Abstract
Because of its relatively recent evolution, Homo sapiens exhibits relatively little within-species genomic diversity. However, because of genome size, a proportionally small amount of variation creates ample opportunity for both rare mutations that may be disease-causative as well as more common genetic variation that may be important in disease modification or pharmacogenetics. Primarily because of the East African origin of modern humans, individuals of African ancestry (AA) exhibit greater degrees of genetic diversity than more recently established populations, such as those of European ancestry (EA) or Asian ancestry. These population effects extend to differences in the frequency of common gene variants that may be important in heart failure natural history or therapy. For cell-signaling mechanisms important in heart failure, we review and present new data on genetic variation between AA and EA populations. The data indicate that 1) neurohormonal signaling mechanisms frequently (16 of the 19 investigated polymorphisms) exhibit racial differences in the allele frequencies of variants comprising key constituents, 2) some of these differences in allele frequency may differentially affect the natural history of heart failure in AA vs. EA individuals, and 3) in many cases these differences likely play a role in observed racial differences in drug or device response.
Keywords: Genetic polymorphisms, pharmacogenetics, racial ancestry, heart failure
Introduction
Variability in drug response is typical of cardiovascular therapies (1) and may be related to common genetic variation (1–3), operationally defined as a polymorphism with a minor allele frequency of ≥ 1% (4). Race-associated differences in cardiovascular disease natural history and drug response are well known, and may be associated with common genetic variation in drug targets or pharmacokinetics (2). Such pharmacogenetic variation can potentially explain variability in racially based response to therapies for heart failure, a phenomenon that is becoming more widely appreciated.
We review and provide new information on race-associated differences in heart failure natural history and therapeutic responses that are associated with common genetic variation. The presented information is confined to heart failure with reduced left ventricular ejection fraction (HFrEF, or simply HF), and the race comparisons are between Sub-Saharan African-ancestry (AA) vs. European-ancestry (EA) populations regardless of geographic location. The overall aim is to explore the hypothesis that reported racial differences in HF natural history or therapeutic responses are influenced by differences in genetic variation in relevant cardiac myocyte neurohormonal signaling mechanisms.
Race and Genetic Variation
An argument for a genetic basis for race-associated differences in disease natural history or therapeutic response is predicated on the idea that different frequencies of genetic variants between racial groups may lead to differentiated outcomes between those groups. Genetic alteration of a disease or therapeutic phenotype within a racially defined population is dependent on a) the frequency of the polymorphism in that population, b) the difference in the allele frequency of the polymorphism between the indicated population and a general, non-racially selected population, and c) the extent of the biological effect of the polymorphism on the phenotype.
Extrapolating racial identity from common attributes such as skin color can be problematic due to the assumption that physical characteristics are a proper proxy for variation in biologic phenotype-modifying, non-anthropomorphic genes. Nevertheless, extensive clinical experience supports that phenotype heterogeneity in disease risk, prognosis, and response to therapy can be attributed to self-identified race, and it is appropriate to explore to what extent genetic variation contributes to these differences (5,6). Understanding the underlying genetic basis of such heterogeneity will help devise new drugs, devices and diagnostics that improve outcomes.
Genetic diversity in humans is relatively low compared with other species, including other primates (7). This is generally attributed to Homo sapiens' relatively recent evolution in East Africa approximately 200,000 years ago, and the subsequent immigration of modern populations from Africa in the past 100,000 years (6). Based on the first detailed single-nucleotide polymorphism (SNP) map of the human genome encompassing 1.42 million variants occurring every 1.9 Kb, humans were estimated to be 99.6% to 99.8% identical at the nucleotide level (6,8). The more recent 1000 Genomes Project, which has the goal of identifying pan-genomic and coding region variations down to respective allele frequencies of 1% and 0.1%, identified in its recently published pilot phase (9) about 15 million SNPs, 1 every 800 bases, from whole-genome sequencing of 179 individuals in 3 racial categories. The average number of SNPs per individual was about 3 million, and the variation from the reference genome was 0.125% (9). Thus, although the most recent estimate of single-nucleotide variation is about 0.1%, the 3 million SNPs per individual plus other types of genetic variation provide ample potential for genomic diversity within and between populations. Despite the notion that the vast majority of SNPs represent silent (synonymous) variation or an amino acid change (non-synonymous) with no clear biological function effects, substantial effort has been invested in identifying the small fraction of SNPs and other variants that associate with human phenotypes and disease risks.
African-ancestry populations exhibit greater degrees of genetic variation compared with non-African cohorts (10,11). Given that modern European and Asian populations descended from founder groups that diverged from ancestral African populations, it is expected that genetic diversity in non-African groups would be lower since ancestral founder populations would contain only a subset of the total ancestral African variation. However, most of the genetic variation in African populations can also be found in non-African populations. Overall, 10% to 15% of all human genetic variation is explained by differences between Sub-Saharan Africans, Northern Europeans, and East Asians. Stated another way, approximately 85% to 90% of known variation is captured by studying any 1 of the 3 "major" population groups (Africa, Asia, and Europe), and only an additional 10% to 15% can be ascertained by inclusion of the other 2 groups (12). Thus, genetic variation between populations is only slightly more different than variation within a given population (13).
These data have relevance for the evaluation of genetic variation related to health and disease. A priori, for any given variant there is an increased probability of it being represented in an AA vs. a non-AA population. Furthermore, for any variant locus shared between AA and non-AA populations, the observed allele frequencies may differ, sometimes widely, between racial populations.
In the example of the ADRA2C 322-325 insertion (Ins)-deletion (Del) polymorphism (rs2234888), various studies have noted a 7- to 10-fold increase in the prevalence of the Del variant in AA populations (14–16). Some of the difference in allele frequency is likely due to the lower frequency of the Del allele in the founder population(s) that immigrated to Northern Europe. Presumably, there may be differential allele frequency across Africa, with lower Del frequencies in East African populations. This question has not been extensively investigated, although one analysis of black South Africans, far removed from the migration point, noted the Del allele to be present in more than 50% of individuals (17). The same logic and arguments apply to other variants that exhibit marked racial differences in frequencies, such as GRK5 Gln41Leu (rs2230345) (18) and SCN5A Ser1103Tyr (rs7626962) (19), both of which have minor alleles of demonstrated functional importance with frequencies that are ≥ 10-fold higher in AA vs. EA populations. However, even in these highly minor allele-enriched examples, the major allele has a frequency >0.5. Thus there is a non-trivial percentage of AA individuals that do not have the minor allele in the heterozygous or homozygous state. Therefore, one can readily appreciate that skin color would be a poor method of determining whether a person carries the minor allele for ADRA2C Ins322-325Del, GRK5 Gln41Leu, or SCN5A Ser1103Tyr.
In spite of widespread variation in the frequency of various genetic markers, the associated odds ratios for disease risk in different racial groups appears to be less variable (20). However, as will be presented, in HF therapeutics there is good evidence that racially based pharmacogenetic differences exist for both effectiveness and safety.
Common Genetic Variation in Signaling Mechanisms Important in HF
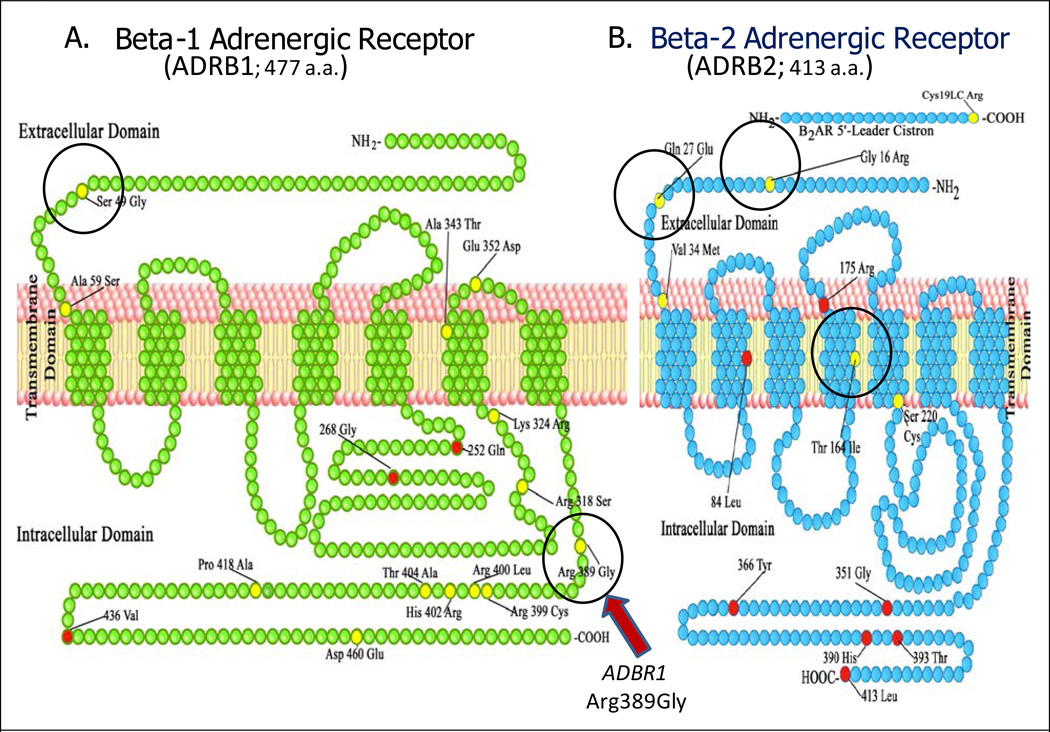
Numerous reports have documented substantial polymorphic variation in neurohormonal and associated signaling mechanisms important in HF therapeutics. For example, the β1-adrenergic receptor (ADRB1, Figure 1A)—the initiating signal transduction protein in a system that can cause a dilated cardiomyopathy when overexpressed in transgenic mice and is also the primary drug target for a major class of HF therapeutics (β-blockers) (21)—exhibits about 5 times the average SNP percentage detected in the ENCODE regions in the human genome (22) and 2 times that detected in exons in the 1000 Genomes Project (9), with 26 SNPs in the 1431 coding region nucleotides, half of which result in an amino acid change (23). Most genomic common variation has no impact on protein function, so the focus of this study is on variants with well-characterized biologic or pharmacologic effects.
Figure 1.
Consensus coding sequence (CCDS, http://www.ensembl.org/info/genome/genebuild/ccds.html) for ADRB1 (A) and ADRB2 (B). Amino acid (a.a.) changes resulting from nonsynonymous SNPs are in yellow, while unchanged amino acids at the site of synonymous SNPs are in red. Polymorphisms discussed in the text are circled, and the ADRB1 Arg389gly locus is highlighted by an arrow. Reproduced and modified from reference (23), with the permission of Le Jacq Communications, Inc and Congestive Heart Failure.
Table 1 is a compilation of allele frequency data from non-synonymous SNPs, insertion-deletions (InDels), or other polymorphisms that have been shown to have functional significance in selected genes encoding signaling pathway proteins of documented or potential importance in HF natural history or therapeutics. The methods for generating the data in Table 1 are given in the Supplement and in the Table 1 footnotes. The basis for combining AA populations from Sub-Saharan Africa and elsewhere is that this alignment forms a distinct genetic group compared with EA populations from the United States, Europe, and elsewhere (44).
Table 1.
