Abstract
Upon endoplasmic reticulum Ca2+ store depletion, Orai channels in the plasma membrane are activated directly by endoplasmic reticulum-resident STIM proteins to generate the Ca2+-selective, Ca2+ release-activated Ca2+ (CRAC) current. After the molecular identification of Orai, a plethora of functional and biochemical studies sought to compare Orai homologs, determine their stoichiometry, identify structural domains responsible for the biophysical fingerprint of the CRAC current, identify the physiological functions, and investigate Orai homologs as potential therapeutic targets. Subsequently, the solved crystal structure of Drosophila Orai (dOrai) substantiated many findings from structure-function studies, but also revealed an unexpected hexameric structure. In this review, we explore Orai channels as elucidated by functional and biochemical studies, analyze the dOrai crystal structure and its implications for Orai channel function, and present newly available information from molecular dynamics simulations that shed light on Orai channel gating and permeation.
Main Text
In the ion channel field, store-operated channels have a relatively recent history, and have been investigated at the molecular level only within the past decade. First observed as a calcium signal within cells (1), then detected as an ionic current in the late 1980s (2,3), and much later associated with a pair of gene products in 2005–2006 (4–8), STIM and Orai proteins have captured wide interest as a two-component channel that links the endoplasmic reticulum (ER) to the plasma membrane (PM), with broad implications for channel biophysics, cell biology, physiology, and drug development. In store-operated calcium entry, extracellular calcium ions (Ca2+) enter into cells in response to ER Ca2+ store depletion. Once inside the cell, Ca2+ serves as a second messenger to regulate signaling cascades that result in cytoskeletal rearrangement, secretion, gene transcription, alterations in motility, and cell proliferation. The best-characterized store-operated channel to date is the Ca2+ release-activated Ca2+ (CRAC) channel, which is endogenously expressed in T lymphocytes and other cells of the immune system. Sustained or oscillatory Ca2+ signaling mediated by CRAC current influx over a period of hours after T-cell receptor engagement with antigen initiates the immune response by triggering downstream changes in gene expression that are required for cytokine production, cell proliferation, and differentiation of T cells into effector subsets (9,10).
A series of RNAi screens—four using Drosophila S2 cells and one using HeLa cells—led to the identification of the two key proteins, STIM and Orai, that together form the CRAC channel (4–8). STIM proteins had been identified previously and named stromal interacting molecules (STIM), but the link to Ca2+ signaling was uncovered by these RNAi screens in combination with functional studies. Orai (also known as CRACM), however, was completely novel. RNAi knockdown of either STIM or Orai completely suppressed functional CRAC current, establishing a functional requirement for each protein. Overexpression of STIM and Orai together yielded greatly increased currents with the same biophysical properties as native CRAC current, indicating that no other protein component is limiting up to a very high channel density (8,11–13). STIM proteins are single-pass ER transmembrane proteins that sense ER luminal Ca2+ store depletion and physically relay the message by translocating to ER-PM junctions (4,14,15). Orai proteins in the PM then bind to ER STIM and accumulate in puncta by a diffusion trap (16–18). Orai is a four-transmembrane (TM)-spanning, pore-forming subunit of the CRAC channel that opens to permit Ca2+ influx upon physical interaction with STIM. There are two mammalian homologs of STIM, i.e., STIM1 and STIM2; and three mammalian homologs of Orai, namely, Orai1, Orai2, and Orai3; these are distributed widely throughout the tissues of the body, with selected tissues exhibiting elevated expression (9,19).
With the STIM and Orai proteins identified, the Ca2+ signaling field exploded into activity with work on several fronts including channel structure-function, cell physiology, mouse knockouts, biomedical studies on patients, and drug development. After the initial description of STIM as the missing link between the ER and the PM, and Orai as the pore-forming subunit of store-operated Ca2+ signaling (4–8,14,20–22) in 2005 and 2006, respectively, more than 850 articles on STIM1 and more than 750 on Orai1 have appeared. Fig. 1 A shows a diagram of a human Orai1 subunit. Within its 301 amino-acid primary sequence, several functionally important residues are highlighted. The vast majority of structure-function studies have focused on Orai1; however, due to high sequence homology, many of the resulting structural insights have been applied to other Orai homologs. Within the core domain of four TM-spanning segments, the pore-lining TM1 helix is identical in all three Orais, with the extracellular loop between TM3 and TM4 being the most divergent; and the 3-4 loop of Orai1 is uniquely glycosylated. Here, we review a subset of structure-function studies and note the contributions they have made toward a molecular understanding of the biophysical properties of Orai channel gating and ion selectivity that underlie the CRAC current.
Figure 1.
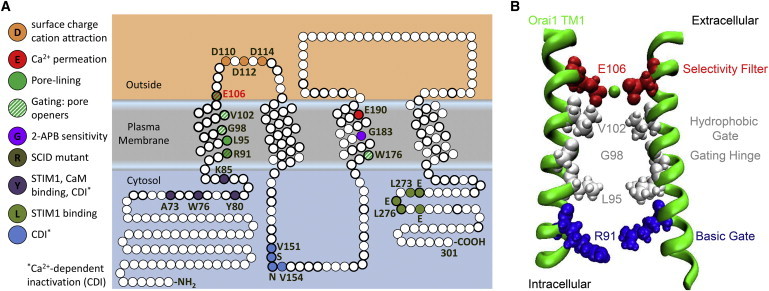
Orai1 structure-function mapping. (A) Annotated sequence of Orai1. (Circles) Residues; (bold) conservation in the three human Orai channels. Color-coded channel functions defined by mutational analysis (discussed in text) are highlighted from N- to C-terminus: N-terminal STIM1 and CaM binding; Ca2+-dependent inactivation (CDI); mutation that causes human SCID; constitutively active channel mutants; Ca2+ permeation; cation electrostatic attraction; second CDI site; TM3 residues that contribute to permeation and gating; and C-terminal STIM1 binding. (B) TM1 residues lining the Orai1 store-operated pore elucidated by functional analysis: selectivity filter E106, hydrophobic gate V102, gating hinge G98, L95, and basic gate R91. For clarity, only two TM1 domains, from two Orai1 monomers, are represented.
Structure-function studies
Before the Orai family was identified and cloned, the endogenous CRAC channel had already been shown to have unusual properties, including gating by ER Ca2+ store depletion, a very low (for an ion channel) single-channel conductance of ∼10 fS, a very high degree of Ca2+ selectivity (>1000-fold over Na+), conduction of small monovalent cations in the absence of divalents, and inward rectification (16,17). Identification of the Orai gene family opened the door to classic electrophysiological approaches with mutagenesis in expression systems, aimed at identifying the molecular basis for the CRAC channel’s biophysical fingerprint. Orai is unrelated to previously identified ion channels, and its activation by STIM proteins in the ER is highly unusual. At the level of channel function, site-directed mutagenesis and thiol-reactive probes in the context of patch-clamp and fluorometric Ca2+ imaging experiments have identified amino-acid residues that contribute to ion selectivity and gating of Orai1 and other Orai homologs (Fig. 1).
