Abstract
A recent discovery in membrane research is the ability of styrene-maleic acid (SMA) copolymers to solubilize membranes in the form of nanodisks allowing extraction and purification of membrane proteins from their native environment in a single detergent-free step. This has important implications for membrane research because it allows isolation as well as characterization of proteins and lipids in a near-native environment. Here, we aimed to unravel the molecular mode of action of SMA copolymers by performing systematic studies using model membranes of varying compositions and employing complementary biophysical approaches. We found that the SMA copolymer is a highly efficient membrane-solubilizing agent and that lipid bilayer properties such as fluidity, thickness, lateral pressure profile, and charge density all play distinct roles in the kinetics of solubilization. More specifically, relatively thin membranes, decreased lateral chain pressure, low charge density at the membrane surface, and increased salt concentration promote the speed and yield of vesicle solubilization. Experiments using a native membrane lipid extract showed that the SMA copolymer does not discriminate between different lipids and thus retains the native lipid composition in the solubilized particles. A model is proposed for the mode of action of SMA copolymers in which membrane solubilization is mainly driven by the hydrophobic effect and is further favored by physical properties of the polymer such as its relatively small cross-sectional area and rigid pendant groups. These results may be helpful for development of novel applications for this new type of solubilizing agent, and for optimization of the SMA technology for solubilization of the wide variety of cell membranes found in nature.
Introduction
A recent finding in membrane research is the ability of styrene maleic acid (SMA) copolymers to solubilize membranes into lipid nanodisks without the assistance of detergents (1–6). This has two implications: 1) it allows direct solubilization and purification of membrane proteins while maintaining their lipid environment (5,6), thereby avoiding cumbersome, detergent-based procedures (7) with concomitant risk of protein aggregation and/or denaturation; and 2) the embedding into nanodisks ensures a stable lipid environment for membrane proteins, yet with a small particle size that allows characterization by a wide range of biophysical approaches (1–6).
Other advantages of the use of nanodisks are that the embedded proteins are accessible to both sides of the membrane, facilitating functional studies (8), and that a defined oligomerization state of the proteins is maintained, which may be important for understanding mechanistic processes (9). A disadvantage of nanodisks, however, is that they do not allow the study of vectorial function such as solute transport or signal transduction, which requires a membrane that separates two distinct aqueous compartments.
Most of the studies on nanodisks described in literature so far involve nanodisks that are stabilized by amphipathic membrane scaffolding proteins (MSPs), as developed by Bayburt and Sligar (10) and Denisov et al. (11). In these systems, many new applications of nanodisks for studies of membrane proteins have been developed (9,12–14). However, reconstitution of membrane proteins in MSP-bounded nanodisks typically requires several steps, including prior purification of the protein in detergent (10,11). The SMA technology thus combines the advantages of a straightforward protocol for detergent-free purification of membrane proteins with embedding of the proteins into nanodisks as a stable and native host in which they maintain their activity.
Despite this huge potential of SMA for membrane research, very little is known about the molecular mechanism of SMA-induced nanodisk formation. In this study, we investigate the physico-chemical properties of membranes that regulate the kinetics and efficiency of nanodisk formation by using model membrane systems. The main advantage of these model systems is that they allow systematic modulation of a wide range of physical and chemical properties of the membrane simply by variation of lipid composition. By using complementary biophysical techniques, we will show that the SMA copolymer is a very efficient membrane-solubilizing agent and that lipid bilayer physico-chemical properties, such as lipid packing and electrostatic interactions, play an important role in determining the efficiency of the solubilization process. We present a model for the mode of action of SMA copolymers and compare this to the action of similar amphipathic polymers as well as MSPs. The fresh insights obtained in this study will be helpful to exploit the full potential of the SMA copolymer for research on membranes and membrane proteins.
Materials and Methods
Materials
Lipids were purchased from Avanti Polar Lipids (Alabaster, AL). The following lipids were used:
di-13:0 PC (1,2-ditridecanoyl-sn-glycero-3-phosphocholine); di-14:0 PC (1,2-dimyristoyl-sn-glycero-3-phosphocholine); di-14:0 PG (1,2-dimyristoyl-sn-glycero-3-phosphoglycerol); di-16:0 PC (1,2-dipalmitoyl-sn-glycero-3-phosphocholine); di-16:0 PG (1,2-dipalmitoyl-sn-glycero-3-phosphoglycerol); di-18:0 PC (1,2-distearoyl-sn-glycero-3-phosphocholine); di-18:0 PG (1,2-distearoyl-sn-glycero-3-phosphoglycerol); 16:0-18:1 PC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine); di-14:1 PC (1,2-dimyristoleoyl-sn-glycero-3-phosphocholine); di-16:1 PC (1,2-dipalmitoleoyl-sn-glycero-3-phosphocholine); di-18:1 PC (1,2-dioleoyl-sn-glycero-3-phosphocholine); di-20:1 PC (1,2-dieicosenoyl-sn-glycero-3-phosphocholine); di-22:1 PC (1,2-dierucoyl-sn-glycero-3-phosphocholine); di-18:2 PC (1,2-dilinoleoyl-sn-glycero-3-phosphocholine); lyso-18:1 PC (1-oleoyl-2-hydroxy-sn-glycero-3-phosphocholine); di-18:1 PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine); di-18:1 Rh-PE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-n-lissamine rhodamine B sulfonyl-Rhodamine PE); and TLE (total polar lipid extract) of Escherichia coli membranes (catalog No. 100600P).
SMAn (Styrene maleic anhydride) copolymer was obtained as a kind gift from Cray Valley (Exton, PA). Membrane scaffold protein MSP1D1 was kindly provided by the NMR Department, Bijvoet Center for Biomolecular Research, Utrecht University, where MSP1D1 was expressed and purified as described by Bayburt and Sligar (15). Amphipol A8-35 was purchased from Affymetrix (Santa Clara, CA), and TRIS (tris-hydroxymethyl-aminomethane), NaCl, chloroform, and methanol were purchased from Sigma Aldrich (St. Louis, MO).
SMA copolymer preparation
The SMA2000 copolymer used throughout this study was prepared in exactly the same way as described in Swainsbury et al. (6). Briefly, a 5% (w/v) SMAn suspension in 1 M KOH was refluxed for at least 4 h after which the SMA was recovered by acid precipitation in 1.1 M HCl. The precipitated SMA was washed at least four times with 10 mM HCl. Finally, the SMA was lyophilized and stored at room temperature until required. 5% (w/v) SMA solutions used for experiments were prepared by dissolving lyophilized SMA in 50 mM Tris-HCl, after which the solution was adjusted to pH 8.0.
Large unilamellar vesicle preparation
Phospholipid stock solutions were prepared by dissolving dry lipid powder in 2:1 (v/v) chloroform/methanol at a concentration of 10 mM. Multilamellar vesicles (MLVs) at a concentration of 10 mM phospholipid were prepared by hydration of vacuum-dried lipid films using 50 mM Tris-HCl pH 8.0 buffer with 150 mM NaCl or without NaCl. After hydration, at least 10 freeze-thaw cycles were performed using liquid nitrogen and a water bath set at a temperature at least 10°C above the gel-to-liquid crystalline phase transition temperature (Tm) of the respective phospholipids. Subsequently, MLV dispersions were extruded through either a 200-nm or 400-nm Whatman polycarbonate filter using the Avanti mini-extruder (Sigma Aldrich) at least 15 times >Tm to prepare large unilamellar vesicles (LUVs) of 200-nm or 400-nm diameter. Phospholipid concentrations were determined by the phosphate assay according to Rouser et al. (16).