Racial distribution of selected signaling pathway alleles in patients with and without HF
| Gene, amino acid or nucleotide position, SNP reference (rs) number, cohort, clinical trial |
*MAF, AAa,b populations |
*MAF, EAc,d populations |
Subject Ns |
†P-value AA vs EA |
|
|---|---|---|---|---|---|
| AA | EA | ||||
| 24–26ADRB1 Arg389Gly (rs1801253), (NF) | a,b0.43 Gly | c0.28 Gly | 269 | 399 | <0.0001 |
| ADRB1 Arg389Gly (HF, BEST) | a0.43 Gly | c0.28 Gly | 207 | 762 | <0.0001 |
| 14,24ADRB1 Ser49Gly (rs1801252), (NF) | a,b0.22 Gly | c0.12 Gly | 119 | 147 | 0.002 |
| ADRB1 Ser49Gly (HF, BEST) | a0.24 Gly | c0.14 Gly | 205 | 756 | <0.0001 |
| 24ADRB2 Gly16Arg (rs1042713), (dbSNP) | a,b0.49 Arg | c0.35 Arg | 210 | 160 | 0.0001 |
| ADRB2 Gly16Arg (HF, BEST) | a0.47 Arg | c0.39 Arg | 207 | 762 | 0.003 |
| 24ADRB2 Gln27Glu (rs1042714), (dbSNP) | a,b0.16 Glu | c0.45 Glu | 107 | 107 | <0.0001 |
| ADRB2 Gln27Glu (HF, BEST) | a0.19 Glu | c0.39 Glu | 207 | 762 | <0.0001 |
| 24ADRB2 Thr164Ile (rs1800888), (dbSNP) | a,b0.00 Ile | c0.011 Ile | 166 | 272 | 0.09 |
| ADRB2 Thr164Ile (HF, BEST) | a0.002 Ile | c0.014 Ile | 207 | 762 | 0.053 |
| 14,15ADRA2C Ins322-325Del (rs61767072), (NF) | a0.40 Del | c0.036 Del | 132 | 179 | <0.0001 |
| 16ADRA2C Ins322-325Del (HF, BEST) | a0.43 Del | c0.04 Del | 207 | 762 | <0.0001 |
| 24,27GNB3 C825T (rs5443), (NF) | b,e0.86 T | d0.30 T | 159 | 1914 | <0.0001 |
| 28GNB3 C825T (HF, A-HeFT) | a0.72 T | c0.34 T | 350 | 424 | <0.0001 |
| 24,29NOS3 Glu298Asp (rs1799983), (NF) | ab,f0.11 Asp | cg0.35 Asp | 277 | 295 | <0.0001 |
| 30NOS3 Glu298Asp (HF, A-HeFT) | a0.11 Asp | c0.37 Asp | 352 | 424 | <0.0001 |
| 24,31CYP11B2 T-344C (rs1799998), (dbSNP), (NF) | ab,h0.20 C | c,d0.45 C | 624 | 592 | <0.0001 |
| 32CYP11B2 T-344C (HF, A-HeFT) | a0.22 C | c0.43 C | 354 | 424 | <0.0001 |
| 24NR3C2 Ile180Val (rs5522), (dbSNP) | a,b0.083 Val | c0.103 Val | 241 | 195 | 0.38 |
| 33NR3C2 Ile180Val (HF) | – | c0.12Val | – | 156 | – |
| 34–36ACE Del/intron16/Ins (rs1799752), (NF) | a0.41 Ins | d0.43 Ins | 467 | 196 | 0.48 |
| 37,38ACE Del/intron16/Ins (HF) | a0.37 Ins | ci0.36 Ins | 145 | 324 | 0.97 |
| AUIKI A1166C (3 UTR), (rs5186), (dbSNP), (NF) | a0.051 C | c0.26 C | 242 | 242 | <0.0001 |
| AUIKI A1166C (3 UTR), (HF) | a0.10 C | ci0.32 C | 145 | 384 | <0.0001 |
| 24,35,40AGT Thr174Met (rs4762), (NF) | a,b0.05 Met | c,d0.096 Met | 316 | 469 | 0.001 |
| 41AGT Thr174Met (Htn) | a0.05 Met | – | 187 | – | – |
| 38AGT Thr174Met (HF) | – | i0.18 Met | – | 58 | NA |
| 24,35,40AGT Met235Thr (rs699), (NF) | a,b0.86 Thr | c,d0.38 Thr | 299 | 450 | <0.0001 |
| 41AGT Met235Thr (Htn) | a0.83 Thr | c0.41 Thr | 187 | 611 | <0.0001 |
| 38AGT Met235Thr (HF) | – | i0.48 Thr | – | 58 | – |
| 24EDN1 Lys198Asn (rs5370), (dbSNP), (NF) | a,b0.18 Asn | c0.22 Asn | 266 | 220 | 0.120 |
| 42EDN1 Lys198Asn (HF, BEST) | a0.23Asn | c0.23Asn | 69 | 212 | 0.99 |
| 24ECE1 Thr341Ile (rs1076669), (dbSNP), (NF) | a,b0.00 Ile | c0.076 Ile | 157 | 216 | <0.0001 |
| 42ECE1 Thr341Ile (HF, BEST) | a0.007 Ile | c0.09 Ile | 69 | 212 | 0.0009 |
| 24GRK5 Gln41Leu (rs2230345), (dbSNP), (NF) | a,b0.31 Leu | c0.025 Leu | 141 | 141 | <0.0001 |
| 18GRK5 Gln41Leu (rs2230345), (HF) | a0.23 Leu | c0.017 Leu | 711 | 1749 | <0.0001 |
| 19,24SCN5A Ser1103Tyr (rs7626962), (dbSNP), (NF) | a0.065 Tyr | c0.001 Tyr | 527 | 493 | <0.0001 |
| 43SCN5A Ser1103Tyr (rs7626962), (dbSNP), (HF) | a0.09 Tyr | – | 112 | – | – |
Minor allele frequency.
Chi-square 2-sided test (Fisher exact test substituted for low cell counts) performed on the number of alleles.
AA = African ancestry; A-HeFT, African-American Heart Failure Trial; BEST = β-Blocker Evaluation of Survival Trial; dbSNP = Database of Single-Nucleotide Polymorphisms; EA = European ancestry; HF = heart failure with reduced left ventricular ejection fraction; Htn, hypertension; NF = nonfailing; UTR = untranslated region; SNP = single-nucleotide polymorphism.
African-American;
Sub-Saharan African;
European-American (Caucasian);
European (Caucasian);
South African or Zimbabwean;
Brazilian-African;
Brazilian-European (Caucasian);
United Kingdom-European;
French-Canadian European.
As can be seen in Table 1, of the 19 selected polymorphisms in 15 genes in non-HF (NF) populations, 16 variants exhibit evidence of racial differences in minor allele frequency (MAF). Moreover, in every case the difference in allele frequency is maintained in HF populations (i.e., there is good agreement in MAFs between HF and NF populations). The data in Table 1 illustrate that 1) racially based genetic variation is common in genes encoding proteins that are important in neurohormonal signaling systems; and 2) the racial differences in distribution of polymorphic variants are generally not altered in HF.
For ADRB2 Gln27Glu, GNB3 C825T, NOS3 Glu298Asp, and AGTR1 A1166C, the differences in MAF allele frequencies are very different (i.e., ≥ 2.5-fold between races, Table 1); and for ADRB2 Thr164Ile, ADRA2C Ins322-325Del, ECE1 Thr341Ile, GRK5 Gln41Leu, and SCN5A Ser1103Tyr, the MAF frequencies are extremely different (i.e., >5-fold). Having established that AA and EA racial differences are common in polymorphisms of signaling systems important in HF, does this have any practical importance? We will first consider potential impacts on HF natural history.
Does Common Genetic Variation Influence HF Natural History in Either AA or EA Populations?
Table 2 lists the Table 1 polymorphisms with a racial difference in MAF, the pharmacologic effects of their minor vs. major alleles, the predicted biologic or pharmacologic effects of the frequency difference in AA vs. EA individuals, and whether the projected relative signaling effect would be cytoprotective or cytopathic in failing human cardiac myocytes based on the assumption that increases in signaling are harmful and decreases are beneficial. In 13 of the 14 polymorphisms listed in Table 2 (exception is ECE1 Thr341Ile, where functional effects are unknown), the minor alleles have effects that differ from the major allele and could influence HF risk or progression. Race-associated differential genetic effects on HF natural history could in turn affect therapeutic responses.
Table 2.
Impact of AA vs. EA MAF differences on cardiac myocyte cell signaling* and biologic effect on cardiac myocytes, adrenergic nerve terminals, vascular smooth muscle or endothelium
| Gene polymorphism |
Effect of minor vs major allele |
AA vs. EA MAF | AA vs. EA cardiac myocyte relative effect (cytoprotective or cytopathic/harmful)* |
Pharmacogenetic effect in AA vs. EA* (Actual Possible or Predicted) |
|---|---|---|---|---|
| ADRB1 Arg389Gly | ↓NE† affinity ↓ signal transduction ↓ constitutive activity |
~50% ↑ in Gly | Protective (↓ ADRB1 signaling) |
↓ response to β-blockers (Actual for bucindolol, Predicted for standard β-blockers but not supported by empirical data) |
| ADRB1 Ser49Gly | ↑ internalization, downregulation |
~70% ↑ in Gly | Protective (↓ ADRB1 signaling) |
↓ response to β-blockers (Predicted but not supported by empirical data) |
| ADRB2 Gly16Arg | ↑ internalization, downregulation |
~20% ↑ in Arg | Protective | ↓ response to nonselective β- blockers (Predicted, not supported by empirical data) |
| ADRB2 Gln27Glu | ↓ internalization, downregulation |
~ 50% ↓ in Glu | Protective (↓ ADRB2 signaling) |
↓ response to carvedilol, CRT (Possible, no data in AA) |
| ADRB2 Thr164Ile | ↓ signal transduction ↓ NE, EPI‡ affinity ↑ vasoconstriction |
Ile ↓, rare in AA | Harmful (restoration of cardiac ADRB2 signaling); Protective (↓ afterload) |
↑ response to nonselective β- blockers (Predicted, no reported data) |
| GRK5 Gln41Leu | ↑ ADRB1,2 phosphorylation, uncoupling |
~10-fold ↑ in Leu |
Protective (↑ ADRB1,2 phosphorylation and desensitization |
↓ response to carvedilol, metoprolol (Actual) |
|
ADRA2C Ins322- 325Del |
↑ sympatholysis ↑ adrenergic drive |
~10-fold ↑ in Del | Harmful (↑ adrenergic drive) |
↓ response to bucindolol, ↑ CV adverse events to bucindolol (Actual) |
| GNB3 C825T | ↑ vasoconstriction | ~2.5-fold ↑ in T | Harmful (↑ afterload) | ↑ response to BiDil (Possible based on AA data, no data in EA) |
| NOS3 Glu298Asp | ↓ NOS3 activity | ~70% ↓ in Asp | Protective§ (↑ NOS3 activity) |
↑ response to BiDil (Possible based on AA data, no data in EA) |
| CYP11B2 T-344C | ↑ aldosterone synthesis |
~50% ↓ in C | Harmful (↑ aldosterone) |
↑ response to BiDil (Possible based on AA data, no data in EA) ↑ response to RAAS inhibition (Predicted, no data in AA or EA) |
| AGTR1 A1166C | ↑ AGTR1 density | ~70% ↓ in C | Protective (↓ AGTR1 density) |
↓ response to ARBs (Possible based on data in EAs, no AA data); ↓ response to ACEIs (Possible, based on non- genotyped empirical data in AAs) |
| AGT Met235Thr | ↑ AGT levels | ~2-fold ↑ in Thr | Harmful (↑ AGT) | ↑ response to ACEIs, ARBs (Predicted, for ACEIs not supported by non-genotyped empirical data in AAs; no support for ARBs) |
| ECE1 Thr341Ile | ? | ~10-fold ↓ in Ile | ? | ↑ Response to bucindolol (Possible, based on EA data) |
| SCN5A Ser1103Tyr | ↑ late INa; Ca2+ overload |
>10-fold ↑ in Tyr | Harmful; pro- arrhythmic |
↑ indication for prevention of sudden cardiac death by ICDs (Actual) |
Based on biologic or pharmacologic effects between genotypes, and allele frequency differences between AA (African-ancestry) and EA (European-ancestry) patients.
Norepinephrine (NE).
Epinephrine (EPI).
Minor allele frequency (MAF) difference may be an adaptive response to NOS3 uncoupling in AA; Actual means by-genotype differences in response in AA and EA, which would imply a differential response in AA vs. EA based on the differences in MAF. Possible means 1) by-genotype response differences for drug class in AA, EA, or EA + AA populations, or 2) decreased response in AA predicted by MAF signaling effects. Predicted means no clinical data, based entirely on cell-signaling effects; ACEI = angiotensin converting enzyme inhibitors; ARB = angiotensin-receptor blockers; CRT = cardiac resynchronization therapy; ICD = implantable cardioverter defibrillator; RAAS = renin-angiotensin-aldosterone system; CV = cardiovascular
In contrast to the low frequency (MAF <1%) mutations in contractile protein, cytoskeletal, and other genes that cause inherited dilated cardiomyopathies (45), there is no good evidence that any of the cell-signaling polymorphisms listed in Tables 1 or 2 predispose to the risk of developing HF. The fact that the MAF distributions are quite similar in HF and NF populations indicates that HF populations are not generally enriched in these signaling molecule minor alleles. Although earlier, smaller studies did report enrichment in HF for some of these MAFs (46–48), most subsequent larger studies (49–51), including meta-analyses (52,53), have not confirmed the findings. In general there is a need for well-controlled, prospective studies with large numbers of patients and clinical endpoints to address this issue, and whether any of the variants listed in Table 1 could influence the risk of developing HF is not settled.
In terms of possible effects on disease progression, of the HF studies with enough patients and events to provide adequate statistical power, the adrenergic receptor polymorphism substudy (1040 patients, 193 deaths, and 426 deaths or first HF hospitalizations [HFHs]) (16,25) of the placebo-controlled β-Blocker Evaluation of Survival Trial (BEST) and a large observational study from the Universities of Cincinnati and Pennsylvania (2460 patients and 765 deaths) (18), each with >20% of AA patients, provide the most robust data. Effects of polymorphic variants on natural history of established HF can be assessed in BEST in the 525 placebo-treated patients, and in the Cincinnati/Pennsylvania study by effects in patients untreated with β-blockers, as described in the Supplement.
Among the potential candidate gene variants listed in Tables 1 and 2, perhaps the most likely to influence the progression of HF is the ADRB1 Arg389Gly polymorphism (Figure 1A). This is because in human ventricular preparations, compared with its 389Gly counterpart the 389Arg version of the receptor has 3-to-4 times the signal transduction capacity (25), a higher proportion of receptors that are constitutively active as inferred from inverse agonist activity (25), and a much higher affinity for norepinephrine (54) (Table 2). As a result of this marked difference in agonist affinity and the lower affinity of β2 receptors for norepinephrine, the protein product of ADRB1 389Arg can be considered the norepinephrine receptor in the human heart (21,54). Increased release and cardiac concentrations of the β1-selective neurotransmitter norepinephrine produce and lead to the progression of ventricular chamber remodeling and HF (21). Table 3 summarizes the ADRB1 Arg389Gly data from placebo-treated patients in BEST (16,25) and MERIT-HF (Metoprolol CR/XL Randomized Intervention Trial in Congestive Heart Failure, 55), as well data from the Cincinnati/Pennsylvania β-blocker untreated patients (18).
