SUMMARY
Inappropriate inflammasome activation contributes to multiple human diseases, but the mechanisms by which inflammasomes are suppressed are poorly understood. The NFκB inhibitor A20 is a ubiquitin-modifying enzyme that may prevent human inflammatory diseases and lymphomas. Here, we report that A20-deficient macrophages, unlike normal cells, exhibit spontaneous NLRP3 inflammasome activity to LPS alone. The kinase RIPK3, but not the adaptor MyD88, is required for this response. In normal cells, A20 constitutively associates with caspase-1 and pro-IL-1β, and NLRP3 activation further promotes A20 recruitment to the inflammasome. Pro-IL-1β also co-immunoprecipitates with RIPK1, RIPK3, caspase-1 and caspase-8 in a complex that is modified with K63-linked and unanchored polyubiquitin. In A20-deficient macrophages, this pro-IL-1β-associated ubiquitination is markedly increased in a RIPK3-dependent manner. Mass spectrometric and mutational analyses reveal that K133 of pro-IL-1β is a physiological ubiquitination site that supports processing. Our study reveals a novel mechanism by which A20 prevents inflammatory diseases.
INTRODUCTION
Secretion of the pro-inflammatory cytokines, interleukin-1β (IL-1β) and IL-18, is normally regulated by at least two distinct signals (Martinon et al., 2002). First, expression of the inactive pro-proteins requires nuclear factor-κ B (NF-κB) activity transduced by pattern recognition receptors (PRRs) such as toll-like receptors (TLRs). Subsequently, the proteolytic processing of these cytokines usually requires caspase-1 (Casp1) activation by cytosolic PRRs possessing a pyrin domain and/or caspase activation and recruitment domain (CARD) (Davis et al., 2011; Martinon et al., 2009). Upon detecting the presence of microbes or “danger signals”, these proteins recruit Casp1 either directly or via the adaptor protein, ASC (apoptosis-associated speck-like protein containing a C-terminal CARD), to form a signaling complex called the inflammasome. Though critical for efficient pathogen clearance, exaggerated inflammasome functions can play pathogenic roles in various autoimmune, allergic, and inflammatory disorders (Lamkanfi and Dixit, 2012; Strowig et al., 2012). The mechanisms by which inflammasomes are regulated are incompletely understood.
The NLRP3 (nucleotide-binding domain and leucine-rich repeats containing pyrin domain 3) inflammasome is normally induced by large particulates such as uric acid crystals, cholesterol crystals, amyloid or misfolded protein, and asbestos (Leemans et al., 2011). Exaggerated activities of NLRP3 to these particulates are implicated in the pathogenesis of gout, type-2 diabetes, atherosclerosis, neurodegenerative diseases, and asthma (Davis et al., 2011). How NLRP3 responds to such diverse stimuli is incompletely understood but may involve lysosomal rupture, reactive oxygen species (ROS), or mitochondrial damage (Leemans et al., 2011). The loss of intracellular K+ ions appears to be required, as blocking K+ efflux suppresses NLRP3 activities (Muñoz-Planillo et al., 2013). NLRP3 can also sense cellular damage from within or nearby cells by the release of ATP, which causes K+ efflux through the P2X7 channel (Mariathasan et al., 2006). In response to Gram-negative bacteria, NLRP3 activation also requires the upregulation of Casp11 by type I interferons (IFNs) (Kayagaki et al., 2011; Rathinam et al., 2012). While several molecules required for inflammasome activation have been described, inhibitors of these protein complexes have not been completely defined.
Covalent conjugation of ubiquitin (Ub) molecules to target proteins, termed ubiquitination, regulates protein stability and protein-protein interactions in signaling complexes. Poly-Ub chains can be linked via N-terminal amino groups on Met1 (termed linear chains) or via ε-amino groups on any of the seven lysines of Ub. These distinct poly-Ub conformations are recognized by specific Ub sensor proteins that facilitate the localization, activity, and interacting partners of ubiquitinated proteins (Corn and Vucic, 2014). In particular, K48-linked poly-Ub chains generally target proteins for proteasomal degradation, whereas linear and K63-linked chains primarily serve as a scaffolding network for the formation of signaling complexes.
