Abstract
Purpose
Recent advances in DNA sequencing have led to the development of breast cancer susceptibility gene panels for germline genetic testing of patients. We assessed the frequency of mutations in 17 predisposition genes, including BRCA1 and BRCA2, in a large cohort of patients with triple-negative breast cancer (TNBC) unselected for family history of breast or ovarian cancer to determine the utility of germline genetic testing for those with TNBC.
Patients and Methods
Patients with TNBC (N = 1,824) unselected for family history of breast or ovarian cancer were recruited through 12 studies, and germline DNA was sequenced to identify mutations.
Results
Deleterious mutations were identified in 14.6% of all patients. Of these, 11.2% had mutations in the BRCA1 (8.5%) and BRCA2 (2.7%) genes. Deleterious mutations in 15 other predisposition genes were detected in 3.7% of patients, with the majority observed in genes involved in homologous recombination, including PALB2 (1.2%) and BARD1, RAD51D, RAD51C, and BRIP1 (0.3% to 0.5%). Patients with TNBC with mutations were diagnosed at an earlier age (P < .001) and had higher-grade tumors (P = .01) than those without mutations.
Conclusion
Deleterious mutations in predisposition genes are present at high frequency in patients with TNBC unselected for family history of cancer. Mutation prevalence estimates suggest that patients with TNBC, regardless of age at diagnosis or family history of cancer, should be considered for germline genetic testing of BRCA1 and BRCA2. Although mutations in other predisposition genes are observed among patients with TNBC, better cancer risk estimates are needed before these mutations are used for clinical risk assessment in relatives.
INTRODUCTION
Triple-negative breast cancer (TNBC), defined by little or no expression of estrogen receptor (ER), progesterone receptor (PR), or human epidermal growth factor receptor 2 (HER2) in tumor material, accounts for 12% to 15% of all breast cancers.1,2 TNBC occurs most frequently in young or premenopausal women and African Americans. Patients with TNBC often have a worse outcome than patients with other breast cancer subtypes,3,4 with 5-year survival estimated at 70% for those with TNBC compared with > 80% for all other subtypes.5 Germline mutations in the BRCA1 breast and ovarian cancer susceptibility gene have been associated with TNBC, with 60% to 80% of breast tumors from BRCA1 mutation carriers displaying a TNBC phenotype.6 Additional studies have identified BRCA1 mutations in up to 29% of patients of Ashkenazi Jewish ethnicity presenting with TNBC,7 20% of those with TNBC diagnosed at a young age and/or with a family history of breast cancer,8 and 8% to 14% of those with TNBC unselected for family history.9–11 In addition, three (3.9%) of 77 patients with TNBC with a median age at diagnosis of 51 years,12 six (9%) of 64 patients with TNBC of Ashkenazi Jewish ancestry,13 and 5.2% of patients with TNBC without a significant family history of breast or ovarian cancer14 have been shown to carry germline BRCA2 mutations. Although a substantial proportion of TNBCs arise as a result of inherited mutations in BRCA1 and BRCA2, the contribution of mutations in these genes to TNBC, not specifically selected for age at diagnosis or enriched family history of breast or ovarian cancer, remains to be established. Furthermore, although the development of panel-based testing has revealed that 10% of high-risk patients with no BRCA1 or BRCA2 mutation may carry inherited deleterious mutations in other breast cancer predisposition genes,15 the frequency of inherited mutations in the non-BRCA1/2 predisposition genes among patients with TNBC has not been determined. In this study, we conducted panel-based mutation screening of breast cancer predisposition genes in a large cohort of patients with TNBC in an effort to better understand the contribution of inherited mutations in moderate- and high-risk predisposition genes to TNBC and determine the best parameters for selection of patients with TNBC for BRCA testing.
PATIENTS AND METHODS
Study Populations
The Triple-Negative Breast Cancer Consortium has access to DNA and phenotypic information from consecutive patients with TNBC recruited through oncology clinics from 11 clinical centers in the United States (Mayo Clinic Breast Cancer Study, Dana-Farber Cancer Institute, Ohio State University, Roswell Park Cancer Institute, Kansas University Medical Center, and Fox Chase Cancer Center), Germany (Bavarian Breast Cancer Cases and Controls and Gene Environment Interaction and Breast Cancer in Germany), Finland (Helsinki Breast Cancer Study), Greece (Demokritos), and the United Kingdom (Sheffield Breast Cancer Study; Table 1; Data Supplement). Patients with TNBC from the POSH (Prospective Study of Outcomes in Sporadic Versus Hereditary Breast Cancer) multicenter United Kingdom trial of women diagnosed at age < 40 years were also included (Data Supplement). Selection of patients with TNBC was independent of family history of breast or ovarian cancer and age at diagnosis. All 1,824 patients with TNBC were recruited to institutional review board–approved studies.
Table 1.
Sample Demographics
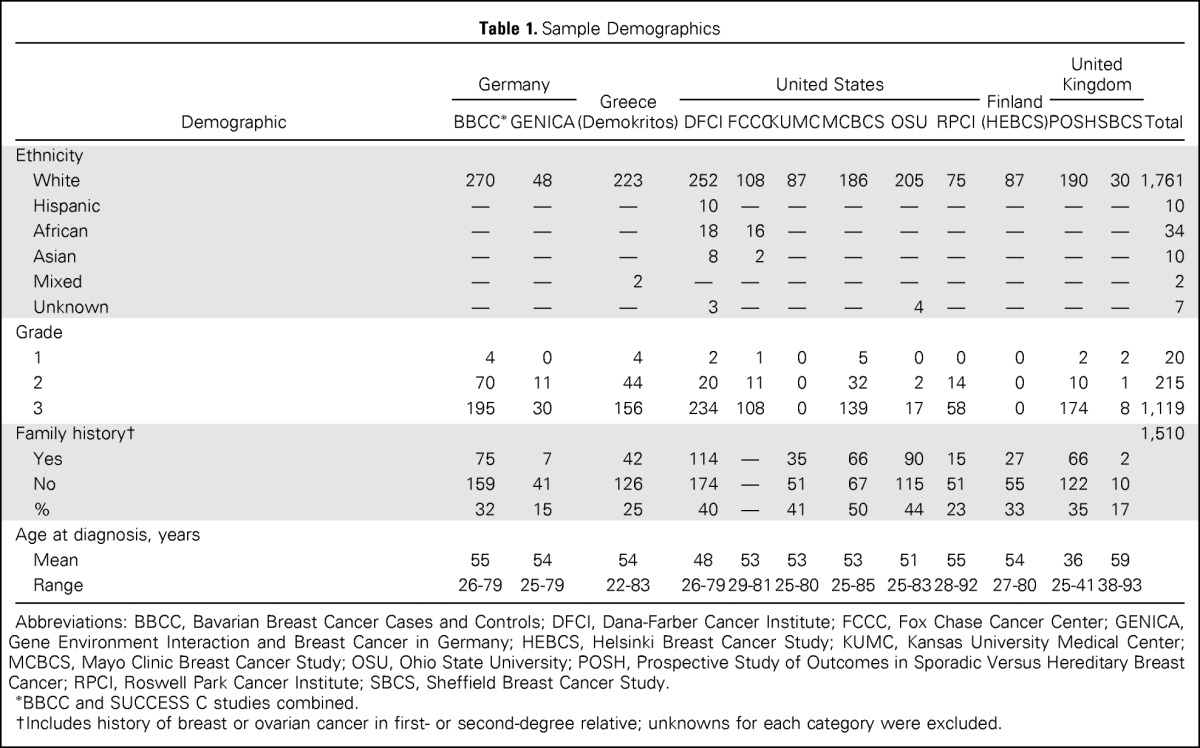
| Demographic | Germany |
Greece (Demokritos) | United States |
Finland (HEBCS) | United Kingdom |