Table 3.
Impact of adrenergic signaling genetic variation on outcomes in placebo-treated HF patients in the BEST and MERIT-HF clinical trials or β-blocker-untreated patients in the Cincinnati/Pennsylvania observational study. HRs are expressed as minor allele carriers vs. major allele homozygotes
| Gene polymorphism | HR [no. events] (95% CI), ACM* or ACM/Tx† |
HR [no. events] (95% CI), ACM/HFH‡ |
||||
|---|---|---|---|---|---|---|
| EA | AA | EA + AA | EA | AA | EA + AA | |
|
ADRB1 Arg389Gly, BEST* |
1.25 [84] (0.81,1.93) p=0.32 |
0.85 [15] (0.27,2.63) p=0.77 |
1.14 [97] (0.76,1.69) p=0.79 |
1.03 [174] (0.76,1.39) p=0.84 |
0.77 [41] (0.38,1.54) p=0.45 |
1.02 [215] (0.79,1.32) p=0.88 |
| Interaction p=0.51 | Interaction p=0.42 | |||||
|
ADRB1 Arg389Gly, MERIT-HF55 |
NA | NA | NA | NA | NA | 1.00§ [NA] (0.61,1.64) p=0.99 |
|
ADRB1 Arg389Gly, † Cincinnati/Pennsylvania18 |
1.98 [NA] (1.07,3.65) p=0.03 |
unadjusted p=0.55¶ |
1.76 [NA] (1.09,2.85) p=0.02 |
NA | NA | NA |
|
ADRB1 Ser49Gly, BEST* |
0.64 [84] (0.37,1.12) p=0.11 |
0.94 [15] (0.32,2.76) p=0.91 |
0.68 [99] (0.42,1.11) p=0.12 |
0.78 [174] (0.54,1.13) p=0.19 |
0.61 [41] (0.31,1.22) p=0.16 |
0.74 [215] (0.54,1.03) p=0.07 |
| Interaction p=0.49 | Interaction p=0.53 | |||||
|
ADRB2 Gly16Arg, BEST* |
1.05 [84] (0.68,1.63) p=0.82 |
0.62 [15] (0.21,1.82) p=0.38 |
0.95 [99] (0.63,1.42) p=0.80 |
1.06 [174] (0.78,1.43) p=0.72 |
0.85 [41] (0.42,1.69) p=0.63 |
1.02 [215] (0.77,1.35) p=0.89 |
| Interaction p=0.38 | Interaction p=0.56 | |||||
|
ADRB2 Gln27Glu, BEST* |
1.11 [84] (0.71,1.74) p=0.65 |
2.91 [15] (1.03,8.19) p=0.035 |
1.37 [99] (0.91,2.07) p=0.13 |
1.21 [174] (0.88,1.67) p=0.23 |
1.72 [41] (0.92,3.21) p=0.085 |
1.27 [215] (0.97,1.68) p=0.085 |
| Interaction p=0.086 | Interaction p=0.34 | |||||
|
ADRB2 Thr164Ile, BEST* |
1.34 [84] (0.42,4.24) p=0.62 |
n=1 164Ile |
1.29 [99] (0.41,4.09) p=0.66 |
1.20 [174] (0.53,2.70) p=0.67 |
n=1 164Ile |
1.06 [215] (0.47,2.40) p=0.88 |
| Interaction p=NA | Interaction p=NA | |||||
|
ADRA2C Ins322-325Del, BEST* |
0.96 [84] (0.44,2.07) p=0.91 |
1.13 [15] (0.36,3.60) p=0.83 |
0.86 [99] (0.51,1.43) p=0.55 |
0.79 [174] (0.45,1.39) p=0.41 |
0.98 [41] (0.51,1.89) p=0.95 |
0.91 [215] (0.65,1.76) p=0.58 |
| Interaction p=0.74 | Interaction p=0.62 | |||||
|
GRK5 Gln41Leu, Cincinnati/Pennsylvania18 (adjusted for age, sex) |
NA [3] (Too few events) |
0.32 [NA] (0.13,0.80) p=0.01 |
0.76 [NA] (0.40,1.32) p=0.39 |
NA | NA | NA |
All-cause mortality (ACM).
ACM or cardiac transplantation.
ACM or heart failure hospitalization (HFH).
All races.
Events too few for Cox modeling.
CI = confidence interval; HR = hazard ratio; NA = not available; Tx = cardiac transplantation.
In BEST as well as in MERIT-HF (which did not report pharmacogenetic data by race), in placebo-treated patients there was no evidence of an effect of ADRB1 Arg389Gly genotypes on the outcomes of time to all-cause mortality (ACM) or the combined endpoint of ACM or HFH (Table 3). In contrast, in the Cincinnati/Pennsylvania study, EA (but not AA) patients who were untreated with β-blocking agents had improved transplant-free survival from the time of HF diagnosis if they were ADRB1 389Arg homozygotes, compared with 389Gly genotypes (18). One possible difference between the clinical trial/placebo groups data from BEST and MERIT-HF as compared with the Cincinnati/Pennsylvania study is much shorter mean or median follow-up times in BEST (2 years) (25) and MERIT-HF (1 year) (55), compared with 46 months in the observational study (18). It is conceivable that a longer follow-up time in HF is necessary to detect genotype-based differences in outcomes for common genetic variation in neurohormonal cell signaling systems. However, the Cincinnati/Pennsylvania data imply a protective effect of the ADRB1 389Arg allele, which is contrary to work in transgenic cardiac overexpressor mice as well as multiple cell biology studies that demonstrate that heightened ADRB1 signaling is cytopathic to cardiac myocytes (21). Thus, natural history and biologic plausibility data do not consistently support an effect of the ADRB1 Arg389Gly polymorphism on clinical HF outcomes. However, larger prospective studies with relatively long follow-up periods, of greater duration than is usually the case in therapeutic clinical trials, are needed before reaching firm conclusions. This principle also applies to the other genetic variants listed in Table 1.
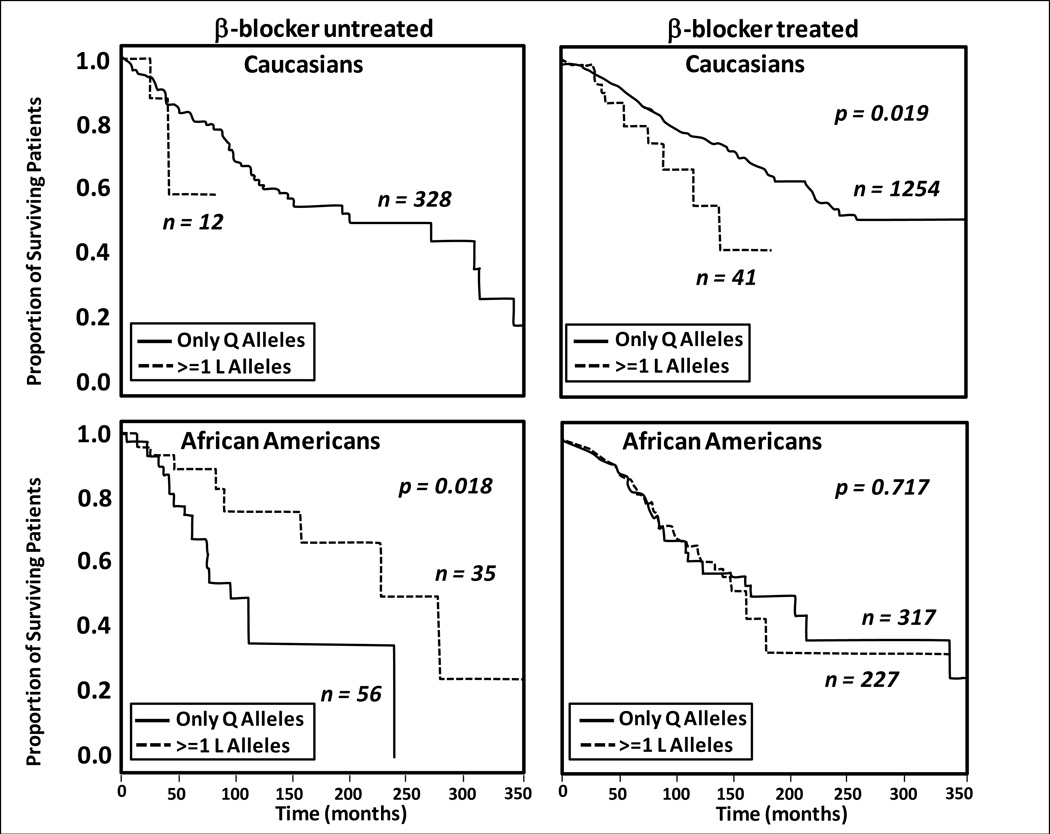
The Cincinnati/Pennsylvania study produced a very interesting result in β-blocker-untreated patients regarding the GRK5 Gln41Leu polymorphism: AA patients (but not EA patients or the entire EA + AA cohort) had improved transplant-free survival (Table 3, Figure 2 L-panels) in 41Leu genotypes compared with 41Gln homozygotes (18). The 41Leu variant of GRK5 is a gain-of-function polymorphism whose protein product increases β-adrenergic receptor phosphorylation, which produces receptor desensitization and a genetic anti-adrenergic effect (18,56). The approximate 10-fold higher prevalence of GRK5 41Leu genotypes in AA patients compared with EA patients may therefore provide a significant amount of "endogenous β-blockade" (18,56), with the effect detectable in patients untreated with β-blocking agents.
Figure 2.
Kaplan-Meier curves for time to all-cause mortality or cardiac transplantation from the time of heart failure diagnosis of patients with GRK5 Gln(Q)41Leu(L) genotypes (18). P-values are from an unadjusted model log-rank statistic. Reproduced with permission of Elsevier and the Journal of the American College of Cardiology.
Table 3 also gives the BEST placebo-treated patient data for adrenergic receptor polymorphisms listed in Tables 1 and 2. For the entire EA + AA cohort, there were no differences between major allele homozygotes and MAF carriers for the ADRB1 Ser49Gly, ADRB2, or ADRA2C Ins322-325Del polymorphisms, for either the ACM or ACM/HFH endpoints. In AA patients the ACM hazard ratio p-value is 0.035 (above the 0.01 critical value, see Supplement) and the interaction test is p<0.10 for ADRB2 27Glu carriers having an increased mortality risk, but this is based on only 15 total events Although the EA ACM and ACM/HFH hazard ratios of respectively 1.11 and 1.21 are substantially lower than AA values, the race-treatment interaction p-values are not significant for either endpoint. There are no other differences with p<0.05 in the BEST receptor polymorphism data in Table 3.
In addition to pump dysfunction, the other major clinical component of HF disease progression is the development of serious arrhythmias. The SCN5A 1103Tyr variant of the Nav1.5 sodium channel is found in 6% to 9% of AA individuals but is extremely rare in EA subjects (19,44, Table 1), and it has been associated with a 4-fold increase in implantable cardioverter defibrillator (ICD) discharges for sustained ventricular arrhythmias in AA HF patients (43). The SCN5A Ser1103Tyr polymorphism clearly has the potential to influence HF progression disproportionally in AA vs. EA populations, and also has implications for device and even drug therapy as discussed below.
Therefore, the cell-signaling polymorphisms denoted in Tables 1 and 2, some of which are capable of major effects on cardiac myocyte function, do not appear to fundamentally affect the risk of developing HF. One likely reason for this is that neurohormonal cell-signaling mechanisms only modulate cardiac myocyte function and are not critical for cell integrity. In addition, when neurohormonal activation occurs in HF, it is typically ontogenetically beyond the reproductive years, obviating any possibility of natural selection eliminating deleterious alleles. In terms of polymorphism effects on HF disease progression, the most compelling evidence is for GRK5 Gln41Leu producing a beneficial effect and for SCN5A Ser1103Tyr producing an adverse effect in AA individuals, due to markedly higher (≥10-fold) MAF frequencies.
Genetic Variation and HF Therapy: Interaction of Race, Signaling Polymorphisms, and Therapeutic Responses
There is considerable evidence that several of the polymorphisms in Table 1 exert an influence on drug or device therapeutic response. Table 4 summarizes pharmacogenetic effects on HF clinical, left ventricular remodeling, or biomarker endpoints that have been reported for some of the polymorphisms listed in Table 1, independent of race. Many of these studies were underpowered, but some (16,18,25,54) had adequate prespecified statistical design. In most cases the results are reinforced by biologic plausibility, and in some instances there are large differences in functional activity between the polymorphic variants that have been shown to interact with the therapeutic agent or drug class (15,16,25,54–56).
Table 4.
