Abstract
NF-κB proteins play a central and subunit-specific role in the response to DNA damage. Previous work identified p50/NF-κB1 as being necessary for cytotoxicity in response to DNA alkylation damage. Given the importance of damage-induced cell death for maintenance of genomic stability, we examined whether Nfkb1 acts as a tumor suppressor in the setting of alkylation damage. Hprt mutation analysis demonstrates that Nfkb1−/− cells accumulate more alkylator-induced, but not ionizing radiation (IR)-induced, mutations than similarly treated wildtype cells. Subsequent in vivo tumor induction studies reveal that following alkylator treatment, but not IR, Nfkb1−/− mice develop more lymphomas than similarly treated Nfkb1+/+ animals. Heterozygous mice develop lymphomas at an intermediate rate and retain functional p50 in their tumors indicating that Nfkb1 acts in a haploinsufficient manner. Analysis of human cancers, including therapy-related myeloid neoplasms, demonstrates that NFKB1 mRNA expression is down regulated compared to control samples in multiple hematological malignancies. These data indicate that Nfkb1 is a haploinsufficient, pathway-specific tumor suppressor that prevents the development of hematologic malignancy in the setting of alkylation damage.
Keywords: NF-κB, tumor suppression, DNA damage, lymphoma
INTRODUCTION
Damage to DNA is a central factor in the acquisition of genomic errors and development of cancer. Chemotherapy represents an exogenous source of DNA damage that is highly genotoxic. Therapy-related myeloid neoplasia (t-MN) is a devastating complication of cytotoxic chemotherapy whereby a secondary malignancy is induced following treatment of an initial malignant or non-malignant disease.1 The importance of DNA alkylation damage to secondary malignancy formation is underlined by the observation that in the largest study of t-MN reported to date, 78 % of patients received an alkylating agent for their primary tumor.2
NF-κB represents a family of proteins comprised of five subunits: p50 (NF-κB1, p105), p52 (NF-κB2, p100), p65 (relA), c-rel, and relB that play a complex role in tumor formation. While the oncogenic effects of this transcription factor are consistent with the propensity of NF-κB to mediate survival signaling,3, 4 tumor suppressive actions are evident in the finding that various subunits can indirectly mediate the tumor suppressive effects of p53 or ARF.5–9 Despite indirect tumor suppression, targeted deletion of individual NF-κB proteins has not yet revealed a direct tumor suppressive role for any specific subunit.10
We recently reported that p50/NF-κB1 is necessary for cytotoxicity by alkylating agents such as temozolomide (TMZ) or N-methyl-N-nitrosourea (MNU).11 These agents induce cell death by forming O6-methylguanine (O6-MeG) lesions that mispair with deoxythymidine (T) residues and induce cytotoxicity in a mismatch repair-dependent fashion.12 In cells where damage is tolerated, O6-MeG:T mismatches lead to mutations that are potentially oncogenic.13 Given that depletion of p50, or deletion of Nfkb1, renders cells tolerant of alkylation damage without affecting damage repair,11 we hypothesized that this NF-κB subunit acts to maintain genomic stability specifically in the setting of DNA alkylation damage. In this report, we demonstrate that Nfkb1 protects mice against alkylator-induced, but not ionizing radiation (IR)-induced, lymphoma formation in a haploinsufficient manner.
RESULTS AND DISCUSSION
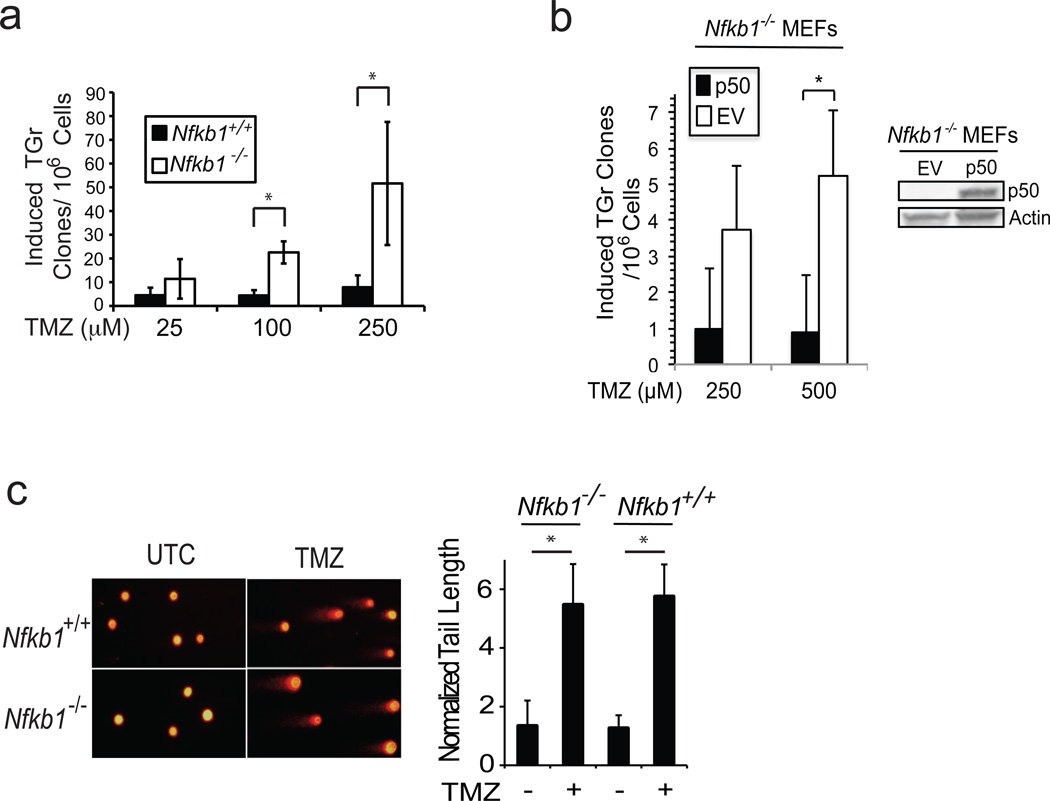
