Abstract
Posttranslational modification by small ubiquitin-like modifiers (SUMOs), known as SUMOylation, is a key regulatory event in many eukaryotic cellular processes in which SUMOs interact with a large number of target proteins. SUMO binding motifs (SBMs) are small peptides derived from these target proteins that interact noncovalently with SUMOs and induce conformational changes. To determine the effect of SBMs on the mechanical properties of SUMO1 (the first member of the human SUMO family), we performed single-molecule force spectroscopy experiments on SUMO1/SBM complexes. The unfolding force of SUMO1 (at a pulling speed of 400 nm/s) increased from ∼130 pN to ∼170 pN upon binding to SBMs, indicating mechanical stabilization upon complexation. Pulling-speed-dependent experiments and Monte Carlo simulations measured a large decrease in distance to the unfolding transition state for SUMO1 upon SBM binding, which is by far the largest change measured for any ligand binding protein. The stiffness of SUMO1 (measured as a spring constant for the deformation response along the line joining the N- and C-termini) increased upon SBM binding from ∼1 N/m to ∼3.5 N/m. The relatively higher flexibility of ligand-free SUMO1 might play a role in accessing various conformations before binding to a target.
Introduction
Protein-protein/ligand interactions are ubiquitous in biology and they are an important aspect of biological function of many proteins (1). They play an important role in many cellular functions, and an increasing number of biochemical and biophysical methods are being employed to understand these interactions and their role in protein function. There is a plethora of biophysical methods for determining the effect of ligand binding on the thermodynamic stability of proteins, and it has been observed generally that the thermodynamic stability of the protein is enhanced upon ligand binding (1). Recently, with the development of single-molecule force spectroscopy techniques, the effect of ligand binding on mechanical stability has been elucidated for many proteins (2). Single-molecule atomic force microscopy (SM-AFM) demonstrated that ligand binding may affect the mechanical stability of proteins or modulate their unfolding pathways (2–12).
Posttranslational modification by small ubiquitin-like modifiers (SUMOs), known as SUMOylation, is an important regulatory event in many cellular processes, such as gene transcription, cell cycle progression, signal transduction, DNA repair, and transport of nuclear bodies. During SUMOylation, SUMOs interact with various enzymes and target proteins either covalently or noncovalently. SUMO-modified proteins act as a platform for binding of other proteins, and hence, SUMO plays a key role in protein interaction networks (13,14). SUMO interacts with many enzymes and proteins, for example, Ubc9, RanGAP1, Thymine-DNA glycolase, E2-25K, RanBP2, and PIASX, with varied consequences (15). The covalent attachment of SUMO to its target proteins is by formation of an isopeptide bond through the C-terminal glycine of SUMO. A consensus binding motif with amino acid sequence Ф-K-X-D/E, where Ф is any large hydrophobic residue and X is any residue, has been identified in the target proteins for covalent attachment (16). SUMO binding motifs (SBMs) that are necessary for the target protein to interact with SUMO noncovalently have also been identified, and they generally have a consensus site with a stretch of hydrophobic amino acids (V/I)-X-(V/I)-(V/I) flanked by acidic residues (13,14,16). The SBMs orient in either a parallel or an antiparallel β-strand conformation that extends the SUMO β-sheet, allowing the interaction of hydrophobic residues of the SBM and the hydrophobic core between the β-sheet and the α-helix of SUMO (14). SBMs have been found in various proteins, including SUMOylating enzymes, SUMO targets, and other SUMO-binding proteins.