Summary of signaling molecule polymorphism effects by drug or device class, irrespective of races
| Drug or class | Polymorphism (effects on response in HF) |
|---|---|
| Standard β-blockers |
GRK5 Gln41Leu (↓ by Leu carriers);18ADRB2 Gln27Glu (carvedilol ↑ in Glu genotypes)57,58ACE Del/intron16/Ins (↑ in Del homozygotes)47 |
| Bucindolol (β-blocker/sympatholytic) |
ADRB1 Arg389Gly (↑in Arg homozygotes,25 ↓ in {389Gly + ADRA2C 322-325Del} genotypes;54EDN1 Lys198Asn (gene dose- related ↓ in Asn genotypes);42ECE1 Thr341Ile (↓ in Ile genotypes) |
| Angiotensin-converting enzyme inhibitors |
ACE Del/intron16/Ins (↑ in Del homozygotes)59 |
| Angiotensin AT-1 receptor blockers | ↑ biomarker (NT-proBNP) response in AGTR1 1166C genotypes60 |
| Mineralocorticoid receptor blockers | No effects reported |
| Hydralazine/isosorbide dinitrate |
NOS3 Glu298Asp (↑ in Glu homozygotes);30CYP11B2 T-344C (↑ in −344T homozygotes);32GNB3 825T(↑ in TT homozygotes)28 |
| Cardiac resynchronization therapy |
ADRB2 Gln27Glu (↑ in Glu homozygotes);61NR3C2 Ile180Val(↑ in Ile homozygotes)62 |
| ICD appropriate discharge for VT/VF | SCN5A Ser1103Tyr (↑ events in Tyr carriers)43 |
HF = heart failure; ICD = implantable cardioverter defibrillator; NT-proBNP = N-terminal pro-brain natriuretic peptide; VF = ventricular fibrillation; VT = ventricular tachycardia.
Table 5 gives some of the evidence for racial effects on specific HF therapeutic responses that may be influenced by the products of genes listed in Table 1. Because HF clinical response-racial data are limited in several circumstances, antihypertensive response is included for some drug classes. Decreased effectiveness in AA HF patient populations has been reported for 3 drug classes: standard β-blockers (18,65); the β-blocker/sympatholytic agent bucindolol (67); and angiotensin-converting enzyme inhibitors (68,69). Blunted antihypertensive responses to β-blockers (66) and angiotensin-converting enzyme inhibitors (70) have also been reported in AA patients. In contrast, the response to fixed-dose hydralazine/isosorbide dinitrate appears to be enhanced in AA HF patients (68), and ICD therapy appears to be more effective in a genetically defined subset of AA HF patients (43).
Table 5.
Racial effects (AA vs. EA) on HF or hypertension response by drug or device class
| Drug or class | Racial effect in HF (AA or EA) | Racial effect in hypertension (AA or EA) |
|---|---|---|
| β-blockers | Cresci et al,18↓ in AA (carvedilol, metoprolol) Yancy et al, 63 ↔ in AA (carvedilol) Goldstein et al, 64 ↔ ↓ in AA (metoprolol) Lanfear et al, 65↓ in AA (all standard P-blockers) |
Cubeddu et al,66 ↓ in AA (propranolol) |
| Bucindolol (β-blocker/sympatholytic) | BEST,67 ↓ in AA | No data |
| Angiotensin-converting enzyme inhibitors |
Carson et al,68 ↓ in AA Exner et al,69 ↓ in AA |
Weir et al,70 ↓ in AA |
| Angiotensin AT-1 receptor blockers | Prisant et al, ↔ in AA | Ofili et al, 72 ↔ in AA |
| Mineralocorticoid receptor blockers | No data | No data |
| Hydralazine/isosorbide dinitrate | Carson et al,68 ↑ in AA | No data |
| Cardiac resynchronization therapy | No data | No data |
| ICD | ↑appropriate discharge in a subset of AA, by virtue of markedly higher SCN5A 1103Tyr frequency43 |
No data |
AA = African ancestry; EA = European ancestry; HF = heart failure; BEST = β-Blocker Evaluation of Survival Trial.
There is clearly the potential for some of the polymorphisms listed in Tables 1 and 2 to account for the Table 5 racial effects. The potential impact of race-related differences in MAF on therapeutic response based on estimations of the predicted, possible, or actual observed effects of therapeutic agents or classes is given in the last column of Table 2. The estimates follow the general paradigm that increased cardiac myocyte neurohormonal signaling is harmful, decreased signaling is beneficial, and the predicted therapeutic effect of an inhibitor is directly related to the degree of signaling. Most of the pharmacogenetic interactions are assigned to the possible (some clinical evidence in support) or predicted (no clinical data in support) categories. Some of the the pharmacogenetic interactions listed in Table 2 that are supported by clinical outcomes data will now be discussed by therapeutic class.
β-blockers, including the β-blocker/sympatholytic bucindolol
ADRB1 Arg389Gly, Ser49Gly
The higher-functioning 389Arg allele of ADRB1 present at lower frequency in AA patients (Tables 1 and 2) was originally hypothesized to confer a greater therapeutic HF response to β-blockers (25) (Table 2). However, thus far for clinical outcomes this seems to be the case only for the β-blocker/sympatholytic agent bucindolol (25,54), an experimental compound in development for HF and prevention of atrial fibrillation. There is no evidence of a differential ADRB1 Arg389Gly clinical therapeutic effect for standard HF-approved β-blockers (18,55,73), which are devoid of β1 389Arg adrenergic receptor inverse agonist effects in isolated preparations of a failing human heart (25) and lack systemic sympatholytic properties in HF patients. The latter property likely contributes to a favorable therapeutic effect of bucindolol on the ADRB1 389Arg "norepinephrine receptor" (54). Thus, the ADRB1 Arg389Gly polymorphism could explain the reduced response to bucindolol in BEST AA patients (66) and the perhaps smaller racial interaction with carvedilol (63) or metoprolol (64). However, the Cincinnati/Pennsylvania study (18), which contained the largest cohort of β-blocker-treated AA HF patients studied to date, found reduced efficacy with carvedilol and metoprolol (18). In addition, in a large (1094 patients, 56% AA, 1368 primary events) well-controlled health care system retrospective study evaluating the effects of all U.S. marketed β-blockers, the reduction in ACM and first re-HFH in AA HF patients, although statistically significant (p = 0.024 compared to the 20% of patients not treated with β-blockers), was 40–50% less than in EA patients (65).
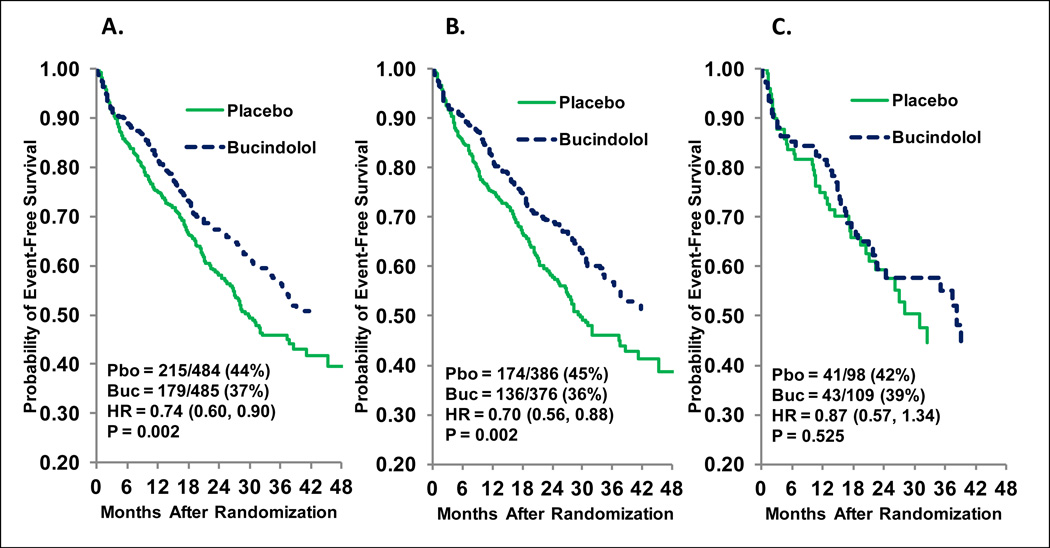
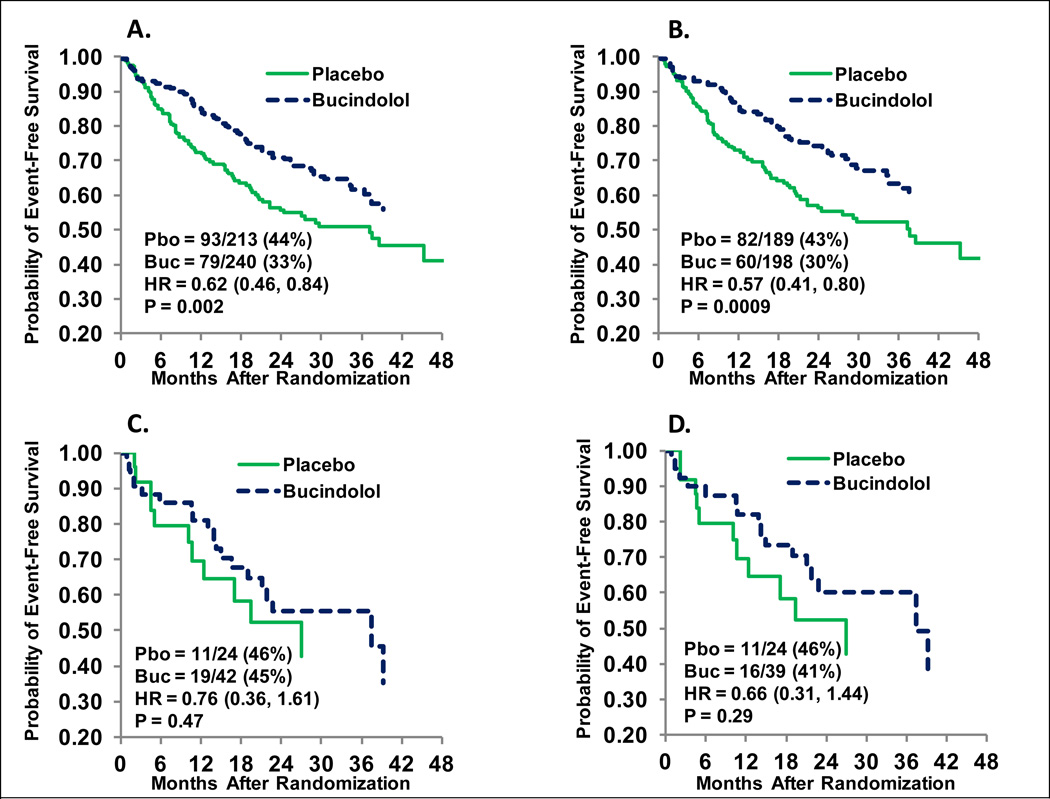
Thus the weight of the evidence favors a blunted effectiveness of β-blockers in AA compared to EA HF patients. Figures 3 and 4 address the question of whether a lower frequency of the favorable response ADRB1 389Arg homozygous (Arg389Arg) genotype explains the attenuated response of bucindolol in AA patients (i.e., whether pharmacogenetics alone can explain lower efficacy in AAs) (67). Baseline characteristics for EA or AA patients in the BEST 1040-patient adrenergic receptor polymorphism substudy (16,25,54) are given in the Supplement (Table S1), and the differences are typical of other HF studies (68). For the primary ACM endpoint of BEST, as in the entire cohort (67), there was a statistically significant (p=0.023) test for interaction between non-AA and AA patients in the substudy population, with AA patients showing no evidence of a favorable treatment effect. There were relatively few (n=36) ACM events in the 207-member AA subgroup, and in order to assess the interaction of race and ADRB1 Arg389Gly genotype, time to ACM/HFH was evaluated. Figure 3 gives the Kaplan-Meier curves for AA (84 events) and EA (310 events) patients for ACM/HFH in the substudy parent (all genotypes) population. The AA group has an attenuated hazard ratio (0.87, p=0.52) vs. the EA group (0.70, p=0.002; interaction p=0.39), a pattern similar to the entire cohort (data not shown). In Figure 4 the treatment effects in the ADRB1 Arg389Arg genotype subgroup are enhanced in both racial groups, but attenuation relative to EA persists in AA patients (hazard ratio 0.76 in AA [C] vs. 0.57 in EA [B]). However, when class-IV HF subjects, who in BEST tended to be volume overloaded/unstable at the time of randomization (74) are excluded, the AA hazard ratio improves further to 0.66 (Figure 4D), approaching the EA hazard ratio in the same patient population (0.58 [95% confidence interval 0.41–0.82], data not shown). Compared with all genotypes (Figure 3C), the 32% of patients who were ADRB1 Arg389Arg (Figure 4C) have an approximate 2-fold increase in effect size (using the relative effect size [RES] calculation [Supplement]), and then restriction to class-III (Figure 4D) results in an approximate 3-fold increase compared with Figure 3C. These data suggest that differences in β-blocker/sympatholytic response exist between AA and EA patients beyond the 37% lower (32% AA, 51% EA) ADRB1 Arg389Arg genotype in AA individuals, but that strategies in addition to pharmacogenetics such as more precise clinical phenotyping (3)—in this case excluding volume overloaded, unstable class-IV patients consistent with most HF guidelinescould be used to to bring the AA therapeutic response in line with that of EA individuals. In the example presented in Figures 3 and 4, the largest increment in effectiveness (2-fold) was achieved by treating patients with a preferred genotype, followed by an additional 1.5-fold achieved by patient selection. The result is an effect size (Supplement) increasing from a negligible 13% to a quite therapeutic 34%, albeit achieved in only 30% of the original AA population.
