization dynamics of SFB in jejunum, ileum, cecum and colon of SW mice. Our data showed that the ileum is a primary niche for SFB colonization, which is consistent with previous observations [21]. The colonization levels of SFB in the ileum were not influenced significantly by aging, gender or H. hepaticus infection. In addition, SFB were also detected, to lesser extent, in the jejunum, cecum and colon. Interestingly, the colonization efficiency of SFB (both presence and colonization levels) in the large intestine was significantly increased during aging from 8 to 16 WPI. Colonization levels of SFB in the jejunum, cecum and colon of the females trended to be higher than those in the males at 8 WPI but the opposite was true at 16 WPI.
H. hepaticus infection altered colonization dynamics of SFB in the cecum and colon in a gender-dependent manner. At 8WPI, the H. hepaticus infection increased SFB levels in the males but did not impact its levels in the females. However, there was a decrease of the SFB levels in the infected females compared to the controls, but no significant change was noted for males at 16 WPI. We previously reported that H. hepaticus infection in these SW mice increased the mRNA levels of a proinflammatory Th1 cytokine Ifn-γ in the ileum and cecum of the females but not the males compared to the sham controls [19]. Increased Ifn-γ production plays a pivotal role in clearing Yersinia enterocolitica infection in C57BL/6 mice [22]. Thus, the enhanced Ifn-γ production in the H. hepaticus-infected females likely contributes to the reduced colonization levels of both SFB (compared to the sham female controls, this study) and H. hepaticus (compared to the infected males) [19]. In contrast, the H. hepaticus infection in the males increased the mRNA levels of ileal Il-10 with concomitant down-regulation of the mRNA levels of the ileal Ifn-γ in the infected males compared to the infected females [19, 23]. Therefore, the enhanced expression of Il-10 at least partially contributed to the higher levels of SFB in the large intestine compared to the sham controls at 8 WPI.
The finding that higher Il-17A mRNA levels were positively correlated with higher SFB levels further supports SFB as an inducer of Th17 cell response. In addition, higher Il-17A mRNA levels in the ileum of females than males could partially contribute to the lower levels of H. hepaticus colonization in the SW females compared to the SW males at 16 WPI demonstrated in our previous study, since Th17 cell response plays an important role in resistance to bacterial infection [3, 16, 19]. Our data also indicated that the relative mRNA levels of Il-17A decreased from the jejunum, ileum, cecum, and colon, particularly at 16 WPI. In contrast, the mRNA levels of Foxp3 were relatively comparable in these intestinal regions of SW mice and were not influenced by age, gender and H. hepaticus infection. The results were consistent with the previous findings that Il-17 mRNA levels decreased from ileum, cecum and colon of SFB-positive C3H/HeN mice [4, 24]. In addition, there was no significant difference in Foxp3-postitive Treg cell populations between ileum and colon of C3H/HeN [4, 24]. Intriguingly, alteration of colonization dynamics of SFB associated with H. hepaticus infection appeared not to have effects on the mRNA of Il-17A and Foxp3. This may be due to the fact that SW mice are out-bred, immune-competent and resistant to the development of overt intestinal or hepatic pathology induced by H. hepaticus infection [19].
Our previous study demonstrated that colonization dynamics of select species of the altered Schaedler flora (ASF) is changed by aging and gender [25]. ASF are comprised of 8 intestinal commensal bacterial species, 2 aerotolerant Lactobacillus strains (ASF360 and ASF361), 2 Clostridium sp. strains (ASF356, ASF502), Eubacterium sp. (strain ASF492), Bacteroides sp. strain (ASF519), a low-G+C-content gram-positive bacterial strain (ASF500), and Mucispirillum schaedleri (ASF457) [26]. During aging from 8 to 16 WPI, ASF457 and ASF500 in the cecum and of colon of SW mice of both sexes are increased, whereas ASF492 colonization levels are decreased in the colon of males but not females [25]. H. hepaticus infection altered colonization levels of ASF360 in the jejunum of both males (increased) and females (decreased) as well as in the ileum of males (increased) at 16 WPI. Cecal ASF502 colonization levels are also decreased in the H. hepaticus-infected male mice compared to the sham controls at 8 WPI [25]. These lines of evidence indicate that changes in colonization dynamics of the select commensal bacteria such as SFB and ASF strains could potentially play an important role in health and disease of the host.
Taken together, we demonstrated that colonization dynamics of SFB in the large intestine is altered by gender, during aging and with H. hepaticus infection. The ability of SFB to shape the host’s immunity via induction of the skewed differentiation of proinflammatory Th17 cells and mucosal IgA production highlights the importance of the in vivo interactions between SFB and pathogenic infections [3, 4, 17, 18]. In addition, recent identification of SFB 16S rRNA gene in the ileum of human subjects further emphasizes the potential effects of these bacteria on human health and disease [6–8]. Further investigations are needed to ascertain how changes in colonization dynamics of SFB influence intestinal mucosal immune responses in the host. Conventional and quantitative PCR assays of SFB established in this study will serve as useful tools to detect SFB in clinical and environmental samples or murine models of metabolic and intestinal disease [24].