Total | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BBCC* | GENICA | DFCI | FCCC | KUMC | MCBCS | OSU | RPCI | POSH | SBCS | ||||
| Ethnicity | |||||||||||||
| White | 270 | 48 | 223 | 252 | 108 | 87 | 186 | 205 | 75 | 87 | 190 | 30 | 1,761 |
| Hispanic | — | — | — | 10 | — | — | — | — | — | — | — | — | 10 |
| African | — | — | — | 18 | 16 | — | — | — | — | — | — | — | 34 |
| Asian | — | — | — | 8 | 2 | — | — | — | — | — | — | — | 10 |
| Mixed | — | — | 2 | — | — | — | — | — | — | — | — | — | 2 |
| Unknown | — | — | — | 3 | — | — | — | 4 | — | — | — | — | 7 |
| Grade | |||||||||||||
| 1 | 4 | 0 | 4 | 2 | 1 | 0 | 5 | 0 | 0 | 0 | 2 | 2 | 20 |
| 2 | 70 | 11 | 44 | 20 | 11 | 0 | 32 | 2 | 14 | 0 | 10 | 1 | 215 |
| 3 | 195 | 30 | 156 | 234 | 108 | 0 | 139 | 17 | 58 | 0 | 174 | 8 | 1,119 |
| Family history† | 1,510 | ||||||||||||
| Yes | 75 | 7 | 42 | 114 | — | 35 | 66 | 90 | 15 | 27 | 66 | 2 | |
| No | 159 | 41 | 126 | 174 | — | 51 | 67 | 115 | 51 | 55 | 122 | 10 | |
| % | 32 | 15 | 25 | 40 | — | 41 | 50 | 44 | 23 | 33 | 35 | 17 | |
| Age at diagnosis, years | |||||||||||||
| Mean | 55 | 54 | 54 | 48 | 53 | 53 | 53 | 51 | 55 | 54 | 36 | 59 | |
| Range | 26-79 | 25-79 | 22-83 | 26-79 | 29-81 | 25-80 | 25-85 | 25-83 | 28-92 | 27-80 | 25-41 | 38-93 | |
Abbreviations: BBCC, Bavarian Breast Cancer Cases and Controls; DFCI, Dana-Farber Cancer Institute; FCCC, Fox Chase Cancer Center; GENICA, Gene Environment Interaction and Breast Cancer in Germany; HEBCS, Helsinki Breast Cancer Study; KUMC, Kansas University Medical Center; MCBCS, Mayo Clinic Breast Cancer Study; OSU, Ohio State University; POSH, Prospective Study of Outcomes in Sporadic Versus Hereditary Breast Cancer; RPCI, Roswell Park Cancer Institute; SBCS, Sheffield Breast Cancer Study.
BBCC and SUCCESS C studies combined.
Includes history of breast or ovarian cancer in first- or second-degree relative; unknowns for each category were excluded.
Panel-Based Mutation Analysis
Germline DNA samples from 1,824 patients with TNBC underwent custom capture (eArray; Agilent, Santa Clara, CA) of all coding sequences and intron/exon boundaries of coding exons from 122 DNA repair genes, including 17 breast cancer predisposition genes (BRCA1, BRCA2, PALB2, BARD1, BRIP1, RAD51C, RAD51D, RAD50, NBN, MRE11A, XRCC2, ATM, CHEK2, TP53, PTEN, STK11, and CDH1). Products from each capture reaction were sequenced on a HiSeq 2000 (Illumina, San Diego, CA; Data Supplement), and all likely deleterious mutations were validated by Sanger sequencing.
Bioinformatic Analysis
Paired end reads (100 bp) were aligned to the hg19 reference human genome using Novoalign (Novocraft Technologies, Selangor, Malaysia). Realignment and recalibration were performed using GATK software (version 1.6-7; https://www.broadinstitute.org/gatk). Germline variations were called with a combination of GATK Unified Genotyper16 and Samtools (version 0.1.18; http://www.htslib.org).17 Annotations were defined using SnpEFF (version 3.0c; http://snpeff.sourceforge.net/index.html)18 and ANNOVAR (http://www.openbioinformatics.org/annovar).19 Population allele frequencies were extracted from the Exome Variant Server (http://evs.gs.washington.edu/EVS), 1000 Genomes (http://www.1000genomes.org), and dbSNP (version 137; http://www.ncbi.nlm.nih.gov/projects/SNP). Known deleterious missense mutations in BRCA1, BRCA2, and TP53 were included in all analyses (Data Supplement). Predicted deleterious missense mutations were selected using algorithms in ANNOVAR (eg, SIFT, PolyPhen2, LRT, MutationTaster, PhyloP, GERP)19,20 and AlignGVGD (http://agvgd.iarc.fr).21
Statistical Analysis
Likely deleterious mutations from genes other than BRCA1 or BRCA2 were combined in an “other” category. Patients with TNBC without mutations were categorized as wild type. The t test, χ2 test, and Fisher's exact test were used for evaluating associations with mutation status. P values < .05 were considered statistically significant.
RESULTS
Study Population
The 1,824 female patients with TNBC in this study were recruited from 12 centers (Table 1). Of the 1,817 patients with established ethnicity, 1,762 (97%) were white, and 34 (1.9%), 10 (0.6%), and 10 (0.6%) were of African, Asian, and Hispanic ethnicities, respectively. Age at diagnosis ranged from 22 to 93 years, with an average age of 51 years. This was similar to the average age of patients with TNBC in the METABRIC (Molecular Taxonomy of Breast Cancer International Consortium) study (52.7 years; P = .2477)22 but younger than that of patients with TNBC in the Cancer Genome Atlas Network (54.2 years; P = .027).23 Of the 1,510 patients with available family history information, 514 (34%) had at least one first- or second-degree relative with breast cancer, and 4% had a relative with ovarian cancer. The TNBCs were predominantly grade 3 (81%; Table 1).
Germline Mutations
All coding exons and consensus splice sites of 17 known cancer predisposition genes were screened for mutations in the 1,824 patients with TNBC. Overall, 271 deleterious mutations were identified in 267 patients (14.6%; Table 2; Data Supplement). Of these, 155 (57%) occurred in BRCA1, 49 (18%) in BRCA2, and 67 (25%) in 12 of 15 other predisposition genes (Table 1; Fig 1). The frequency of mutations by center ranged from 4% to 24% (Data Supplement). The elevated mutation frequency (24%) in the Dana-Farber Cancer Institute study resulted from 21 Ashkenazi Jewish founder mutations in BRCA1 and BRCA2.
Table 2.