Nanodisk preparation for transmission electron microscopy and dynamic light-scattering experiments
Nanodisks of saturated and unsaturated phospholipids used for transmission electron microscopy (TEM) and dynamic light-scattering (DLS) experiments were prepared by the addition of SMA copolymer (5% w/v) in a 3:1 (w/w) SMA/phospholipid (∼1:3.7 molar ratio) to lipid vesicles of either 200 nm or 400 nm in diameter at a phospholipid concentration of 10 mM in 50 mM Tris pH 8.0 and 150 mM NaCl at Tm for saturated PC lipids and at 25°C for unsaturated lipids. The only exception was di-22:1 PC, which required a 9:1 (w/w) SMA/phospholipid and a temperature of 60°C for complete solubilization. All nanodisk samples were equilibrated overnight.
Turbidity experiments
The kinetics of solubilization of vesicles (400-nm diameter) by SMA copolymers was monitored by turbidity measurements at 350 nm using a Lambda 18 spectrophotometer (PerkinElmer, Waltham, MA) equipped with a Peltier element. In all experiments a quartz cuvette was used holding a total volume of 700 μL. The phospholipid vesicle dispersions (0.5 mM phospholipid in 50 mM Tris-HCl pH 8.0 with either 150 mM or without NaCl) were temperature-equilibrated for at least 15 min before addition of 15 μL of a 5% (w/v) SMA 2000 solution yielding a final SMA copolymer concentration of 1.05 mg/mL (∼0.1% w/v). This corresponds to a SMA/phospholipid of 3:1 (w/w) (1:3.7 molar ratio), and unless otherwise noted this ratio was used throughout this study. SMA addition was directly followed by quickly mixing the contents of the cuvette using a 200-μL pipette after which the starting point was set to zero (t = 0). Stirring the solutions yielded similar time traces but was avoided due to higher noise levels. Similar conditions were used in experiments with membrane scaffold protein MSP1D1. More specifically, MSP1D1 in 100 mM Tris pH 7.4 and 200 mM NaCl buffer was added to LUV dispersions (0.5 mM phospholipid, total volume of 700 μL) in either a 1:1 (w/w) or 3:1 (w/w) phospholipid/MSP1D1. Time traces of optical absorbance at 350 nm for all experiments were reproducible. The curves displayed are representative, and from a single experiment.
Analysis of lipid composition by thin layer chromatography (TLC) and gas chromatography (GC)
MLVs of Escherichia coli total polar lipid extract (Avanti Polar Lipids) were prepared as described above at a lipid concentration of 0.5 mM in 50 mM Tris-HCl pH 8 and 150 mM NaCl buffer. Vesicles were treated with an amount of SMA that is insufficient for complete solubilization (1.4:1 (w/w) SMA/lipid) for 15 min at 25°C while shaking gently. Nanodisks (supernatant) and unsolubilized lipid material (pellet) were separated by ultracentrifugation at 115,000 g for 1 h at 4°C. Lipids were purified from the supernatant, the pellet, and intact vesicles by the Bligh and Dyer extraction method (17). Purified lipid samples were applied to an HPTLC Silica gel 60 plate (Macherey Nagel, Düren, Germany) using a Linomat 5 sample applicator (CAMAG, Muttenz, Switzerland) that was developed with chloroform/methanol/ammonia/water (68:28:3:1 by volume) in a ADC2 developing chamber (CAMAG). The plate was preincubated in a 1.2% (w/v) boric acid solution in water/ethanol 1:1 (v/v), which was dried for 2 h at 160°C. Lipids were visualized by dipping the plate into a methanol solution of 10% copper(II) sulfate (w/v) in 8% sulfuric acid (98%), and 8% phosphoric acid (85%) and then drying the plate by heating at 130°C for 15 min. Relative intensities were determined by densitometry using QUANTITY ONE software (BioRad, Hercules, CA). Molar percentages were determined from a concentration calibration of the TLE of E. coli membranes.
The fatty acid composition of the samples was analyzed as follows: After extraction, a part of the lipid material was esterified by methyl esterification of the lipids in 2.5% volume sulfuric acid (98%) for 2 h at 70°C. The reaction was stopped by the extra addition of water/methanol (1:1 v/v) and the methylated fatty acids were captured in the organic phase by the addition of hexane (2:1 v/v aqueous phase to hexane). The hexane phase was washed with water twice after which 0.2 mL of isopropanol was added. Samples were then dried under a nitrogen flow and the purified methylated fatty acids, redissolved in hexane, were analyzed by gas chromatography on a Trace GC Ultra (InterScience, Troy, NY) using a Stabilwax polar column (Thermo Fisher Scientific, Waltham, MA) of 30 min with a 0.31-mm internal diameter and a 0.25-μm film thickness. Fatty acid assignment was facilitated by using two standards: GLC 63b (Nu-Check Prep, Elysian, MN), and BAME (Bacterial Acid Methyl Ester) Mix (Sigma-Aldrich).
Size exclusion chromatography
LUVs of 200 nm in diameter of di-14:0 PC containing 0.5% mol Rh-PE (di-18:1 phosphatidylethanolamine rhodamine sulfonyl, 10 mM phospholipid) in 50 mM Tris-HCl pH 8.0 treated with different concentrations of SMA copolymer were applied after 15 min of incubation with SMA at 30°C to a Sephacryl S300 10/300 column (Pharmacia, Uppsala, Sweden) that was preequilibrated with Tris-HCl 50 mM pH 8.0 buffer at room temperature. Sample loading (200 μL) and elution were controlled manually using a model No. P-6000 pump (Pharmacia). The flow rate was set to 0.5 mL/min and at least 30 fractions of 200 μL were collected. Elution profiles were determined by measuring the Rh-PE absorption at 572 nm for each collected fraction using a Lambda 18 spectrophotometer (PerkinElmer).
TEM
Di-14:0 PC vesicles (untreated and treated with SMA) were visualized using cryo-TEM. A 3-μL aliquot (10 mM phospholipid) was pipetted onto a glow-discharged holey carbon copper grid in the environmental chamber of a Vitrobot (FEI, Hillsboro, OR) at 26°C with a relative humidity kept >98%. The copper grids were blotted once for 2 s and quickly plunged into liquid ethane. A No. 20 Philips Tecnai electron microscope (FEI) was used, operating at an acceleration voltage of 200 kV. Di-14:0 PC intermediate vesicle structures were obtained by the addition of SMA copolymers in the SMA/lipid 1:4 (limiting concentration, i.e., insufficient amount of SMA to induce complete solubilization) to vesicles of di-14:0 PC at 30°C.
Nanodisks of di-14:0 PC and of other lipid composition were visualized with negative-staining TEM. Aliquots (5 μL) of nanodisk dispersions (10 mM phospholipid) were adsorbed to glow-discharged carbon-coated copper grids for 120 s. Subsequently, the grids were washed twice with water for 15 s and negatively stained with 2% (w/v) uranyl acetate for 45 s. Excess solution on the grids was removed with filter paper. The grids were air-dried and examined using a model No. 10 Philips Tecnai electron microscope (FEI) operating at an acceleration voltage of 100 kV.
DLS
DLS experiments were performed on a Zetasizer Nano ZS (Malvern Instruments, Worcestershire, United Kingdom) at 25°C. Nanodisks of different lipid composition (10 mM phospholipid) and vesicle dispersions (5 × 10−4 mM phospholipid) in 50 mM Tris-HCl, 150–250 mM NaCl pH 8.0 buffer were measured for at least 15 times each measurement, being an average of 20 subruns of 15 s. Size-intensity distributions were generated using ZETASIZER softwares Ver. 6.20 and Ver. 7.03 and analyzed using the multiple narrow distribution. Hydrodynamic diameters (DH) were calculated from the intensity distributions with the assumption that nanodisks are spherically shaped.