Earlier studies have shown that small peptides with the consensus sequence of the SBM can also interact with SUMO1 (first member of the human SUMO family) in vitro without any enzymatic cascade reactions (13,16). The SBMs used in this study, henceforth called S10 and S12 based on the number of residues in their amino acid sequences (DDDVLIVYEL and KVDVIDLTIESS, respectively), have been shown to interact with SUMO1 (17). S10 and S12 bind to SUMO1 with dissociation constants (Kd) of ∼70 μM and ∼6 μM, respectively (16,17). S10 and S12 peptides have been derived from the interacting regions of SUMO binding proteins RanBp2 and PiasX, respectively. RanBP2 catalyzes SUMO E3 ligase activity and is required to localize SUMO1-modified RanGAP1 at the nuclear pore complex to facilitate nucleocytoplasmic trafficking (18). The SBM of PiasX plays an important role in its interaction with SUMOylated transcription factor Elk-1, which favors transcriptional activation (13). Both S10 and S12 interact with SUMO1 noncovalently at the same binding site. S10 forms an antiparallel β-strand with the β2-strand of SUMO1 and interacts with the residues in the β2-strand as well as with those in the α-helix of SUMO1. S12 binds in the same region of SUMO1 but in a reverse orientation to the β2-strand of SUMO1 (13,16,17). The binding groove of SUMO1 consists of hydrophobic and aromatic residues that help in forming strong hydrophobic interactions with the SBMs. The residues in the folded region of SUMO1 that are involved in interaction with the SBMs are Glu-33, Ile-34, His-35, Phe-36, Val-38, Leu-47, and Tyr-51 (13). The structure and topology diagrams of the ligand-free and ligand-bound forms of SUMO1 are shown in Fig. 1. Here, we investigate the effect of SBMs S10 and S12 on the mechanical stability of SUMO1 when these complexes are pulled along the line joining the N-C termini of SUMO1.
Figure 1.
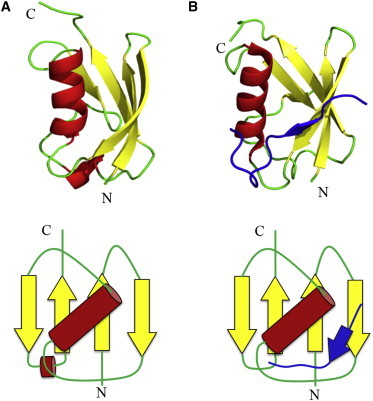
Structure and topology map of SUMO1 in ligand-free (A) and ligand-bound (B) forms. N- and C-termini, along which the stretching force is applied in the pulling experiments, are labeled in the figure. Protein Data Bank IDs of the structures of SUMO1 and S12-bound SUMO1 are 1A5R and 2ASQ, respectively. Another SBM peptide, S10, also binds in the same region of SUMO1 but in the reverse orientation. The unstructured N-terminus of SUMO1 (i.e., 21 residues) is not shown in the figure. To see this figure in color, go online.
Materials and Methods
Protein expression and purification
The octameric protein (SUMO1)8 was expressed and purified using the protocol described earlier (19). Briefly, human SUMO1 was modified to insert the restriction sites of BamHI at the 5′ end and those of BglII and KpnI at the 3′ end. The gene was then cloned into the pQE80L vector between the BamHI and KpnI sites. The gene of (SUMO1)8 was constructed by iterative cloning method as described by Carrion-Vazquez et al. (20). (SUMO1)8 protein was overexpressed in the BLR(DE3) strain of E. coli by induction with 1 mM isopropyl β-D-1 thiogalactopyranoside for 6 h after the OD600 of the cell culture has reached 0.6. The harvested cells were suspended in phosphate-buffered saline (PBS) (pH 7.4) containing 1 mM dithiothreitol and 0.1 mM phenylmethylsulfonyl fluoride. The cells were lysed by sonication and centrifuged and the supernatant was applied to a column of Ni-NTA-coated agarose beads. The beads were washed with PBS containing 20 mM imidazole and the protein was eluted with PBS containing 250 mM imidazole at pH 7.4. The protein was further purified by size-exclusion chromatography using a superdex200 column (Amersham Biosciences, Chennai, India). The purity of the protein was checked using sodium dodecyl sulfate polyacrylamide gel electrophoresis. The purified protein was stored at 4°C. SBM peptides S10 and S12 were purchased from USV (Mumbai, India).
Sample preparation for single-molecule pulling experiments
Solid SBM peptides were weighed and added to 2 μM (SUMO1)8 solutions in PBS (pH 7.4) such that the final concentration of the SBM peptide was 1 mM. For concentration-dependent studies, small amounts from the stock solutions of SBM peptides were mixed with the protein solution to get the final desired peptide concentration. The protein and the SBM peptide were mixed and equilibrated on a rotary shaker overnight (or for at least 3 h) before the experiment. The solutions were centrifuged at 13,000 rpm for 5 min to remove any undissolved SBM peptide and the supernatant was used for performing pulling experiments by SM-AFM.