Alkylating agents are mutagenic and loss of p50/Nfkb1 leads to increased survival following alkylator treatment.11 These findings suggest that loss of Nfkb1 may affect alkylator-induced mutagenesis. To examine mutation formation, the hypoxanthine-guanine phospho-ribosyltransferase (hprt) assay was used in Nfkb1−/− and Nfkb1+/+ MEFs. This assay accurately reports alkylator-induced mutations. Primary, low passage MEFs were treated with alkylating agent and mutation induction assessed. Significantly more 6-TG resistant clones develop following treatment of Nfkb1−/− MEFs than Nfkb1+/+ (P < 0.03, Figure 1a), suggesting that Nfkb1 maintains genomic integrity in the setting of alkylation damage. To examine whether it is specifically p50, the mature protein product of Nfkb1, that prevents mutation induction, Hprt assay was performed following reexpression of p50. Nfkb1−/− MEFs stably expressing p50 acquire less 6-TG resistant clones following alkylator treatment than isogenic cells expressing empty vector (EV) (Figure 1b). Next, to examine DNA strand break induction, alkaline comet assay was performed following treatment. Equal amounts of strand breaks are induced in Nfkb1+/+ and Nfkb1−/− MEFs within 3 hours of treatment (Figure 1c). This finding suggests that the difference in the mutation frequency following alkylator exposure is not due to quantitative differences in the level of induced DNA damage, a finding supported by previous data showing that loss of Nfkb1 does not affect damage repair.11 The above observations, coupled with the finding that loss of Nfkb1 results in a decrease in alkylation-induced apoptosis,11 suggest that the difference in mutagenesis is likely due to increased survival of damaged Nfkb1−/− compared to Nfkb1+/+ MEFs.
Figure 1.
Nfkb1 prevents alkylator-induced mutation. (a) Hprt mutation assay in primary Nfkb1+/+ and Nfkb1−/− MEFs treated with increasing concentrations of TMZ and selected in 5 µg/ml 6-TG. Data show mean number of 6-TG resistant (TGr) clones ± SEM of 4 independent experiments performed at least in duplicate. * P < 0.05. (b) Hprt assay in Nfkb1−/− MEFs stably expressing p50 or empty vector (EV) following treatment with TMZ. * P < 0.05. (c) Alkaline comet assay in Nfkb1+/+ and Nfkb1−/− MEFs treated with vehicle (UTC) or 100 µM TMZ for 3 hours. Graph demonstrates quantification of average tail length normalized to nucleus diameter ± SD. * P < 1×10−5. N = 50 cells. Experiment was repeated. Hprt assay in (a) was performed using early passage primary MEFs, harvested from day 14 embryos, treated with TMZ and maintained in exponential growth for 7 days. Cells were subcultured in growth medium alone (plating efficiency) or in the presence of 5 µg/ml 6-TG for selection of mutants. Induced Hprt mutations were calculated as the number of 6-TG resistant colonies per 106 cells plated after correction for plating efficiency. Importantly, 5 ug/ml 6-TG is 100 % lethal to un-mutated MEFs of both genotypes. For re-expression of p50, immortal Nfkb1−/− MEFs were infected with the retroviral vector MigR1-p50 or MigR1-EV. MigR1-p50 was created by liberating p50 from pcDNA3.1-p5033 by digestion with PmeI and XhoI, and ligated into the BglII site of pMSCV-MigR1 containing an IRESGFP insert. Retrovirus was produced with Platinum-GP packaging cells following transfection of MigR1-p50 or MigR1-EV using XtremeGENE transfection reagent (Roche). MEFs were then spinoculated with virus/polybrene-containing supernatant and colonies sorted by FACS. Alkaline single cell gel electrophoresis (Comet assay) was performed as described.34 Briefly, MEFs were treated with TMZ for 3 hours, trypsinized and resuspended in low-melting point agarose. A single cell suspension in the agarose was layered on glass slides and following cell lysis, submerged in alkaline (pH > 13) buffer and electrophoresis performed until slight migration pattern observed in the untreated nuclei. Dried slides were stained with ethidium bromide and visualized by fluorescence microscopy. Tail length was quantified as tail length normalized to nuclear diameter.