Gene-Based Age at Diagnosis and Family History of Cancer
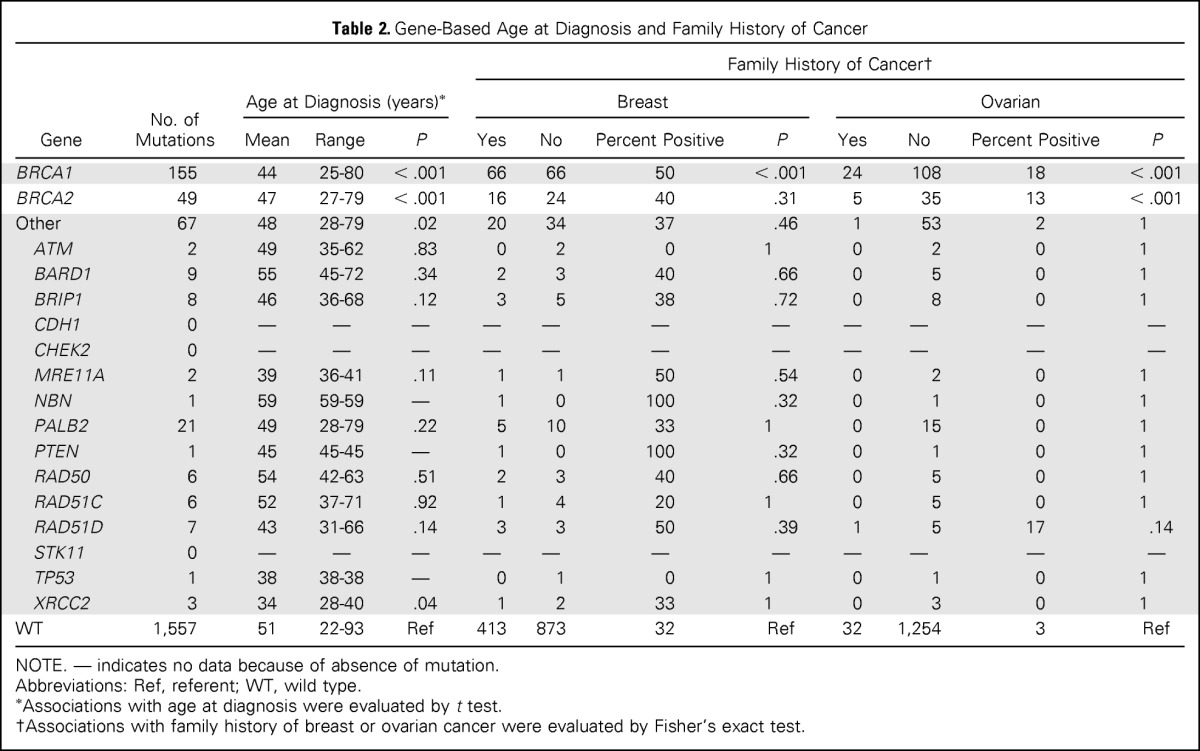
| Gene | No. of Mutations | Age at Diagnosis (years)* |
Family History of Cancer† |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Breast |
Ovarian |
|||||||||||
| Mean | Range | P | Yes | No | Percent Positive | P | Yes | No | Percent Positive | P | ||
| BRCA1 | 155 | 44 | 25-80 | < .001 | 66 | 66 | 50 | < .001 | 24 | 108 | 18 | < .001 |
| BRCA2 | 49 | 47 | 27-79 | < .001 | 16 | 24 | 40 | .31 | 5 | 35 | 13 | < .001 |
| Other | 67 | 48 | 28-79 | .02 | 20 | 34 | 37 | .46 | 1 | 53 | 2 | 1 |
| ATM | 2 | 49 | 35-62 | .83 | 0 | 2 | 0 | 1 | 0 | 2 | 0 | 1 |
| BARD1 | 9 | 55 | 45-72 | .34 | 2 | 3 | 40 | .66 | 0 | 5 | 0 | 1 |
| BRIP1 | 8 | 46 | 36-68 | .12 | 3 | 5 | 38 | .72 | 0 | 8 | 0 | 1 |
| CDH1 | 0 | — | — | — | — | — | — | — | — | — | — | — |
| CHEK2 | 0 | — | — | — | — | — | — | — | — | — | — | — |
| MRE11A | 2 | 39 | 36-41 | .11 | 1 | 1 | 50 | .54 | 0 | 2 | 0 | 1 |
| NBN | 1 | 59 | 59-59 | — | 1 | 0 | 100 | .32 | 0 | 1 | 0 | 1 |
| PALB2 | 21 | 49 | 28-79 | .22 | 5 | 10 | 33 | 1 | 0 | 15 | 0 | 1 |
| PTEN | 1 | 45 | 45-45 | — | 1 | 0 | 100 | .32 | 0 | 1 | 0 | 1 |
| RAD50 | 6 | 54 | 42-63 | .51 | 2 | 3 | 40 | .66 | 0 | 5 | 0 | 1 |
| RAD51C | 6 | 52 | 37-71 | .92 | 1 | 4 | 20 | 1 | 0 | 5 | 0 | 1 |
| RAD51D | 7 | 43 | 31-66 | .14 | 3 | 3 | 50 | .39 | 1 | 5 | 17 | .14 |
| STK11 | 0 | — | — | — | — | — | — | — | — | — | — | — |
| TP53 | 1 | 38 | 38-38 | — | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 |
| XRCC2 | 3 | 34 | 28-40 | .04 | 1 | 2 | 33 | 1 | 0 | 3 | 0 | 1 |
| WT | 1,557 | 51 | 22-93 | Ref | 413 | 873 | 32 | Ref | 32 | 1,254 | 3 | Ref |
NOTE. — indicates no data because of absence of mutation.
Abbreviations: Ref, referent; WT, wild type.
Associations with age at diagnosis were evaluated by t test.
Associations with family history of breast or ovarian cancer were evaluated by Fisher's exact test.
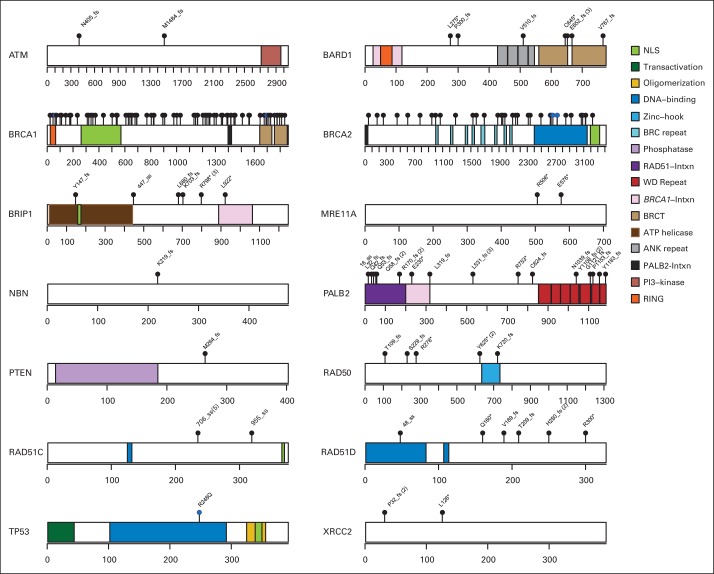
Fig 1.
Germline likely deleterious mutations from 14 breast cancer predisposition genes in unselected patients with triple-negative breast cancer. Locations of likely deleterious mutations and domains in proteins encoded by predisposition genes are shown by lollipop structures. Mutations are labeled, other than in BRCA1 and BRCA2. Protein domain patterns are shown in key. Scales on genes reflect number of amino acid residues.
Deleterious BRCA1 mutations were detected in 8.5% of patients with TNBC, including 145 truncating (frameshift, nonsense, and splice) mutations and 10 known deleterious missense mutations.24 The 185delAG (c.68_69delAG) Ashkenazi Jewish founder mutation was identified in 18 patients with TNBC, and the 5382insC (c.5266dupC) Eastern European founder mutation was found in 19 patients with TNBC (Data Supplement). Another 21 recurrent BRCA1 mutations were observed in 64 other patients with TNBC (Data Supplement). The 49 deleterious BRCA2 mutations (2.7%) included 41 truncating mutations, of which six were the 6174delT (c.5946delT) Ashkenazi Jewish founder mutation, three were splice mutations, and five were known deleterious missense mutations in the BRCA2 DNA binding domain.24 Three recurrent mutations in BRCA2 accounted for 16 patient cases of TNBC (Data Supplement). Likely deleterious mutations in non-BRCA1/2 predisposition genes were identified in 3.7% of all unselected patients with TNBC. In particular, 21 patients (1.2%) with TNBC had deleterious PALB2 truncating mutations, including 15 diagnosed at age ≤ 50 years. Three patients from Finland were found to carry the PALB2 c.1592delT founder mutation.25 In addition, deleterious mutations were detected in BARD1 (n = 9), BRIP1 (n = 8), RAD51D (n = 7), RAD50 (n = 6), and RAD51C (n = 6; Data Supplement). In contrast, no mutations were observed in CHEK2, consistent with an association between CHEK2 mutations and ER-positive breast cancer,23,26 or in the CDH1 and STK11 genes. Four patients with TNBC carried > 1 deleterious mutation (Data Supplement), including an individual with BRCA1:c.68_69delAG and BRCA2:c.5946delT mutations diagnosed at age 68 years. None had a family history of breast or ovarian cancer.