A20, encoded by the tumor necrosis factor-α (TNFα)-induced protein 3 (TNFAIP3) gene, is a potent regulator of Ub-dependent signaling (Shembade and Harhaj, 2012; Verstrepen et al., 2010). Polymorphisms in the human TNFAIP3 gene locus are associated with a number of inflammatory diseases, suggesting that A20 prevents the incidence and/or severity of these diseases (Catrysse et al., 2014; Ma and Malynn, 2012). Correspondingly, A20-deficient mice spontaneously develop a systemic inflammatory syndrome that culminates in premature death (Lee et al., 2000), while cell-specific deletions of A20 cause diseases in mice reminiscent of those in humans (Chu et al., 2011; Hammer et al., 2011; Heger et al., 2014; Kool et al., 2011; Matmati et al., 2011; Tavares et al., 2010; Vereecke et al., 2010). Deficiency in MyD88 (myeloid differentiation primary response 88) or commensal depletion abrogates the severity of several of these conditions, indicating that A20 restricts TLR and IL-1-receptor (IL-1R) family signaling (Boone et al., 2004; Turer et al., 2008). Hence, A20 appears critically positioned at the interface of host responses to commensal microbes. Given the genetic links between inflammasomes and A20 with human inflammatory diseases, we have investigated the potential role of A20 in regulating inflammasomes.
RESULTS
A20 prevents spontaneous secretion of IL-1β and IL-18
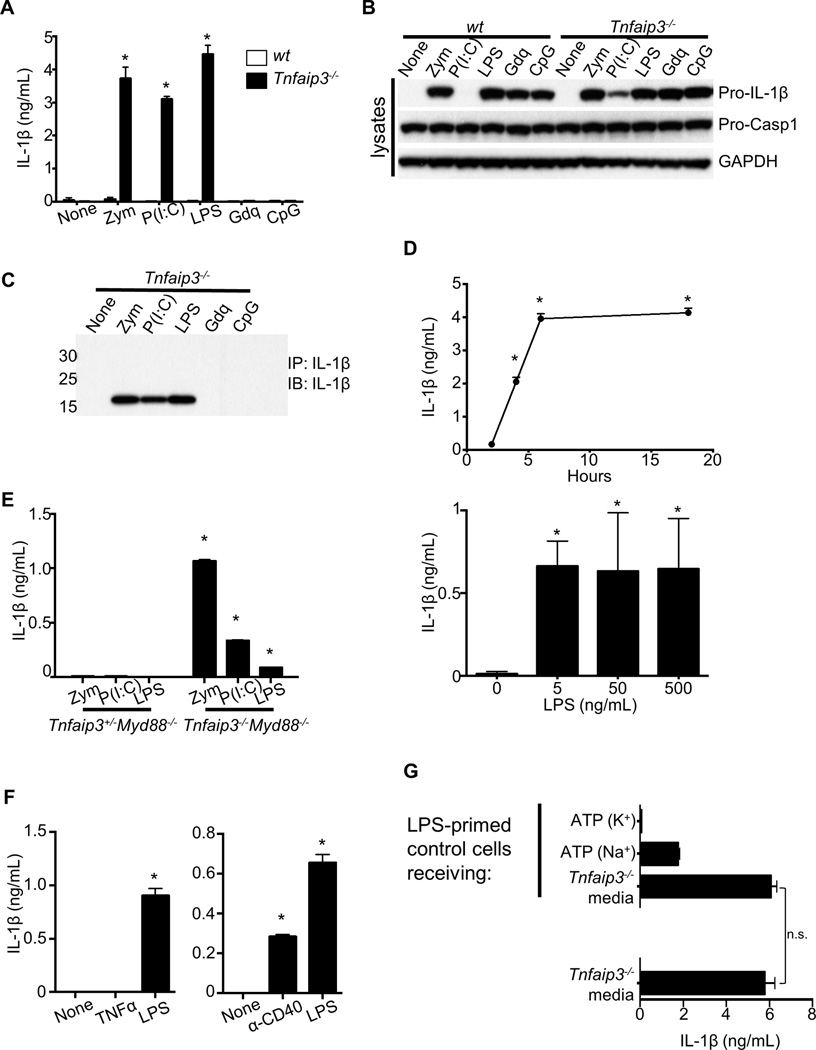
To determine whether A20 regulates inflammasome activity, we stimulated bone marrow-derived macrophages (BMDMs) from Tnfaip3−/− and control mice with various TLR ligands. Consistent with the two-signal requirement for IL-1β secretion (Martinon et al., 2002), TLR ligands alone induced only the intracellular expression of pro-IL-1β in wild-type (wt) cells (Figures 1A, 1B). No mature IL-1β was detected in the supernatants (Figure 1A). Surprisingly, even without a second signal for inflammasome activation, Tnfaip3−/− BMDMs secreted IL-1β in response to zymosan (Zym), poly(I:C), and LPS (Figure 1A). Similar results were obtained for the secretion of IL-18 (not shown). Other TLR ligands induced the expression of pro-IL-1β in both Tnfaip3−/− and wt cells but failed to induce its processing and secretion (Figures 1B and 1A). To determine whether the secreted IL-1β was proteolytically mature, we immunoprecipitated (IP’d) IL-1β from the supernatants of