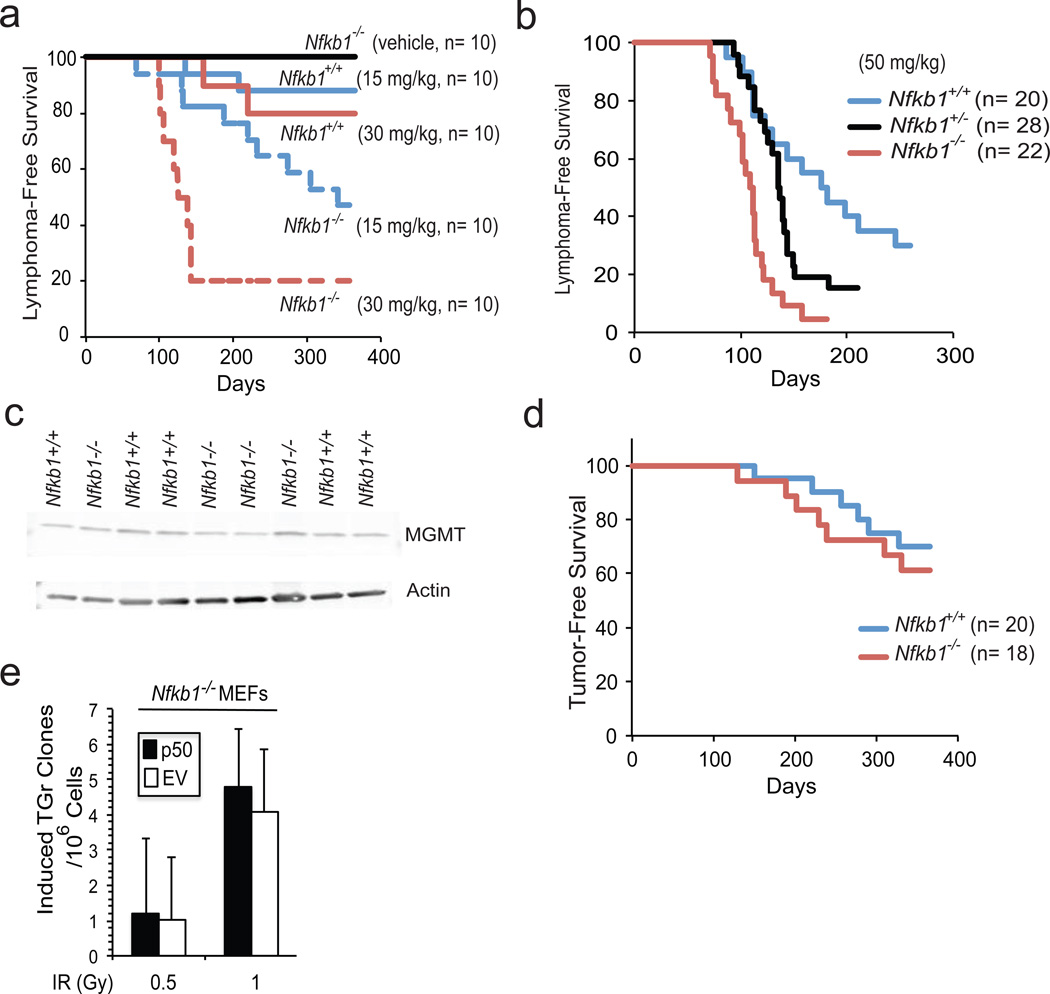
Increase in mutation formation suggests that Nfkb1 may be tumor suppressive in the setting of DNA damage. To examine this hypothesis, we attempted to transform MEFs using TMZ or MNU but found that primary cells of both genotypes are resistant to alkylator-induced transformation in vitro. Therefore, in vivo tumor formation was examined using Nfkb1+/+ and Nfkb1−/− mice. MNU was used as the inducing carcinogen due to its well-described tumor-forming dose response profile.14, 15 6-week old Nfkb1−/− and Nfkb1+/+ mice were injected with either vehicle or MNU and followed for 1 year. At each MNU concentration, significantly more Nfkb1−/− than Nfkb1+/+ mice develop thymic lymphomas (P < 0.002 at 30 mg/kg and P < 0.02 at 15 mg/kg) (Figure 2a). Next, to examine potential Nfkb1 gene-dosage effects, Nfkb1−/− animals, backcrossed for 12 generations with C57BL/6 mice, were obtained and bred with wildtype C57BL/6 animals. Subsequently, Nfkb1−/−, Nfkb1+/− and Nfkb1+/+ mice were injected with 50 mg/kg MNU. At this higher MNU concentration, Nfkb1−/− mice also form significantly more thymic lymphomas than Nfkb1+/+ animals (P < 0.0001) (Figure 2b and Supplementary Figure 1) while heterozygotes have an intermediate tumor induction rate (P ≤ 0.05 relative to Nfkb1+/+ and Nfkb1−/−). FACS analysis demonstrates that the lesions are CD4−/CD8− and CD4+/CD8+ T-cell tumors (data not shown).
Figure 2.
Nfkb1 prevents alkylator-induced tumor formation. (a, b and d) Kaplan-Meier survival curves in mice following a single i.p. injection of MNU. (a) B6/129 Nfkb1+/+ and Nfkb1−/− animals treated with two concentrations of MNU. Statistical significance: P < 0.002 at 30 mg/ kg MNU and P < 0.02 at 15 mg/kg. (b) C57BL/6 mice treated with 50 mg/kg MNU. Statistical significance: P < 0.0001: Nfkb1+/+ vs. Nfkb1−/−; P = 0.05: Nfkb1+/+ vs. Nfkb1+/−; and P = 0.002: Nfkb1+/− vs. Nfkb1−/−. (c) Western blot with anti-MGMT antibody in thymus extract of untreated C57BL/6 Nfkb1+/+ and Nfkb1−/− mice. (d) C57BL/6 Nfkb1+/+ and Nfkb1−/− animals treated with whole body IR. Statistical significance: P > 0.5. (e) Hprt assay in Nfkb1−/− MEFs stably expressing p50 or empty vector (EV) following treatment with IR. Animal experiments were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the University of Chicago. Nfkb1−/− mouse strains used include B6;129P-Nfkb1tm1Bal/J and B6.Cg-Nfkb1tm1Bal/J that have been backcrossed for 12 generations with C57BL/6J mice (Jackson Laboratories, Bar Harbor, Maine, USA). The respective Nfkb1+/+ controls include: B6;129PF1/J and C57BL/6J. For tumorigenesis in the B6;129P strain, male mice were used and for tumorigenesis in C57BL/6J mice, equal ratios of male to female animals of all genotypes were used. Animals were housed in a pathogen-free, biosafety level II facility. MNU was dissolved in DMSO and diluted in PBS within 10 min of injection. The final preparation was delivered in a total volume of 500 µl per 25 g mouse. Six-week old mice were given a single i.p. injection of MNU or vehicle. For radiation carcinogenesis, equal ratios of male to female animals were treated with four weekly doses of 1 Gy total body irradiation using a Phillips RT250 x-ray generator. Animals were observed at least 2 times per week and sacrificed when they exhibited signs of cachexia, hunched position, respiratory distress or a visible mass. Following euthanasia, autopsy was performed. For evaluation of thymic MGMT expression, 5– 8 week old male C57BL/6 mice were sacrificed and the thymus harvested. Immunoblot was performed on cell lysates with anti-MGMT antibody (sc-8828, Santa Cruz Biotechnology, Dallas, Texas, USA). For histological analysis, tumors were removed and either snap frozen, disaggregated for FACS analysis or immersed in paraformaldyhyde prior to sectioning. Statistical analysis was performed using one-sided Mantel-Cox Log rank analysis.