Lipid monolayers
Surface pressure isotherms versus time were recorded for lipid monolayers of di-14:0 PC, di-14:0 PG, and mixtures of both (4:1 mol PC/PG) while adding 20 μL 5% (w/v) SMA (1 mg/20 mL, which is an excess amount). Lipid monolayers were spread at the air-water interface by spreading a phospholipid solution (0.5 mM phospholipid in 9:1 v/v chloroform/methanol) until the required initial surface pressure was reached. Equilibration of the initial surface pressure and solvent evaporation was allowed for 15 min before each experiment. All measurements were performed in a 6 × 5.5 cm compartment of a homemade Teflon trough filled with 20 mL buffer of 50 mM Tris-HCl pH 8.0 with 150 mM or without NaCl at 25 ± 1°C and constant area. The surface pressure in time was recorded using a commercially available monolayer system (MicroTroughXS; Kibron, Helsinki, Finland). The reproducibility of the surface pressure increase in each experiment was within the maximum error of ±1 mN/m.
Results
The SMA copolymer is a highly efficient membrane-solubilizing agent
LUVs scatter light efficiently, but nanodisks are too small to do so. Therefore the kinetics of vesicle solubilization upon addition of SMA copolymers can be followed simply by monitoring the decrease in optical density.
Initially, the effect was investigated of SMA addition to vesicles of saturated di-14:0 PC lipids, which undergo a gel to liquid-crystalline phase transition at 23°C (17). Fig. 1 A shows that the kinetics are temperature-dependent, with slow solubilization below the phase transition temperature (Tm), slightly faster solubilization when Tm is approached, and very fast solubilization within seconds at and above Tm. In all experiments, the SMA/lipid was kept constant at 3:1 (w/w). This represents an excess of SMA, based on the observation that the kinetics of solubilization for di-14:0 PC vesicles did not change when the ratio was lowered to 1:1 (w/w). At lower SMA/lipid, slower kinetics was observed at all temperatures (data not shown).
Figure 1.
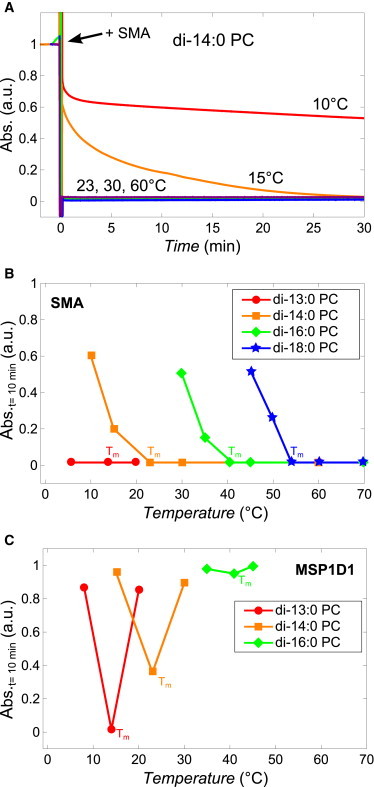
Kinetics of solubilization of lipid vesicles (400 nm) induced by the SMA copolymer and membrane scaffold protein MSP1D1. (A) Normalized time trace of absorbance at 350 nm showing the kinetics of solubilization of di-14:0 PC vesicles induced by the SMA copolymer at different temperatures. (B) Normalized absorbance values at 350 nm of saturated lipid vesicle dispersions after 10 min of incubation with SMA copolymer (3:1 w/w SMA/lipid) at different temperatures (solid lines are added to guide the eye). Tm values of the different lipids are indicated. (C) Normalized absorbance values at 350 nm of saturated lipid vesicle dispersions after 10 min of incubation with the membrane scaffold protein MSP1D1 (1:1 w/w MSP1D1/lipid) at different temperatures (solid lines are added to guide the eye). To see this figure in color, go online.
Similar solubilization trends were observed for vesicles of saturated lipids with different acyl-chain length (see Fig. S1 in the Supporting Material for full traces). Fig. 1 B shows the relative optical density value of the LUV dispersions after 10 min of SMA incubation as function of temperature. For all lipids, rapid solubilization occurs at and above Tm. In case of the shortest lipid, di-13:0 PC (red line), it was found that also at <Tm the LUV dispersion clarifies rapidly, even within seconds. For the longer lipids, at <Tm, the rate of solubilization is relatively slow but becomes faster when the temperature approaches Tm. Upon overnight incubation all lipid dispersions clarified except for those for which the temperature was set to 15°C <Tm or more. In those cases, the rate of solubilization was so slow that no complete solubilization could be observed overnight (data not shown).
The SMA copolymer solubilizes lipid vesicles much more efficiently than the membrane scaffolding protein MSP1D1
For comparison, the ability of the membrane scaffold protein MSP1D1 to solubilize lipid vesicles was also tested. MSP1D1 is a widely used recombinant form of apolipoprotein A-I designed by Bayburt and Sligar (10,15), Denisov et al. (11), and Nath et al. (18) to reconstitute membrane proteins into nanodisks that are bounded by MSP1D1. The relative optical density was recorded as function of incubation time of MSP1D1 (1:1 w/w protein/lipid) in di-13:0 PC, di-14:0 PC, and di-16:0 PC lipid dispersions at different temperatures (see Fig. S2 for full traces). Fig. 1 C shows that after 10 min of incubation, a significant effect occurs only for di-13:0 PC and di-14:0 PC at Tm. In the case of di-16:0 PC even at Tm, no significant solubilization is observed. Similar results were obtained for all lipids when the experiments at Tm were performed at an increased protein/lipid of 3:1 (w/w) (data not shown), demonstrating that the inability of MSP1D1 to solubilize vesicles was most likely not due to an insufficient amount of protein being present. These results are in agreement with results from previous studies on MSPs and model membranes under similar conditions as described, e.g., apoA-1 (19) and apoLp-III (20).
The SMA copolymer induces a complete conversion of vesicles into nanodisks
The solubilization process of lipid vesicles by the SMA copolymer was further characterized using di-14:0 PC as model lipid. Fig. 2 A shows size-exclusion chromatograms of 200-nm lipid vesicles treated with different concentrations of SMA. In the absence of SMA, the vesicles elute at ∼1.5 mL (red line). When the vesicles are mixed with a subsolubilizing amount of SMA, corresponding to a 1:4 (w/w) SMA/lipid (green line), the elution profile shows a decreased signal at 1.5 mL while a broad peak appears at higher retention volumes indicating the elution of smaller particles in addition to intact vesicles. When mixed with an excess amount of SMA, corresponding to a 3:1 (w/w) SMA/lipid (blue line), a single peak at ∼3.0 mL is observed suggesting conversion of vesicles into even smaller particles.
Figure 2.
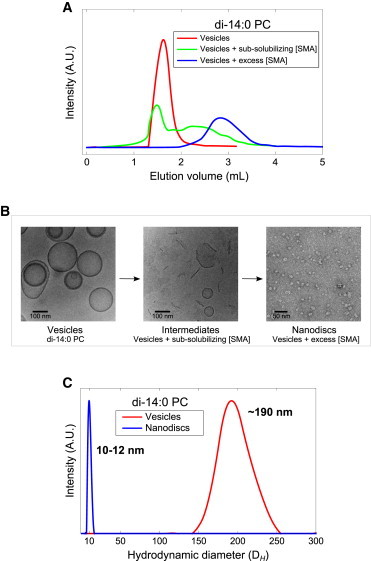
Structural characterization of di-14:0 PC lipid vesicle (200 nm) solubilization by the SMA copolymer. (A) Size-exclusion chromatograms of di-14:0 PC lipid vesicles (red), di-14:0 PC lipid vesicles incubated with a subsolubilizing amount of SMA corresponding to a 1:4 (w/w) SMA/phospholipid (green), and di-14:0 PC lipid vesicles incubated with excess SMA, which corresponds to a 3:1 (w/w) SMA/phospholipid (blue). (B) Cryo- (left, middle) and negative staining (right) transmission electron microscopy images of particles obtained from the fractions corresponding to the three chromatograms from Fig. 2A showing vesicles (left), intermediate vesicular structures and open bilayer fragments (middle), and nanodisks (right). (C) Size distributions from DLS experiments on vesicles (red) and nanodisks obtained by incubating vesicles with excess SMA (blue). To see this figure in color, go online.