SM-AFM
Single-molecule pulling experiments were performed on a custom-built atomic force microscope as described elsewhere (21). Approximately 50 μL of (SUMO1)8 protein solution (or protein/SBM peptide mixture solution) was placed on a gold-coated glass coverslip and let it for 30 min before starting a pulling experiment. Gold-coated reflective cantilevers with spring constants of ∼35 pN/nm were purchased from Bruker (Goleta, CA). Calibration of cantilevers was done using the equipartition theorem before each pulling experiment (22). All the experiments were performed at room temperature.
Data analysis
In the data analysis, force peaks in force-versus-extension (FX) traces were fitted to the worm-like chain (WLC) model of polymer elasticity (23) using Eq. 1:
| (1) |
where p and Lc denote the persistence length and contour length, respectively, kB is Boltzmann’s constant and T is the absolute temperature. The persistence-length values used for fitting varied between 0.3 and 0.7. This range of persistence length for the WLC model was found to best fit the FX traces.
The spontaneous rate of unfolding (ku0) and distance to the unfolding transition state (Δxu) are calculated using Monte Carlo simulations as described elsewhere (19). The transition-state activation energy was calculated using the Arrhenius equation (Eq. 2) and the spring constant was calculated using harmonic approximation (Eq. 3) as described earlier by Rief and co-workers (24,25).
| (2) |
| (3) |
where ΔG‡ is the activation energy, kA is Arrhenius frequency factor, and ks is the spring constant for deformation along the N-C-termini pulling direction.
Results and Discussion
Mechanical stability of SUMO1 increases upon peptide binding
SM-AFM experiments were performed on (SUMO1)8 and its complexes with SBM peptides S10 and S12 at a constant pulling speed of 400 nm/s. Representative FX traces are shown in Fig. 2. The FX curves were fitted to the WLC model of polymer elasticity and the change in contour length (ΔLC) upon unfolding was observed to be ∼24 nm, corresponding to the complete unraveling of SUMO1 (i.e., 76 residues in its fold), as reported previously (19). This also indicates that SUMO1 unfolds in a two-state pathway upon binding S10 or S12. The featureless spacer before the first unfolding force peak in the FX traces corresponds to the unraveling of the long unstructured peptide at the N-terminus of SUMO1 (19). The maximum of each peak corresponds to the force at which the corresponding SUMO1 unfolded, and the last peak with very high force corresponds to detachment of the protein either from the cantilever tip or from the surface. The FX traces show that SUMO1 unfolds at a higher force upon binding to S10 and S12, clearly indicating enhancement of the mechanical stability of the protein.
Figure 2.
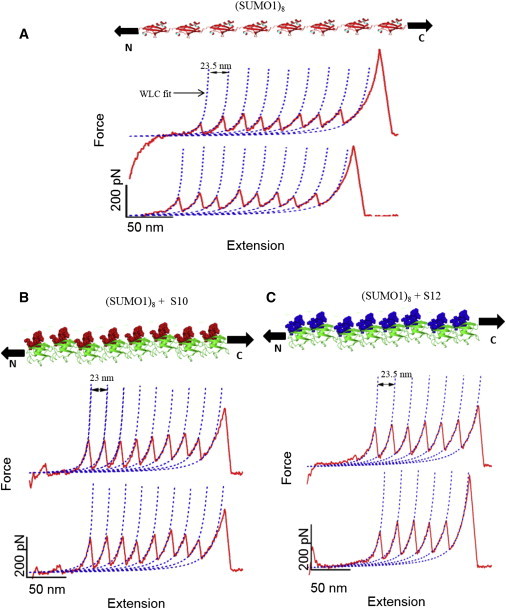
SUMO binding peptides (SBMs) alter the mechanical properties of SUMO1. Force-extension (FX) traces of (SUMO1)8 (A), (SUMO1)8 in the presence of S10 (B), and (SUMO1)8 in presence of S12 (C) are shown. Cartoons of octameric SUMO1 and its complex with the SBM peptides appear above the corresponding FX traces. All force peaks in the FX traces are fitted to the WLC model (dotted lines) to obtain ΔLC. SUMO1 has a ΔLC of ∼24 nm and is not altered upon binding to the peptides. The unfolding force of SUMO1 increases upon binding to the SBMs, as can be seen from the FX traces and the force scale. SUMO1 is being pulled along the line joining the N- and C-termini in the experiment, as indicated by the arrows. To see this figure in color, go online.