The carcinogenic effect of MNU is blocked primarily by O6-methylguanine-DNA methyltransferase (MGMT), a protein that specifically removes alkyl groups from the O6 position of guanine.15 Therefore, MGMT expression level was examined in untreated mice. Equal MGMT protein expression is seen in thymic tissue from Nfkb1−/− and Nfkb1+/+ animals (Figure 2c) suggesting that the difference in tumor formation rate is not due to differences in O6-MeG repair. In addition, given that Nfkb1−/− animals have defects in innate and adaptive immunity,10, 16 we examined whether tumors from Nfkb1−/− animals can be transplanted into mice of both genotypes. Nfkb1−/− and Nfkb1+/+ recipient mice develop disseminated cancer at equal rates following injection of tumor cells from Nfkb1−/− animals (Supplementary Figure 2) indicating that Nfkb1−/− tumors are equally transplantable.
It was previously noted that loss of Nfkb1 does not affect the cytotoxic response to IR.11 We therefore examined tumor formation in response to whole-body IR. Remarkably, at the IR dose used, no significant difference in tumor formation rate is noted between Nfkb1−/− and Nfkb1+/+ animals (P > 0.5) (Figure 2d). While these data suggest that Nfkb1 does not protect mice from IR-induced tumor formation, it is possible that at a different dose of IR, or if greater numbers of animals are used, a difference in tumor formation may be noted. Nevertheless, given that the dose of IR used results in approximately 30% tumor formation in both genotypes, a rate comparable to that induced by 30 mg/kg MNU in Nfkb1+/+ mice (Figure 2a), it is evident that loss of Nfkb1 renders animals significantly more sensitive to tumor formation by MNU than IR. Given the findings in response to IR, we also examined mutation induction following IR. In contrast to TMZ, expression of p50 does not affect mutation frequency in response to IR (Figure 2e). Together, these findings indicate that loss of Nfkb1 leads to an increase in the susceptibility of mice to alkylation-induced tumor formation and moreover, suggest that Nfkb1 acts as a pathway-specific tumor suppressor.
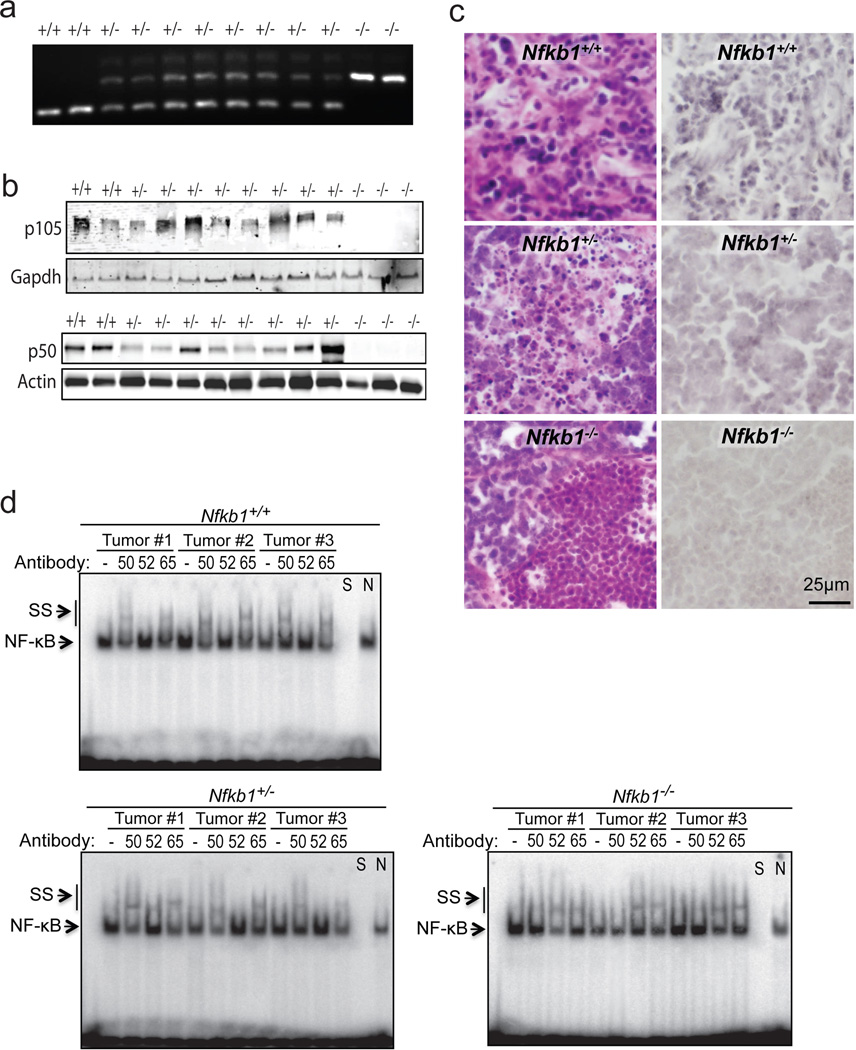
To examine whether Nfkb1 functions in a haploinsufficient manner, tumors from mice were harvested. All the tumors from Nfkb1+/− animals retain an Nfkb1 allele (Figure 3a). As loss of heterozygosity can also occur via post-translational mechanisms, expression of p105 and p50 was examined. In tumors from Nfkb1+/− animals, both p105 and p50 expression are universally retained (Figure 3b). Moreover, immunohistochemical analysis demonstrates that p50 is found in tumor cells from Nfkb1+/− mice and not solely in the stroma (Figure 3c). Finally, to examine the functional status of p50, binding to κB consensus DNA was evaluated using EMSA. Protein from three tumors of each genotype was harvested and examined for subunit binding by supershift analysis. In addition to p50 binding, p52 was examined as this subunit most often cross-compensates when p50 is lost,11,17 and p65 was studied as this is the primary interacting partner of p50. Supershift analysis demonstrates that while tumors from Nfkb1+/+ animals contain p50/p65 dimers, in Nfkb1−/− mice although p65 remains, p50 binding is lost and replaced by p52 (Figure 3d). Most significantly, tumors from Nfkb1+/− animals retain p50 that binds DNA. These findings indicate that functional p50, i.e. p50 that does not contain mutations in critical DNA binding regions, is maintained in tumors from heterozygous mice and, when considered with the intermediate tumor formation rate in heterozygote animals (Figure 2b), suggest that Nfkb1 is a haploinsufficient tumor suppressor.