The eluted samples were characterized by electron microscopy. As illustrated in Fig. 2 B, left, intact vesicles were found to have an average diameter of 175 nm. Treatment of vesicles with a subsolubilizing amount of SMA resulted in the appearance of open vesicular intermediates and membrane fragments (Fig. 2 B, middle). Vesicles treated with an excess amount of SMA reveal disk-like particles of ∼10–12 nm in diameter (Fig. 2 B, right), consistent with previous observations by Jamshad et al. (21), who also used the SMA 2000 copolymer, and Orwick et al. (3), who used the more hydrophobic SMA 3000 copolymer.
DLS experiments confirmed the transition of lipid vesicles into nanodisks (Fig. 2 C). Lipid vesicles were found to have a hydrodynamic diameter of ∼190 nm and nanodisks were found to have a fairly homogeneous apparent diameter of ∼10–12 nm, both in good agreement with the electron microscopy experiments. Characterization of the intermediate structures by DLS led to irreproducible size-intensity distributions due to the formation of differently sized and shaped intermediate vesicular particles, and are, for that reason, omitted.
The size of nanodisks does not significantly change upon variation of acyl-chain length
We next investigated whether fatty acid chain length of the lipids affects the size of the nanodisks. Vesicles of saturated PC lipids with varying length of acyl chains were treated with an excess amount of SMA at Tm in order to form nanodisks (see Materials and Methods). TEM and DLS analysis showed that the particle shapes as well as the average diameter of the nanodisks (12 ± 2 nm) are very similar, regardless of the acyl-chain length. Similar particle shapes and nanodisk diameters were also observed for vesicles of unsaturated lipids with different chain lengths (see Fig. S3). However, the presence of unsaturated chains in the lipids resulted in very different kinetics of vesicle solubilization, as will be discussed next.
Lipid packing strongly influences the efficiency of solubilization
Based on the fast solubilization of saturated lipids in the liquid-crystalline phase, one would also expect that addition of SMA to unsaturated lipids would lead to fast solubilization, because these lipids are in the fluid phase around room temperature. Surprisingly, this was not the case. Fig. 3 A shows the changes in relative optical density in time for di-18:1 PC vesicles. At 10°C and 20°C, solubilization is relatively slow and the curves display more complex kinetics than saturated PC lipids, possibly due to differences in the kinetics of formation and size of intermediate vesicular structures. At increased temperatures, solubilization becomes faster, with complete solubilization occurring within 10 min at 30°C and above. A similar trend was observed for unsaturated lipids with other acyl-chain lengths (see Fig. S4 for full traces). Fig. 3 B shows the relative optical density after 10 min of incubation with SMA. At 30°C and above, complete solubilization is obtained for all unsaturated lipids, except for the longest lipid di-22:1 PC. From this figure, two trends are clearly visible: 1) the rate of solubilization decreases with increasing acyl-chain length and 2) solubilization becomes faster at higher temperature.
Figure 3.
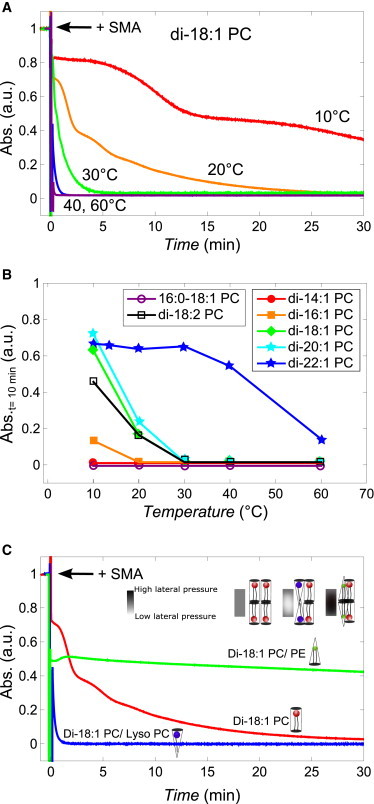
(A) Kinetics of solubilization of unsaturated lipid vesicles (400 nm) induced by the SMA copolymer and the effect of lateral chain pressure. In all cases, a 3:1 (w/w) SMA/lipid solubilization kinetics of di-18:1 PC vesicles is induced by the SMA copolymer at different temperatures. (B) Normalized absorbance values at 350 nm of unsaturated lipid vesicle dispersions after 10 min of incubation with SMA copolymer at different temperatures (solid lines are added to guide the eye). (C) Normalized absorbance time trace of di-18:1 PC vesicles (red), di-18:1 PC/di-18:1 PE (1:1 molar ratio, green), and di-18:1-PC/lyso-18:1-PC (4:1 molar ratio, blue) after the addition of SMA copolymer, all at 20°C. (Inset) Schematic representation of the expected changes in the lateral chain pressure. To see this figure in color, go online.
Fig. 3 B also shows that bilayers of 16:0-18:1 PC are solubilized much faster than di-18:1 PC vesicles, while the effect of an extra double bond per acyl chain (di-18:2 PC vesicles) does not show significantly slower kinetics.
To further investigate the effect of lipid packing on solubilization, PE lipids were incorporated into PC lipid bilayers. This results in an increase of the lateral pressure in the acyl-chain region and an increase of the order of the lipid acyl chains due to the negative intrinsic curvature of PE lipids (22–25). Fig. 3 C shows that the presence of di-18:1 PE in di-18:1 PC vesicles results in incomplete solubilization, with a quick initial drop in optical density but no further change. By contrast, incorporation of lyso-PC lipids, which tend to decrease the lateral chain pressure due to their positive intrinsic curvature (22–24), results in immediate solubilization. Together, these data show that lipid packing has a strong influence on the kinetics of solubilization by SMA.
Electrostatic interactions affect membrane insertion and solubilization
At physiological pH, SMA copolymers carry multiple negative charges due to the partial deprotonation of maleic acid groups (26,27). In biological membranes, which often contain anionic lipids, this might lead to repulsive electrostatic interactions during membrane solubilization. Such electrostatic interactions would be particularly relevant when the polymer interacts with the membrane surface and inserts into the membrane. This step can be conveniently monitored by monolayer experiments.
Fig. 4 A shows that the addition of SMA copolymers to lipid monolayers of di-14:0 PC in the presence of salt results in an increase in surface pressure of 17 mN/m at an initial surface pressure of 25 mN/m. This increase is strong, because reported increases in surface pressure from other surface active molecules such as proteins are usually between 5 and 15 mN/m at similar initial surface pressure (28,29). Fig. 4 A also shows that the addition of 20% mol anionic di-14:0 PG lipids has very little effect on the insertion of SMA copolymers into the lipid monolayer. However, a significantly lower increase in surface pressure is observed when monolayers of pure anionic lipids are used and when salt is absent, implying that electrostatic repulsions are no longer screened. This shows that repulsive electrostatic interactions between SMA copolymers and anionic lipids can inhibit membrane insertion to a certain extent. Fig. 4 B displays the increase in surface pressure at different initial surface pressures for di-14:0 PC and di-14:0 PG lipid monolayers in the presence of NaCl. From the linear relationship between the surface pressure increase and the initial surface pressure, the maximal insertion pressure can be extrapolated to be ∼48 mN/m for both PC and PG lipid monolayers. This demonstrates that SMA readily inserts into lipid monolayers far above the surface pressure of ≈30 mN/m estimated for biological membranes (30). The observation that this happens even when the monolayer is completely negatively charged further highlights the strong insertion capability of the polymer.
Figure 4.