Age at Diagnosis
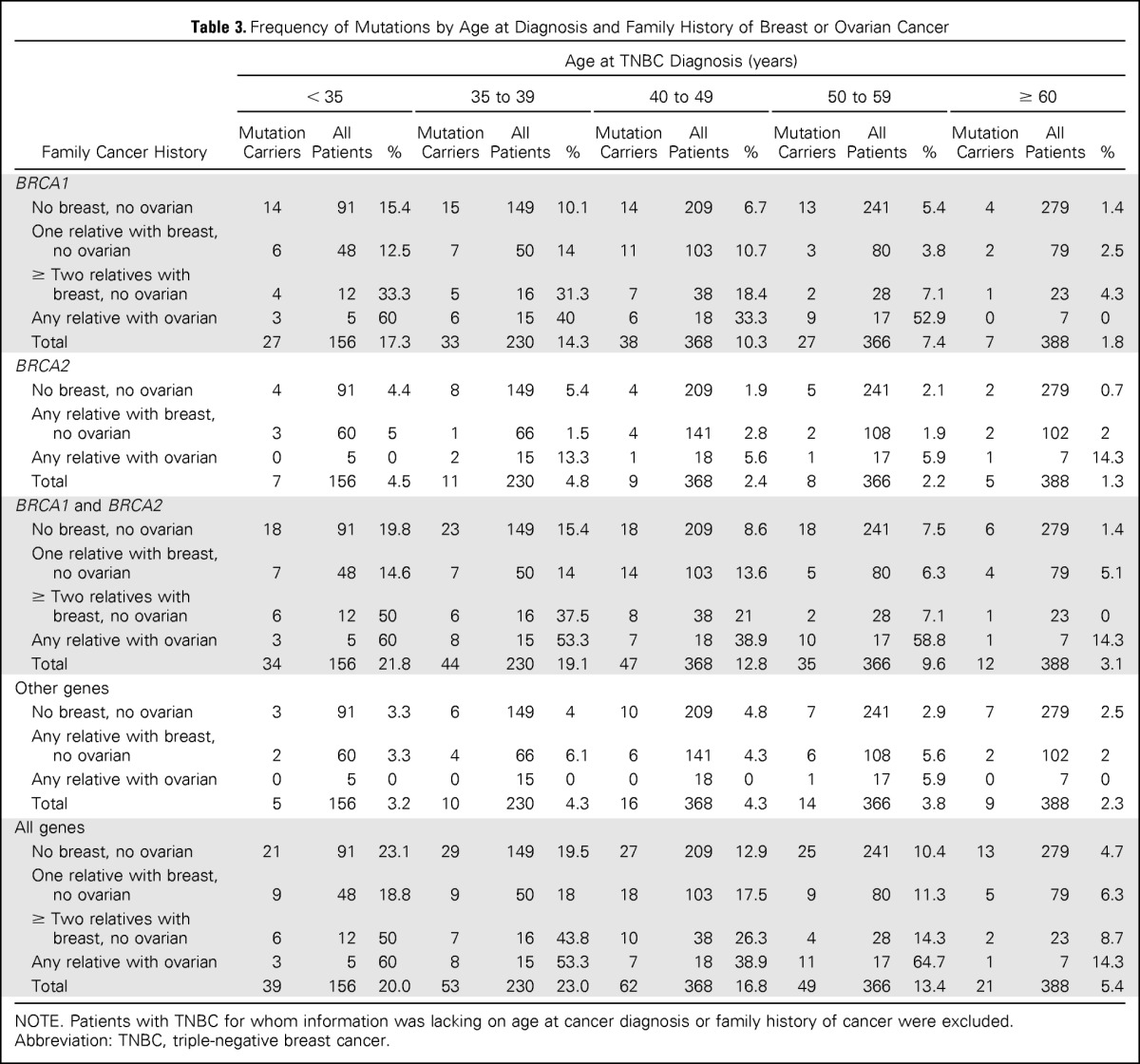
In this study, 38% of all deleterious mutations were detected in patients with TNBC diagnosed at age < 40 years. The average age at diagnosis of TNBC was significantly younger for patients with deleterious (45 years; P < .001), BRCA1 (44 years; P < .001), BRCA2 (47 years; P < .001), and non-BRCA1/2 gene mutations (48 years; P = .02), relative to those with TNBC with no mutations (ie, wild type; 52 years; Table 2; Fig 2). Consistent with this, the distribution of age at TNBC diagnosis for BRCA1, BRCA2, and non-BRCA1/2 gene mutation carriers differed from that among noncarriers (Fig 2). However, 10% (n = 27) of all mutation carriers and 5.5% of all patients with TNBC were diagnosed at age ≥ 60 years. Of these, 37% (n = 10) carried mutations in non-BRCA1/2 genes, and 50% (n = 13) had no family history of breast or ovarian cancer (Table 3).
Fig 2.
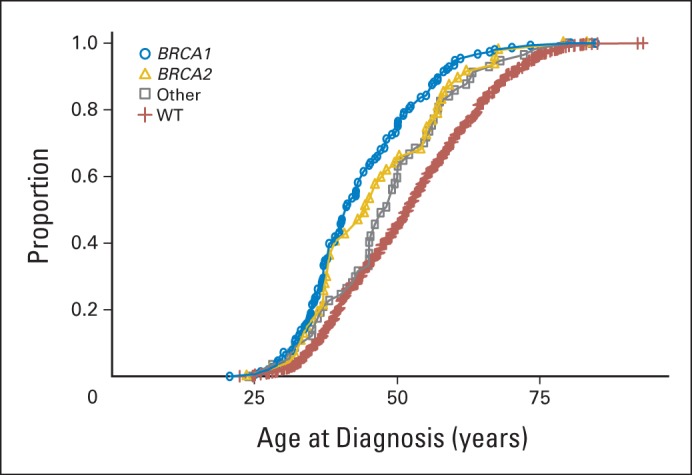
Age at onset of triple-negative breast cancer by mutation status. Distribution shown for patients with triple-negative breast cancer with BRCA1, BRCA2, and other non-BRCA1/2 mutations and no mutations (ie, wild type [WT]).
Table 3.