Figure 3.
Time to all-cause mortality or first heart failure hospitalization in the BEST “Adrenergic Receptor Polymorphism Substudy.” A. European-ancestry (EA) + African-ancestry (AA) patients; B. EA patients; C. AA patients. Pbo = placebo events/patients; buc = bucindolol events/patients; HR = hazard ratio. Cox modeling was unadjusted.
Figure 4.
Time to all-cause mortality or first heart failure hospitalization in the BEST “Adrenergic Receptor Polymorphism Substudy,” ADRB1 Arg389Arg cohort. A. European-ancestry (EA) + African-ancestry (AA) New York Heart Association Class III and IV patients; B. EA patients Class III and IV; C. AA patients Class III and IV; D. AA patients Class III only. Pbo = placebo events/patients; Buc = bucindolol events/patients; HR = hazard ratio. Cox modeling was unadjusted.
The 49Gly minor allele of ADRB1 (Figure 1A) has an approximate 70% higher MAF in AA vs. EA patients (Table 1), and this would lead to a greater degree of cardiac myocyte ADRB1 receptor down-regulation in AA HF patients (Table 2) that could attenuate β-blocker treatment effects (Table 2) by reducing ADRB1 signaling. There are conflicting reports of effects of the ADRB1 Ser49Gly polymorphism on β-blocker effect. A recent report found that open-label carvedilol-treated, Brazilian, AA patients who were 49Gly carriers had improved hospitalization-free survival compared with 49Ser homozygotes, while non-black patients treated with the same 25 mg bi-daily target dose showed no difference between genotypes (75). However, in a large, 75%-EA HF patient population studied prospectively and treated with carvedilol or metoprolol, no effect of ADRB1 Ser49Gly was found, including no racial interaction (73). In BEST, there was no effect of the ADRB1 Ser49Gly polymorphism on bucindolol vs. placebo treatment effects in either AA or EA patients, with respective interaction p-values of 0.23 and 0.74 for ACM/HFH events. In AA patients, the hazard ratio was numerically higher in 49Gly carriers (1.26 [95% confidence interval 0.60–2.63]) compared with 49Ser homozygotes (0.72 [0.42–1.26]), indicating a trend for AA ADRB1 49Gly carriers to do worse on bucindolol, the opposite of the result with carvedilol in the Brazilian study (75). A Swedish study that did not report racial demographics but was presumably conducted in predominately EA patients found a therapeutic advantage to the the 49Gly genotype in patients treated with low doses of open-label metoprolol, which disappeared on higher doses (76). However, the possible enhanced effects of β-blockers in ADRB1 49Gly genotypes (75,76) are not supported by biologic plausibility, which would predict an attenuated effect (Table 2), at least in placebo-controlled studies.
GRK5 Gln41Leu
The other good candidate for a racial-genetic interaction to β-blocker therapy is the GRK5 Gln41Leu polymorphism investigated in the large (n=711 AA HF patients, n=1749 EA HF patients) Cincinnati/Pennsylvania observational study (18). Similar to bucindolol in BEST (67, Figure 3), carvedilol or metoprolol exhibited no evidence of a beneficial treatment effect in the all-genotype AA population (Figure 5). In AA patients with GRK5 41Leu genotypes who were not treated with β-blocking agents, transplant-free survival from the time of HF diagnosis was better (age/sex-adjusted hazard ratio 0.325, p=0.01, Figure 2) than in 41Gln homozygotes. However, in the β-blocker-treated cohort, AA patients with 41Leu genotypes did equally well compared with 41Gln homozygotes, while EA patients with 41Leu genotypes (limited by only 10 events) may have done worse than their Gln homozygous counterparts (p=0.019 in an unadjusted model, upper right panel Figure 2). These data were interpreted as the GRK5 41Leu polymorphism conferring endogenous β-blockade preferentially in AA patients by virtue of their approximate 10-fold higher prevalence of 41Leu genotypes (18), which then obviated potentially favorable effects of β-blockade.
Figure 5.
Effect of race on transplant-free survival in the presence or absence of P-blocking agents in the Cincinnati/Pennsylvania study (18). A. Combined European-ancestry (EA) and African-ancestry (AA) heart failure cohort (18), stratified by +/− P-blocker (carvedilol 65%, metoprolol 25%, other 10%) use; (top inset, EA [“Caucasians”]; bottom inset, AA (“African-Americans”]). Solid line = P-blocker-untreated; dashed line = P-blocker-treated. B. Transplant-free survival of P-blocker-untreated heart failure subjects, stratified by race. C. Transplant-free survival of P-blocker-treated heart failure subjects, stratified by race. Solid line = EA; dashed line = AA. P-values are from an unadjusted model log-rank statistic. Reproduced with permission of Elsevier and the Journal of the American College of Cardiology. BB = β-blocker.
ADRB2 Gln27Glu, Gly16Arg, Thr164Ile
These 3 polymorphisms are depicted in Figure 1B. Two studies have reported that the minor allele of ADRB2 Gln27Glu predicts a positive reverse remodeling response to the β1/β2/α1 receptor blocker carvedilol: 27Glu carriers in one (57) and Glu homozygotes (58) in the other. The ADRB2 27Glu variant is resistant to agonist-induced receptor down-regulation and actually up-regulates in response to agonist exposure (77), which may make the human β2 adrenergic receptor and its subsequent blockade more important in 27Glu genotypes. Although ADRB2 genotype data relative to race and β-blocker response have not been reported, the allele frequency data in Tables 1 and 2 would predict that AA patients may be at a possible disadvantage to treatment with nonselective β-blockers (Table 2) due to their lower ADRB2 27Glu allele frequency.
There is no evidence that ADRB2 Gly16Arg, which exhibits only a slight difference (increase by about 20%, Table 2) in MAF in AA vs. EA HF patients, affects β-blocker response (Table 2). The ADRB2 164Ile allele is rare and was detected in only one AA patient in BEST (Table 1); hence, there are no data on race-associated pharmacogenetic effects (Table 2). Also, the ADRB2 facilitates norepinephrine release, but in BEST there were no effects of ADRB2 polymorphisms on baseline norepinephrine or 3-month change, for either race or treatment group (data not shown).
ADRA2C Ins322-325Del
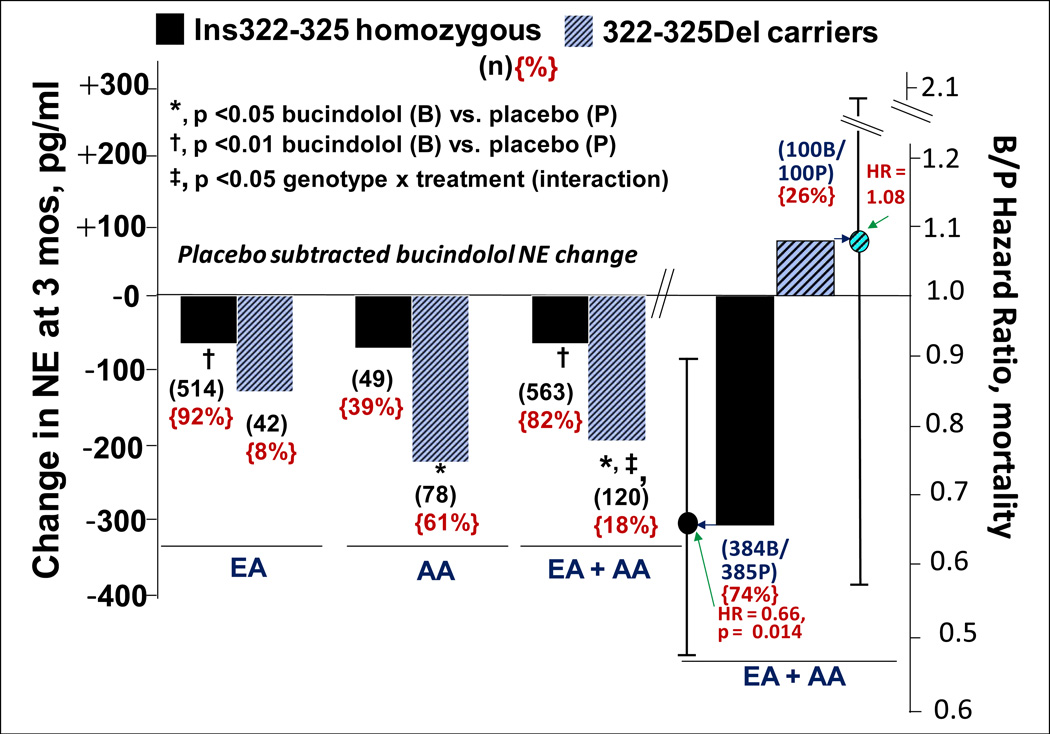
In HF patients, the ADRA2C Ins322-325Del polymorphism affects the cardiac sympatholytic response to bucindolol (16), which is a secondary pharmacodynamic property of this nonselective β-blocker (78). The ADRA2C receptor is located prejunctionally on cardiac adrenergic neurons, where it provides tonic inhibition to norepinephrine release (79), and a 4-amino acid deletion in the third intracytoplasmic loop at positions 322-325 completely abolishes functionality (15). In patients with advanced HF and elevated adrenergic activity, the ADRA2C Ins322-325Del polymorphism does not appreciably affect circulating norepinephrine levels, but influences bucindolol's sympatholytic effects (16). The norepinephrine-lowering effects of bucindolol, and potentially other sympatholytic agents, are markedly enhanced in HF patients with 322-325Del genotypes (Figure 6) (16).
Figure 6.
Bucindolol-associated norepinephrine (NE)-lowering and effects on all-cause mortality by ADRA2C Ins322-325Del genotype. L-y axis and panel, systemic venous NE changes (pg ± SEM) at 3 months, placebo subtracted in European-ancestry (EA) or African-ancestry (AA) patients or the combination in BEST. R-y axis and panel, bucindolol:placebo hazard ratio for all-cause mortality in EA + AA patients. There were no differences in baseline NE, which ranged from 436 pg/mL to 557 pg/mL in the various groups. The treatment × genotype interaction p-value for all-cause mortality is 0.29.
Similar to GRK5 41Leu genotypes, in AA vs. EA subjects the 322-325Del or minor allele genotypes are present at a 10-fold higher frequency (14–16, Tables 1 and 2). In advanced HF, marked sympatholysis can be associated with increased cardiovascular adverse events, due to adrenergic support being reduced to levels below what are required to support the failing heart (16,78). Because of the markedly higher ADRA2C 322-325Del frequency, AA patients treated with bucindolol are potentially more prone to this type of adverse event, manifested as increased mortality from sudden death or precipitation of HFH (78).
Figure 6 shows the large effect on bucindolol-associated systemic norepinephrine lowering conferred by ADRA2C 322-325 genotypes, and that the effect is present in both EA and AA patients. Systemic plasma norepinephrine is a biomarker for cardiac norepinephrine, which directly participates in the pathophysiology of adverse remodeling as well as in the support of cardiac function (21). As can be seen in Figure 6, the sympatholytic effect of bucindolol is similar in AA and EA patients, but the clinical impact would be much greater in the former population because of the ~10-fold higher frequency of ADRA2C 322-325Del genotypes. The exaggerated sympatholysis in ADRA2C 322-325Del genotypes in advanced HF patients is associated with a loss in favorable effects of bucindolol on mortality (16) (Figure 6), which is especially pronounced in the presence of ADRB1 389Gly genotypes and is not evident in patients who are ADRB1 Arg389Arg (54). Since AA populations have a much higher frequency of ADRA2C 322-325Del as well as ADRB1 389Gly genotypes, this may unfavorably impact the response of bucindolol, a situation that can be avoided by confining treatment to patients with an ADRB1 Arg389Arg genotype (54). The data presented in Figure 6 and their clinical implications underscore how gene variant differences in allele frequency may impact drug response (2), and how a second gene variant's pharmacologic effects can impact outcomes associated with a functionally important polymorphism (54).
Standard HF β-blockers devoid of sympatholytic activity such as carvedilol or metoprolol do not have potential for adverse interactions with ADRA2C 322-325Del genotypes, which was borne out in a large clinical trial (73).
ACE Del/intron16/Ins, EDN1 Lys198Asn, ECE1 Thr341Ile
A significant effect on β-blocker therapy was shown for the ACE Del/intron16/Ins polymorphism (47), which is discussed in the renin-angiotensin-aldosterone system (RAAS) inhibitor section below. In a 309-patient, nonischemic cardiomyopathy subgroup of the BEST DNA bank population, 9 polymorphisms in 6 genes within the endothelin system were measured and tested against the outcome of ACM and HFH (42). A haplotype including an exon 5, non-synonymous SNP that is correlated with elevated endothelin-1 levels, EDN1 Lys198Asn was highly related to outcome in the bucindolol group, with the minor allele associated with an adverse treatment effect (42). The finding appeared to be independent of any ADRB1 effects, but the data were generated in only 30% of the total number of DNA substudy patients. In addition, since the EDN1 Lys198Asn allele frequencies don't differ by race (Table 1), differential effects in AA vs. EA patients would not be expected. However, another endothelin system polymorphism, ECE1 Thr341Ile does exhibit substantial racial differences, with an approximate 10-fold higher MAF in AA patients (Tables 1 and 2). Both the ACE Del/Ins association with standard β-blocker responses (47) and the EDN1 Lys198Asn or ECE1 Thr341Ile pharmacogenetic interactions with bucindolol are likely examples of indirect (from the primary target[s]) pharmacogenetic interactions, based on inhibitory effects of β-blockers on the renin-angiotensin system and cross-regulation of the endothelin and β-adrenergic systems.