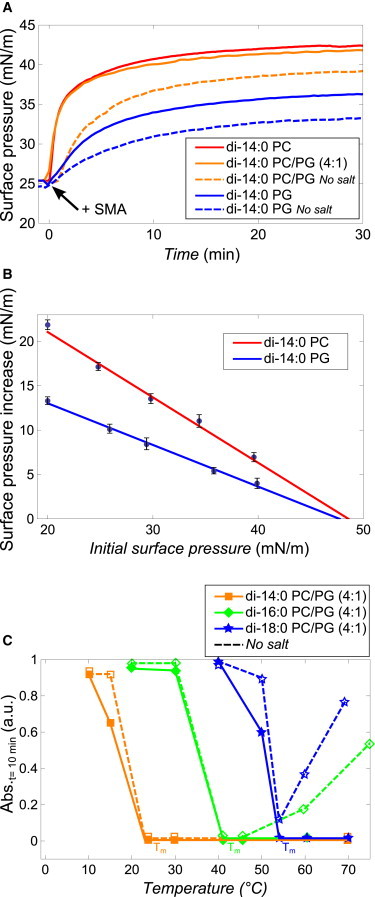
Effect of electrostatic interactions on membrane insertion and solubilization by SMA copolymers. (A) Surface pressure increase in time upon the addition of excess SMA to monolayers of di-14:0 PC (red), di-14:0 PC/PG (4:1) (orange), and di-14:0 PG (blue) in 50 mM Tris-HCl pH 8.0 with 150 mM NaCl (solid) or without NaCl (dashed). (B) Surface pressure increase as function of initial surface pressure for di-14:0 PC and di-14:0 PG lipid monolayers in 50 mM Tris-HCl pH 8.0 with 150 mM NaCl as subphase. (Solid lines) Linear fit with the maximum insertion pressure extrapolated to be ∼48 mN/m. Error bars are standard errors based on at least two independent measurements. (C) Normalized absorbance values at 350 nm of lipid vesicle dispersions after 10 min of incubation with the SMA copolymer at different temperatures. The diameter of the vesicles was 400 nm and a 3:1 (w/w) SMA/lipid was used. (Solid lines) Data recorded in Tris-HCl pH 8.0 buffer with 150 mM NaCl; (dashed lines) without NaCl. To see this figure in color, go online.
We next investigated to what extent electrostatic interactions influence the kinetics of solubilization by analyzing the effects of incorporation of anionic lipids on solubilization kinetics of vesicles in the presence and absence of salt (see Fig. S5 for full traces). Fig. 4 C shows the optical density values after 10 min of incubation of SMA with di-14:0 PC/PG, di-16:0 PC/PG, and di-18:0 PC/PG vesicles in the presence of salt (solid lines). The trends are similar to those observed with pure PC lipids (Fig. 1 B), but with solubilization rates that are slightly slower at >Tm. This is particularly true for the case where salt is absent (dashed lines). Above Tm, solubilization is fast for di-14:0 PC/PG but it becomes significantly slower for the longer chain lipids di-16:0 PC/PG and di-18:0 PC/PG in the absence of salt. Rather surprisingly, in this case solubilization is getting more difficult with increasing temperature. Possibly, this effect is related to changes in the intrinsic membrane curvature, as will be discussed later. In any case, these results indicate that increased repulsive electrostatic interactions slow down the overall rate and efficiency of solubilization.
The SMA copolymer does not preferentially solubilize specific lipid species
Finally, we tested the effect of SMA copolymers on vesicles of a native extract of E. coli membranes. This system was chosen because E. coli is often used for the expression of membrane proteins, and therefore efficient solubilization is required for extraction of these proteins from the membrane by using SMA. The acyl-chain composition of E. coli lipids is governed by c16:0 and c18:1 fatty acids (Fig. 5 D). Based on the results of solubilization of 16:0–18:1 PC vesicles (Fig. 3 B), there is no reason to expect that the acyl-chain composition would significantly slow down solubilization in this case. However, the overall lipid composition of E. coli membranes may make it difficult to solubilize: the TLE of E. coli membrane consists for a large part of the nonbilayer lipid PE (∼65 mol %) and furthermore contains the anionic lipids PG (∼25 mol %) and cardiolipin (∼10 mol %). As shown in Fig. 5 A, solubilization of vesicles of E. coli lipids is indeed inefficient at the standard 3:1 (w/w) SMA/lipid. On the other hand, solubilization could be significantly improved by increasing the SMA/lipid to 9:1 (w/w) and raising the salt concentration to 450 mM. Apparently, by increasing the concentrations of SMA and/or salt, polymer binding to the membrane is promoted and thereby solubilization. Also in this case, TEM and DLS analysis of the solubilized E. coli vesicles showed that nanodisks are formed with an average diameter of 8–10 nm (Fig. 5 B), similar to that found for pure PC lipids (see Fig. S3).
Figure 5.
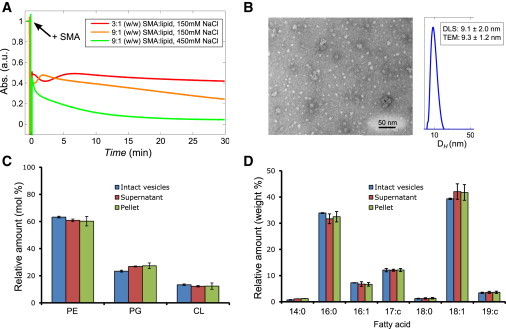
Characterization of the solubilization of vesicles (400 nm) of an E. coli native lipid extract by the SMA copolymer. (A) Time traces of absorbance at 350 nm of E. coli vesicle solubilization at different SMA copolymer and salt concentrations, all at 20°C. (B) Negative staining TEM image and DLS size distribution of E.coli nanodisks as prepared by the addition of SMA in a 9:1 (w/w) SMA/lipid in the presence of 450 mM NaCl. (Inset) Average diameters and standard deviations that were found. Analysis by TEM was based on 20 individual nanodisk particles and analysis by DLS was based on multiple measurements on a single independent sample as described in Materials and Methods. (C) Lipid composition (mol %) and (D) fatty acid composition (weight %) of intact vesicles of E. coli lipids, and of pellet and supernatant obtained after partial solubilization by SMA. Error bars represent the standard deviation of three independent experiments. To see this figure in color, go online.
The observation that the efficiency of solubilization is modulated by lipid packing properties and electrostatic interactions raises the question whether SMA may preferentially solubilize specific lipids. To test this, a subsolubilizing amount of SMA (1.4:1 w/w SMA/lipid) was added to vesicles of the native extract to induce only partial solubilization (see Fig. S6). The samples were then centrifuged to separate pellet (nonsolubilized vesicles) and supernatant (nanodisks) and their lipid compositions were determined from TLC analysis. As shown in Fig. 5 C, the lipid composition in the supernatant is not significantly different from that of the pellet or that of the intact vesicles. Similarly, GC analysis revealed that the fatty acid composition of the lipids is similar in all cases (Fig. 5 D). Hence, SMA copolymers do not seem to selectively solubilize phospholipid species in this system.
Discussion
Insight into the mode of action of SMA copolymers is important to optimally exploit its use in the purification and characterization of membrane proteins but also for development of novel applications in membrane research. Here, it was found that the SMA copolymer is a very efficient membrane solubilizing agent but that the kinetics of solubilization are modulated by physico-chemical properties of the lipids in very distinct ways.
How does the SMA copolymer exert its action and how can we understand the effects of lipid composition? Below, we present a model for the molecular mechanism of membrane solubilization by SMA and discuss how it is related to our experimental findings. We then discuss the physico-chemical properties of SMA that are responsible for its high solubilizing efficiency and we will compare this with the mode of action of other amphiphilic polymers that are used in membrane research.
Model for the mode of action of the SMA copolymer
The effect of lipid composition on vesicle solubilization by SMA copolymers can be understood based on the model presented in Fig. 6.
Figure 6.