Ion selectivity and Orai1 pore-lining residues
Three independent studies identified a glutamate residue situated at the extracellular end of TM1 as the most important determinant of Ca2+ selectivity (20–22). All three studies showed that a conservative point mutation, glutamate to aspartate (E180 in Drosophila Orai, E106 in Orai1 and E81 in Orai3; see Fig. 1), drastically altered ion selectivity as determined by patch-clamp analysis, strongly implicating Orai as the pore-forming subunit of the CRAC channel. The normal inwardly rectifying, highly selective Ca2+ channel instead conducted outwardly rectified, nonselective, primarily monovalent currents. Furthermore, these studies also showed that substitution of the critical glutamate by an uncharged amino acid resulted in a nonconducting, dominant-negative subunit capable of strongly suppressing endogenous CRAC current. Negatively charged aspartates in the first extracellular loop near the glutamate filter residue may fine-tune Ca2+ selectivity by locally attracting external cations toward the pore (21,22).
In addition, a glutamate situated at the extracellular side of TM3, Orai1 E190, was also shown to increase selectivity for Ca2+ over monovalent cations (20,21). Based on these three studies (20–22), it was hypothesized that the selectivity filter consists of a ring of glutamates (E106 in Orai1) that form a pore with diameter close to 3.5 Å, i.e., near the size cutoff for Cs+. In this view, mutation of the key glutamate residue (E106 in Orai1) to aspartate (one carbon shorter than glutamate) would widen the pore and increase permeability to larger monovalent cations such as Cs+. Subsequent studies have confirmed that Orai3 residue E81, homologous to Orai1 E106, also controls Ca2+ selectivity in Orai3 (23,24).
Two comprehensive cysteine-scanning mutagenesis studies, each focused on Orai1, defined the entire pore to be lined by TM1 residues. McNally et al. (25) systematically mutated every residue in TM1, the extracellular TM1-TM2 loop, and TM3 of Orai1 to cysteine and tested the mutants for reactivity with cadmium ions (Cd2+), reported by current block during patch-clamp experiments. They found that particular TM1 residues, when mutated to cysteine, were sensitive to Cd2+ block. These residues followed a periodic pattern that, when mapped to a generic α-helical projection, aligned to one face of the α-helix. In the second study, Zhou et al. (26) mutated every residue in TM1 and TM3 of Orai1 to cysteine and took a biochemical approach to identifying residues involved in pore formation, with formation of intersubunit disulfide bridges as a readout. Taken together, the two cysteine-scanning studies predicted TM1 to be α-helical, with residues E106, V102, G98, L95, and R91 all on one side of the α-helix, lining the pore formed by multiple TM1 domains gathered about a central axis.
Gating by STIM1
STIM1 directly activates Orai1 by binding to C- and N- terminal cytosolic domains. Deletions and truncations of STIM1 zeroed in on a region of ∼100 amino acids as the essential CRAC-activation domain (CAD; also known as STIM-Orai activating region, SOAR) (27,28). As shown by FRET and patch-clamp studies, the CAD domain allows STIM1 to bind and activate Orai1 through the C-terminal cytosolic domain of Orai1 by interactions within a leucine-rich, acidic coiled-coil motif (27,29–32). A second site at the conserved portion of the N-terminus adjacent to TM1 is also required for CAD/STIM1 binding and Orai1 channel activity (27,33–36). STIM interacts strongly with the C-termini of Orai proteins and weakly with the N-termini (27,37). Although it is clear that both STIM1-binding sites are required for channel opening, the precise sequence of events involving first the Orai1 C-terminus and then the N-terminus is still under investigation.
A human Orai1 channelopathy opened the door for further functional analysis of residues lining the Orai1 pore and the possible location of channel gates. Arginine 91 of Orai1 mutated to tryptophan causes a lethal severe combined immune deficiency (SCID) (6), and is localized toward the intracellular side of Orai1 TM1. Implicated in pore formation, R91 may also be involved in gating because mutation to bulky hydrophobic residues, including the channelopathy mutation R91W, blocked the channel, whereas mutation to hydrophilic residues allowed the channel to function normally (38,39). R91 is one of several arginines that collectively may form an electrostatic gate toward the cytosolic portion of TM1. The glycine at position 98 in the middle of TM1 was proposed to be a gating hinge (39), permitting the channel’s STIM1-induced conformational changes to open the channel by widening the cytosolic gate. This notion was supported by the first-known activating mutations G98D and G98P, which resulted in constitutively preactivated, nonselective Orai1 channels. Substitution of alanine at G98, but not in adjacent residues, resulted in an appropriately expressed but nonfunctional channel.
The cysteine-oxidizing agent diamide blocked the R91C mutant through an intersubunit disulfide bond, but the double mutant R91C/G98D was not capable of being blocked. In addition, the double mutant R91W/G98D was constitutively open, suggesting that widening induced by mutation of G98 overcame block that would normally be induced by R91W. These results suggested that the flexibility of glycine permits the channel to open and close in response to STIM1 binding and unbinding, and that pore-lining R91 residues from adjacent subunits function as a physical gate that widens the inner pore as basic residues pivot away from each other during channel opening (39). The valine at position 102 of Orai1, situated between the glycine hinge and the selectivity filter glutamate, was also proposed to function as a gate (40), in part because mutating it to more polar residues resulted in preactivated, nonselective channels. Furthermore, STIM1 binding to the constitutively open nonselective V102C restored selectivity for Ca2+.
Although the picture remains uncertain, the conclusion of these studies is that Orai1 TM1 not only lines the conducting pore but includes two activation gates that flank a central glycine hinge: a hydrophobic gate at V102, and an inner gate at R91 (Fig. 1 B).
As has been previously noted (41), TM1 and TM3 have an internal homology suggestive of evolution by gene duplication. Whereas cysteine-scanning studies excluded TM3 from pore-formation (25), other functional studies found that mutating certain TM3 residues also could alter permeation and gating properties of Orai channels. The conserved TM3 glutamates (E190 in Orai1) were shown to contribute to Ca2+ selectivity; neutralizing mutations to alanine or glutamine resulted in increased Cs+ permeability (in Orai1) or decreased sensitivity to the channel modulator 2-APB (in Orai3) (20,21,23,42). The conserved TM3 glycines (G183 in Orai1) are also necessary for proper channel gating (43,44). Mutating the conserved Orai1 TM3 tryptophan (W176 in Orai1) to cysteine also results in preactivated channels (44).