TLR-stimulated Tnfaip3−/− cells and assessed the molecular weight of the secreted IL-1β by immunoblotting (IB). These experiments confirmed the IL-1β secretion in each case to be the processed 17 kDa form (Figure 1C). The secretion of IL-1β from Tnfaip3−/− cells was detected as early as 2 h after LPS stimulation and at LPS doses as low as 5 ng/mL (Figure 1D). Thus, A20 prevents spontaneous pro-IL-1β processing and secretion in cells stimulated with Zym, poly(I:C), and LPS.
Figure 1. A20 deficiency causes secretion of mature IL-1β in the absence of an exogenous second signal for inflammasome activation.
(A–C) BMDMs cultured from wt or Tnfaip3−/− mice were stimulated with the following ligands for 15 h: 10 µg/mL Zym, 100 µg/mL poly(I:C) (P(I:C)), 500 ng/mL LPS, 3 µg/mL Gdq, and 5 µM CpG. (A) Supernatants were analyzed by ELISA for IL-1β secretion. (B) Whole cell lysates were analyzed by IB for the indicated proteins after 6 h stimulation of each ligand as in A. (C) IL-1β was IP’d from supernatants and analyzed by IB to confirm proteolytic processing. (D) IL-1β secretions by Tnfaip3−/− BMDMs in response to 500 ng/mL LPS for 0–18 h, or 0–500 ng/mL LPS for 18 h, were quantitated by ELISA. (E) IL-1β secretions in response to Dectin-1, TLR3, and TLR4 ligands by A20-sufficient and -deficient BMDMs on Myd88−/− background were measured after ~15 h. Note TLR2 cannot respond to Zym without MyD88. (F) IL-1β secretions by A20-deficient cells after ~15 h of TNFα or anti-CD40 stimulation were measured. (G) Supernatant from A20-deficient cells stimulated with LPS for ~15 h (bottom of graph) was transferred to LPS-primed control cells. After overnight incubation, the supernatant was measured again for any additional IL-1β secretion by control cells. As controls for NLRP3 responsiveness, IL-1β responses to ATP by control cells in the presence or absence K+ efflux were measured. To eliminate any potential complications by autocrine TNFα signaling, both control and A20-deficient cells used here were on Tnf−/− background. Each bar in graphs represents mean of triplicate wells + s.d. * denotes statistical significance by student t test (p < 0.05) when comparison is made between control and A20-deficient cells for each condition. n.s. = not statistically significant. All data presented here and hereafter are representative of at least 3 independent experiments.