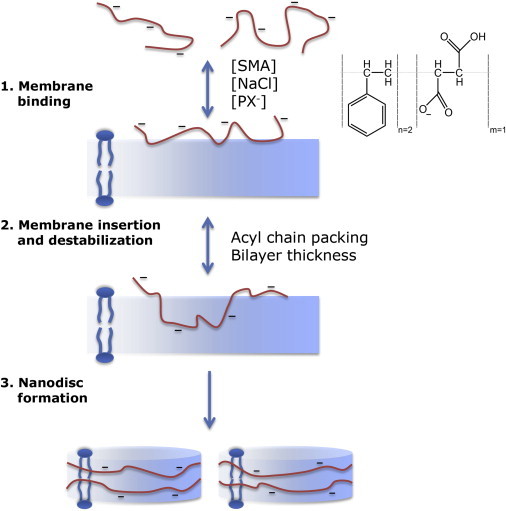
Schematic illustration of membrane solubilization by the SMA copolymer. (Inset) Schematic representation of the SMA copolymer used in this study. Step 1: anionic SMA copolymers bind to the lipid membrane, a process that is driven mainly by hydrophobic interactions and modulated by electrostatic interactions. Binding increases when the salt concentration increases and the number of anionic lipids (PX−) decreases. Step 2: insertion of the SMA copolymer in the membrane hydrophobic core, a process mainly determined by acyl-chain packing. A high lateral pressure in the acyl-chain region will decrease the efficiency of insertion. Step 3: insertion into the hydrophobic core leads to membrane destabilization and to the formation of vesicular intermediates. Further solubilization will lead to the formation of nanodisks, which are stabilized by a SMA copolymer belt around the nanodisk. The kinetics and efficiency of membrane destabilization and nanodisk formation is determined by acyl-chain packing properties and bilayer thickness. To see this figure in color, go online.
Step 1: membrane binding
In a first step, SMA copolymers bind to the membrane. In our model, the extent of binding depends on the SMA concentration. It is modulated by electrostatic interactions between SMA copolymers and the copolymers and anionic lipids at the membrane surface. Despite these repulsive electrostatic interactions, SMA has a strong interaction with membranes. This is most clearly illustrated by the lipid monolayer experiments, which showed a fast and exceptionally strong insertion of SMA in films of zwitterionic as well as anionic lipids. In both cases, the polymers are able to insert even at surface pressures far above those estimated for biological membranes, suggesting that SMA copolymers will insert into any biological membrane.
What is the driving force for the interaction of SMA with membranes? We propose that binding is mainly driven by the hydrophobic effect via the polymer styrene moieties and the lipid acyl chains (21,31) and that this driving force is sufficiently strong to overcome any repulsive electrostatic interactions. Nevertheless, electrostatic repulsion does modulate the extent of binding, as illustrated by the effects of increasing the salt concentration or decreasing the amount of anionic lipids in the membrane. Both changes would lead to increased binding, and hence to an increased efficiency of solubilization in the next step of the process.
Step 2: membrane insertion and destabilization
In this step, SMA copolymers insert more deeply into the hydrophobic core of the membrane. We envision that SMA will need packing defects in the membrane for efficient insertion. This is supported by the observation that in all systems of saturated lipids, maximal solubilization of SMA occurs at Tm of the lipids, where gel-phase domains coexist with domains of lipids in the liquid-crystalline phase, leading to large packing defects (32). However, also in the absence of large packing defects, lipid packing is important for solubilization, as evident from the observation that at >Tm, solubilization takes place much faster than at <Tm, when the lipids are in the gel phase and are thus more tightly packed. Interestingly, unsaturated lipids were found to be more difficult to solubilize than saturated lipids in the fluid phase. At first glance this is surprising, because unsaturated chains are more disordered and thus are more loosely packed. On the other hand, due to their double bonds, these membranes have an increased lateral pressure in the acyl-chain region (24,33,34), and this might lead to a less efficient insertion of the polymers. The observations that incorporation of PE, with its negative intrinsic curvature, inhibits solubilization, while incorporation of lyso-PC, with its positive intrinsic curvature, has the opposite effect, are consistent with the notion that the acyl-chain pressure plays an important role in solubilization.
In most cases, solubilization was improved by increasing temperature. This can be expected because increasing temperature will lead to a general increase in reaction kinetics and hence to faster solubilization. However, there is one exception: solubilization does not increase as a function of temperature upon SMA addition to anionic saturated lipids that have long acyl chains in the liquid-crystalline phase in the absence of salt (Fig. 4 C). Most likely this is a direct consequence of the low membrane binding of SMA, due to strong repulsive electrostatic interactions in this particular case. We propose that this low binding allows an opposing temperature-dependent effect to become dominant, namely the increase of lipid chain dynamics. This would lead to an increased negative curvature of the membrane at higher temperatures and hence, based on the result discussed above, to slower and inefficient solubilization. Such an increase in negative curvature will be stronger for longer lipids and would explain the trend of less efficient solubilization for increasing bilayer thickness.
In general, we suggest that the bilayer thickness plays an important role especially at the stage where the vesicles disintegrate and fall apart into intermediate vesicular structures and membrane fragments. This is simply because it will be more difficult to break up thicker membranes due to an increase of the forces that hold the membrane together, such as van der Waals interactions and the hydrophobic effect. This view is in line with the observations that short-chained saturated lipids are easily solubilized, even in the gel phase, and that unsaturated lipids at lower temperature are less efficiently solubilized when the effective length of the acyl chains increases (Fig. 4 B).
Together, these results show that the rate and yield of solubilization by SMA copolymers depends greatly on the lipid composition of the model membranes. Despite this, we found that the lipid composition of nanodisks largely reflects that of the initial membrane. This has also been observed recently for nanodisks that contain a membrane protein purified from the purple bacterium Rhodobacter sphaeroides (6). These findings thus imply that it is the physical properties of lipid membranes, rather than those of individual lipid constituents, that determine the solubilization kinetics and efficiency.
Step 3: nanodisk formation
In the last step, membrane fragments are further solubilized and the formation of nanodisks is facilitated. Recently, Jamshad et al. (21) and Orwick et al. (3) structurally characterized the nanodisks of di-14:0 PC bounded by SMA. In these studies it was shown that nanodisks are indeed shaped like a disk and that the SMA copolymer places its phenyl groups between the lipid acyl chains, thereby stabilizing the nanodisks. The reported disk-like shape and average diameter of the nanodisks is very similar to what we observed here. In addition, we found that the average nanodisk diameter of 10–12 nm is completely independent of lipid chain length and unsaturation. However, the parameters that control this nanodisk diameter are still unclear (see discussion below).
The SMA copolymer forms nanodisks efficiently because of its chemical structure
The hydrophobic effect drives the interaction between SMA copolymers and membranes. But what sets this polymer apart from other amphiphilic polymers? We argue that the properties of the phenyl and carboxylic acid pendant groups are responsible for this in several ways (Fig. 6).
-
1)
These residues are small, thereby giving SMA a small cross-sectional area. This makes it relatively easy for the polymer to insert into membranes even when no packing defects are present.
-
2)
Insertion of the short residues, and in particularly the rigid phenyl groups, in between the acyl chains (22) leads to a minimal loss in conformational entropy of the residues when the polymer is wrapped around a nanodisk compared to when the polymer is in solution.
-
3)
Because there is minimal steric hindrance, the short side-groups allow the polymer to adopt a curvature that is required to encircle a nanodisk.
An additional helpful property of the SMA copolymer might be its ability to interact with membranes via the polar moments of the side-groups. It was recently suggested that polyacids interact with the membrane based on the favorable interaction between the dipole moment of their carboxylic acid groups and the dipole potential of membranes (35). The same would hold not only for the carboxylic acid groups of SMA but for the phenyl side-groups as well. Due to the orientation of the lipid fatty acid carbonyls and the preferential orientation of water molecules that hydrate the lipid headgroups, a positive potential arises in the center of the bilayer (36). Because the phenyl side-groups have a large quadrupolar moment with a negative potential on both sides of the aromatic ring (37), this would favor their insertion into the interior of the bilayer.