Based upon these findings, we propose the following picture for how TM1 and TM3 may coordinate: TM1-2 loop aspartates create a ring of negative charges that attract a cloud of Ca2+ ions toward the critical glutamate E106 ring that forms the selectivity filter. Flanking TM1 hydrophobic residues form a large energetic barrier that needs to be overcome for ions to permeate. Glycines in the middle of TM1 form a gating hinge; and arginine (TM1) or tryptophan (TM3) may form an intracellular gate toward the inside. The parallels in TM1 and TM3 mutagenesis results suggest coordinated movement during channel activation by STIM1.
Calcium-dependent inactivation
Classic CRAC current is in part defined by fast Ca2+-dependent inactivation (CDI), a process that occurs in tens of milliseconds and is mediated by Ca2+ influx into the cytosol (45,46). Of the three human Orai homologs, Orai3 exhibits the greatest degree of CDI, and Orai1 the least (47). CDI is thought to be mediated by cytosolic domains of both STIM and Orai and modulated by the overall expression ratio of the two proteins. Neutralization of several C-terminal STIM1 acidic residues (475–483) outside of the CAD diminished or completely abolished CDI (48–50). The N-terminal, PM-proximal region of Orai1 and Orai3 channels has been identified as a calmodulin-binding domain, which mediates interactions with calmodulin and thereby affects CDI. Mutations within the Orai1 (e.g., A73E; W76A, E, or S; and Y80E) or Orai3 calmodulin-binding domain eliminate CDI (49,51). Mutations introduced into the cytosolic TM2-TM3 loop of Orai1 (residues 151–154) abolish CDI (52), and decreasing the ratio of STIM1 to Orai1, diminishes CDI (53,54).
STIM-independent gating of Orai3 by 2-APB
2-APB (2-aminoethyl diphenylborinate) has been previously described as a CRAC channel modulator that augments channel activity at low micromolar concentrations and blocks at higher concentrations (55). Of the three human Orai channel homologs, Orai3 and, to a lesser extent, Orai1 can be activated by high concentrations (>50 μM) of 2-APB (56–58). 2-APB-activation of Orai3 occurs without STIM1-Orai3 interaction or store-depletion and is thought to be mediated by residues within the Orai3 TM2-TM3 region (43,58). The mechanism by which 2-APB activates Orai3 involves pore dilation, resulting in nonselective cation currents, a biphasic I-V shape, larger single-channel conductance, and wider pore diameters (23,24,43,56–58). TM3 residues are likely to be critical for 2-APB-induced pore dilation. Mutation G183A in TM3 of Orai1 results in Orai1 channels that can also be activated by 2-APB; mutating the homologous G158 in TM3 of Orai3 to cysteine results in delayed 2-APB activation and washout due to intrasubunit disulfide bridge formation between the introduced G158C and endogenous TM2 C101 (43).
Finally, a very recent targeted cysteine mutagenesis study on 2-APB-activated Orai3 showed that TM1 residues Q83, V77, and L70 line the channel pore. Other cysteine mutants of TM1 residues (E81, G73, and R66) homologous to Orai1 pore-lining residues made Orai3 channels insensitive to 2-APB-activation. Orai3 TM3 residue E165 was found to only assist in pore formation of the 2-APB-activated, but not store-operated, Orai3 channel (59). These data suggest that the store-operated (i.e., STIM-activated) Orai channel conduction pathway differs from that of the 2-APB-activated Orai3 channel pore. The STIM-activated Orai channel conduction pathway is solely composed of TM1 residues. However, the 2-APB-activated Orai3 channel pore forms with assistance from TM3 residues, namely E165 and G158. The wider pore of 2-APB-activated Orai3 might facilitate the interaction between E165 and G158 with the conduction pathway and the ions that fill it.
Orai subunit stoichiometry
Ion channels are homomeric or heteromeric assemblies of protein subunits that form ion conduction pathways through the otherwise very hydrophobic barrier of cell membranes. Examples of channel dimers, trimers, tetramers (or domain tetramers in the case of single-subunit voltage-gated Na+ and Ca2+ channels), pentamers, and hexamers have been described. After the identification of genes encoding members of the Orai family, researchers began to employ biochemical, functional, and imaging techniques to determine the stoichiometric makeup of the Orai channel pore. To investigate Orai channel stoichiometry, tandem dimeric, trimeric, or tetrameric Orai1 constructs with or without pore-blocking dominant-negative mutations were coexpressed together with STIM1 (60). Current suppression in tetramers with a single subunit pore mutation suggested that store-operated Orai1 channels are tetrameric.
Other studies utilized fluorescently labeled Orai multimers or monomers, and in conjunction with total internal reflection fluorescence microscopy, counted numbers of photobleaching steps of single molecular entities in individually resolved diffraction-limited spots. Three studies using this technique, termed subunit counting, agreed that Drosophila Orai (dOrai), Orai1, and Orai3 each formed tetrameric complexes when activated by STIM proteins (61–63). However, there was disagreement about the resting state. One group observed dimeric complexes of dOrai, Orai1, and Orai3 when expressed alone (61,63). A different optical approach tracked single molecules, and analyzed the brightness of EGFP-Orai diffusing into prebleached regions of live HEK cells. That study found support for a tetrameric Orai1 channel complex at rest (64). All the above studies are in agreement on a tetrameric active form of Orai, although reports of Orai dimers and heteromeric Orai channels suggested a broader versatility in subunit organization. Arachidonate-regulated Ca2+ (ARC) channels were described as Ca2+-selective, store-independent, pentameric assemblies of three Orai1 and two Orai3 subunits (65), and complexes of Orai1 with TRPC channels have been reported (66).
The tetrameric subunit stoichiometry determined by concatemers and optical approaches was called into question by publication of the crystal structure of dOrai, which clearly showed a hexameric closed state with three Orai dimeric units organized around a central pore (67). A subsequent study compared the biophysical properties of tetrameric and hexameric concatenated Orai1 subunits, and reported that only the tetrameric assembly recapitulated the Ca2+-selectivity of endogenous CRAC channels, whereas the hexameric complex generated nonselective cationic currents (68). The authors interpreted this to imply that Orai channels may natively exist as tetramers, forming CRAC channels with high selectivity for Ca2+. However, another very recent study (69) utilized hexameric concatemers of Orai1 and introduced mutations to prevent STIM1 binding into individual subunits, concluding that Orai1 hexamers recapitulate the Ca2+-selectivity of native CRAC current.