Zymosan can activate MyD88-dependent as well as independent pathways through TLR2 and Dectin-1, respectively (Brown et al., 2002; Underhill et al., 1999), and both MyD88 and TRIF (TLR/IL-1R domain-containing adaptor inducing IFN) are involved in TLR4 responses to LPS (Hoebe et al., 2003). We therefore assessed the contribution of MyD88 to the spontaneous IL-1β secretion induced by these ligands in Tnfaip3−/− cells. Tnfaip3−/−Myd88−/− cells retained the ability to process and secrete IL-1β upon Zym or LPS stimulation (Figure 1E). Predictably, the IL-1β responses of A20-deficient cells to poly(I:C) were unaffected by MyD88 deficiency, as TLR3 uses only TRIF to signal (Figure 1E). Hence, TRIF-dependent signals and other MyD88-independent signals are sufficient to drive pro-IL-1β processing in the absence of A20. To determine whether other MyD88-independent signals beside TLR3 and Dectin-1 could induce IL-1β secretion, we stimulated Tnfaip3−/− cells with TNFα or agonistic anti-CD40 antibody. We found that anti-CD40, but not TNFα, triggered IL-1β secretion in these cells (Figure 1F). Additionally, TNFα deficiency had no effects on IL-1β secretion by A20-deficient cells in response to all of these stimuli (not shown), ruling out involvement of TNFα autocrine signaling.
TLR3 and TLR4 can trigger programmed necrosis under certain conditions (Feoktistova et al., 2011; He et al., 2011; Kaiser et al., 2013). Thus, secretion of IL-1β by Tnfaip3−/− cells could result from the release of danger-associated molecular patterns (DAMPs) by dying cells (e.g., ATP) to activate inflammasomes. To test whether Tnfaip3−/− cells might release such ligands, we stimulated LPS-primed control BMDMs with supernatants from LPS-stimulated Tnfaip3−/− cells and assayed for additional IL-1β production by control cells. We found that Tnfaip3−/− BMDMderived supernatant failed to stimulate additional IL-1β secretion from Tnfaip3+/+ cells (Figure 1G). In contrast, addition of ATP to these cells induced robust IL-1β secretion that could be inhibited by preventing K+ efflux, as predicted for NLRP3 activation (Figure 1G). Taken together, these experiments suggest that A20 suppresses spontaneous IL-1β production in a cell autonomous fashion.
A20 suppresses NLRP3 inflammasome functions
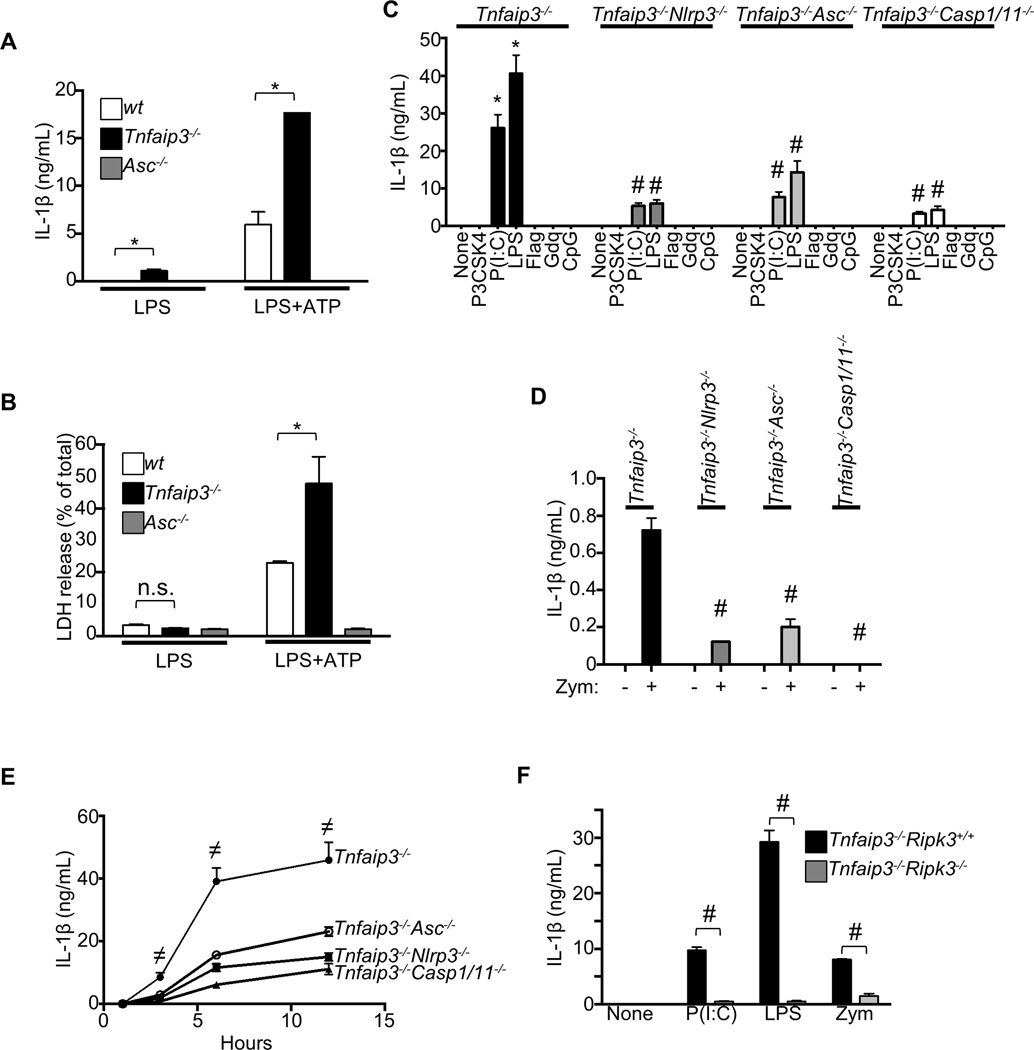