Figure 3.
Thymic tumors from Nfkb1+/− animals retain heterozygosity. (a) Tumors from Nfkb1−/−, Nfkb1+/− and Nfkb1+/+ mice were examined by pcr using genotype specific primers. (b) Immunoblot of tumor lysate from the indicated mice with anti-p105 (C-terminal) antibody (upper) and anti-p50 antibody (lower). (c) Immunohistochemical analysis, using anti-p50 antibody, of tumors harvested from Nfkb1+/+, Nfkb1+/− and Nfkb1−/− mice. Hematoxylin and eosin sections from the same tumors are also shown (left). (d) EMSA using the Ig-κB probe of tumor lysates from three separate tumors from mice of each genotype. Supershift (SS) was performed with anti-p50 (50), anti-p52 (52) or anti-p65 (65) antibody where shown. To confirm the band specificity, competition with specific (S) and nonspecific (N) DNA was performed as indicated.
Genotyping was performed using snap frozen tumor samples following thawing and lysis at 55°C for 18 hours. Nucleic acid was extracted and DNA amplified by PCR using primers described for Nfkb1−/− animals (Jackson Laboratory). Amplified DNA was run through 2 % agarose gel. For tumor protein, snap frozen samples were thawed, homogenized in 700 ml RIPA lysis buffer and incubated on ice for 5 minutes. Subsequently, samples were centrifuged (14000g × 5 minutes) and the supernatant used for immunobloting and EMSA. Immunoblots were performed with anti-p105 C-terminal (#4717, Cell Signaling Technologies), anti-p50 (sc-8414, Santa Cruz Biotechnology), anti-actin (sc-7210) or anti-gapdh (sc-137179) antibodies. EMSA was performed as previously described11 using a probe bearing the decameric Ig-κB consensus sequence (5’-GGGACTTTCC-3’). Supershift was performed by pre-incubation with anti-p50 (sc-1190x), anti-p52 (sc-298x) or anti-p65 (sc-8008) antibodies for 30 min on ice.
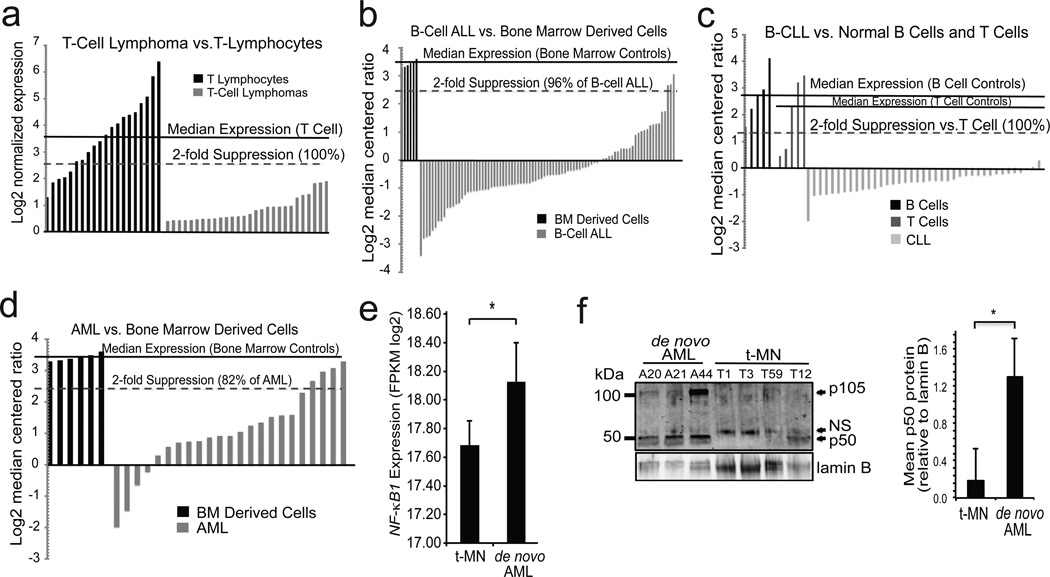
Tumor suppression by Nfkb1 raises the question of whether this subunit acts in a similar fashion in man. Although NFKB1 is rarely mutated in human tumors, the haploinsufficient nature of this factor suggests that reduced expression may be sufficient to facilitate tumor formation. We therefore analyzed NFKB1 expression data in T-cell lymphomas and noted that 100 % of tumors have at least 2-fold decreased expression relative to CD8+ and CD4+ T-cells from normal human peripheral blood (Figure 4a).18 Next, a series of other tumors were studied and a highly significant decrease in NFKB1 expression noted in multiple hematological malignancies (P < 1×10−7), including B-cell ALL (Figure 4b), T-cell ALL (Supplementary Figure 3a),19 B-cell CLL (Figure 4c and Supplementary Figure 3b),20, 21 diffuse large B-cell lymphoma (DLBCL) (Supplementary Figure 3c)22 and AML (Figure 4d).19 In addition, P65 expression level was examined. Contrary to NFKB1, P65 expression is up-regulated in T-cell ALL (2.3-fold), B-cell ALL (1.6-fold) and AML (1.5-fold) (Supplementary Figure 4a), decreased in T-cell lymphoma (Supplementary Figure 4b) and unchanged in DLBCL (Supplementary Figure 4c) suggesting that a general decrease in NF-κB pathway mRNA is not present. The above data demonstrate that in multiple different human hematological malignancies a striking decrease in NFKB1 mRNA expression is apparent.