The unfolding force (Fu) distributions are shown in Fig. 3. The ΔLC distributions are given in Fig. S1 in the Supporting Material. Although the ΔLC (∼24 nm) remains unchanged upon ligand binding, the overlay of unfolding force histograms shows a clear increase in the mechanical stability upon ligand binding. Table 1 shows the average Fu and ΔLC along with their standard deviations. By measuring the unfolding force of SUMO1 in the presence of a peptide that does not bind to it, we have also confirmed that the increase in unfolding force is not due to the high concentration of the peptide in solution but rather to the ligand binding of SUMO1. The unfolding force of SUMO1 remains unchanged in the presence of this peptide in solution (Fig. S2). The unfolding force of SUMO1 increases from 132 pN to 166 pN upon binding to S10 and to 179 pN upon binding to S12. The unfolding force of apo SUMO1 is consistent with our earlier report (19). The increase in unfolding force upon binding to SBM peptides is significant, as revealed by the statistical p-test, with p < 0.0001 for both S10 and S12. We further studied the concentration dependence of S10 and S12 on the mechanical stability of SUMO1 (Fig. 4 and Fig. S3). The unfolding force of SUMO1 saturates at high concentrations of SBM peptides, and the concentrations of S10 and S12 at which 50% saturation occurs are 140 μM and 33 μM, respectively. These values are in the range expected from their dissociation constants, i.e., 6 μM for SUMO1 and S12 complex and 70 μM for SUMO1 and S10 complex, obtained from bulk studies reported previously (16).
Figure 3.
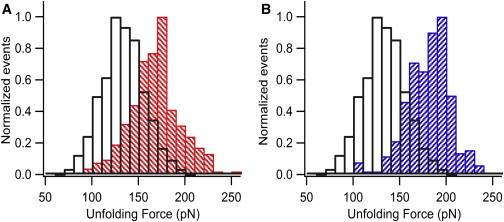
The unfolding force of SUMO1 increases upon binding the SBM peptides. The overlay of unfolding force histograms of SUMO1 (open bars) and SUMO1 bound to S10 (red-hatched bars) (A) and S12 (blue-hatched bars) (B) are shown. The histograms clearly show that the average unfolding force of SUMO1 increased from 132 pN to 166 pN upon binding to S10 and to 179 pN upon binding to S12. To see this figure in color, go online.
Table 1.
Mechanical properties of SUMO1 and SUMO1 in complex with SBM peptides
| Protein | ΔLC (nm) | Unfolding force (pN) |
|---|---|---|
| SUMO1 | 23.8 ± 1.2 | 132 ± 23 |
| SUMO1 + S10 | 23.6 ± 1.1 | 166 ± 26 |
| SUMO1 + S12 | 23.7 ± 0.8 | 179 ± 23 |
Values are represented as the mean ± SD.
Figure 4.
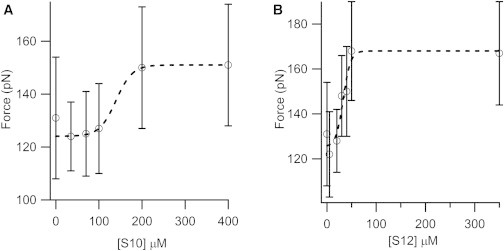
Dependence of the unfolding force of SUMO1 on SBM peptide concentration. The unfolding force of SUMO1 increases with increasing concentrations of S10 (A) and S12 (B), with a sudden change at concentrations close to the Kd of the complex of the protein and the SBM peptide. Sigmoidal fits are shown by dotted lines (see text for more details). The unfolding force histograms of SUMO1 at various concentrations of S10 and S12 are shown in Fig. S3.
Although proteins such as cadherin, dihydrofolate reductase, acylphosphatase, GB1, and M-crystallin have shown an increase in mechanical stability upon binding to metal ions or small molecules, there are many other proteins, for example, maltose-binding protein, leucine-binding protein, ribose-binding protein, and von Willebrand factor, whose mechanical stability is not altered by ligand binding (2,26). This might be due to the anisotropic nature of the stretching force. The orientation of the ligand-binding region with respect to the direction of applied force plays a prominent role in determining the ligand effect on mechanical properties, as has been illustrated previously for maltose-binding protein and titin-telethonin complex (7,27).