Each method of determining subunit stoichiometry has limitations that could lead to disparate results. Tandem concatemers with artificial linkers between subunits might not assemble as anticipated. Photobleaching and dark states of fluorescent tags may result in undercounting of subunits, and subunit counting studies are done at low expression levels. At the opposite extreme of very high protein concentration, crystal structures are static representations determined, of necessity, under nonphysiological conditions. One way to reconcile some of the findings would be to suppose that newly synthesized Orai subunits are dimeric and are organized by the STIM protein into tetramers and hexamers (trimers of dimers). Subunit stoichiometry and assembly of Orai channels remain topics of lively debate; the weight of recent evidence is shifting to favor a hexameric state for the functional channel.
The dOrai crystal structure
The culmination of functional and biochemical studies discussed above painted a picture of what the pore looks like—a concentric arrangement of Orai1 TM1 domains, with residues E106, V102, G98, L95, R91, and A88 protruding into the central cavity, which forms a narrow conduction pathway traversed mainly by Ca2+ ions as they enter the cell (Fig. 1 B). These studies also unveiled the functional identities of these residues: E106 as the selectivity filter, V102 as the hydrophobic gate, G98 as the gating hinge, and R91 as the basic gate. These associations, based on functional analysis of site-directed mutants, were largely substantiated by the solved crystal structure of dOrai (67). The crystal structure of dOrai (in a closed state), although validating some functional conclusions, also contained surprises.
Drosophila dOrai shares 73% sequence identity with Orai1 and has nearly identical biophysical properties (70). The four TM domains of dOrai are almost identical to the three human homologs—Orai1, Orai2, and Orai3. Therefore, the closed state crystal structure of truncated (residues 132–341) and mutated (C224S, P276R, P277R, and C283T) dOrai, solved to 3.35 Å resolution, is the best structural representation presently available (67). The structure lacks substantial parts of the N-and C termini, the extracellular TM1-TM2 loop, and the intracellular TM2-TM3 loop. However, all four TM domains are resolved and the conduction pathway is clearly defined (Fig. 2). Therefore, the dOrai crystal structure gives insight into possible gating mechanisms, hypothetical conformational changes that might occur as the channel opens/closes, and possible STIM binding sites.
Figure 2.
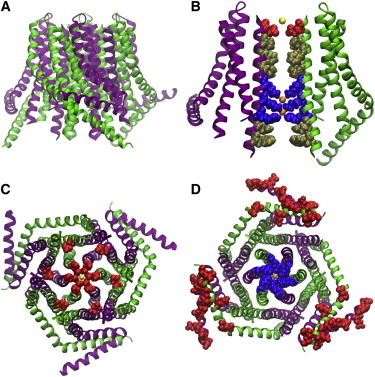
The dOrai crystal structure. (A) Side view showing hexameric assembly composed three α-subunits (green) with TM4 extended and three β-subunits (purple) with TM4 bent. (B) Pore view showing nine residues that line the conduction pathway, 55 Å in length. For clarity, only two of the six subunits are represented. (Red) Selectivity filter residue E178; (yellow) bound Ca2+; (gold) five hydrophobic residues; and (blue) basic residues, which bind two unidentified anions (orange). (C) View from the extracellular side showing concentric circles of α-helices with two rings of glutamates (red): the inner ring of TM1 E178 residues (corresponding to Orai1 E106) surrounding a Ca2+ ion at the center of the pore, and the outer ring formed by TM3 E262 residues (corresponding to Orai1 E190). (D) View from the intracellular side showing TM1 residues with positive charge (blue) at the center surrounding the anion (orange) in the pore, flanked by M4 extensions with three red stripes of negative charge at the C-terminal STIM-binding sites.
The dOrai crystal structure reveals a hexameric assembly of subunits. The subunits are symmetrically arranged as a trimer of dimers around the central axis of the channel that forms the pore (Fig. 2, A and B). Each subunit is composed of four α-helical TM domains (M1–M4) and an M4 extension that protrudes into the cytosol. The disposition of the M4 extension defines the two conformations available to each of the subunits in the structure; α-subunits have M4 extensions that extend deep into the cytosol, whereas β-subunits have bent M4 extensions that hover just beneath the intracellular side of the PM. Each one of the three dimers that comprise the hexamer is composed of one α- and one β-subunit; α- and β-subunits alternate within the hexameric assembly. The M4 extensions of each α- and β-subunit within a dimer are paired in an antiparallel, coiled-coil helical arrangement.
The view of the structure from the extracellular side is defined by three concentric rings of TM domains (Fig. 2 C). The innermost ring, composed of M1 domains, forms the conduction pathway of the channel. The middle ring, composed of M2 and M3 domains, provides rigidity and thus structural integrity to the hexameric complex. The outermost concentric ring, composed of M4 and M4 extension domains, is the most peripheral ring and therefore likely to be the most mobile. Both the inner and middle concentric circles of the structure exhibit sixfold symmetry, while the outer ring exhibits threefold symmetry.
Orai pore
The six M1 domains are surprisingly long; at ∼55 Å apiece, each protrudes ∼20–25 Å deep into the cytosol, forming what appears to be an extended pore. All six of the M1 domains contribute residues to the formation of the conduction pathway along the central axis of the channel. The side chains of nine residues homologous to Orai1 E106, V102, F99, L95, R91, K87, R83, Y80, and W76 protrude into the pore (Fig. 2 B). The pore can be subdivided into four distinct regions: 1) the selectivity filter—a ring of glutamates; 2) the hydrophobic region spanning three α-helical turns; 3) the basic region spanning three α-helical turns; and 4) the cytosolic region spanning two α-helical turns.
At ∼6 Å in diameter, the selectivity filter glutamate ring (homologous to Orai1 E106) is the narrowest part of the pore, and is surrounded by a ring of TM3 glutamate residues, homologous with Orai1 E190 (Fig. 2 C). Moreover, the unresolved TM1-TM2 loop with three negatively charged residues likely contributes to a negative electrostatic potential at the external surface of the pore. The hydrophobic region is rigid, stabilized by an extensive network of van der Waals interactions, and contains V174 (homologous to the Orai1 hydrophobic V102 gate). It measures 8–10 Å in diameter and is ∼18 Å long. The basic region of the pore contributes 18 basic residues to the conduction pathway, including K163 (homologous to the aforementioned basic R91 gate of Orai1 channels). These basic residues surround what appears to be an anion bound to the crystal (Fig. 2 D).