Figure 4.
NFKB1 mRNA is decreased in hematological malignancy. (a–d) Waterfall plots of data from human tumor databases. Median expression in the control samples (solid line) and % below 2-fold suppression (dashed line) are indicated for each dataset. (a) T-cell lymphoma samples vs. circulating T cells of healthy volunteers. P = 8×10−13. (b) Pediatric B-cell ALL bone marrow (BM) samples vs. BM of healthy volunteers. P = 1.1×10−11. (c) Adult B-cell CLL peripheral blood samples vs. B cells isolated from healthy adults, or T cells isolated from 1 neonate, 2 fetal and 2 adult samples. P < 1×10−12 (CLL vs. B or T cells). (d) Pediatric AML BM vs. BM of healthy volunteers. P = 2.7×10−7. (e) NFKB1 expression level assessed by RNA sequencing in t-MN (N =18) compared to de novo AML (N = 10). NFKB1 expression is output as Fragments per Kilobase per Million mapped reads (FPKM). * P = 0.026. RNA sequencing data are extracted from our previous study,35 plus five previously unpublished t-MN samples. (f) Immunoblot (left) with anti-p50 antibody in bone marrow samples from patients with de novo AML (n=3) and t-MN (n= 4). Loading was examined with anti-lamin B. Quantification of p50 protein level (right) is shown relative to lamin B. * P < 0.05. NFKB1 expression level in human lymphomas and leukemias was determined from published datasets. Oncomine (Compendia Bioscience, Ann Arbor, MI, USA) was initially used for screening of data and preliminary analysis. Subsequently, identified datasets [GEO accession: GSE7186;19 GSE2466;20 and GSE633818] were downloaded from the Gene Expression Omnibus using the Bioconductor package GEOquery.36, 37 Data for DLBCL by Rosenwald et. al.22 was downloaded from the Lymphoma/Leukemia Molecular Profiling Project Gateway (http://llmpp.nih.gov/DLBCL/). Data was quantile normalized and log2 transformed as-needed using the limma and lumi packages.38 The median NFKB1 expression level of control samples was calculated, and the percentage of tumor samples with 2-fold lower expression determined. Means and standard deviations of NFKB1 log2 normalized expression or log2 median centered ratios for patient samples and controls in each dataset were calculated using Graphpad Prism (San Diego, CA, USA). Five t-MN samples were processed for next-generation RNA sequencing on the Illumina platform as previously described.35 Patient characteristics and read depth are provided (Supplemental Table 1). Data analysis and expression level estimates were performed as previously described.35 Raw sequencing data will be deposited in the NCBI Short Read Archive. Two-tailed Student’s t tests were used with an alpha of < 0.05. For t-MN, one-sided Wilcoxon rank sum test was used. For immunoblotting, bone marrow samples were thawed, washed in PBS and lysed with RIPA buffer prior to SDS-PAGE and immunoblotting. Anti-lamin B antibody (sc-6216). p50 quantification was performed by densitometry by measuring band intensity with ImageJ freeware (W. S. Rasband, National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/) and the data plotted as ratio over loading after compensating for background intensity taken in a non-immunoreactive region of the blot.
Given that Nfkb1 is tumor suppressive specifically following alkylator treatment and that over 75 % of t-MN patients receive an alkylating agent for their primary tumor,2 NFKB1 expression was examined in t-MN. We performed next-generation RNA sequencing on a series of patients with t-MN and NFKB1 expression level was compared to that in samples from patients with de novo AML, which occurs without a history of prior malignancy. t-MN samples have significantly reduced NFKB1 relative to that found in de novo AML (P = 0.026, Figure 4e). While t-MN has a higher frequency of adverse-risk cytogenetics (e.g. −5/del(5q) and −7/del(7q)),2 NFKB1 expression in patients with adverse-risk cytogenetics is also decreased compared to de novo AML with similar karyotypes (17.68 ± 0.18 sterr, n = 14 vs. 18.17 ± 0.20, n = 5). Although this smaller subset does not achieve statistical significance (P = 0.19), the median values in the subgroups are comparable to those in the complete sample sets. Finally, to examine whether mRNA differences in de novo AML and t-MN patients are reflected by differences in protein expression, immunoblotting was performed on the original bone marrow samples from patients with both types of disease. A general trend for less p50 protein is seen in t-MN samples compared to de novo AML, an observation confirmed by densitometry following normalization to loading (Figure 4f and Supplementary Figure 4d). Importantly, t-MN samples also have less p105, supporting the observation that NFKB1 expression is reduced at the mRNA level in these tumors. In addition, it is notable that in myeloid neoplasms decreased NFKB1 expression is associated with significantly reduced expression of death pathway genes (Supplementary Figure 5), supporting the association of reduced NFκB1 with increased damage tolerance and cell survival. In sum, the human data are consistent with the animal findings and suggest that chemotherapy exposure and reduced NF-κB1 cooperate to promote malignancy.