Hydralazine/isosorbide dinitrate and NOS3 Glu298Asp, CYP11B2 T-344C, or GNB3 C825T
In a 354-patient DNA bank substudy of the African-American Heart Failure Trial (A-HeFT, 80), McNamara and associates have published data indicating that polymorphisms in NOS3 (endothelial nitric oxide synthase) (30), CYP11B2 (aldosterone synthase) (32), and GNB3 (guanine nucleotide β3 subunit) (28) influence the response to fixed-dose hydralazine/isosorbide dinitrate (trade name BiDil) in AA patients. The more favorable response genotypes are the major allele homozygotes in NOS3 and CYP11B2 and the minor allele homozygote in GNB3 that is the major allele in AA populations (Tables 1 and 2). In each case, the favorable response genotype is present at a higher frequency in AA populations as compared with EA populations, and there is evidence that BiDil is more effective in AA compared with EA patients (68). Although the A-HeFT DNA/pharmacogenetic substudy was relatively small, these data are highly intriguing and suggest that at least for one type/class of HF therapy, AA patients are at a pharmacogenetic advantage (81). However, because there are no comparable EA data, these findings are classified as possible in Table 2.
NOS3 Glu298Asp
The biologic/pharmacologic plausibility for the pharmacogenetic interactions of BiDil are not as obvious as for β-blockers, as the modulating polymorphisms are more remote from (28,30) or even unrelated to (32) the drug target. However, the exon 7 NOS3 Glu298Asp polymorphism results in a loss of function (82) in an enzyme that may partially mediate (83) or at least modulates NO donor pharmacologic action, and the more favorable effect in the higher-function NOS3 Glu298Glu genotype that is enriched in AAs (Tables 1 and 2) provides biologic plausibility for the pharmacogenetic interaction observed in the A-HeFT DNA substudy (30). The observed pharmacogenetic effect was an improvement in the primary endpoint composite score (mortality, HFH, and change in quality of life at 6 months) by BiDil vs. placebo in patients with a Glu298Glu genotype but not in 298Asp carriers (30).
GNB3 C825T
This GNB3 polymorphism affects a gene product in vascular smooth muscle contraction signal transduction pathways (84). A synonymous C→T transition in exon 10 of GNB3 in some unknown way leads to the formation of a splice variant that has a 41-amino acid deletion, and is a gain-of-function change (84). Thus the GNB3 825T favorable response polymorphism for BiDil is for a gene variant that would lead to greater vasoconstriction, resulting in biologic plausibility for the observed favorable effects (28). In the A-HeFT DNA substudy, BiDil vs. placebo improved the composite score and HF hospitalization-free survival in 825T homozygotes, but not in C825 carriers (28).
CYP11B2 T-344C
Although there is no known mechanism of action for BiDil modulating aldosterone synthase, the −344T homozygous promoter polymorphism in CYP11B2 is related to increased aldosterone urinary excretion (85), and it was associated with deterioration in the composite score in the placebo arm of the A-HeFT DNA substudy (32). This is potentially an example of a pharmacogenetic effect being manifested through effects on increasing event rate in placebo-treated patients, making drug effectiveness easier to detect (3). In the A-HeFT DNA substudy, BiDil vs. placebo improved the composite score in −344T homozygotes but not in −344C carriers (32).
ACEIs, ARBs, or MRAs and polymorphisms in the RAAS
The RAAS is of undeniable importance in the natural history of myocardial remodeling and HF, and inhibitors of angiotensin II formation (ACEIs), angiotensin AT-1 receptor blockers (ARBs), and mineralocorticoid receptor antagonists (MRAs) all produce substantial favorable effects on HF natural history. With the exception of AGT Thr174Met (40,86), all the RAAS polymorphisms listed in Table 1 have been associated with effects on cognate gene expression or hormone levels (36,40,85–89), and AGT Thr174Met has been associated with hypertension (40). The AGTR1 A1166T and the 2 AGT polymorphisms exhibit racial differences in allele frequencies, while ACE Del/intron16/Ins and NR3C2 Ile180Val do not. However, in racially unselected HF populations, relatively few of these gene variants have been shown to be associated with differential responses to RAAS inhibitors.
In 2 medium-sized observational studies conducted at the University of Pittsburgh, McNamara et al reported that either β-blockers (n=328) (47) or higher doses of ACEIs (n=479) (59) were required to reduce an elevated risk of mortality or cardiac transplantation in HF patients who had an ACE Del homozygous genotype. These findings are consistent with the ACE Del allele being associated with higher circulating (36) angiotensin-II levels and higher cardiac ACE gene expression (87), and with β-blockers inhibiting renin release (90). There weren't enough (about 10%) AA subjects in the Pittsburgh studies (47,59) to assess a racial difference in response, and the ACE Del/Ins polymorphism does not exhibit a racial difference in allele frequencies (Table 1). However, a racial difference in ACEI response could result from different allele frequencies in AGT Met235Thr, AGT Thr174Met, AGTR1 A1166C, or CYP11B2 T-344C polymorphisms (Tables 1 and 2).
The Veterans Affairs Vasodilator-Heart Failure (V-HeFT) II study provided evidence of a lesser response to enalapril in AA vs. EA HF patients (68), but the larger Valsartan Heart Failure Trial (Val-HeFT) trial did not observe differences between captopril responses in AA vs. EA patients (71). The definitive information on race and ACEI response in HF comes from a 1196 patient matched-cohort design report from the Studies of Left Ventricular Dysfunction (SOLVD) trial, where compared to EA 800 AA patients had a reduction in enalapril efficacy for prevention of HFHs but not mortality (69). RAAS polymorphisms were not measured in any of these multicenter, placebo-controlled HF trials, so a possible relationship between race-associated RAAS genetic variation and blunted response can only be inferred. However, assuming that the effects of ACEIs are attenuated in HF just as they are in blood pressure responses in hypertension (70), the polymorphism that best predicts such an effect is AGTR1 A1166C, whose 70% lower 1166C MAF in AAs would predict lower AGTR1 receptor density (88) and RAAS drug response (60) (listed as possible in Table 2).
Val-HeFT also reported no effect of race on the response to the ARB valsartan (71), but support for a possible pharmacogenetic interaction between ARBs and the AGTR1 A1166C polymorphism does come from a small Canadian study in EA HF patients (60). This study demonstrated that 1166C genotypes had a more favorable N-terminal pro-brain natriuretic peptide response to candesartan compared with A1166 homozygotes (Table 4) (60). Therefore, Table 2 lists a racial interaction of ARBs via AGTR1 A1166C as possible. Surprisingly, there are no reports of racial effects of MRAs in HF, but the potential for such heterogeneity exists via the approximate 2-fold difference in MAF of the promoter region CYP11B2 T-344C polymorphism (Table 1).
Device therapies and signaling polymorphisms
Improvement in left ventricular chamber function by synchronizing contraction with biventricular pacing is expectedly accompanied by decreases in neurohormonal biomarkers (91,92), and so polymorphic variation in signaling mechanisms could affect cardiac resynchronization therapy (CRT) responses. There are 2 polymorphisms, ADRB2 Gln27Glu (61) and NR3C2 Ile180Val (62), whose minor allele genotypes have been reported to be associated with a CRT reverse remodeling response (Table 4). As seen in Table 1, there is no obvious racial difference in MAF for NR3C2 Ile180Val, but there is a modest ≥ 2-fold lower frequency of the higher response ADRB2 27Glu allele in AA patients that could affect CRT response (Tables 1 and 2). However, thus far only CRT-treated EA patients have been genetically investigated for either polymorphism (61,62). CRT responses by racial subgroup have been reported from the Multicenter Automatic Defibrillator Implantation Trial–Cardiac Resynchronization Therapy (MADIT-CRT) (93) study in the absence of any genetic information, where no differences in effectiveness were observed.
The other device that has proved to be invaluable in HF therapy is the ICD. AA compared with EA HF patients may have a disproportionately higher rate of ventricular arrhythmias and sudden cardiac death (94), which is likely due in part to genetic factors. The nearly exclusive presence in AAs of the Ser1103Tyr variant of SCN5A (Tables 1 and 2)—the gene that encodes the α subunit of the human Nav1.5 sodium channel and plays a central role in cardiac conduction and repolarization as well as in regulation of [Ca2+]i through downstream effects on Na+-Ca2+ exchange (which is also under β-adrenergic regulation)—is especially intriguing. It has been proposed that the metabolic abnormalities and cellular derangements accompanying HF may serve as proarrhythmic substrates for 1103Tyr minor allele carriers, and the variant has been shown to increase the “late” or sustained Na+ current under certain conditions (95). The SCN5A 1103Tyr variant is associated with a 4-fold increase in sustained ventricular arrhythmias in HF patients with ICDs (43), essentially making ICDs more clinically effective (more lives saved from sudden cardiac death) in AA patients with an SCN5A 1103Tyr genotype (43). Moreover, these observations suggest that a pharmacogenetic approach to sudden cardiac death prevention in HF may be indicated, with agents such as ranolazine (96) that diminish the sustained Na+ current potentially having a role. In HF there is precedent for major antiarrhythmic pharmacogenetic effects via the ADRB1 Arg389Gly polymorphism, for prevention of ventricular tachycardia/fibrillation (97) and atrial fibrillation (98).
Non-Cardiac disease common genetic variation
This review does not address common genetic variation important in non-cardiac pathophysiologic settings, but based on population genetics it is likely that at least some of the polymorphism differences between AA and EA extend to affecting non-cardiac disease mechanisms or their treatment. It would be expected that some signaling systems that were shown to have AA vs. EA differences in nonfailing/presumably healthy populations, such as those involving ADRB1 and ADRB2, would exhibit similar genetic variation in non-cardiac diseases in which they may be important, for example in CNS disorders.
Conclusions
Due to population genetics, AA vs. EA HF patients exhibit substantial differences in frequencies of functionally important alleles that code for important constituents of neurohormonal cell-signaling pathways. For at least two polymorphisms this may differentially affect the progression of HF in AA vs. EA patients. Importantly, the effectiveness or safety of several drugs and a device may be influenced by AA vs. EA self-identified race, and it is likely that genetic variation in signaling pathways that are important in HF contribute to this therapeutic heterogeneity. Compared to EA, AA race can be associated with allele frequency-based genetic profiles that 1) can lead to therapeutic response attenuation (standard β-blockers and the experimental β-blocker/sympatholytic bucindolol); 2) possibly decrease therapeutic response (ACEIs and ARBs); 3) enhance effectiveness (fixed-dose hydralazine/isosorbide dinitrate); or 4) increase event rate, making the therapy more clinically effective (ICDs). In order to better understand the basis for heterogeneous therapeutic responses, race, pharmacogenetics and their interaction need to be considered in the development of drugs and devices for the treatment of HF. As demonstrated, identifying genetic subgroups with an enhanced response is a key to delivering optimal therapies for racial cohorts, regardless of the net effect of the therapeutic agent in the parent population and frequency of the preferred genotype.
Supplementary Material
Acknowledgments
The authors thank Laura Hofstatter, Rachel Rosenberg and Morgan Deblecourt for their assistance in manuscript preparation.
Funding sources: NHLBI, VA Cooperative Studies Program (BEST trial substudy "Pharmacogenomics of Beta-adrenergic Receptor Polymorphisms and Response to Beta-Blockers in Heart Failure," S. Liggett, PI; M. Bristow, Substudies Committee Chairman); R01 HL7701 (S. Liggett, PI); 2R01 HL48013 (M. Bristow, PI).
Abbreviations and Acronyms
- AA
African ancestry
- ACEI
angiotensin-converting enzyme inhibitor
- ACM
all-cause mortality
- A-HeFT
African-American Heart Failure Trial
- ARB
angiotensin-receptor blocker
- BEST
β-Blocker Evaluation of Survival Trial
- CCDS
Consensus coding sequence
- CRT
cardiac resynchronization therapy
- Del
deletion
- EA
European ancestry
- ENCODE
Encyclopedia of DNA Elements (Project)
- HF
heart failure
- HFH
heart failure hospitalization
- HFrEF
heart failure with reduced left ventricular ejection fraction
- ICD
implantable cardioverter defibrillator
- InDel
insertion-deletion
- Ins
insertion
- MADIT-CRT
Multicenter Automatic Defibrillator Implantation Trial–Cardiac Resynchronization Therapy
- MAF
minor allele frequency
- MERIT-HF
Metoprolol CR/XL Randomized Intervention Trial in Congestive Heart Failure
- MRA
mineralocorticoid receptor antagonists
- NF
non-heart failure
- RAAS
renin-angiotensin-aldosterone system
- RES
relative effect size
- SNP
single-nucleotide polymorphism
- SOLVD
Studies of Left Ventricular Dysfunction
- Val-HeFT
Valsartan Heart Failure Trial
- V-HeFT
Veterans Affairs Vasodilator-Heart Failure
Footnotes
Disclosures: GD and MRB are employees of and have equity in ARCA biopharma, which is developing bucindolol for prevention of atrial fibrillation in heart failure patients. SBL has equity in ARCA biopharma and has served as a consultant.