Frequency of Mutations by Age at Diagnosis and Family History of Breast or Ovarian Cancer
| Family Cancer History | Age at TNBC Diagnosis (years) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| < 35 |
35 to 39 |
40 to 49 |
50 to 59 |
≥ 60 |
|||||||||||
| Mutation Carriers | All Patients | % | Mutation Carriers | All Patients | % | Mutation Carriers | All Patients | % | Mutation Carriers | All Patients | % | Mutation Carriers | All Patients | % | |
| BRCA1 | |||||||||||||||
| No breast, no ovarian | 14 | 91 | 15.4 | 15 | 149 | 10.1 | 14 | 209 | 6.7 | 13 | 241 | 5.4 | 4 | 279 | 1.4 |
| One relative with breast, no ovarian | 6 | 48 | 12.5 | 7 | 50 | 14 | 11 | 103 | 10.7 | 3 | 80 | 3.8 | 2 | 79 | 2.5 |
| ≥ Two relatives with breast, no ovarian | 4 | 12 | 33.3 | 5 | 16 | 31.3 | 7 | 38 | 18.4 | 2 | 28 | 7.1 | 1 | 23 | 4.3 |
| Any relative with ovarian | 3 | 5 | 60 | 6 | 15 | 40 | 6 | 18 | 33.3 | 9 | 17 | 52.9 | 0 | 7 | 0 |
| Total | 27 | 156 | 17.3 | 33 | 230 | 14.3 | 38 | 368 | 10.3 | 27 | 366 | 7.4 | 7 | 388 | 1.8 |
| BRCA2 | |||||||||||||||
| No breast, no ovarian | 4 | 91 | 4.4 | 8 | 149 | 5.4 | 4 | 209 | 1.9 | 5 | 241 | 2.1 | 2 | 279 | 0.7 |
| Any relative with breast, no ovarian | 3 | 60 | 5 | 1 | 66 | 1.5 | 4 | 141 | 2.8 | 2 | 108 | 1.9 | 2 | 102 | 2 |
| Any relative with ovarian | 0 | 5 | 0 | 2 | 15 | 13.3 | 1 | 18 | 5.6 | 1 | 17 | 5.9 | 1 | 7 | 14.3 |
| Total | 7 | 156 | 4.5 | 11 | 230 | 4.8 | 9 | 368 | 2.4 | 8 | 366 | 2.2 | 5 | 388 | 1.3 |
| BRCA1 and BRCA2 | |||||||||||||||
| No breast, no ovarian | 18 | 91 | 19.8 | 23 | 149 | 15.4 | 18 | 209 | 8.6 | 18 | 241 | 7.5 | 6 | 279 | 1.4 |
| One relative with breast, no ovarian | 7 | 48 | 14.6 | 7 | 50 | 14 | 14 | 103 | 13.6 | 5 | 80 | 6.3 | 4 | 79 | 5.1 |
| ≥ Two relatives with breast, no ovarian | 6 | 12 | 50 | 6 | 16 | 37.5 | 8 | 38 | 21 | 2 | 28 | 7.1 | 1 | 23 | 0 |
| Any relative with ovarian | 3 | 5 | 60 | 8 | 15 | 53.3 | 7 | 18 | 38.9 | 10 | 17 | 58.8 | 1 | 7 | 14.3 |
| Total | 34 | 156 | 21.8 | 44 | 230 | 19.1 | 47 | 368 | 12.8 | 35 | 366 | 9.6 | 12 | 388 | 3.1 |
| Other genes | |||||||||||||||
| No breast, no ovarian | 3 | 91 | 3.3 | 6 | 149 | 4 | 10 | 209 | 4.8 | 7 | 241 | 2.9 | 7 | 279 | 2.5 |
| Any relative with breast, no ovarian | 2 | 60 | 3.3 | 4 | 66 | 6.1 | 6 | 141 | 4.3 | 6 | 108 | 5.6 | 2 | 102 | 2 |
| Any relative with ovarian | 0 | 5 | 0 | 0 | 15 | 0 | 0 | 18 | 0 | 1 | 17 | 5.9 | 0 | 7 | 0 |
| Total | 5 | 156 | 3.2 | 10 | 230 | 4.3 | 16 | 368 | 4.3 | 14 | 366 | 3.8 | 9 | 388 | 2.3 |
| All genes | |||||||||||||||
| No breast, no ovarian | 21 | 91 | 23.1 | 29 | 149 | 19.5 | 27 | 209 | 12.9 | 25 | 241 | 10.4 | 13 | 279 | 4.7 |
| One relative with breast, no ovarian | 9 | 48 | 18.8 | 9 | 50 | 18 | 18 | 103 | 17.5 | 9 | 80 | 11.3 | 5 | 79 | 6.3 |
| ≥ Two relatives with breast, no ovarian | 6 | 12 | 50 | 7 | 16 | 43.8 | 10 | 38 | 26.3 | 4 | 28 | 14.3 | 2 | 23 | 8.7 |
| Any relative with ovarian | 3 | 5 | 60 | 8 | 15 | 53.3 | 7 | 18 | 38.9 | 11 | 17 | 64.7 | 1 | 7 | 14.3 |
| Total | 39 | 156 | 20.0 | 53 | 230 | 23.0 | 62 | 368 | 16.8 | 49 | 366 | 13.4 | 21 | 388 | 5.4 |
NOTE. Patients with TNBC for whom information was lacking on age at cancer diagnosis or family history of cancer were excluded.
Abbreviation: TNBC, triple-negative breast cancer.
Family History
We also evaluated whether patient cases of TNBC with mutations in the 17 predisposition genes were associated with a greater family history of breast and/or ovarian cancers than nonmutated patient cases (Table 2). Patient cases of TNBC with BRCA1 mutations were enriched for a family history of breast (50%; P < .001) and ovarian cancers (18%; P < .001), whereas patient cases of TNBC with BRCA2 mutations were only enriched for a family history of ovarian cancer (Table 2). However, patient cases of TNBC with mutations in the non-BRCA1/2 genes were not significantly associated with an enriched family history for either breast or ovarian cancer (Table 2). In particular, only five of 21 PALB2 mutation carriers and 12 of 36 BARD1, BRIP1, RAD50, RAD51C, or RAD51D mutation carriers had a family history of breast or ovarian cancer. Thus, many patients with TNBC with mutations in predisposition genes may not be identified by a family history of cancer.
Association With Tumor Pathology
Overall, patients with TNBC with mutations presented with higher-grade tumors than noncarriers (P < .001; Fig 3; Data Supplement). Although 83% (1,117 of 1,351) of all TNBCs with pathology data were grade 3, this increased to 94% (105 of 112) for BRCA1 carriers (P < .001) and 90% (35 of 39) for BRCA2 carriers (P = .35); however, this decreased slightly to 82% (37 of 45) for TNBCs in non-BRCA1/2 mutation carriers (P = .84).
Fig 3.
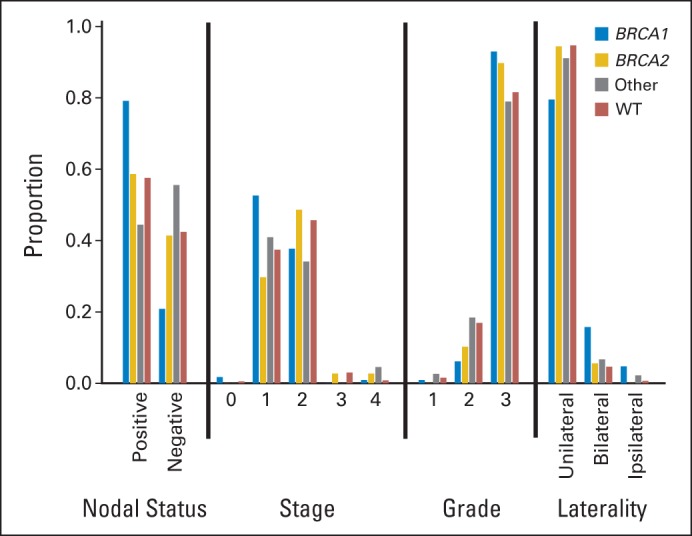
Tumor characteristics of patients with triple-negative breast cancer by mutation status. Proportion of those with BRCA1, BRCA2, and other non-BRCA1/2 mutations and no mutations (ie, wild type [WT]) exhibiting specified nodal status, tumor stage, tumor grade, and bilateral breast cancer status.
Missense Mutations
A total of 66 unique missense mutations from 91 patients were predicted deleterious by at least six of seven methods, or five of six methods when AlignGVGD was not available (Data Supplement). This included 21 unique variants in BRCA1 and BRCA2, accounting for 31 patients. Of 10 BRCA1 and BRCA2 known deleterious variants identified, only p.Ile2627Phe,27–30 which alters splicing of BRCA2, and p.Leu22Ser in BRCA1 were not predicted as deleterious. These findings suggest that a high proportion (40% to 80%) of the missense mutations predicted as deleterious in this study are likely to predispose individuals to TNBC (Data Supplement) and that an additional 1% to 3% of patient cases of TNBC may be associated with deleterious missense mutations in the predisposition genes.
Mutation Prediction
Patients with TNBC are often considered for breast cancer predisposition gene panel testing, because mutations in predisposition genes are common,14 and both carriers of mutations and their family members may benefit from informed cancer risk management. Although criteria for testing based on age at diagnosis and presence of family history have been suggested, detailed predisposition gene mutation rates for patients with TNBC based on these phenotypic categories are not currently available. Here we combined mutation results with phenotypic characteristics of patients with TNBC to provide estimates of mutation frequency by categories of age at diagnosis and family history of cancer (Table 3). When including all genes, > 18% of patients with TNBC diagnosed at age < 60 years with or without a family history of cancer carried deleterious mutations. Conversely, 5% (n = 13) diagnosed at age ≥ 60 years and with no family history of cancer carried mutations.
DISCUSSION
We present results from the largest series to date, to our knowledge, of patients with TNBC analyzed for germline mutations in a panel of known breast cancer predisposition genes. We found that 14.6% of 1,824 patients with TNBC unselected for family history of cancer carried germline deleterious mutations in 14 of 17 predisposition genes tested. BRCA1 and BRCA2 mutations were found in 11.2% of patients, consistent with other studies of TNBC, whereas mutations in 12 other genes were found in 3.7% of patients. In addition, 1% to 3% of patients carried missense mutations predicted by in silico methods to be deleterious (Data Supplement). Furthermore, the detection of BRCA1 and BRCA2 large exonic deletions or duplications in six (2%) of 294 patients with TNBC from the GeparSixto study31 suggests that approximately 2% of the patients with TNBC in our study may have also carried this type of mutation. Thus, between 14.6% and 20% of patients with TNBC may have deleterious germline mutations in these genes.