Pulling-speed-dependent mechanical unfolding properties and estimation of kinetic parameters
Mechanical unfolding of proteins is a kinetic process, and the pulling speed affects the unfolding force of proteins. Further pulling experiments on SUMO1 bound to the SBM peptides were performed by varying the pulling speed. It is found that the ΔLC is independent of pulling speed, but the unfolding force varies with the speed. The histograms of unfolding forces of SUMO1 complexes with S10 and S12 at various pulling speeds are shown in Fig. S4. When the speed is varied from 100 nm/s to 2000 nm/s, the unfolding force increases from 138 pN to 195 pN for SUMO1 bound to S10 and from 153 pN to 213 pN for SUMO1 bound to S12. These speed-dependent unfolding forces can be used to calculate kinetic parameters such as the spontaneous rate of unfolding (ku0) and the distance to the unfolding transition state (Δxu) using Monte Carlo simulations (28,29). The semilogarithmic plot of unfolding forces versus pulling speed is shown in Fig. 5. The plot clearly shows that the unfolding force of SUMO1 in the presence of the SBM peptides is always higher than that of SUMO1 alone. In addition, SUMO1 bound to S12 shows greater stability than SUMO1 bound to S10 at all the pulling speeds. The plot also underlines that the dependence of unfolding force on the pulling speed is more prominent after binding to the SBM peptides.
Figure 5.
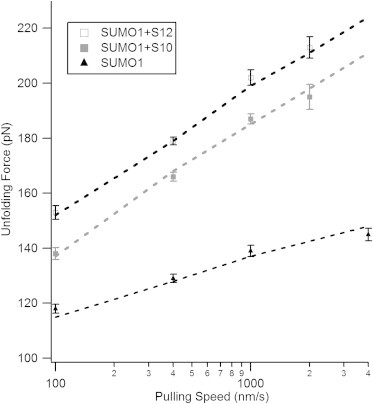
The distance to the unfolding transition state (Δxu) for SUMO1 becomes smaller upon binding to the SBM peptides. Shown are the pulling-speed-dependent unfolding forces of SUMO1 alone (triangles) and in the presence of S10 (solid squares) and S12 (open squares). The graph shows that SUMO1 is more dependent on the pulling speed in the presence of the SBM peptides. Dashed lines are Monte Carlo fits to the experimental data. SUMO1 in the presence of the SBM peptides has a Δxu of 0.25 nm, which is smaller than that of the ligand-free SUMO1 (0.51 nm). Values are expressed as the mean ± SE. The ku0and Δxu values are given in Table 2. The speed dependence data for ligand-free SUMO1 is taken from our earlier report (19).
In Monte Carlo simulations, the values of ku0 ∼ 3 × 10−3 s−1 and Δxu ∼ 0.25 nm best fit the experimental data of SUMO1 bound to S10 and ku0 ∼ 1.15 × 10−3 s−1 and Δxu ∼ 0.25 nm best fit that of SUMO1 bound to S12. In general, the ku0 depends on the magnitude of the unfolding force and Δxu depends on the variation of unfolding force with the pulling speed. Although the magnitude of the unfolding force differed for S10 and S12, the variation of unfolding force with pulling speed is similar, as revealed by the fact that Δxu is the same in both cases, but ku0 values are different. Δxu of SUMO1 complexed to the SBM peptides (0.25 nm) is much smaller compared to that of the ligand-free SUMO1 (0.51 nm). All values are listed in Table 2.
Table 2.
Kinetic parameters of SUMO1 and its complexes
| Protein | Δxu(nm) | ku0(s−1) | ΔG‡ (kBT)a | ks(N/m)a |
|---|---|---|---|---|
| SUMO1 | 0.51 | 1.15 × 10−5 | 32.1 | 1.0 |
| SUMO1+ S10 | 0.25 | 3 × 10−3 | 26.5 | 3.5 |
| SUMO1 + S12 | 0.25 | 1.5 × 10−3 | 27.2 | 3.6 |
Calculated using values obtained from Monte Carlo simulations.
A schematic energy landscape constructed from the values obtained through Monte Carlo simulations is shown in Fig. 6. The native state of SUMO1 bound to SBM peptides S10 and S12 is stabilized with respect to the ligand-free state, as revealed by the enthalpy values (16). The Δxu value decreases from 0.51 nm to 0.25 nm upon ligand binding, and interestingly, this decrease is the same for both the SBM peptides. The activation energy of unfolding is higher in the ligand-free state than in the bound state. The activation energies of SUMO1 bound to S10 (26.5 kBT) and S12 (27.2 kBT) are similar.
Figure 6.