References
- 1.Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med. 2001;7:201–204. doi: 10.1016/s1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- 2.Johnson JA. Ethnic differences in cardiovascular drug response: potential contribution of pharmacogenetics. Circulation. 2008;118:1383–1393. doi: 10.1161/CIRCULATIONAHA.107.704023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bristow MR. Pharmacogenetic targeting of drugs for heart failure. Pharmacol Ther. 2012;134:107–115. doi: 10.1016/j.pharmthera.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Twyman R. [Accessed April 18, 2014];Mutation or polymorphism? Wellcome Trust, The Human Genome website. Available at: http://genome.wellcome.ac.uk/doc_WTD020780.html.
- 5.Smedley BD, Stith AY, Nelson AR, editors. Unequal Treatment: Confronting Racial and Ethnic Disparities in Health Care. Washington, DC: National Academies Press; 2003. [PubMed] [Google Scholar]
- 6.Tishkoff SA, Kidd KK. Implications of biogeography of human populations for ‘race’ and medicine. Nat Genetics. 2004;36:S21–S27. doi: 10.1038/ng1438. [DOI] [PubMed] [Google Scholar]
- 7.Li WH, Sadler Low nucleotide diversity in man. Genetics. 1991;129:513–523. doi: 10.1093/genetics/129.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachidanandam R, Weissman D, Schmidt SC, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 9.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Ann Rev Genomics Hum Genet. 2003;4:293–340. doi: 10.1146/annurev.genom.4.070802.110226. [DOI] [PubMed] [Google Scholar]
- 11.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 12.Jorde LB, Wooding SP. Genetic variation, classification and ‘race’. Nat Gen. 2004;36:S28–S33. doi: 10.1038/ng1435. [DOI] [PubMed] [Google Scholar]
- 13.Rosenberg NA, Pritchard JK, Weber JL, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 14.Kurnik D, Li C, Sofowora GG, et al. Beta-1-adrenoceptor genetic variants and ethnicity independently affect response to beta-blockade. Pharmacogenet Genomics. 2008;18:895–902. doi: 10.1097/FPC.0b013e328309733f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small KM, Forbes SL, Rahman FF, Bridges KM, Liggett SB. A four amino acid deletion polymorphism in the third intracellular loop of the human alpha 2C–adrenergic receptor confers impaired coupling to multiple effectors. J Biol Chem. 2000;275:23059–23064. doi: 10.1074/jbc.M000796200. [DOI] [PubMed] [Google Scholar]
- 16.Bristow MR, Murphy GA, Krause-Steinrauf H, et al. An alpha2C–adrenergic receptor polymorphism alters the norepinephrine-lowering effects and therapeutic response of the beta-blocker bucindolol in chronic heart failure. Circ Heart Fail. 2010;3:21–28. doi: 10.1161/CIRCHEARTFAILURE.109.885962. [DOI] [PubMed] [Google Scholar]
- 17.Du Preez J, Matolweni LO, Greenberg J, Mntla P, Adeyemo AA, Mayosi BM. The alpha 2C Del322-325 adrenergic receptor polymorphism is not associated with heart failure due to idiopathic dilated cardiomyopathy in black Africans. Cardiovasc J Afr. 2008;19:15–16. [PMC free article] [PubMed] [Google Scholar]
- 18.Cresci S, Kelly RJ, Cappola TP, et al. Clinical and genetic modifiers of long-term survival in heart failure. J Am Coll Cardiol. 2009;54:432–444. doi: 10.1016/j.jacc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackerman MJ, Splawski I, Makielski JC, et al. Spectrum and prevalence of cardiac sodium channel variants among black, white, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/long QT syndrome genetic testing. Heart Rhythm. 2004;1:600–607. doi: 10.1016/j.hrthm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 20.Ioannidis JP, Ntzani EE, Trikalinos TA. ‘Racial’ differences in genetic effects for complex diseases. Nat Genet. 2004;36:1312–1318. doi: 10.1038/ng1474. [DOI] [PubMed] [Google Scholar]
- 21.Bristow MR. Treatment of chronic heart failure with β-adrenergic receptor antagonists: a convergence of receptor pharmacology and clinical cardiology. Circ Res. 2011;109:1176–1194. doi: 10.1161/CIRCRESAHA.111.245092. [DOI] [PubMed] [Google Scholar]
- 22.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor MR, Bristow MR. The emerging pharmacogenomics of the beta-adrenergic receptors. Congest Heart Fail. 2004;10:281–288. doi: 10.1111/j.1527-5299.2004.02019.x. [DOI] [PubMed] [Google Scholar]
- 24.National Center for Biotechnology Information (NCBI) Database of Single Nucleotide Polymorphisms (dbSNP) website. [Accessed April 18, 2014]; Available at: http://www.ncbi.nlm.nih.gov/snp/.
- 25.Liggett SB, Mialet-Perez J, Thaneemit-Chen S, et al. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci U S A. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie HG, Dishy V, Sofowora G, et al. Arg389Gly beta 1-adrenoceptor polymorphism varies in frequency among different ethnic groups but does not alter response in vivo. Pharmacogenetics. 2001;11:191–197. doi: 10.1097/00008571-200104000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Rosskopf D, Manthey I, Siffert W. Identification and ethnic distribution of major haplotypes in the gene GNB3 encoding the G-protein beta3 subunit. Pharmacogenetics. 2002;12:209–220. doi: 10.1097/00008571-200204000-00005. [DOI] [PubMed] [Google Scholar]
- 28.McNamara DM, Taylor AL, Tam SW, et al. G protein beta 3 subunit (GNB3) genotype predicts enhanced benefit of fixed-dose isosorbide dinitrate and hydralazine: results of the A-HeFT trial. JACC: Heart Fail; in press. [DOI] [PubMed] [Google Scholar]
- 29.Marroni AS, Metzger IF, Souza-Costa DC, et al. Consistent interethnic differences in the distribution of clinically relevant endothelial nitric oxide synthase genetic polymorphisms. Nitric Oxide. 2005;12:177–182. doi: 10.1016/j.niox.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 30.McNamara DM, Tam SW, Sabolinksi ML, et al. Endothelial nitric oxide synthase (NOS3) polymorphisms in African Americans with heart failure: results from the A-HeFT trial. J Card Fail. 2009;15:191–198. doi: 10.1016/j.cardfail.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 31.Barbato A, Russo P, Siani A, et al. Aldosterone synthase gene (CYP11B2) C-344T polymorphism, plasma aldosterone, renin activity and blood pressure in a multi-ethnic population. J Hypertens. 2004;22:1895–1901. doi: 10.1097/00004872-200410000-00011. [DOI] [PubMed] [Google Scholar]
- 32.McNamara DM, Tam SW, Sabolinski ML, et al. Aldosterone synthase promoter polymorphism predicts outcome in African Americans with heart failure: results from the A-HeFT Trial. J Am Coll Cardiol. 2006;48:1277–1282. doi: 10.1016/j.jacc.2006.07.030. [DOI] [PubMed] [Google Scholar]
- 33.De Maria R, Landolina M, Gasparini M, et al. Genetic variants of the renin-angiotensin-aldosterone system and reverse remodeling after cardiac resynchronization therapy. J Card Fail. 2012;18:762–768. doi: 10.1016/j.cardfail.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 34.Rutledge DR, Browe CS, Ross EA. Frequencies of the angiotensinogen gene and angiotensin I converting enzyme (ACE) gene polymorphisms in African Americans. Biochem Mol Biol Int. 1994;34:1271–1275. [PubMed] [Google Scholar]
- 35.Henderson SO, Haiman CA, Mack W. Multiple polymorphisms in the renin- angiotensin-aldosterone system (ACE, CYP11B2, AGTR1) and their contribution to hypertension in African Americans and Latinos in the multiethnic cohort. Am J Med Sci. 2004;328:266–273. doi: 10.1097/00000441-200411000-00006. [DOI] [PubMed] [Google Scholar]
- 36.Tiret L, Rigat B, Visvikis S, et al. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am J Hum Gen. 1992;51:197–205. [PMC free article] [PubMed] [Google Scholar]
- 37.Blanco RR, Austin H, Vest RN, 3rd, et al. Angiotensin receptor type 1 single nucleotide polymorphism 1166A/C is associated with malignant arrhythmias and altered circulating miR-115 levels in patients with chronic heart failure. J Card Fail. 2012;18:717–723. doi: 10.1016/j.cardfail.2012.06.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zakrzewski-Jakubiak M, de Denus S, Dubé MP, Bélanger F, White M, Turgeon J. Ten renin-angiotensin system-related gene polymorphisms in maximally treated Canadian Caucasian patients with heart failure. Br J Clin Pharmacol. 2008;65:742–751. doi: 10.1111/j.1365-2125.2007.03091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gainer JV, Hunley TE, Kon V, Nadeau JH, Muldowney JA3rd, Brown NJ. Angiotensin II type I receptor polymorphism in African Americans lower frequency of the C1166 variant. Biochem Mol Biol Int. 1997;43:227–231. doi: 10.1080/15216549700204001. [DOI] [PubMed] [Google Scholar]
- 40.Jeunemaitre X, Soubrier F, Kotelevtsev YV, et al. Molecular basis of human hypertension: role of angiotensinogen. Cell. 1992;71:169–180. doi: 10.1016/0092-8674(92)90275-h. [DOI] [PubMed] [Google Scholar]
- 41.Rotimi C, Morrison L, Cooper R, Oyejide C, Effiong E, Ladipo M, Osotemihen B, Ward R. Angiotensinogen gene in human hypertension Lack of an association of the 235T allele among African Americans. Hypertension. 1994;24:591–594. doi: 10.1161/01.hyp.24.5.591. [DOI] [PubMed] [Google Scholar]
- 42.Taylor MR, Slavov D, Humphrey K, et al. Pharmacogenetic effect of an endothelin-1 haplotype on response to bucindolol therapy in chronic heart failure. Pharmacogenet Genomics. 2009;19:35–43. doi: 10.1097/FPC.0b013e328317cc57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun AY, Koontz JI, Shah SH, et al. The S1103Y cardiac sodium channel variant is associated with implantable cardioverter-defibrillator events in blacks with heart failure and reduced ejection fraction. Circ Cardiovasc Genet. 2011;4:163–168. doi: 10.1161/CIRCGENETICS.110.958652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Siu H, Jin L, Xiong M. Manifold learning for human population structure studies. PLoS One. 2012;7:e29901. doi: 10.1371/journal.pone.0029901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hershberger RE, Hedges DJ, Morales A. Dilated cardiomyopathy: the complexity of a diverse genetic architecture. Nat Rev Cardiol. 2013;10:531–547. doi: 10.1038/nrcardio.2013.105. [DOI] [PubMed] [Google Scholar]
- 46.Raynolds MV, Bristow MR, Bush EW, et al. Angiotensin-converting enzyme DD genotype in patients with ischaemic or idiopathic dilated cardiomyopathy. Lancet. 1993;342:1073–1075. doi: 10.1016/0140-6736(93)92061-w. [DOI] [PubMed] [Google Scholar]
- 47.McNamara DM, Holubkov R, Janosko K, et al. Pharmacogenetic interactions between beta-blocker therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. Circulation. 2001;103:1644–1648. doi: 10.1161/01.cir.103.12.1644. [DOI] [PubMed] [Google Scholar]
- 48.Small KM, Wagoner LE, Levin AM, Kardia SL, Liggett SB. Synergistic polymorphisms of beta1- and alpha2C–adrenergic receptors and the risk of congestive heart failure. N Engl J Med. 2002;347:1135–1142. doi: 10.1056/NEJMoa020803. [DOI] [PubMed] [Google Scholar]
- 49.Covolo L, Gelatti U, Metra M, et al. Angiotensin-converting-enzyme gene polymorphism and heart failure: a case-control study. Biomarkers. 2003;8:429–436. doi: 10.1080/13547500310001599052. [DOI] [PubMed] [Google Scholar]
- 50.Metra M, Zani C, Covolo L, et al. Role of beta1- and alpha2C–adrenergic receptor polymorphisms and their combination in heart failure: a case-control study. Eur J Heart Fail. 2006;8:131–135. doi: 10.1016/j.ejheart.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 51.Canham RM, Das SR, Leonard D, et al. Alpha2cDel322-325 and beta1Arg389 adrenergic polymorphisms are not associated with reduced left ventricular ejection fraction or increased left ventricular volume. J Am Coll Cardiol. 2007;49:274–277. doi: 10.1016/j.jacc.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 52.Yang J, Zhao Y, Hao P, et al. Impact of angiotensin I converting enzyme insertion/deletion polymorphisms on dilated cardiomyopathy and hypertrophic cardiomyopathy risk. PLoS One. 2013;8:e63309. doi: 10.1371/journal.pone.0063309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kitsios G, Zintzaras E. Genetic variation associated with ischemic heart failure: a HuGE review and meta-analysis. Am J Epidemiol. 2007;166:619–633. doi: 10.1093/aje/kwm129. [DOI] [PubMed] [Google Scholar]