We next determined whether A20 could also inhibit NLRP3 responses to a second signal. Addition of ATP to A20-deficient cells after 4 h of LPS stimulation induced massive IL-1β secretion compared to wt cells (Figure 2A). Hence, A20 limits the magnitude of NLRP3 inflammasomes triggered by LPS plus ATP, as well as preventing spontaneous IL-1β secretion. Beside cleaving pro-IL-1β/18, overt Casp1 activity can also cause pyroptosis, an inflammatory form of lytic cell death (Davis et al., 2011). Accordingly, considerably more Tnfaip3−/− cells died in response to LPS plus ATP than control cells (Figure 2B). These results indicate that A20 also restricts pyroptosis induced by NLRP3 inflammasome activation. Of note, the amounts of cell death did not differ between wt and Tnfaip3−/− cultures in response to LPS alone (Figure 2B), confirming that Tnfaip3−/− cells did not die necrotically and release DAMPs as second signals for inflammasome activation (Figure 1G).
Figure 2. Spontaneous NLRP3 activity in Tnfaip3−/− cells depends on RIPK3.
(A–B) (A) IL-1β responses by wt or Tnfaip3−/− BMDMs to ATP were measured by ELISA. Cells were prestimulated with LPS for 4 h (IL-1β secretions displayed on left of graph), followed by 30-min pulse with 5 mM ATP. 3 h after addition of fresh media, IL-1β responses were measured (right of graph). (B) Pyroptotic responses to ATP were measured by LDH release assays. (C–E) Effects of NLRP3, ASC, or Casp1/Casp11 deficiency on IL-1β secretion by Tnfaip3−/− cells were assessed. Single and compound mutants were stimulated with the indicated ligands for ~15 h (CD) or with 500 ng/mL LPS for the indicated time (E). The ligands used in C and D were 1 µg/mL Pam3CSK4 (P3CSK4), 100 µg/mL poly(I:C), 500 ng/mL LPS, 100 ng/mL flagellin (Flag), 3 µg/mL Gdq, and 5 µM CpG. (F) Effects of RIPK3 deficiency on IL-1β responses by A20-deficient cells to poly(I:C), LPS, and Zym were assessed by ELISA. Each bar in graphs represents mean of triplicate wells + s.d. * denotes statistical significance (p < 0.05) when comparison is made between control and A20-deficient cells for each condition. # denotes statistical significance by student t test (p < 0.05) when comparison is made between A20 deficiency alone with the indicated compound mutations. ≠ denotes statistically significant contribution by NLRP3, ASC, or Casp1 and Casp11 functions to spontaneous IL-1β secretion by A20-deficient cells.