Support for the special role of these side-groups in SMA comes from comparison with the chemistry and mode of action of other amphiphilic polymers. The A8-35 amphipol, for example, is a polymer that is often used as replacement for detergents to stabilize membrane proteins (38,39). Its pendant groups are mainly carboxylic acid and large alkyl groups (see Fig. S7 A). These alkyl chains give the polymer a large cross-sectional area, which would interfere with membrane solubilization and nanodisk formation due to steric hindrance, while their flexibility would cause a large loss in conformational entropy when it hypothetically would be wrapped around a nanodisk. Instead, it is much more favorable for the polymer to wrap around membrane proteins directly. Another interesting example is PEAA (poly(ethyl)acrylic acid; see Fig. S7 B). This polymer consists of relatively small pendant carboxylic acid and ethyl groups. Thomas et al. (40) and Chung et al. (41) reported that PEAA is able to solubilize saturated di-14:0 PC and di-16:0 PC vesicles. This situation leads to the formation of structures similar in shape and size to SMA-bounded nanodisks. However, the solubilization kinetics is much slower, and range of lipid composition able to be solubilized much more limited than for SMA. This is in line with the notion that the presence of small pendant groups such as phenyl groups rather than the alkyl group is helpful for efficient solubilization of membrane into nanodisks.
Finally, we address MSPs, which are amphiphilic biopolymers that, like SMA, are able to assemble nanodisks. They form α-helices with a relatively large cross-sectional area, which would require the presence of large surface defects for membrane insertion. Indeed, as described here and previously, MSPs can only efficiently solubilize PC vesicles at Tm (19,20) when there are large surface defects, and then only in the case of short-chained lipids, when lipid-lipid interactions are less strong and solubilization occurs more easily.
The SMA copolymer versus membrane scaffold proteins
These fresh insights into the mode of action of SMA allow us to compare some aspects of the assembly of nanodisks by MSPs and SMA. Due to the efficient solubilizing properties of SMA, it can solubilize a wide variety of lipids with different acyl-chain lengths and headgroups. Thus SMA, in contrast to MSPs, is able to directly solubilize a wide variety of biological membranes into nanodisks without the help of detergents (5,6,8). Importantly, we showed that SMA does not solubilize lipids in a preferential way. This implies that the native lipid environment is retained for membrane proteins isolated in nanodisks by using SMA (5,6). In apparent contrast, for MSPs it is possible to choose which lipids are used for reconstitution. However, it should be realized that for SMA-bounded nanodisks it is also possible to capture membrane proteins in a specific lipid composition; this can be accomplished by first reconstituting the protein in vesicles of the desired lipids by conventional methods, and then solubilizing these reconstituted vesicles with SMA.
An advantage of the use of MSP-bounded nanodisks is that the diameter can be conveniently controlled between 7 and 20 nm by choosing an MSP of a certain length (42). The nanodisk diameter is an important parameter because, in principle, it determines the size of a membrane protein (complex) that can be captured in a nanodisk. For the SMA copolymer, it is not (yet) clear what controls the nanodisk diameter and how this can be modulated (43). Here, we found that the SMA 2000 copolymer always yielded nanodisks with a diameter of ∼10 nm, regardless of the actual lipid used. Interestingly, it has been reported that both SMA 2000 and the more hydrophobic SMA 3000 yield larger nanodisks when a membrane protein is embedded. Incorporation of a photosynthetic reaction center (6), the mitochondrial respiratory complex IV (5), and bacteriorhodopsin (4) all led to nanodisks of a diameter between 12 and 17 nm. The capturing of the staphylococcal penicillin-binding protein complex PBP2/PBP2a even led to nanodisks up to 24 nm in diameter (44). Apparently, the protein content, and most likely specifically its shape and diameter, are key parameters in determining the size of nanodisks.
Conclusions
We found that that SMA is a very efficient membrane solubilizing agent, displaying kinetics that are strongly influenced by lipid packing properties and electrostatic interactions. Its small phenyl and carboxylic acids pendant groups both account for the high solubilizing efficiency of the polymer as well as for its ability to stabilize nanodisks. The results will be helpful for optimization of procedures involving the use of SMA for isolation of membrane proteins from different membranes, the extraction of more complex lipid compositions that might include sphingolipids and sterols, and for development of new applications of this polymer in membrane research. From a broader perspective, they will contribute to the understanding of the mode of action of polymers as membrane-active agents to modify membrane properties with possible implications for drug delivery systems, among other uses.
Acknowledgments
We thank Charles Mateer from Cray Valley (USA) for providing the SMAn copolymer; Mark Daniels and Sylke Lievestro from the NMR spectroscopy group, Utrecht University, for the expression and purification of MSP1D1 membrane scaffold protein; Bonny Kuipers for his assistance during dynamic light scattering experiments; and Ruud Cox for his assistance during gas chromatography experiments. We thank Francisco Calderón Celis for his contributions as master student in our group.
Financial support via S.S. and J.A.K. of the research program of the Foundation for Fundamental Research on Matter and the support from a grant from the Netherlands Organization for Scientific Research (NWO-ECHO grant No. 711-013-005 to J.D.P.) is gratefully acknowledged.
Supporting Material
References
- 1.Knowles T.J., Finka R., Overduin M. Membrane proteins solubilized intact in lipid containing nanoparticles bounded by styrene maleic acid copolymer. J. Am. Chem. Soc. 2009;131:7484–7485. doi: 10.1021/ja810046q. [DOI] [PubMed] [Google Scholar]
- 2.Jamshad M., Lin Y.-P., Dafforn T.R. Surfactant-free purification of membrane proteins with intact native membrane environment. Biochem. Soc. Trans. 2011;39:813–818. doi: 10.1042/BST0390813. [DOI] [PubMed] [Google Scholar]
- 3.Orwick M.C., Judge P.J., Watts A. Detergent-free formation and physicochemical characterization of nanosized lipid-polymer complexes: Lipodisq. Angew. Chem. Int. Ed. Engl. 2012;51:4653–4657. doi: 10.1002/anie.201201355. [DOI] [PubMed] [Google Scholar]
- 4.Orwick-Rydmark M., Lovett J.E., Watts A. Detergent-free incorporation of a seven-transmembrane receptor protein into nanosized bilayer Lipodisq particles for functional and biophysical studies. Nano Lett. 2012;12:4687–4692. doi: 10.1021/nl3020395. [DOI] [PubMed] [Google Scholar]
- 5.Long A.R., O’Brien C.C., Alder N.N. A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs. BMC Biotechnol. 2013;13:41. doi: 10.1186/1472-6750-13-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Swainsbury D.J.K., Scheidelaar S., Jones M.R. Bacterial reaction centers purified with styrene maleic acid copolymer retain native membrane functional properties and display enhanced stability. Angew. Chem. Int. Ed. 2014 doi: 10.1002/anie.201406412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seddon A.M., Curnow P., Booth P.J. Membrane proteins, lipids and detergents: not just a soap opera. Biochim. Biophys. Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Gulati S., Jamshad M., Rothnie A.J. Detergent-free purification of ABC (ATP-binding-cassette) transporters. Biochem. J. 2014;461:269–278. doi: 10.1042/BJ20131477. [DOI] [PubMed] [Google Scholar]