- 54.O’Connor CM, Fiuzat M, Carson PE, et al. Combinatorial pharmacogenetic interactions of bucindolol and β1, α2C adrenergic receptor polymorphisms. PLoS One. 2012;7:e44324. doi: 10.1371/journal.pone.0044324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White HL, de Boer RA, Maqbool A, et al. An evaluation of the beta-1 adrenergic receptor Arg389Gly polymorphism in individuals with heart failure: a MERIT-HF sub-study. Eur J Heart Fail. 2003;5:463–468. doi: 10.1016/s1388-9842(03)00044-8. [DOI] [PubMed] [Google Scholar]
- 56.Liggett SB, Cresci S, Kelly RJ, et al. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaye DM, Smirk B, Williams C, Jennings G, Esler M, Holst D. Beta-adrenoceptor genotype influences the response to carvedilol in patients with congestive heart failure. Pharmacogenetics. 2003;13:379–382. doi: 10.1097/00008571-200307000-00002. [DOI] [PubMed] [Google Scholar]
- 58.Metra M, Covolo L, Pezzali N, et al. Role of beta-adrenergic receptor gene polymorphisms in the long-term effects of beta-blockade with carvedilol in patients with chronic heart failure. Cardiovasc Drugs Ther. 2010;24:49–60. doi: 10.1007/s10557-010-6220-5. [DOI] [PubMed] [Google Scholar]
- 59.McNamara DM, Holubkov R, Postava L, et al. Pharmacogenetic interactions between angiotensin-converting enzyme inhibitor therapy and the angiotensin-converting enzyme deletion polymorphism in patients with congestive heart failure. J Am Coll Cardiol. 2004;44:2019–2026. doi: 10.1016/j.jacc.2004.08.048. [DOI] [PubMed] [Google Scholar]
- 60.de Denus S, Zakrzewski-Jakubiak M, Dubé MP, et al. Effects of AGTR1 A1166C gene polymorphism in patients with heart failure treated with candesartan. Ann Pharmacother. 2008;42:925–932. doi: 10.1345/aph.1K657. [DOI] [PubMed] [Google Scholar]
- 61.Pezzali N, Curnis A, Specchia C, et al. Adrenergic receptor gene polymorphism and left ventricular reverse remodelling after cardiac resynchronization therapy: preliminary results. Europace. 2013;15:1475–1481. doi: 10.1093/europace/eut136. [DOI] [PubMed] [Google Scholar]
- 62.De Maria R, Landolina M, Gasparini M, et al. Genetic variants of the renin-angiotensin-aldosterone system and reverse remodeling after cardiac resynchronization therapy. J Card Fail. 2012;18:762–768. doi: 10.1016/j.cardfail.2012.07.008. [DOI] [PubMed] [Google Scholar]
- 63.Yancy CW, Fowler MB, Colucci WS, et al. Race and the response to adrenergic blockade with carvedilol in patients with chronic heart failure. N Engl J Med. 2001;344:1358–1365. doi: 10.1056/NEJM200105033441803. [DOI] [PubMed] [Google Scholar]
- 64.Goldstein S, Deedwania P, Gottlieb S, Wikstrand J MERIT-HF Study Group. Metoprolol CR/XL in black patients with heart failure (from the Metoprolol CR/XL randomized intervention trial in chronic heart failure) Am J Cardiol. 2003;92:478–480. doi: 10.1016/s0002-9149(03)00674-x. [DOI] [PubMed] [Google Scholar]
- 65.Lanfear DE, Hrobowski TN, Peterson EL, et al. Association of β-blocker exposure with outcomes in heart failure differs between African American and white patients. Circ Heart Fail. 2012;5:202–208. doi: 10.1161/CIRCHEARTFAILURE.111.965780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cubeddu LX, Aranda J, Singh B, et al. A comparison of verapamil and propranolol for the initial treatment of hypertension Racial differences in response. JAMA. 1986;256:2214–2221. [PubMed] [Google Scholar]
- 67.Beta-Blocker Evaluation of Survival Trial Investigators. A trial of the beta-blocker bucindolol in patients with advanced chronic heart failure. N Engl J Med. 2001;344:1659–1667. doi: 10.1056/NEJM200105313442202. [DOI] [PubMed] [Google Scholar]
- 68.Carson P, Ziesche S, Johnson G, Cohn JN. Racial differences in response to therapy for heart failure: analysis of the vasodilator-heart failure trials Vasodilator-Heart Failure Trial Study Group. J Card Fail. 1999;5:178–187. doi: 10.1016/s1071-9164(99)90001-5. [DOI] [PubMed] [Google Scholar]
- 69.Exner DV, Dries DL, Domanski MJ, Cohn JN. Lesser response to angiotensin-converting-enzyme inhibitor therapy in black as compared with white patients with left ventricular dysfunction. N Engl J Med. 2001;344:1351–1357. doi: 10.1056/NEJM200105033441802. [DOI] [PubMed] [Google Scholar]
- 70.Weir MR, Gray JM, Paster R, Saunders E. Differing mechanisms of action of angiotensin-converting enzyme inhibition in black and white hypertensive patients The Trandolapril Multicenter Study Group. Hypertension. 1995;26:124–130. doi: 10.1161/01.hyp.26.1.124. [DOI] [PubMed] [Google Scholar]
- 71.Prisant LM, Thomas KL, Lewis EF, et al. Racial analysis of patients with myocardial infarction complicated by heart failure and/or left ventricular dysfunction treated with valsartan, captopril, or both. J Am Coll Cardiol. 2008;51:1865–1871. doi: 10.1016/j.jacc.2007.12.050. [DOI] [PubMed] [Google Scholar]
- 72.Ofili EO, Ferdinand KC, Saunders E, et al. Irbesartan/HCTZ fixed combinations in patients of different racial/ethnic groups with uncontrolled systolic blood pressure on monotherapy. J Natl Med Assoc. 2006;98:618–626. [PMC free article] [PubMed] [Google Scholar]
- 73.Sehnert AJ, Daniels SE, Elashoff M, et al. Lack of association between adrenergic receptor genotypes and survival in heart failure patients treated with carvedilol or metoprolol. J Am Coll Cardiol. 2008;52:644–651. doi: 10.1016/j.jacc.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 74.Anderson JL, Krause-Steinrauf H, Goldman S, et al. Failure of benefit and early hazard of bucindolol for Class IV heart failure. J Card Fail. 2003;9:266–277. doi: 10.1054/jcaf.2003.42. [DOI] [PubMed] [Google Scholar]
- 75.Pereira SB, Velloso MW, Chermont S, et al. β-adrenergic receptor polymorphisms in susceptibility, response to treatment and prognosis in heart failure: implication of ethnicity. Mol Med Rep. 2013;7:259–265. doi: 10.3892/mmr.2012.1120. [DOI] [PubMed] [Google Scholar]
- 76.Magnusson Y, Levin MC, Eggertsen R, et al. Ser49Gly of beta1-adrenergic receptor is associated with effective beta-blocker dose in dilated cardiomyopathy. Clin Pharmacol Ther. 2005;78:221–231. doi: 10.1016/j.clpt.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 77.Green SA, Turki J, Innis M, Liggett SB. Amino-terminal polymorphisms of the human beta 2-adrenergic receptor impart distinct agonist-promoted regulatory properties. Biochemistry. 1994;33:9414–9419. doi: 10.1021/bi00198a006. [DOI] [PubMed] [Google Scholar]
- 78.Bristow MR, Krause-Steinrauf H, Nuzzo R, et al. Effect of baseline or changes in adrenergic activity on clinical outcomes in the beta-blocker evaluation of survival trial. Circulation. 2004;110:1437–1442. doi: 10.1161/01.CIR.0000141297.50027.A4. [DOI] [PubMed] [Google Scholar]
- 79.Hein L, Altman JD, Kobilka BK. Two functionally distinct alpha2-adrenergic receptors regulate sympathetic neurotransmission. Nature. 1999;402:181–184. doi: 10.1038/46040. [DOI] [PubMed] [Google Scholar]
- 80.Taylor AL, Ziesche S, Yancy C, et al. Combination of isosorbide dinitrate and hydralazine in blacks with heart failure. N Engl J Med. 2004;351:2049–2057. doi: 10.1056/NEJMoa042934. [DOI] [PubMed] [Google Scholar]
- 81.Bristow MR. Polymorphic Variation in the G Protein Beta3 Subunit (GNB3) Gene and Response to BiDil in A-HeFT: Basis for an African-American Pharmacogenetic Advantage to NO Donor Therapy? JACC: Heart Fail; in press. [DOI] [PubMed] [Google Scholar]
- 82.Leeson CP, Hingorani AD, Mullen MJ, et al. Glu298Asp endothelial nitric oxide synthase gene polymorphism interacts with environmental and dietary factors to influence endothelial function. Circ Res. 2002;90:1153–1158. doi: 10.1161/01.res.0000020562.07492.d4. [DOI] [PubMed] [Google Scholar]
- 83.Bonini MG, Stadler K, Silva SO, et al. Constitutive nitric oxide synthase activation is a significant route for nitroglycerin-mediated vasodilation. Proc Natl Acad Sci U S A. 2008;105:8569–8574. doi: 10.1073/pnas.0708615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Siffert W, Rosskopf D, Siffert G, et al. Association of a human G-protein beta3 subunit variant with hypertension. Nat Genet. 1998;18:45–48. doi: 10.1038/ng0198-45. [DOI] [PubMed] [Google Scholar]
- 85.Davies E, Holloway CD, Ingram MC, et al. Aldosterone excretion rate and blood pressure in essential hypertension are related to polymorphic differences in the aldosterone synthase gene CYP11B2. Hypertension. 1999;33:703–707. doi: 10.1161/01.hyp.33.2.703. [DOI] [PubMed] [Google Scholar]
- 86.Brand E, Chatelain N, Paillard F, et al. Detection of putative functional angiotensinogen (AGT) gene variants controlling plasma AGT levels by combined segregation-linkage analysis. Eur J Hum Genet. 2002;10:715–723. doi: 10.1038/sj.ejhg.5200874. [DOI] [PubMed] [Google Scholar]
- 87.Danser AH, Schalekamp MA, Bax WA, et al. Angiotensin-converting enzyme in the human heart Effect of the deletion/insertion polymorphism. Circulation. 1995;92:1387–1388. doi: 10.1161/01.cir.92.6.1387. [DOI] [PubMed] [Google Scholar]
- 88.Martin MM, Buckenberger JA, Jiang J, et al. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microRNA-155 binding. J Biol Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.van Leeuwen N, Bellingrath S, de Kloet ER, et al. Human mineralocorticoid receptor (MR) gene haplotypes modulate MR expression and transactivation: implication for the stress response. Psychoneuroendocrinology. 2011;36:699–709. doi: 10.1016/j.psyneuen.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 90.Blumenfeld JD, Sealey JE, Mann SJ, et al. Beta-adrenergic receptor blockade as a therapeutic approach for suppressing the renin-angiotensin-aldosterone system in normotensive and hypertensive subjects. Am J Hypertens. 1999;12:451–459. doi: 10.1016/s0895-7061(99)00005-9. [DOI] [PubMed] [Google Scholar]
- 91.Erol-Yilmaz A, Verberne HJ, Schrama TA, et al. Cardiac resynchronization induces favorable neurohumoral changes. Pacing Clin Electrophysiol. 2005;28:304–310. doi: 10.1111/j.1540-8159.2005.09508.x. [DOI] [PubMed] [Google Scholar]
- 92.Tarquini R, Guerra CT, Porciani MC, et al. Effects of cardiac resynchronization therapy on systemic inflammation and neurohormonal pathways in heart failure. Cardiol J. 2009;16:545–552. [PubMed] [Google Scholar]
- 93.Elanchenny M, Moss AJ, McNitt S, et al. Effectiveness of cardiac resynchronization therapy with defibrillator in at-risk black and white cardiac patients. Ann Noninvasive Electrocardiol. 2013;18:140–148. doi: 10.1111/anec.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Armstrong D, Wing S, Tyroler HA. Race differences in estimates of sudden coronary heart disease mortality, 1980–1988: the impact of ill-defined death. J Clin Epidemiol. 1996;49:1247–1251. doi: 10.1016/s0895-4356(96)00217-x. [DOI] [PubMed] [Google Scholar]
- 95.Splawski I, Timothy KW, Tateyama M, et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297:1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- 96.Undrovinas AI, Belardinelli L, Undrovinas NA, Sabbah HN. Ranolazine improves abnormal repolarization and contraction in left ventricular myocytes of dogs with heart failure by inhibiting late sodium current. J Cardiovasc Electrophysiol. 2006;(17 Suppl 1):S169–S177. doi: 10.1111/j.1540-8167.2006.00401.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aleong RG, Sauer WH, Robertson AD, Liggett SB, Bristow MR. Adrenergic receptor polymorphisms and prevention of ventricular arrhythmias with bucindolol in patients with chronic heart failure. Circ Arrhythm Electrophysiol. 2013;6:137–143. doi: 10.1161/CIRCEP.111.969618. [DOI] [PubMed] [Google Scholar]
- 98.Aleong RG, Sauer WH, Davis G, et al. Prevention of atrial fibrillation by bucindolol is dependent on the beta-1389 Arg/Gly adrenergic receptor polymorphism. Vol. 1. JACC: Heart Fail; 2013. pp. 338–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.