The basic region is enriched with residues that are associated with helix flexibility (serine, threonine, and glycine). G170, a residue homologous to the G98 gating hinge identified in Orai1 channels, is also nearby. The rigidity of the selectivity filter and hydrophobic regions, and the relative flexibility of the basic region and proposed gating hinge, suggest a plausible mechanism for channel gating: as the channel goes from the closed to open state, M1 helices in the basic region move apart from one another, thereby opening the channel pore. Additionally, the anion bound within the basic region may act in gating as well. A complementary dOrai K163W structure (a mutation homologous to Orai1 SCID mutation R91W) exhibits altered anionic binding within the basic region. This further suggests that the basic region of the channel is in part responsible for channel gating and that an unidentified physiological anionic species such as phosphate might be responsible for stabilizing the closed state of the channel.
STIM interactions and gating
The dOrai crystal structure also offers insight into the plausible binding sites for STIM molecules. As mentioned, the M4 extensions from adjacent α- and β-subunits within each of the three dimers in the hexameric structure are paired by antiparallel coiled-coil helix associations. The coiled-coil interactions occur between residues I316 and L319. These residues are homologous to Orai1 L273 and L276; mutation of these two residues disrupts STIM1-Orai1 interaction and inhibits Orai1 channel opening (31,71,72). Therefore, Hou et al. (67) proposed that STIM binds the M4-extensions. The multimerization state of STIM is uncertain. STIM is likely to be a dimer in resting cells when ER Ca2+ stores are full (17). After store depletion, STIM1 forms higher-order oligomers that physically migrate laterally within the ER membrane to ER/PM junctions; STIM binding then causes Orai channels to open. An NMR structure of a STIM1 fragment bound to Orai1 suggests a 1:1 molecular interaction (73). However, functional experiments in which the ratio of STIM/Orai is varied suggest that binding stoichiometry is variable, and that channel activity is graded accordingly (53,54,72). Increasing the ratio of STIM to Orai first increases and then drastically inhibits channel activity. A ratio of two STIM1 monomers per Orai1 subunit most closely recapitulates properties of endogenous CRAC currents and results in optimal channel activity.
Thus, to achieve maximal channel activation, it appears that an Orai hexamer may require binding of up to 12 STIM monomers. It is likely that graded activation takes place through a complex gating sequence involving varying numbers of STIM proteins bound, but the vanishingly small single-channel current has precluded direct measurement. The C-terminal Orai interaction with STIM then results in a conformational change that allows M4-extensions to protrude further into the cytosol and enables STIM to secondarily interact with cytosolic portions of the M1 helices. STIM interaction with the M1 helices in turn allows the M1 helices to move outward, thereby opening the channel pore.
Molecular dynamics
The dOrai crystal structure has enabled atomic-level computational studies of interactions among water molecules, ions, and the membrane-embedded Orai channel. The first atomistic molecular dynamics (MD) simulation study of dOrai revealed a possible gating mechanism in which water molecules within the pore contribute to channel activation in both the wild-type (WT) and constitutively active V174A mutant, homologous to Orai1 V102A (74). The authors of the study characterize the binding of Na+ (in the absence of Ca2+) by calculating the potential of mean force for the translocation of a single Na+ through the pore along the transmembrane direction. The potentials of mean force suggested that the V174A mutant channel has a more favorable permeation pathway than the WT channel, consistent with functional data showing that V174A channels are preactivated (67).
The V174A mutation alters the number of water molecules present in the pore, thereby reshaping the local electrostatic field in the pore region from A174 to R155 (74), suggesting that channel activation may be regulated by internal waters. However, the authors acknowledged that this water-regulated pore model may not represent the general mechanism for Orai channel activation, and that the role of pore waters in Orai channel gating has yet to be confirmed experimentally. The study also did not focus on STIM gating of Orai channels, leaving unclear the specific mechanism by which STIM molecules open the Orai pore. Finally, although Cl− counterions were observed to bind exclusively in the basic region of the pore, the role of the putative anion binding site revealed by the dOrai crystal structure in channel gating and conduction remains elusive.
Hou et al. (67) suggested that phosphate or pyrophosphate may be the counterion bound in the anion binding region. We speculate that phosphates assist in neutralizing the basic region of the pore, thereby stabilizing the closed state of the channel. To explore this possibility, we (M.L.W., J.A.F., and D.J.T.) ran atomistic MD simulations of the WT and V174A mutant channels embedded in a POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) bilayer with excess water and either Gd3+ or Ca2+ in the selectivity filter (SF) (75). Snapshots of the system are shown in Fig. 3, A and B. We used PHYRE2 (76) to build in the extracellular TM1-TM2 and intracellular TM2-TM3 loops that were missing in the crystal structure. Additionally, we reverted crystallization-related mutations to ensure that our simulations were representative of the WT dOrai channel. The titratable residues K159, K163, K170, E178, and D184 were neutralized based on pKa values calculated using PROPKA 3.1 (77). Finally, we included phosphates in the anion binding region suggested by the crystal structure.
Figure 3.
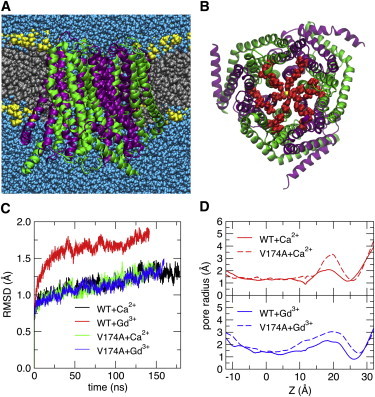
MD simulation of wild-type dOrai. (A) Configuration snapshot showing dOrai in secondary structure representation colored by subunit as in Fig. 2, with lipid tails (silver), lipid headgroups (yellow), and water (blue) shown in solid-sphere representation. (B) Extracellular view of dOrai with the water and lipids removed for clarity. In addition to the glutamate rings in TM1 (E178) and TM3 (E262) observed in the crystal structure, the full sequence shows another set of concentric acidic side chains formed by D182 and D184 in the TM1-TM2 connecting loop. (C) Cα RMSD from the initial configuration. Most simulation systems reached a steady state after ∼120 ns; WT+Gd3+ required only ∼50 ns. (D) Pore radius profiles of WT dOrai and the V174A mutant with Ca2+ or Gd3+ bound in the selectivity filter. The coordinate Z is defined as the distance (along the TM direction) from the center-of-mass for TM1 residues 144–180. The local minimum at Z ∼25 Å corresponds to the position of the E178 carboxyl groups, and the maximum at Z ∼20 Å corresponds to the position of the residue 174 side chain.
We found that one cation (either Ca2+ or Gd3+) in the SF and two phosphate ions in the basic region of the pore yielded a stable structure in all four systems; the Cα-root-mean-squared deviations (RMSDs) (with respect to the crystal structure) appear to be converging to values <2 Å within the first ∼120 ns of each simulation (Fig. 3 C). This is in contrast with the simulations by Dong et al. (74) of WT and V174A dOrai channels, which required 300 and 500 ns to reach Cα-RMSD plateau values of ∼3 and ∼3.5 Å, respectively, with Na+ as the permeant ion and Cl– as the anion. The fact that the structural integrity of the channel depends upon the choice of counteranion and protonation state of titratable residues in the pore region suggests that changes in the local pH around the channel may play an important role in stabilizing or destabilizing the closed state by changing the charge distribution in the pore region, and lends support to the possibility, mentioned above, that STIM binding may aid in releasing the anion from the basic region of the Orai pore.