The current work uncovers a previously unidentified tumor suppressive role for Nfkb1. Specifically, loss of Nfkb1 increases the susceptibility of animals to alkylator-induced lymphoma formation. Interestingly, tumor formation in response to IR is not increased suggesting that Nfkb1 mediates its tumor suppressive effects in a pathway-specific manner. Pathway-specific tumor suppression is consistent with the prior finding that the mature product of Nfkb1, p50, facilitates cytotoxicity specifically in response to certain types of DNA damage.11 Moreover, the observation that Nfkb1−/− animals do not have increased susceptibility to tumor formation by the carcinogen, diethylnitrosamine,23 an agent that induces tumors by forming O4-ethylthymidine adducts, further emphasizes the damage-specific nature of tumor suppression by Nfkb1. Despite tumor suppression, it is notable that Nfkb1−/− animals are not tumor prone at baseline.16 While lack of spontaneous tumor formation in animals deficient for a tumor suppressor is not uncommon,24 the absence of increased tumorigenesis may be related to the fact that Nfkb1−/− animals are housed in a pathogen-free environment. The primary phenotype of Nfkb1−/− mice is their deficiency in innate and adaptive immunity.10, 16 Although alterations in the inflammatory environment likely contribute to tumor formation, the damage-specific nature of tumor suppression, coupled with the increased mutagenesis in vitro, argues against tumor suppression by Nfkb1 being solely an immune-mediated phenomenon.
The data suggest that tumor suppression occurs in a haploinsufficient manner indicating that reduced expression, without complete loss, is sufficient to facilitate oncogenesis. This observation is supported by prior work indicating that even partial decrease in p50/p105 is sufficient to compromise the damage response.11 From a mechanistic prospective, a decrease in p50 level results in formation of compensatory p52/p65 heterodimers, as noted on EMSA in the tumor tissue (Figure 3d). While p52 can cross compensate for p50 in certain respects,17 it cannot functionally compensate for p50 in the response to DNA damage.11 Haploinsufficiency is particularly relevant to man because complete loss of NFKB1/p50 in human tumors is rare, likely because of the critical role of NF-κB in normal cellular physiology. The lack of significant NFKB1 gene deletion or mutation in cancer raises the question of how NFKB1 expression is reduced in human malignancy. While decreased expression may be a result of epigenetic factors such as promoter methylation, several oncogenes implicated in hematological malignancy, including tal1, lmo1, bcl-6 and myc negatively regulate Nfkb1 expression.25–27 On the other hand, low NFKB1 expression may be a normal variant found in the general population, an observation supported by the description of a functional NFKB1 promoter polymorphism, −94ins/delATTG, that is associated with reduced NFKB1 expression.28
Therapy-related malignancy is a particularly severe complication of cancer treatment that is becoming more prevalent as patients survive for longer time.1 In fact, up to one in six cancers is thought to be therapy-related.29 Many chemotherapeutic agents that induce t-MN have mechanisms of action that involve the NF-κB1/p50 pathway. From a clinical perspective, NFKB1 expression, like other predisposing factors, may be especially important in patients with non-malignant lesions where the risk/benefit ratio for treatment with a cytotoxic agent can be unclear. In patients that have a higher risk of secondary tumor formation, caution should be taken prior to the use of a cytotoxic agent whose beneficial effects may be slim. In this regard, therapy-related lymphoma was recently reported in several patients with low-grade gliomas that were treated with an alkylator.30
The ability to maintain genomic integrity in the face of DNA damage is a feature of ‘caretaker’ genes;31 however, NF-κB1 does not appear to be directly involved in damage repair. Nevertheless, Nfkb1 is tumor suppressive in the setting of DNA damage. These observations suggest that NF-κB1 may belong to the growing group of low penetrance cancer susceptibility genes that act in combination to determine the overall cellular response to genomic insults.32 Future work will examine the mechanism by which this ubiquitous, yet under-examined NF-κB subunit modulates the response to DNA damage.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to RF de Pooter and B Kee for helpful contributions.
Financial Support: NIH grant 1R01CA136937 (B.Y.), The Ludwig Center for Metastasis Research; Leukemia & Lymphoma Society Fellow award (M.E.M) and the Chicago Cancer Genomes Project.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
REFERENCES
- 1.Allan JM, Travis LB. Mechanisms of therapy-related carcinogenesis. Nat Rev Cancer. 2005;5:943–955. doi: 10.1038/nrc1749. [DOI] [PubMed] [Google Scholar]
- 2.Smith SM, Le Beau MM, Huo D, Karrison T, Sobecks RM, Anastasi J, et al. Clinical-cytogenetic associations in 306 patients with therapy-related myelodysplasia and myeloid leukemia: the University of Chicago series. Blood. 2003;102:43–52. doi: 10.1182/blood-2002-11-3343. [DOI] [PubMed] [Google Scholar]
- 3.Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- 4.Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, et al. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118:285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 5.Perkins ND. NF-kappaB: tumor promoter or suppressor? Trends Cell Biol. 2004;14:64–69. doi: 10.1016/j.tcb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Dajee M, Lazarov M, Zhang JY, Cai T, Green CL, Russell AJ, et al. NF-kappaB blockade and oncogenic Ras trigger invasive human epidermal neoplasia. Nature. 2003;421:639–643. doi: 10.1038/nature01283. [DOI] [PubMed] [Google Scholar]
- 7.Ryan KM, Ernst MK, Rice NR, Vousden KH. Role of NF-kappaB in p53-mediated programmed cell death. Nature. 2000;404:892–897. doi: 10.1038/35009130. [DOI] [PubMed] [Google Scholar]