The selection of patients with TNBC for clinical genetic testing of BRCA1 and BRCA2 is often based on an age-related threshold and the associated probability of finding a mutation. The frequency of mutations in patients presenting with TNBC based on age at diagnosis and family history of breast or ovarian cancer in this study is summarized in Table 3. In our study, only 1.4% of those diagnosed at age > 60 years and with no family history of cancer were found to carry BRCA1 or BRCA2 mutations, supporting the current National Comprehensive Cancer Network guidelines for testing of patients diagnosed with TNBC before age 60 years (Table 3). Similarly, our findings verify that the probability of an underlying pathogenic BRCA1 or BRCA2 mutation exceeds 10% in those diagnosed before age 40 years; however, this probability is < 10% in older age groups in the absence of a family history of cancer (Table 3). This is consistent with the UK National Institute for Clinical Excellence testing guidelines, which do not recommend testing in patients with TNBC diagnosed at age > 40 years and with no family history of cancer. However, on the basis of our data, this latter approach would overlook 24% of all BRCA1 and BRCA2 mutations among TNBCs. Because a relatively high proportion (7.5%) of patients with TNBC with no family history and diagnosed between age 50 and 60 years had mutations, perhaps testing of all patients diagnosed at age < 60 years, or even all patients irrespective of age or family history, should be considered, especially if the cost of mutation screening were to decrease over time.
Deleterious mutations (n = 67) in the non-BRCA1/2 predisposition genes were identified in 3.7% of all patients with TNBC. The frequency of these mutations, especially in PALB2, which has recently been associated with a high lifetime risk of breast cancer,32 was similar to the frequency in high- and moderate-risk breast cancer families,33 suggesting a distinct enrichment for predisposition gene mutations in unselected TNBCs. Furthermore, genes involved in homologous recombination, including PALB2, BARD1, BRIP1, RAD51C, RAD51D, and XRCC2, accounted for 54 (81%) of the 67 mutations in non-BRCA1/2 predisposition genes, suggesting that disruption of homologous recombination repair may be an important event in the development of triple-negative breast tumors. Interestingly, the detection of deleterious mutations in RAD51C and RAD51D, which have been associated with a low to moderate risk of breast cancer but a higher risk of ovarian cancer, also raises the possibility that mutations in these genes confer higher risks of triple-negative and basal subtypes of breast cancer. In contrast, no mutations were observed in CHEK2, CDH1, or STK11, and only one mutation was identified in TP53 or PTEN, indicating that the syndromic breast cancer predisposition genes are rarely involved in predisposition to TNBC.
Clinical testing of predisposition gene panels has recently been developed to improve identification of women at increased risk for breast or ovarian cancer. However, the risks of breast and ovarian cancer associated with mutations in the non-BRCA1/2 predisposition genes are not well defined.34,35 In our study, the prevalence of mutations in the non-BRCA1/2 predisposition genes was stable across all age groups and reported cancer family histories (Table 3), consistent with lower penetrance of disease for mutations in many of these genes. Clinical management guidelines for BRCA1 and BRCA2 mutation carriers have been developed over the 20 years since these genes were identified, with recent studies suggesting that bilateral mastectomy and bilateral oophorectomy in BRCA1/2 mutation carriers may reduce breast cancer– and all cause–related mortalities.36–39 In contrast, management guidelines are not available for carriers of mutations in non-BRCA1/2 predisposition genes.34,35 Thus, the clinical utility of results from a broad gene panel of the type used in our study remains controversial. However, with panel-based genetic testing of BRCA1 and BRCA2 in combination with other genes now well established, and with testing for BRCA1 and BRCA2 mutations likely to increase if poly (ADP-ribose) polymerase inhibitors40,41 are approved for clinical use in patients with breast cancer with BRCA1/2 mutations, or if BRCA1/2 mutations are proven to predict response to platinum-based or other chemotherapies,42 panel testing will likely continue to expand. Clearly, further research with appropriate consent and curation of clinical data from patients receiving panel testing will be needed to establish the most appropriate application of results from the non-BRCA1/2 breast cancer susceptibility genes to patient care.
In conclusion, BRCA1 and BRCA2 mutation testing has a clear role for patients with TNBC, many of whom will meet the current probability threshold guidelines. However, although inclusion of other susceptibility genes in the genetic testing panel is already a widely adopted strategy, it is important that clinical care providers appreciate the current lack of robust estimates of penetrance for many of these genes.
Supplementary Material
Acknowledgment
We thank all the individuals who took part in these studies and all the researchers, clinicians, technicians, and administrative staff who enabled this work to be carried out, in particular the Demokritos participants and research team; the German Cancer Consortium and German Cancer Research Center; Wing-Yee Lo and Christina Justenhoven, Margarete Fischer-Bosch Institute of Clinical Pharmacology, Stuttgart, and University of Tübingen, Tübingen, Germany; Yon-Dschun Ko and Christian Baisc, Evangelische Kliniken Bonn and Johanniter Krankenhaus, Bonn, Germany; Hans-Peter Fischer, Institute of Pathology, University of Bonn, Bonn, Germany; Ute Hamann, Molecular Genetics of Breast Cancer, Deutsches Krebsforschungszentrum, Heidelberg, Germany; Thomas Brüning, Beate Pesch, Sylvia Rabstein, and Anne Lotz, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance, Institute of the Ruhr University Bochum, Bochum, Germany; Volker Harth, Institute of Occupational Medicine and Maritime Medicine, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Johanna Kiiski, Carl Blomqvist, Karl von Smitten, and Irja Erkkilä; Susan M. Gerty, Louise Jones, Linda Haywood, and Nikki Graham and the University of Southampton Faculty of Medicine DNA Bank for sample handling; Malcolm Reed, Sue Higham, Helen Cramp, Sabapathy Balasubramanian, Ian Brock, and Dan Connley; and Karan Rai and the Ohio State University Human Genetics Sample Bank for sample preparation.
Glossary Terms
- BRCA1:
a tumor suppressor gene known to play a role in repairing DNA breaks. Mutations in this gene are associated with increased risks of developing breast or ovarian cancer.
- BRCA2:
a tumor suppressor gene whose protein product is involved in repairing chromosomal damage. Although structurally different from BRCA1, BRCA2 has cellular functions similar to BRCA1. BRCA2 binds to RAD51 to fix DNA breaks caused by irradiation and other environmental agents. Also known as the breast cancer 2 early onset gene.
- sequencing:
a laboratory process that determines the nucleotide sequence of DNA (can involve the whole genome or whole exome or be targeted to as little as one coding sequence). Unlike somatic mutation genotyping, sequencing can detect previously unknown somatic mutations.
Appendix
Part of this work was supported by the European Community's Seventh Framework Programme under grant agreement No. 223175 (Grant No. HEALTH-F2-2009-223175; COGS). The work of Demokritos has been cofinanced by the European Union (European Social Fund) and Greek national funds through the operational program ″Education and Lifelong Learning″ of the National Strategic Reference Framework Research Funding Program: Thales—investing in knowledge society through the European Social Fund. The GENICA network was funded by the Federal Ministry of Education and Research (BMBF) Germany grants 01KW9975/5, 01KW9976/8, 01KW9977/0, and 01KW0114, the Robert Bosch Foundation, Stuttgart, Deutsches Krebsforschungszentrum (DKFZ), Heidelberg, Institute for Prevention and Occupational Medicine of the German Social Accident Insurance (IPA), Bochum, as well as the Department of Internal Medicine, Evangelische Kliniken Bonn gGmbH, Johanniter Krankenhaus, Bonn, Germany. HEBCS was financially supported by the Helsinki University Central Hospital Research Fund, Academy of Finland (266528), the Finnish Cancer Society, the Nordic Cancer Union and the Sigrid Juselius Foundation. RPCI biospecimens and data are from the DataBank and Biorepository (DBBR), a cancer center support grant shared resource supported by National Institutes of Health (P30 CA016056-27). MCBCS was supported by the National Institutes of Health (NIH) Specialized Program of Research Excellence (SPORE) in Breast Cancer to Mayo Clinic (P50 CA116201), the Breast Cancer Research Foundation, and a generous gift from the David F. and Margaret T. Grohne Family Foundation and the Ting Tsung and Wei Fong Chao Foundation. The POSH study was supported by grants from Cancer Research United Kingdom (A7572, A11699, and C22524) and Breast Cancer Campaign L1401. SBCS was supported by Yorkshire Cancer Research S295, S299, and S305PA. OSU was supported by the Stefanie Spielman Breast Fund and the Ohio State University Comprehensive Cancer Center.