To explore the effects of the Gd3+ pore blocker on the protein, we calculated pore radius profiles in the MD simulations of WT and V174A mutant channels with Ca2+ or Gd3+ bound to the E178 residues in the SF. The pore radius profile of the V174A system exhibits an extracellular opening when Ca2+ is in the selectivity filter that is not present in the WT system (Fig. 3 D). This is consistent with functional data showing that V174A channels are constitutively active, while WT channels are closed (as is the dOrai crystal structure). We find that the Gd3+-bound V174A pore has a smaller maximum radius than the Gd3+-bound WT pore, and that Gd3+ results in a smaller pore radius at the WT SF than when Ca2+ is bound (Fig. 3 D) (67). This is consistent with functional data showing that Gd3+ is a strong pore blocker (22,70).
The TM1-TM2 connecting loop, unresolved in the crystal structure but modeled here, positions an outer ring of acidic side chains concentric with the E178 selectivity filter (Fig. 3 B) that may serve as an extracellular vestibule to attract permeating cations or trivalent blockers. Two of the aspartate residues (D182 and D184) were shown in functional studies to be important for fine-tuning ion selectivity and for enhancing block by Gd3+ (21,22). Subtle tuning of the local environment by pH and electrostatics clearly affects the stability of the protein and geometric shape of the pore. Our simulations, in conjunction with those of Dong et al. (74), begin to shed light on the atomistic mechanisms underlying ion conduction, channel block, and pH regulation in the CRAC channel.
Conclusions
The Orai channels are late bloomers in the channel field, with a nearly 30-year discovery time course from store-operated calcium entry and CRAC current to STIM1-operated Orai1 channels. With one crystal structure (dOrai in a closed state) thus far, and a host of studies that probe Orai1 channel function by mutagenesis, the mechanistic picture is rather murky but becoming clearer. The crystal structure and a very recent functional study using concatemers provide strong evidence for a hexameric subunit organization as a trimer of dimers; however, the possibility of a versatile subunit organization should be kept in mind. Gating by STIM1 is a complex process, because varying numbers of STIM1 molecules bind to both C- and N-termini, possibly triggering the channel to open to several conducting substates. The pore-forming TM1 segment of Orai1 is critical for both gating and ion permeation. Selectivity for Ca2+ is determined by the glutamate ring at the outer pore. A gantlet of hydrophobic residues extending through the membrane likely limits the single-channel Ca2+ flux to <10,000 ions per s.
An electrostatic barrier in the inner pore extends into the cytoplasm in the closed state crystal structure; this suggests a major rearrangement is required to widen the inner pore during channel opening, such as TM1 and TM3 pivoting outward from the gating hinge in the middle by a scissoring action of STIM1 dimers exerted from the cytosolic ends of TM1 and TM4. STIM-operated, ligand-gated, and constitutively active Orai channels are proving useful in functional mapping of diverse channel open states with varying permeation properties. A clearer picture will emerge from crystal structures of the open-state channel induced by cocrystallization with STIM1, CAD/SOAR, and/or 2-APB. Complementing this, further MD simulations will reveal the conformational changes that occur as Orai channels close, open, and inactivate, together with atomic-scale details of ion conduction and selectivity.
Taken together, functional studies reaching to single-molecule resolution, x-ray crystallography, and molecular dynamics will ultimately provide a better understanding of the biophysical basis for Orai channel function, as well as the basis for rational drug design of channel blockers and activators.
Acknowledgments
We thank Dr. Joseph Dynes for insightful comments on the manuscript. We apologize to authors whose important contributions could not be covered due to space limitations.
This work used XSEDE high-performance computing resources, provided by the Texas Advanced Computing Center at The University of Texas at Austin, which was supported by the National Science Foundation. This work is supported by National Institutes of Health grant No. NS14609 (to M.D.C.), grant No. GM048071 (to I.P.), and grant No. GM86685 (to D.J.T.). M.L.W. is grateful for support from the University of California-Irvine Medical Scientist Training Program and National Cancer Institute fellowship No. F30CA171717.
References
- 1.Putney J.W., Jr. A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 2.Penner R., Matthews G., Neher E. Regulation of calcium influx by second messengers in rat mast cells. Nature. 1988;334:499–504. doi: 10.1038/334499a0. [DOI] [PubMed] [Google Scholar]
- 3.Lewis R.S., Cahalan M.D. Mitogen-induced oscillations of cytosolic Ca2+ and transmembrane Ca2+ current in human leukemic T cells. Cell Regul. 1989;1:99–112. doi: 10.1091/mbc.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liou J., Kim M.L., Meyer T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roos J., DiGregorio P.J., Stauderman K.A. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feske S., Gwack Y., Rao A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 7.Vig M., Peinelt C., Kinet J.P. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S.L., Yeromin A.V., Cahalan M.D. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc. Natl. Acad. Sci. USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feske S., Skolnik E.Y., Prakriya M. Ion channels and transporters in lymphocyte function and immunity. Nat. Rev. Immunol. 2012;12:532–547. doi: 10.1038/nri3233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cahalan M.D., Chandy K.G. The functional network of ion channels in T lymphocytes. Immunol. Rev. 2009;231:59–87. doi: 10.1111/j.1600-065X.2009.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mercer J.C., DeHaven W.I., Putney J.W., Jr. Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. J. Biol. Chem. 2006;281:24979–24990. doi: 10.1074/jbc.M604589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peinelt C., Vig M., Kinet J.P. Amplification of CRAC current by STIM1 and CRACM1 (Orai1) Nat. Cell Biol. 2006;8:771–773. doi: 10.1038/ncb1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soboloff J., Spassova M.A., Gill D.L. Orai1 and STIM reconstitute store-operated calcium channel function. J. Biol. Chem. 2006;281:20661–20665. doi: 10.1074/jbc.C600126200. [DOI] [PubMed] [Google Scholar]