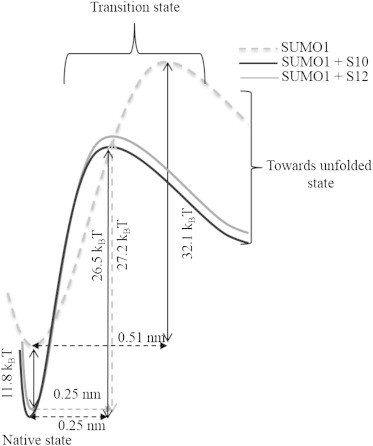
Schematic energy landscape of SUMO1 bound to SBM peptides S10 and S12. SUMO1 in its native state is stabilized by ligand binding. The distance to the unfolding transition state (Δxu) decreases from 0.51 nm to 0.25 nm upon ligand binding. The activation energies of unfolding are similar for SUMO1 in the presence of S10 and S12 but lower than the ligand-free SUMO1. The values of Δxu and ku0 used for this calculation are obtained from Monte Carlo simulations. The ΔG value for the native-state stabilization of SUMO1 upon binding S12 is an estimate from earlier studies (16), and S10 is assumed to have the same value in the above representation.
Effect of SBM peptide binding on the mechanical flexibility of SUMO1
To determine the effect of SBM peptide binding on the mechanical flexibility of SUMO1, the spring constant of the unfolding potential was calculated using harmonic approximation, as described in Materials and Methods. The spring constant (ks) of SUMO1 increased from 1 N/m to 3.5 N/m and 3.6 N/m upon binding to S10 and S12, respectively. This clearly shows that SUMO1 becomes stiffer upon binding to its target SBMs and that its flexibility might play a plausible role in locating these targets. Earlier studies have shown that ligand binding can increase the mechanical stability of proteins. Such increases have been shown, for example, for acylphosphatase upon binding to inorganic phosphate (6), β-adrenergic receptor upon binding to cholesterol (9), and GB1 upon binding to Ni2+ (8). However, in none of these proteins was there a tremendous change in their distance to the unfolding transition state (Δxu). For acylphosphatase and GB1, it has been shown that either the Δxu does not change or the change in Δxu is insignificant, although there were changes in ku0 values. The change in Δxu affects the flexibility of the protein, i.e., the spring constant of the unfolding potential (ks). Although the Δxu value of the β-adrenergic receptor decreases upon binding to cholesterol for certain structural elements, the change is not as significant as in the case of SUMO1 protein. SUMO1 shows a greater decrease in the mechanical flexibility (i.e., an increase in rigidity) upon binding to peptides than do many of the proteins reported in the literature. Serdiuk et al. also observed a greater change in the ks value of E. coli LacY upon ligand binding, but the ks of LacY, unlike that of SUMO1, decreased upon complexation, indicating an increase in its mechanical flexibility (30).
An important question at this stage might be how the enhancement in the mechanical stability and rigidity of SUMO1 is related to the ligand binding position or interface. In the structure of the complex of SUMO1 and S12 (Fig. 1), the peptide ligand binds in the deep hydrophobic groove between the α-helix and the β2-strand. The bound peptide interacts strongly with the hydrophobic and aromatic residues Glu-33, Ile-34, His-35, Phe-36, Val-38, Leu-47, and Tyr-51 present in the binding concave groove of the folded region of SUMO1. The binding groove is far from the experimental pulling geometry of the N- and C-termini and the mechanical clamp (the H-bonded β-strands from the termini). The mechanical clamp geometry is believed to be the major resistance and to contribute to the mechanical resistance in ubiquitin-like proteins (19,31). However, our earlier experiments showed that the residues far from the mechanical clamp would also contribute to the mechanical resistance (19). Hence, we directly see the ligand-binding effect at the hydrophobic interface between the helix and the β-strand through an enhancement of the unfolding force upon ligand binding. The results of this study further support the earlier hypothesis that residues far from the mechanical clamp geometry in proteins may directly influence protein mechanical stability and stiffness.