Footnotes
See accompanying editorial on page 295
Processed as a Rapid Communication manuscript.
Supported by National Institutes of Health Grant No. CA128978, Specialized Program of Research Excellence in Breast Cancer Grant No. P50 CA116201 to Mayo Clinic, Breast Cancer Research Foundation, and a generous gift from David F. and Margaret T. Grohne Family Foundation.
Terms in blue are defined in the glossary, found at the end of this article and online at www.jco.org.
Authors' disclosures of potential conflicts of interest are found in the article online at www.jco.org. Author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Disclosures provided by the authors are available with this article at www.jco.org.
AUTHOR CONTRIBUTIONS
Conception and design: Fergus J. Couch, Steven N. Hart, Xianshu Wang, Susan L. Slager, Celine M. Vachon
Financial support: Fergus J. Couch, Celine M. Vachon
Administrative support: Fergus J. Couch, Emily Hallberg
Provision of study materials or patients: Fergus J. Couch, Steven N. Hart, Priyanka Sharma, Amanda Ewart Toland, Xianshu Wang, Penelope Miron, Janet E. Olson, Andrew K. Godwin, V. Shane Pankratz, Emily Hallberg, Lucia Guidugli, Jaime Davila, Matthias W. Beckmann, Wolfgang Janni, Brigitte Rack, Arif B. Ekici, Dennis Slamon, Irene Konstantopoulou, Florentia Fostira, George Fountzilas, Liisa Pelttari, William J. Tapper, Lorraine Durcan, Simon S. Cross, Robert Pilarski, Charles L. Shapiro, Jennifer Klemp, Song Yao, Judy Garber, Angela Cox, Hiltrud Brauch, Christine Ambrosone, Heli Nevanlinna, Drakoulis Yannoukakos, Susan L. Slager, Celine M. Vachon, Diana M. Eccles, Peter A. Fasching
Collection and assembly of data: Fergus J. Couch, Steven N. Hart, Priyanka Sharma, Xianshu Wang, Penelope Miron, Janet E. Olson, Andrew K. Godwin, Curtis Olswold, Seth Slettedahl, Emily Hallberg, Matthias W. Beckmann, Wolfgang Janni, Brigitte Rack, Arif B. Ekici, Dennis Slamon, Irene Konstantopoulou, Florentia Fostira, Athanassios Vratimos, George Fountzilas, Liisa Pelttari, William J. Tapper, Lorraine Durcan, Simon S. Cross, Robert Pilarski, Charles L. Shapiro, Jennifer Klemp, Song Yao, Judy Garber, Angela Cox, Hiltrud Brauch, Christine Ambrosone, Heli Nevanlinna, Drakoulis Yannoukakos, Susan L. Slager, Celine M. Vachon, Diana M. Eccles, Peter A. Fasching
Data analysis and interpretation: Fergus J. Couch, Steven N. Hart, Amanda Ewart Toland, Xianshu Wang, Janet E. Olson, V. Shane Pankratz, Curtis Olswold, Seth Slettedahl, Lucia Guidugli, Jaime Davila, Wolfgang Janni, Susan L. Slager, Celine M. Vachon, Diana M. Eccles, Peter A. Fasching
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Inherited Mutations in 17 Breast Cancer Susceptibility Genes Among a Large Triple-Negative Breast Cancer Cohort Unselected for Family History of Breast Cancer
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or jco.ascopubs.org/site/ifc.
Fergus J. Couch
Patents, Royalties, Other Intellectual Property: Patent with Myriad Genetics
Steven N. Hart
No relationship to disclose
Priyanka Sharma
Research Funding: Glaxo Smith Kline, Novartis, Celgene, Roche/Genentech, Myriad Genetics, Olympus (I), Cook Medical (I), NinePoint Medical (I)
Travel, Accommodations, Expenses: Novartis, Celgene
Amanda Ewart Toland
No relationship to disclose
Xianshu Wang
No relationship to disclose
Penelope Miron
No relationship to disclose
Janet E. Olson
No relationship to disclose
Andrew K. Godwin
Research Funding: Millennium (Inst), PeptiMed (Inst)
Patents, Royalties, Other Intellectual Property: University of Kansas Research Institute (Inst)
V. Shane Pankratz
No relationship to disclose
Curtis Olswold
No relationship to disclose
Seth Slettedahl
No relationship to disclose
Emily Hallberg
No relationship to disclose
Lucia Guidugli
No relationship to disclose
Jaime Davila
No relationship to disclose
Matthias W. Beckmann
No relationship to disclose
Wolfgang Janni
No relationship to disclose
Brigitte Rack
Honoraria: BiPar/sanofi-aventis, Novartis, Roche, Pfizer, Eli Lilly, Veridex, AstraZeneca
Speakers' Bureau: Roche
Research Funding: BiPar/sanofi-aventis, Novartis, Eli Lilly, AstraZeneca, Veridex
Travel, Accommodations, Expenses: Roche, Novartis, Veridex, Chugai Pharmaceutical
Arif B. Ekici
No relationship to disclose
Dennis Slamon
Leadership: BioMarin
Stock or Other Ownership: Pfizer
Honoraria: Pfizer
Consulting or Advisory Role: Novartis
Irene Konstantopoulou
No relationship to disclose
Florentia Fostira
No relationship to disclose
Athanassios Vratimos
No relationship to disclose
George Fountzilas
No relationship to disclose
Liisa Pelttari
No relationship to disclose
William J. Tapper
No relationship to disclose
Lorraine Durcan
No relationship to disclose
Simon S. Cross
No relationship to disclose
Robert Pilarski
No relationship to disclose
Charles L. Shapiro
No relationship to disclose
Jennifer Klemp
Stock or Other Ownership: Cancer Survivorship Training
Speakers' Bureau: Myriad Genetics, Novartis, Pfizer
Patents, Royalties, Other Intellectual Property: Cancer Survivorship Training: IP and license agreement
Song Yao
No relationship to disclose
Judy Garber
Consulting or Advisory Role: Pfizer, Pfizer (I), Novartis (I)
Other Relationship: Myriad Genetics, Pfizer (I), Novartis (I)
Angela Cox
No relationship to disclose
Hiltrud Brauch
No relationship to disclose
Christine Ambrosone
No relationship to disclose
Heli Nevanlinna
No relationship to disclose
Drakoulis Yannoukakos
No relationship to disclose
Susan L. Slager
No relationship to disclose
Celine M. Vachon
Employment: IMRIS (I)
Patents, Royalties, Other Intellectual Property: Breast density software: patent filed at Mayo Clinic
Diana M. Eccles
Consulting or Advisory Role: AstraZeneca
Research Funding: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca
Peter A. Fasching
Honoraria: Amgen, Novartis, Pfizer, Genomic Health, Roche
Consulting or Advisory Role: Novartis, Genomic Health, Roche
Speakers' Bureau: Novartis, Roche, Amgen, Genomic Health, Pfizer
Research Funding: Novartis, Amgen
REFERENCES
- 1.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Breast Cancer Facts & Figures, 2011-2012. http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-030975.pdf.