- 14.Zhang S.L., Yu Y., Cahalan M.D. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu M.M., Buchanan J., Lewis R.S. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cahalan M.D. STIMulating store-operated Ca2+ entry. Nat. Cell Biol. 2009;11:669–677. doi: 10.1038/ncb0609-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis R.S. Store-operated calcium channels: new perspectives on mechanism and function. Cold Spring Harb. Perspect. Biol. 2011 doi: 10.1101/cshperspect.a003970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu M.M., Covington E.D., Lewis R.S. Single-molecule analysis of diffusion and trapping of STIM1 and Orai1 at endoplasmic reticulum-plasma membrane junctions. Mol. Biol. Cell. 2014;25:3672–3685. doi: 10.1091/mbc.E14-06-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoth M., Niemeyer B.A. The neglected CRAC proteins: Orai2, Orai3, and STIM2. Curr. Top. Membr. 2013;71:237–271. doi: 10.1016/B978-0-12-407870-3.00010-X. [DOI] [PubMed] [Google Scholar]
- 20.Prakriya M., Feske S., Hogan P.G. Orai1 is an essential pore subunit of the CRAC channel. Nature. 2006;443:230–233. doi: 10.1038/nature05122. [DOI] [PubMed] [Google Scholar]
- 21.Vig M., Beck A., Penner R. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr. Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeromin A.V., Zhang S.L., Cahalan M.D. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schindl R., Bergsmann J., Romanin C. 2-aminoethoxydiphenyl borate alters selectivity of Orai3 channels by increasing their pore size. J. Biol. Chem. 2008;283:20261–20267. doi: 10.1074/jbc.M803101200. [DOI] [PubMed] [Google Scholar]
- 24.Yamashita M., Prakriya M. Divergence of Ca2+ selectivity and equilibrium Ca2+ blockade in a Ca2+ release-activated Ca2+ channel. J. Gen. Physiol. 2014;143:325–343. doi: 10.1085/jgp.201311108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNally B.A., Yamashita M., Prakriya M. Structural determinants of ion permeation in CRAC channels. Proc. Natl. Acad. Sci. USA. 2009;106:22516–22521. doi: 10.1073/pnas.0909574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Y., Ramachandran S., Hogan P.G. Pore architecture of the ORAI1 store-operated calcium channel. Proc. Natl. Acad. Sci. USA. 2010;107:4896–4901. doi: 10.1073/pnas.1001169107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park C.Y., Hoover P.J., Lewis R.S. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan J.P., Zeng W., Muallem S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calloway N., Holowka D., Baird B. A basic sequence in STIM1 promotes Ca2+ influx by interacting with the C-terminal acidic coiled coil of Orai1. Biochemistry. 2010;49:1067–1071. doi: 10.1021/bi901936q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frischauf I., Muik M., Romanin C. Molecular determinants of the coupling between STIM1 and Orai channels: differential activation of Orai1–3 channels by a STIM1 coiled-coil mutant. J. Biol. Chem. 2009;284:21696–21706. doi: 10.1074/jbc.M109.018408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muik M., Frischauf I., Romanin C. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J. Biol. Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- 32.Calloway N., Vig M., Baird B. Molecular clustering of STIM1 with Orai1/CRACM1 at the plasma membrane depends dynamically on depletion of Ca2+ stores and on electrostatic interactions. Mol. Biol. Cell. 2009;20:389–399. doi: 10.1091/mbc.E07-11-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng H., Zhou M.H., Zhang S.L. Differential roles of the C and N termini of Orai1 protein in interacting with stromal interaction molecule 1 (STIM1) for Ca2+ release-activated Ca2+ (CRAC) channel activation. J. Biol. Chem. 2013;288:11263–11272. doi: 10.1074/jbc.M113.450254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derler I., Plenk P., Romanin C. The extended transmembrane Orai1 N-terminal (ETON) region combines binding interface and gate for Orai1 activation by STIM1. J. Biol. Chem. 2013;288:29025–29034. doi: 10.1074/jbc.M113.501510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lis A., Zierler S., Penner R. A single lysine in the N-terminal region of store-operated channels is critical for STIM1-mediated gating. J. Gen. Physiol. 2010;136:673–686. doi: 10.1085/jgp.201010484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNally B.A., Somasundaram A., Prakriya M. The C- and N-terminal STIM1 binding sites on Orai1 are required for both trapping and gating CRAC channels. J. Physiol. 2013;591:2833–2850. doi: 10.1113/jphysiol.2012.250456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou Y., Meraner P., Hogan P.G. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat. Struct. Mol. Biol. 2010;17:112–116. doi: 10.1038/nsmb.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Derler I., Fahrner M., Romanin C. Increased hydrophobicity at the N terminus/membrane interface impairs gating of the severe combined immunodeficiency-related ORAI1 mutant. J. Biol. Chem. 2009;284:15903–15915. doi: 10.1074/jbc.M808312200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang S.L., Yeromin A.V., Cahalan M.D. Mutations in Orai1 transmembrane segment 1 cause STIM1-independent activation of Orai1 channels at glycine 98 and channel closure at arginine 91. Proc. Natl. Acad. Sci. USA. 2011;108:17838–17843. doi: 10.1073/pnas.1114821108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNally B.A., Somasundaram A., Prakriya M. Gated regulation of CRAC channel ion selectivity by STIM1. Nature. 2012;482:241–245. doi: 10.1038/nature10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cai X. Molecular evolution and structural analysis of the Ca2+ release-activated Ca2+ channel subunit, Orai. J. Mol. Biol. 2007;368:1284–1291. doi: 10.1016/j.jmb.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 42.Yamashita M., Navarro-Borelly L., Prakriya M. Orai1 mutations alter ion permeation and Ca2+-dependent fast inactivation of CRAC channels: evidence for coupling of permeation and gating. J. Gen. Physiol. 2007;130:525–540. doi: 10.1085/jgp.200709872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amcheslavsky A., Safrina O., Cahalan M.D. Orai3 TM3 point mutation G158C alters kinetics of 2-APB-induced gating by disulfide bridge formation with TM2 C101. J. Gen. Physiol. 2013;142:405–412. doi: 10.1085/jgp.201311030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srikanth S., Yee M.K., Ribalet B. The third transmembrane segment of orai1 protein modulates Ca2+ release-activated Ca2+ (CRAC) channel gating and permeation properties. J. Biol. Chem. 2011;286:35318–35328. doi: 10.1074/jbc.M111.265884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoth M., Penner R. Calcium release-activated calcium current in rat mast cells. J. Physiol. 1993;465:359–386. doi: 10.1113/jphysiol.1993.sp019681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zweifach A., Lewis R.S. Rapid inactivation of depletion-activated calcium current (ICRAC) due to local calcium feedback. J. Gen. Physiol. 1995;105:209–226. doi: 10.1085/jgp.105.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lis A., Peinelt C., Penner R. CRACM1, CRACM2, and CRACM3 are store-operated Ca2+ channels with distinct functional properties. Curr. Biol. 2007;17:794–800. doi: 10.1016/j.cub.2007.