Biological significance of flexibility of SUMO1
The binding mechanism of noncovalent attachment of ligands to SUMO1 is quite different from that of ubiquitin. Ubiquitin binding motifs typically form helices and bind to the β-sheet of ubiquitin (13). On the other hand, in earlier studies, Song et al. discovered a novel mechanism of ligand binding to SUMO1, where the bound orientation of the ligand can reverse depending on the amino acid sequence (13). The SBM peptides S10 and S12 used in this study are two such sequences. Such reversal of orientation in ligand binding is not common in biological systems. This study interestingly shows that the flexibility of SUMO1 decreases upon binding to its target proteins/SBMs. It is likely that the structure of SUMO1 needs to be flexible, with high conformational entropy, to accommodate peptides in such different orientations. The conformational flexibility of SUMO1 is reduced once the ligand is bound, which in our experiments is measured as an increase in stiffness. Measuring the stiffness of overall protein or protein-ligand complexes could serve as a measure of the conformational entropy of the system. Furthermore, we can compare the stiffness of SUMO1 bound to S10 or S12 with that of ubiquitin (∼5.8 N/m), reported previously (19). It shows that after complexation to SBMs S10 and S12, SUMO1 is still not as rigid as ubiquitin and that the complex might become more rigid when it binds to other proteins in the SUMOylation-mediated protein interaction networks. It is also well known that SUMO1 binds to various target proteins and enzymes during the process of SUMOylation and that the binding sites on SUMO are not the same for all the targets. These target binding sites on SUMO are distributed in different regions of the protein surface (32–34). The flexibility and the associated conformational dynamics of SUMO might help it in interacting with its target proteins at various binding sites, and once the target is bound, the complexed SUMO becomes rigid for the subsequent sequence of events that occur during SUMOylation.
Conclusions
This study demonstrates the effect of small peptide binding on the mechanical stability and stiffness of SUMO1. The mechanical stability of SUMO1 increases by ∼35–50 pN upon binding to the SBM peptides. Peptide binding also decreases the flexibility of SUMO1 to a large extent, indicating that flexibility of ligand-free SUMO1 contributes to the protein’s ability to access various conformations before binding to a target for its SUMOylation.
Supporting Material
References
- 1.Sanchez-Ruiz J.M. Ligand effects on protein thermodynamic stability. Biophys. Chem. 2007;126:43–49. doi: 10.1016/j.bpc.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 2.Hu X., Li H. Force spectroscopy studies on protein-ligand interactions: a single protein mechanics perspective. FEBS Lett. 2014;588:3613–3620. doi: 10.1016/j.febslet.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Ramanujam V., Kotamarthi H.C., Ainavarapu S.R. Ca2+ binding enhanced mechanical stability of an archaeal crystallin. PLoS ONE. 2014;9:e94513. doi: 10.1371/journal.pone.0094513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kotamarthi H.C., Sharma R., Ainavarapu S.R. Multiple unfolding pathways of leucine binding protein (LBP) probed by single-molecule force spectroscopy (SMFS) J. Am. Chem. Soc. 2013;135:14768–14774. doi: 10.1021/ja406238q. [DOI] [PubMed] [Google Scholar]
- 5.Ainavarapu S.R., Li L., Fernandez J.M. Ligand binding modulates the mechanical stability of dihydrofolate reductase. Biophys. J. 2005;89:3337–3344. doi: 10.1529/biophysj.105.062034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arad-Haase G., Chuartzman S.G., Reich Z. Mechanical unfolding of acylphosphatase studied by single-molecule force spectroscopy and MD simulations. Biophys. J. 2010;99:238–247. doi: 10.1016/j.bpj.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertz M., Rief M. Ligand binding mechanics of maltose binding protein. J. Mol. Biol. 2009;393:1097–1105. doi: 10.1016/j.jmb.2009.08.066. [DOI] [PubMed] [Google Scholar]
- 8.Cao Y., Yoo T., Li H. Single molecule force spectroscopy reveals engineered metal chelation is a general approach to enhance mechanical stability of proteins. Proc. Natl. Acad. Sci. USA. 2008;105:11152–11157. doi: 10.1073/pnas.0803446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zocher M., Zhang C., Müller D.J. Cholesterol increases kinetic, energetic, and mechanical stability of the human β2-adrenergic receptor. Proc. Natl. Acad. Sci. USA. 2012;109:E3463–E3472. doi: 10.1073/pnas.1210373109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DuVall M.M., Gifford J.L., Herzog W. Altered mechanical properties of titin immunoglobulin domain 27 in the presence of calcium. Eur. Biophys. J. 2013;42:301–307. doi: 10.1007/s00249-012-0875-8. [DOI] [PubMed] [Google Scholar]