- 3.Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J Clin Oncol. 2008;26:1275–1281. doi: 10.1200/JCO.2007.14.4147. [DOI] [PubMed] [Google Scholar]
- 4.Haffty BG, Silber A, Matloff E, et al. Racial differences in the incidence of BRCA1 and BRCA2 mutations in a cohort of early onset breast cancer patients: African American compared to white women. J Med Genet. 2006;43:133–137. doi: 10.1136/jmg.2005.034744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blows FM, Driver KE, Schmidt MK, et al. Subtyping of breast cancer by immunohistochemistry to investigate a relationship between subtype and short and long term survival: A collaborative analysis of data for 10,159 cases from 12 studies. PLoS Med. 2010;7:e1000279. doi: 10.1371/journal.pmed.1000279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Atchley DP, Albarracin CT, Lopez A, et al. Clinical and pathologic characteristics of patients with BRCA-positive and BRCA-negative breast cancer. J Clin Oncol. 2008;26:4282–4288. doi: 10.1200/JCO.2008.16.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Comen EA, Davids M, Kirchhoff T, et al. Prevalence of BRCA1 and BRCA2 mutations in Jewish women with triple negative breast cancer. J Clin Oncol. 2008;26(suppl):749s. abstr 22002. [Google Scholar]
- 8.Robertson L, Hanson H, Seal S, et al. BRCA1 testing should be offered to individuals with triple-negative breast cancer diagnosed below 50 years. Br J Cancer. 2012;106:1234–1238. doi: 10.1038/bjc.2012.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans DG, Howell A, Ward D, et al. Prevalence of BRCA1 and BRCA2 mutations in triple negative breast cancer. J Med Genet. 2011;48:520–522. doi: 10.1136/jmedgenet-2011-100006. [DOI] [PubMed] [Google Scholar]
- 10.Andrés R, Pajares I, Balmaña J, et al. Association of BRCA1 germline mutations in young onset triple-negative breast cancer (TNBC) Clin Transl Oncol. 2014;16:280–284. doi: 10.1007/s12094-013-1070-9. [DOI] [PubMed] [Google Scholar]
- 11.Young SR, Pilarski RT, Donenberg T, et al. The prevalence of BRCA1 mutations among young women with triple-negative breast cancer. BMC Cancer. 2009;9:86. doi: 10.1186/1471-2407-9-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez-Angulo AM, Timms KM, Liu S, et al. Incidence and outcome of BRCA mutations in unselected patients with triple receptor-negative breast cancer. Clin Cancer Res. 2011;17:1082–1089. doi: 10.1158/1078-0432.CCR-10-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comen E, Davids M, Kirchhoff T, et al. Relative contributions of BRCA1 and BRCA2 mutations to “triple-negative” breast cancer in Ashkenazi Women. Breast Cancer Res Treat. 2011;129:185–190. doi: 10.1007/s10549-011-1433-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartman AR, Kaldate RR, Sailer LM, et al. Prevalence of BRCA mutations in an unselected population of triple-negative breast cancer. Cancer. 2012;118:2787–2795. doi: 10.1002/cncr.26576. [DOI] [PubMed] [Google Scholar]
- 15.Laduca H, Stuenkel AJ, Dolinsky JS, et al. Utilization of multigene panels in hereditary cancer predisposition testing: Analysis of more than 2,000 patients. Genet Med. doi: 10.1038/gim.2014.40. [epub ahead of print on April 24, 2014] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H, Handsaker B, Wysoker A, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu X, Jian X, Boerwinkle E. DbNSFP: A lightweight database of human nonsynonymous SNPs and their functional predictions. Hum Mutat. 2011;32:894–899. doi: 10.1002/humu.21517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tavtigian SV, Deffenbaugh AM, Yin L, et al. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet. 2006;43:295–305. doi: 10.1136/jmg.2005.033878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens PJ, Tarpey PS, Davies H, et al. The landscape of cancer genes and mutational processes in breast cancer. Nature. 2012;486:400–404. doi: 10.1038/nature11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindor NM, Guidugli L, Wang X, et al. A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS) Hum Mutat. 2012;33:8–21. doi: 10.1002/humu.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erkko H, Dowty JG, Nikkilä J, et al. Penetrance analysis of the PALB2 c. 1592delT founder mutation. Clin Cancer Res. 2008;14:4667–4671. doi: 10.1158/1078-0432.CCR-08-0210. [DOI] [PubMed] [Google Scholar]
- 26.Weischer M, Nordestgaard BG, Pharoah P, et al. CHEK2*1100delC heterozygosity in women with breast cancer associated with early death, breast cancer-specific death, and increased risk of a second breast cancer. J Clin Oncol. 2012;30:4308–4316. doi: 10.1200/JCO.2012.42.7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farrugia DJ, Agarwal MK, Pankratz VS, et al. Functional assays for classification of BRCA2 variants of uncertain significance. Cancer Res. 2008;68:3523–3531. doi: 10.1158/0008-5472.CAN-07-1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guidugli L, Pankratz VS, Singh N, et al. A classification model for BRCA2 DNA binding domain missense variants based on homology-directed repair activity. Cancer Res. 2013;73:265–275. doi: 10.1158/0008-5472.CAN-12-2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Easton DF, Deffenbaugh AM, Pruss D, et al. A systematic genetic assessment of 1,433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet. 2007;81:873–883. doi: 10.1086/521032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spurdle AB, Whiley PJ, Thompson B, et al. BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J Med Genet. 2012;49:525–532. doi: 10.1136/jmedgenet-2012-101037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Von Minckwitz G, Hahnen E, Fasching PA, et al. Pathological complete response (pCR) rates after carboplatin-containing neoadjuvant chemotherapy in patients with Germline BRCA (gBRCA) mutations and triple-negative breast cancer (TNBC): results from GeparSixto. J Clin Oncol. 2014;(suppl 15s):32. abstr 1005. [Google Scholar]
- 32.Antoniou AC, Casadei S, Heikkinen T, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rahman N, Seal S, Thompson D, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robson M. Multigene panel testing: Planning the next generation of research studies in clinical cancer genetics. J Clin Oncol. 2014;32:1987–1989. doi: 10.1200/JCO.2014.56.0474. [DOI] [PubMed] [Google Scholar]
- 35.Domchek SM, Bradbury A, Garber JE, et al. Multiplex genetic testing for cancer susceptibility: Out on the high wire without a net? J Clin Oncol. 2013;31:1267–1270. doi: 10.1200/JCO.2012.46.9403. [DOI] [PubMed] [Google Scholar]
- 36.Metcalfe K, Gershman S, Ghadirian P, et al. Contralateral mastectomy and survival after breast cancer in carriers of BRCA1 and BRCA2 mutations: Retrospective analysis. BMJ. 2014;348:g226. doi: 10.1136/bmj.g226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metcalfe KA, Lynch HT, Synder CL, et al. The impact of oophorectomy on survival after breast cancer in BRCA1 and BRCA2 mutation carriers. J Clin Oncol. 2014;(suppl 15s):32. doi: 10.1001/jamaoncol.2015.0658. abstr 1507. [DOI] [PubMed] [Google Scholar]
- 38.Tung N, Gaughan E, Hacker MR, et al. Outcome of triple negative breast cancer: Comparison of sporadic and BRCA1-associated cancers. Breast Cancer Res Treat. 2014;146:175–182. doi: 10.1007/s10549-014-2995-6. [DOI] [PubMed] [Google Scholar]
- 39.Finch AP, Lubinski J, Møller P, et al. Impact of oophorectomy on cancer incidence and mortality in women with a BRCA1 or BRCA2 mutation. J Clin Oncol. 2014;32:1547–1553. doi: 10.1200/JCO.2013.53.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tutt A, Robson M, Garber JE, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: A proof-of-concept trial. Lancet. 2010;376:235–244. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- 41.Buisson R, Dion-Côté AM, Coulombe Y, et al. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Byrski T, Gronwald J, Huzarski T, et al. Pathologic complete response rates in young women with BRCA1-positive breast cancers after neoadjuvant chemotherapy. J Clin Oncol. 2010;28:375–379. doi: 10.1200/JCO.2008.20.7019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.