Acknowledgments
This study was supported by the NIH grants (to JGF): P30-ES002109, R01OD11141, R01-CA067529.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 2.Hooper LV, Gordon JI. Commensal host-bacterial relationships in the gut. Science. 2001;292:1115–1118. doi: 10.1126/science.1058709. [DOI] [PubMed] [Google Scholar]
- 3.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Thompson CL, Vier R, Mikaelyan A, Wienemann T, Brune A. ‘Candidatus Arthromitus’ revised: segmented filamentous bacteria in arthropod guts are members of Lachnospiraceae. Environ Microbiol. 2012;14:1454–1465. doi: 10.1111/j.1462-2920.2012.02731.x. [DOI] [PubMed] [Google Scholar]
- 6.Klaasen HL, Koopman JP, Van den Brink ME, Bakker MH, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria in a wide range of vertebrate species. Lab Anim. 1993;27:141–150. doi: 10.1258/002367793780810441. [DOI] [PubMed] [Google Scholar]
- 7.Yin Y, Wang Y, Zhu L, Liu W, Liao N, Jiang M, et al. Comparative analysis of the distribution of segmented filamentous bacteria in humans, mice and chickens. ISME J. 2013;7:615–621. doi: 10.1038/ismej.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonsson H. Segmented filamentous bacteria in human ileostomy samples after high-fiber intake. FEMS Microbiol Lett. 2013;342:24–29. doi: 10.1111/1574-6968.12103. [DOI] [PubMed] [Google Scholar]
- 9.Ericsson AC, Hagan CE, Davis DJ, Franklin CL. Segmented filamentous bacteria: commensal microbes with potential effects on research. Comp Med. 2014;64:90–98. [PMC free article] [PubMed] [Google Scholar]
- 10.Kuwahara T, Ogura Y, Oshima K, Kurokawa K, Ooka T, Hirakawa H, et al. The lifestyle of the segmented filamentous bacterium: a non-culturable gut-associated immunostimulating microbe inferred by whole-genome sequencing. DNA Res. 2011;18:291–303. doi: 10.1093/dnares/dsr022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prakash T, Oshima K, Morita H, Fukuda S, Imaoka A, Kumar N, et al. Complete genome sequences of rat and mouse segmented filamentous bacteria, a potent inducer of th17 cell differentiation. Cell Host Microbe. 2011;10:273–284. doi: 10.1016/j.chom.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 12.Sczesnak A, Segata N, Qin X, Gevers D, Petrosino JF, Huttenhower C, et al. The genome of th17 cell-inducing segmented filamentous bacteria reveals extensive auxotrophy and adaptations to the intestinal environment. Cell Host Microbe. 2011;10:260–272. doi: 10.1016/j.chom.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caselli M, Cassol F, Gentili V, Di Luca D. Genome sequences of segmented filamentous bacteria in animals: implications for human research. Gut Microbes. 2012;3:401–405. doi: 10.4161/gmic.20736. [DOI] [PubMed] [Google Scholar]
- 14.Goto Y, Panea C, Nakato G, Cebula A, Lee C, Diez MG, et al. Segmented filamentous bacteria antigens presented by intestinal dendritic cells drive mucosal Th17 cell differentiation. Immunity. 2014;40:594–607. doi: 10.1016/j.immuni.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lecuyer E, Rakotobe S, Lengline-Garnier H, Lebreton C, Picard M, Juste C, et al. Segmented filamentous bacterium uses secondary and tertiary lymphoid tissues to induce gut IgA and specific T helper 17 cell responses. Immunity. 2014;40:608–620. doi: 10.1016/j.immuni.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 16.Martinez GJ, Nurieva RI, Yang XO, Dong C. Regulation and function of proinflammatory TH17 cells. Ann N Y Acad Sci. 2008;1143:188–211. doi: 10.1196/annals.1443.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki K, Meek B, Doi Y, Muramatsu M, Chiba T, Honjo T, et al. Aberrant expansion of segmented filamentous bacteria in IgA-deficient gut. Proc Natl Acad Sci U S A. 2004;101:1981–1986. doi: 10.1073/pnas.0307317101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohashi Y, Hiraguchi M, Ushida K. The composition of intestinal bacteria affects the level of luminal IgA. Biosci Biotechnol Biochem. 2006;70:3031–3035. doi: 10.1271/bbb.60164. [DOI] [PubMed] [Google Scholar]
- 19.Ge Z, Feng Y, Whary MT, Nambiar PR, Xu S, Ng V, et al. Cytolethal distending toxin is essential for Helicobacter hepaticus colonization in outbred Swiss Webster mice. Infect Immun. 2005;73:3559–3567. doi: 10.1128/IAI.73.6.3559-3567.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouskra D, Brezillon C, Berard M, Werts C, Varona R, Boneca IG, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456:507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 21.Ivanov II, Littman DR. Segmented filamentous bacteria take the stage. Mucosal Immunol. 2010;3:209–212. doi: 10.1038/mi.2010.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Autenrieth IB, Beer M, Bohn E, Kaufmann SH, Heesemann J. Immune responses to Yersinia enterocolitica in susceptible BALB/c and resistant C57BL/6 mice: an essential role for gamma interferon. Infect Immun. 1994;62:2590–2599. doi: 10.1128/iai.62.6.2590-2599.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu Rev Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- 24.Kriegel MA, Sefik E, Hill JA, Wu HJ, Benoist C, Mathis D. Naturally transmitted segmented filamentous bacteria segregate with diabetes protection in nonobese diabetic mice. Proc Natl Acad Sci U S A. 2011;108:11548–11553. doi: 10.1073/pnas.1108924108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ge Z, Feng Y, Taylor NS, Ohtani M, Polz MF, Schauer DB, et al. Colonization dynamics of altered Schaedler flora is influenced by gender, aging, and Helicobacter hepaticus infection in the intestines of Swiss Webster mice. Appl Environ Microbiol. 2006;72:5100–5103. doi: 10.1128/AEM.01934-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robertson BR, O’Rourke JL, Neilan BA, Vandamme P, On SL, Fox JG, et al. Mucispirillum schaedleri gen. nov., sp. nov. a spiral-shaped bacterium colonizing the mucus layer of the gastrointestinal tract of laboratory rodents. Int J Syst Evol Microbiol. 2005;55:1199–1204. doi: 10.1099/ijs.0.63472-0. [DOI] [PubMed] [Google Scholar]