- 8.Rocha S, Garrett MD, Campbell KJ, Schumm K, Perkins ND. Regulation of NF-kappaB and p53 through activation of ATR and Chk1 by the ARF tumour suppressor. Embo J. 2005;24:1157–1169. doi: 10.1038/sj.emboj.7600608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wolff B, Naumann M. INK4 cell cycle inhibitors direct transcriptional inactivation of NF-kappaB. Oncogene. 1999;18:2663–2666. doi: 10.1038/sj.onc.1202617. [DOI] [PubMed] [Google Scholar]
- 10.Gerondakis S, Grumont R, Gugasyan R, Wong L, Isomura I, Ho W, et al. Unravelling the complexities of the NF-kappaB signalling pathway using mouse knockout and transgenic models. Oncogene. 2006;25:6781–6799. doi: 10.1038/sj.onc.1209944. [DOI] [PubMed] [Google Scholar]
- 11.Schmitt AM, Crawley CD, Kang S, Raleigh DR, Yu X, Wahlstrom JS, et al. p50 (NF-kappaB1) is an effector protein in the cytotoxic response to DNA methylation damage. Mol Cell. 2011;44:785–796. doi: 10.1016/j.molcel.2011.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat Rev. 1997;23:35–61. doi: 10.1016/s0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- 13.Margison GP, Santibanez Koref MF, Povey AC. Mechanisms of carcinogenicity/chemotherapy by O6-methylguanine. Mutagenesis. 2002;17:483–487. doi: 10.1093/mutage/17.6.483. [DOI] [PubMed] [Google Scholar]
- 14.Joshi VV, Frei JV. Effects of dose and schedule of methylnitrosourea on incidence of malignant lymphoma in adult female mice. J Natl Cancer Inst. 1970;45:335–339. [PubMed] [Google Scholar]
- 15.Dumenco LL, Allay E, Norton K, Gerson SL. The prevention of thymic lymphomas in transgenic mice by human O6-alkylguanine-DNA alkyltransferase. Science. 1993;259:219–222. doi: 10.1126/science.8421782. [DOI] [PubMed] [Google Scholar]
- 16.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80:321–330. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann A, Leung TH, Baltimore D. Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. Embo J. 2003;22:5530–5539. doi: 10.1093/emboj/cdg534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piccaluga PP, Agostinelli C, Califano A, Rossi M, Basso K, Zupo S, et al. Gene expression analysis of peripheral T cell lymphoma, unspecified, reveals distinct profiles and new potential therapeutic targets. J Clin Invest. 2007;117:823–834. doi: 10.1172/JCI26833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersson A, Ritz C, Lindgren D, Eden P, Lassen C, Heldrup J, et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia. 2007;21:1198–1203. doi: 10.1038/sj.leu.2404688. [DOI] [PubMed] [Google Scholar]
- 20.Haslinger C, Schweifer N, Stilgenbauer S, Dohner H, Lichter P, Kraut N, et al. Microarray gene expression profiling of B-cell chronic lymphocytic leukemia subgroups defined by genomic aberrations and VH mutation status. J Clin Oncol. 2004;22:3937–3949. doi: 10.1200/JCO.2004.12.133. [DOI] [PubMed] [Google Scholar]
- 21.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 23.Glauert HP, Eyigor A, Tharappel JC, Cooper S, Lee EY, Spear BT. Inhibition of hepatocarcinogenesis by the deletion of the p50 subunit of NF-kappaB in mice administered the peroxisome proliferator Wy-14,643. Toxicol Sci. 2006;90:331–336. doi: 10.1093/toxsci/kfj116. [DOI] [PubMed] [Google Scholar]
- 24.Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell. 2006;125:665–677. doi: 10.1016/j.cell.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 25.Chang PY, Draheim K, Kelliher MA, Miyamoto S. NFKB1 is a direct target of the TAL1 oncoprotein in human T leukemia cells. Cancer Res. 2006;66:6008–6013. doi: 10.1158/0008-5472.CAN-06-0194. [DOI] [PubMed] [Google Scholar]
- 26.Li Z, Wang X, Yu RY, Ding BB, Yu JJ, Dai XM, et al. BCL-6 negatively regulates expression of the NF-kappaB1 p105/p50 subunit. J Immunol. 2005;174:205–214. doi: 10.4049/jimmunol.174.1.205. [DOI] [PubMed] [Google Scholar]
- 27.Keller U, Nilsson JA, Maclean KH, Old JB, Cleveland JL. Nfkb 1 is dispensable for Myc-induced lymphomagenesis. Oncogene. 2005;24:6231–6240. doi: 10.1038/sj.onc.1208779. [DOI] [PubMed] [Google Scholar]
- 28.Karban AS, Okazaki T, Panhuysen CI, Gallegos T, Potter JJ, Bailey-Wilson JE, et al. Functional annotation of a novel NFKB1 promoter polymorphism that increases risk for ulcerative colitis. Hum Mol Genet. 2004;13:35–45. doi: 10.1093/hmg/ddh008. [DOI] [PubMed] [Google Scholar]
- 29.Wood ME, Vogel V, Ng A, Foxhall L, Goodwin P, Travis LB. Second malignant neoplasms: assessment and strategies for risk reduction. J Clin Oncol. 2012;30:3734–3745. doi: 10.1200/JCO.2012.41.8681. [DOI] [PubMed] [Google Scholar]
- 30.Neyns B, Cordera S, Joosens E, Pouratian N. Non-Hodgkin's lymphoma in patients with glioma treated with temozolomide. J Clin Oncol. 2008;26:4518–4519. doi: 10.1200/JCO.2008.18.8177. [DOI] [PubMed] [Google Scholar]
- 31.Kinzler KW, Vogelstein B. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–763. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 32.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 33.Yamini B, Yu X, Dolan ME, Wu MH, Darga TE, Kufe DW, et al. Inhibition of nuclear factor-kappaB activity by temozolomide involves O6- methylguanine induced inhibition of p65 DNA binding. Cancer Res. 2007;67:6889–6898. doi: 10.1158/0008-5472.CAN-06-4496. [DOI] [PubMed] [Google Scholar]
- 34.Olive PL, Banath JP. The comet assay: a method to measure DNA damage in individual cells. Nat Protoc. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 35.McNerney ME, Brown CD, Wang X, Bartom ET, Karmakar S, Bandlamudi C, et al. CUX1 is a haploinsufficient tumor suppressor gene on chromosome 7 frequently inactivated in acute myeloid leukemia. Blood. 2013;121:975–983. doi: 10.1182/blood-2012-04-426965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. NCBI GEO: archive for functional genomics data sets--10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics. 2007;23:1846–1847. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 38.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.