- 9.Shi L., Shen Q.-T., Pincet F. SNARE proteins: one to fuse and three to keep the nascent fusion pore open. Science. 2012;335:1355–1359. doi: 10.1126/science.1214984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayburt T.H., Sligar S.G. Self-assembly of single integral membrane proteins into soluble nanoscale phospholipid bilayers. Protein Sci. 2003;12:2476–2481. doi: 10.1110/ps.03267503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Denisov I.G., Grinkova Y.V., Sligar S.G. Directed self-assembly of monodisperse phospholipid bilayer nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 12.Frauenfeld J., Gumbart J., Beckmann R. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 2011;18:614–621. doi: 10.1038/nsmb.2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagn F., Etzkorn M., Wagner G. Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J. Am. Chem. Soc. 2013;135:1919–1925. doi: 10.1021/ja310901f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mazhab-Jafari M.T., Marshall C.B., Ikura M. Membrane-dependent modulation of the mTOR activator Rheb: NMR observations of a GTPase tethered to a lipid-bilayer nanodisc. J. Am. Chem. Soc. 2013;135:3367–3370. doi: 10.1021/ja312508w. [DOI] [PubMed] [Google Scholar]
- 15.Bayburt T.H., Sligar S.G. Membrane protein assembly into nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rouser G., Fleischer S., Yamamoto A. Two-dimensional then layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids. 1970;5:494–496. doi: 10.1007/BF02531316. [DOI] [PubMed] [Google Scholar]
- 17.Silvius D.J.R. Lipid-Protein Interactions. John Wiley; New York: 1982. Thermotropic phase transitions of pure lipids in model membranes and their modifications by membrane proteins. [Google Scholar]
- 18.Nath A., Atkins W.M., Sligar S.G. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 19.Pownall H.J., Massey J.B. Mechanism of association of human plasma apolipoproteins with dimyristoylphosphatidylcholine: effect of lipid clusters on reaction rates. Biophys. J. 1982;37:177–179. doi: 10.1016/S0006-3495(82)84659-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan C.-P.L., Chiu M.H., Weers P.M. Apolipoprotein-induced conversion of phosphatidylcholine bilayer vesicles into nanodisks. Biochim. Biophys. Acta. 2011;1808:606–613. doi: 10.1016/j.bbamem.2010.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jamshad M., Grimard V., Dafforn T.R. Structural analysis of a nanoparticle containing a lipid bilayer used for detergent-free extraction of membrane proteins. Nanotechnology. 2014 doi: 10.1007/s12274-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruner S.M. Intrinsic curvature hypothesis for biomembrane lipid composition: a role for nonbilayer lipids. Proc. Natl. Acad. Sci. USA. 1985;82:3665–3669. doi: 10.1073/pnas.82.11.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsh D. Lateral pressure profile, spontaneous curvature frustration, and the incorporation and conformation of proteins in membranes. Biophys. J. 2007;93:3884–3899. doi: 10.1529/biophysj.107.107938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frolov V.A., Shnyrova A.V., Zimmerberg J. Lipid polymorphisms and membrane shape. Cold Spring Harb. Perspect. Biol. 2011;3:a004747. doi: 10.1101/cshperspect.a004747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Separovic F., Gawrisch K. Effect of unsaturation on the chain order of phosphatidylcholines in a dioleoylphosphatidylethanolamine matrix. Biophys. J. 1996;71:274–282. doi: 10.1016/S0006-3495(96)79223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferry J., Udy D., Fordyce D. Titration and viscosity studies of two copolymers of maleic acid. J. Colloid Sci. 1951;6:429–442. [Google Scholar]
- 27.Garrett E.R., Guile R.L. Potentiometric titrations of a polydicarboxylic acid: maleic acid-styrene copolymer. J. Am. Chem. Soc. 1951;73:4533–4535. [Google Scholar]
- 28.Demel R.A. Monomolecular layers in the study of biomembranes. In: Hilderson H., Ralston G., editors. Physicochemical Methods in the Study of Biomembranes. Vol. 23, Subcellular Biochemistry. Springer; New York: 1994. pp. 83–120. [DOI] [PubMed] [Google Scholar]
- 29.Calvez P., Bussières S., Salesse C. Parameters modulating the maximum insertion pressure of proteins and peptides in lipid monolayers. Biochimie. 2009;91:718–733. doi: 10.1016/j.biochi.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 30.Marsh D. Lateral pressure in membranes. Biochim. Biophys. Acta. 1996;1286:183–223. doi: 10.1016/s0304-4157(96)00009-3. [DOI] [PubMed] [Google Scholar]
- 31.Tonge S.R., Tighe B.J. Responsive hydrophobically associating polymers: a review of structure and properties. Adv. Drug Deliv. Rev. 2001;53:109–122. doi: 10.1016/s0169-409x(01)00223-x. [DOI] [PubMed] [Google Scholar]
- 32.Noordam P.C., Killian A., De Gier J. Comparative study on the properties of saturated phosphatidylethanolamine and phosphatidylcholine bilayers: barrier characteristics and susceptibility to phospholipase A2 degradation. Chem. Phys. Lipids. 1982;31:191–204. doi: 10.1016/0009-3084(82)90044-5. [DOI] [PubMed] [Google Scholar]
- 33.Cantor R.S. Lipid composition and the lateral pressure profile in bilayers. Biophys. J. 1999;76:2625–2639. doi: 10.1016/S0006-3495(99)77415-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mouritsen O.G. Lipids, curvature, and nano-medicine. Eur. J. Lipid Sci. Technol. 2011;113:1174–1187. doi: 10.1002/ejlt.201100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berkovich A.K., Lukashev E.P., Melik-Nubarov N.S. Dipole potential as a driving force for the membrane insertion of polyacrylic acid in slightly acidic milieu. Biochim. Biophys. Acta. 2012;1818:375–383. doi: 10.1016/j.bbamem.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Clarke R.J. The dipole potential of phospholipid membranes and methods for its detection. Adv. Colloid Interface Sci. 2001;89-90:263–281. doi: 10.1016/s0001-8686(00)00061-0. [DOI] [PubMed] [Google Scholar]
- 37.Battaglia M., Buckingham A., Williams J. The electric quadrupole moments of benzene and hexafluorbenzene. Chem. Phys. Lett. 1981;78:421–423. [Google Scholar]
- 38.Tribet C., Audebert R., Popot J.-L. Amphipols: polymers that keep membrane proteins soluble in aqueous solutions. Proc. Natl. Acad. Sci. USA. 1996;93:15047–15050. doi: 10.1073/pnas.93.26.15047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Popot J.-L., Althoff T., Zoonens M. Amphipols from A to Z∗. Annu. Rev. Biophys. 2011;40:379–408. doi: 10.1146/annurev-biophys-042910-155219. [DOI] [PubMed] [Google Scholar]
- 40.Thomas J.L., Devlin B.P., Tirrell D.A. Kinetics of membrane micellization by the hydrophobic polyelectrolyte poly(2-ethylacrylic acid) Biochim. Biophys. Acta. 1996;1278:73–78. doi: 10.1016/0005-2736(95)00192-1. [DOI] [PubMed] [Google Scholar]
- 41.Chung J.C., Gross D.J., Opsahl-Ong L.R. pH-Sensitive, cation-selective channels formed by a simple synthetic polyelectrolyte in artificial bilayer membranes. Macromolecules. 1996;29:4636–4641. [Google Scholar]
- 42.Ritchie T., Grinkova Y., Sligar S. Chapter 11—Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. In: Duzgunes N., editor. Vol. 464, Liposomes, Part F, of Methods in Enzymology. Academic Press; New York: 2009. pp. 211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang R., Sahu I.D., Lorigan G.A. Characterizing the structure of Lipodisq nanoparticles for membrane protein spectroscopic studies. Biochim. Biophys. Acta. 2014 doi: 10.1016/j.bbamem.2014.05.008. www.sciencedirect.com/science/article/pii/S0005273614001801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paulin S., Jamshad M., Taylor P.W. Surfactant-free purification of membrane protein complexes from bacteria: application to the staphylococcal penicillin-binding protein complex PBP2/PBP2a. Nanotechnology. 2014;25:285101. doi: 10.1088/0957-4484/25/28/285101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