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Derler I., Fahrner M., Romanin C. A Ca2+ release-activated Ca2+ (CRAC) modulatory domain (CMD) within STIM1 mediates fast Ca2+-dependent inactivation of ORAI1 channels. J. Biol. Chem. 2009;284:24933–24938. doi: 10.1074/jbc.C109.024083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mullins F.M., Park C.Y., Lewis R.S. STIM1 and calmodulin interact with Orai1 to induce Ca2+-dependent inactivation of CRAC channels. Proc. Natl. Acad. Sci. USA. 2009;106:15495–15500. doi: 10.1073/pnas.0906781106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee K.P., Yuan J.P., Muallem S. Molecular determinants of fast Ca2+-dependent inactivation and gating of the Orai channels. Proc. Natl. Acad. Sci. USA. 2009;106:14687–14692. doi: 10.1073/pnas.0904664106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergsmann J., Derler I., Romanin C. Molecular determinants within N terminus of Orai3 protein that control channel activation and gating. J. Biol. Chem. 2011;286:31565–31575. doi: 10.1074/jbc.M111.227546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Srikanth S., Jung H.J., Gwack Y. The intracellular loop of Orai1 plays a central role in fast inactivation of Ca2+ release-activated Ca2+ channels. J. Biol. Chem. 2010;285:5066–5075. doi: 10.1074/jbc.M109.072736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hoover P.J., Lewis R.S. Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1) Proc. Natl. Acad. Sci. USA. 2011;108:13299–13304. doi: 10.1073/pnas.1101664108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scrimgeour N., Litjens T., Rychkov G.Y. Properties of Orai1 mediated store-operated current depend on the expression levels of STIM1 and Orai1 proteins. J. Physiol. 2009;587:2903–2918. doi: 10.1113/jphysiol.2009.170662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prakriya M., Lewis R.S. Potentiation and inhibition of Ca2+ release-activated Ca2+ channels by 2-aminoethyldiphenyl borate (2-APB) occurs independently of IP3 receptors. J. Physiol. 2001;536:3–19. doi: 10.1111/j.1469-7793.2001.t01-1-00003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DeHaven W.I., Smyth J.T., Putney J.W., Jr. Complex actions of 2-aminoethyldiphenyl borate on store-operated calcium entry. J. Biol. Chem. 2008;283:19265–19273. doi: 10.1074/jbc.M801535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peinelt C., Lis A., Penner R. 2-Aminoethoxydiphenyl borate directly facilitates and indirectly inhibits STIM1-dependent gating of CRAC channels. J. Physiol. 2008;586:3061–3073. doi: 10.1113/jphysiol.2008.151365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S.L., Kozak J.A., Cahalan M.D. Store-dependent and -independent modes regulating Ca2+ release-activated Ca2+ channel activity of human Orai1 and Orai3. J. Biol. Chem. 2008;283:17662–17671. doi: 10.1074/jbc.M801536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Amcheslavsky A., Safrina O., Cahalan M.D. State-dependent block of Orai3 TM1 and TM3 cysteine mutants: insights into 2-APB activation. J. Gen. Physiol. 2014;143:621–631. doi: 10.1085/jgp.201411171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mignen O., Thompson J.L., Shuttleworth T.J. Orai1 subunit stoichiometry of the mammalian CRAC channel pore. J. Physiol. 2008;586:419–425. doi: 10.1113/jphysiol.2007.147249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demuro A., Penna A., Parker I. Subunit stoichiometry of human Orai1 and Orai3 channels in closed and open states. Proc. Natl. Acad. Sci. USA. 2011;108:17832–17837. doi: 10.1073/pnas.1114814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji W., Xu P., Chen L. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc. Natl. Acad. Sci. USA. 2008;105:13668–13673. doi: 10.1073/pnas.0806499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Penna A., Demuro A., Cahalan M.D. The CRAC channel consists of a tetramer formed by STIM-induced dimerization of Orai dimers. Nature. 2008;456:116–120. doi: 10.1038/nature07338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Madl J., Weghuber J., Schütz G.J. Resting state Orai1 diffuses as homotetramer in the plasma membrane of live mammalian cells. J. Biol. Chem. 2010;285:41135–41142. doi: 10.1074/jbc.M110.177881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mignen O., Thompson J.L., Shuttleworth T.J. The molecular architecture of the arachidonate-regulated Ca2+-selective ARC channel is a pentameric assembly of Orai1 and Orai3 subunits. J. Physiol. 2009;587:4181–4197. doi: 10.1113/jphysiol.2009.174193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ong H.L., Ambudkar I.S. The dynamic complexity of the TRPC1 channelosome. Channels (Austin) 2011;5:424–431. doi: 10.4161/chan.5.5.16471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hou X., Pedi L., Long S.B. Crystal structure of the calcium release-activated calcium channel Orai. Science. 2012;338:1308–1313. doi: 10.1126/science.1228757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thompson J.L., Shuttleworth T.J. How many Orai’s does it take to make a CRAC channel? Sci. Rep. 2013;3:1961. doi: 10.1038/srep01961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yen M., Lokteva L.A., Lewis R.S. STIM1 binds to pairs of Orai1 subunits to open the CRAC channel. Biophys. J. 2014;106:314a–315a. [Google Scholar]
- 70.Yeromin A.V., Roos J., Cahalan M.D. A store-operated calcium channel in Drosophila S2 cells. J. Gen. Physiol. 2004;123:167–182. doi: 10.1085/jgp.200308982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Navarro-Borelly L., Somasundaram A., Prakriya M. STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy. J. Physiol. 2008;586:5383–5401. doi: 10.1113/jphysiol.2008.162503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z., Liu L., Xu T. Graded activation of CRAC channel by binding of different numbers of STIM1 to Orai1 subunits. Cell Res. 2011;21:305–315. doi: 10.1038/cr.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stathopulos P.B., Schindl R., Ikura M. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat. Commun. 2013;4:2963. doi: 10.1038/ncomms3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dong H., Fiorin G., Klein M.L. Pore waters regulate ion permeation in a calcium release-activated calcium channel. Proc. Natl. Acad. Sci. USA. 2013;110:17332–17337. doi: 10.1073/pnas.1316969110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wood M.L. Atomistic molecular dynamics simulations of Drosophila orai in a hydrated lipid bilayer. Biophys. J. 2014;106:316a. [Google Scholar]
- 76.Kelley L.A., Sternberg M.J.E. Protein structure prediction on the Web: a case study using the PHYRE server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 77.Rostkowski M., Olsson M.H., Jensen J.H. Graphical analysis of pH-dependent properties of proteins predicted using PROPKA. BMC Struct. Biol. 2011;11:6. doi: 10.1186/1472-6807-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]