- 11.Hann E., Kirkpatrick N., Brockwell D.J. The effect of protein complexation on the mechanical stability of Im9. Biophys. J. 2007;92:L79–L81. doi: 10.1529/biophysj.106.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horejs C., Ristl R., Pum D. Single-molecule force spectroscopy reveals the individual mechanical unfolding pathways of a surface layer protein. J. Biol. Chem. 2011;286:27416–27424. doi: 10.1074/jbc.M111.251322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song J., Zhang Z., Chen Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 2005;280:40122–40129. doi: 10.1074/jbc.M507059200. [DOI] [PubMed] [Google Scholar]
- 14.Gareau J.R., Lima C.D. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat. Rev. Mol. Cell Biol. 2010;11:861–871. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geiss-Friedlander R., Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 16.Song J., Durrin L.K., Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. USA. 2004;101:14373–14378. doi: 10.1073/pnas.0403498101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seu C.S., Chen Y. Identification of SUMO-binding motifs by NMR. Methods Mol. Biol. 2009;497:121–138. doi: 10.1007/978-1-59745-566-4_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gareau J.R., Reverter D., Lima C.D. Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2. J. Biol. Chem. 2012;287:4740–4751. doi: 10.1074/jbc.M111.321141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kotamarthi H.C., Sharma R., Koti Ainavarapu S.R. Single-molecule studies on PolySUMO proteins reveal their mechanical flexibility. Biophys. J. 2013;104:2273–2281. doi: 10.1016/j.bpj.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carrion-Vazquez M., Oberhauser A.F., Fernandez J.M. Mechanical and chemical unfolding of a single protein: a comparison. Proc. Natl. Acad. Sci. USA. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aggarwal V., Kulothungan S.R., Ainavarapu S.R.K. Ligand-modulated parallel mechanical unfolding pathways of maltose-binding proteins. J. Biol. Chem. 2011;286:28056–28065. doi: 10.1074/jbc.M111.249045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Florin E.L., Rief M., Gaub H.E. Sensing specific molecular interactions with the atomic force microscope. Biosens. Bioelectron. 1995;10:895–901. [Google Scholar]
- 23.Bustamante C., Marko J.F., Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 24.Dietz H., Berkemeier F., Rief M. Anisotropic deformation response of single protein molecules. Proc. Natl. Acad. Sci. USA. 2006;103:12724–12728. doi: 10.1073/pnas.0602995103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schlierf M., Rief M. Temperature softening of a protein in single-molecule experiments. J. Mol. Biol. 2005;354:497–503. doi: 10.1016/j.jmb.2005.09.070. [DOI] [PubMed] [Google Scholar]
- 26.Xu A.J., Springer T.A. Calcium stabilizes the von Willebrand factor A2 domain by promoting refolding. Proc. Natl. Acad. Sci. USA. 2012;109:3742–3747. doi: 10.1073/pnas.1121261109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertz M., Wilmanns M., Rief M. The titin-telethonin complex is a directed, superstable molecular bond in the muscle Z-disk. Proc. Natl. Acad. Sci. USA. 2009;106:13307–13310. doi: 10.1073/pnas.0902312106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rief M., Fernandez J.M., Gaub H.E. Elastically coupled two-level systems as a model for biopolymer extensibility. Phys. Rev. Lett. 1998;81:4764–4767. [Google Scholar]
- 29.Rief M., Gautel M., Gaub H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 30.Serdiuk T., Madej M.G., Müller D.J. Substrate-induced changes in the structural properties of LacY. Proc. Natl. Acad. Sci. USA. 2014;111:E1571–E1580. doi: 10.1073/pnas.1404446111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carrion-Vazquez M., Li H., Fernandez J.M. The mechanical stability of ubiquitin is linkage dependent. Nat. Struct. Biol. 2003;10:738–743. doi: 10.1038/nsb965. [DOI] [PubMed] [Google Scholar]
- 32.Capili A.D., Lima C.D. Structure and analysis of a complex between SUMO and Ubc9 illustrates features of a conserved E2-Ubl interaction. J. Mol. Biol. 2007;369:608–618. doi: 10.1016/j.jmb.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baba D., Maita N., Shirakawa M. Crystal structure of thymine DNA glycosylase conjugated to SUMO-1. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 34.Shen L., Tatham M.H., Hay R.T. SUMO protease SENP1 induces isomerization of the scissile peptide bond. Nat. Struct. Mol. Biol. 2006;13:1069–1077. doi: 10.1038/nsmb1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