NLRP3, ASC, and Casp1 are all critical components of inflammasomes. To assess the potential contributions of each of these proteins to the spontaneous IL-1β secretion observed in A20-deficient cells, we interbred Tnfaip3−/− mice with Nlrp3−/−, Asc−/− and Casp1/11−/− mice. BMDMs from all three compound mutant strains secreted 65–100% less IL-1β than Tnfaip3−/− cells after stimulation with poly(I:C), LPS, or Zym (Figures 2C, 2D, 2E). These results indicate that A20 restricts the spontaneous assembly of NLRP3 with ASC and Casp1 in response to these stimuli. Because Casp1-deficient mice also harbor mutant Casp11 allele from 129 strain (Kayagaki et al., 2011), the residual mature IL-1β secretions by Tnfaip3−/−Casp1/11−/− compound mutant cells indicate that A20-deficient cells can additionally process pro-IL-1β through a mechanism that is independent of both Casp1 and Casp11 (Figures 2C, 2E, and not shown).
Depletion of all three baculoviral inhibitor of apoptosis protein (IAP) repeat-containing enzymes (BIRC2, BIRC3, and BIRC4) in BMDMs has been reported to result in spontaneous NLRP3 inflammasome activation by a RIPK3 (receptor-interacting protein kinase 3)-dependent mechanism (Vince et al., 2012). We thus asked whether a similar pathway was responsible for the spontaneous NLRP3 activation in A20-deficient cells. We interbred Tnfaip3−/− mice with Ripk3−/− mice and assayed their BMDMs for inflammasome activity. Tnfaip3−/−Ripk3−/− double mutant BMDMs exhibited drastically reduced IL-1β secretion in response to poly(I:C), LPS, or Zym when compared to Tnfaip3−/− cells (Figure 2F). Thus, RIPK3 is critical for the spontaneous NLRP3 activity in A20-deficient cells.
A20 restricts NLRP3 activity independently of its role in NFκB regulation
The expression of both pro-IL-1β (Figure 1B) and NLRP3 is induced by NFκB activation (Bauernfeind et al., 2009). As A20 restricts TLR-induced NFκB signaling (Boone et al., 2004; Turer et al., 2008), we determined whether the spontaneous NLRP3 inflammasome activity in A20-deficient cells results from increased pro-IL-1β or NLRP3 expression. We noted the amounts of pro-IL-1β expressed in wt and Tnfaip3−/− cells induced by various TLR ligands correlated poorly with the amounts of secreted mature IL-1β (Figures 1A, 1B). For example, poly(I:C) stimulation induced much less pro-IL-1β expression than CpG or gardiquimod (Gdq) from Tnfaip3−/− cells (Figure 1B); yet, poly(I:C) induced robust IL-1β secretion while Gdq and CpG did not (Figures 1A, 1C). More dramatically, poly(I:C) induced much less pro-IL-1β expression in Tnfaip3−/− cells than wt cells stimulated with Zym, LPS, Gdq, or CpG (Figure 1B); yet, poly(I:C)-stimulated Tnfaip3−/− cells readily processed pro-IL-1β for secretion while wt cells failed to do so with all TLR ligands tested (Figures 1A, 1C). We next assayed for NLRP3 expression in control and Tnfaip3−/− cells in response to various TLR ligands. The NLRP3 expression patterns paralleled those of pro-IL-1β (Figure S1A). These data strongly suggest that A20 regulates NLRP3 activity in a manner separate from regulating pro-IL-1β and NLRP3 transcription.
Both TRIF and MyD88 adaptor molecules participate in TLR4 signaling (Hoebe et al., 2003). Hence, we determined to what extent MyD88 deficiency might reduce the NLRP3 and pro-IL-1β expression in A20-deficient cells. Upon LPS stimulation, MyD88 deficiency decreased the amounts of NLRP3 and pro-IL-1β proteins in Tnfaip3−/−Myd88−/− cells to those of wt cells (Figure S1B). Yet, Tnfaip3−/−Myd88−/− cells secreted mature IL-1β in response to LPS (Figure 1E) while wt cells did not (Figure 1A). These data suggest that A20 restrains a TRIF-dependent signal that can otherwise cause aberrant NLRP3 activation.
Unlike Tnfaip3−/− single mutant cells, Tnfaip3−/−Ripk3−/− compound mutant cells secreted little, if any, processed IL-1β (Figure 2F). Although RIPK3 has been reported to play no roles in TLR-induced transcriptional responses (Newton et al., 2004), we entertained the possibility that RIPK3 deficiency might aberrantly affect the NFκB-induced transcriptional responses in A20-deficient cells. We found the kinetics and magnitudes of LPS-induced IκBα phosphorylation and degradation to be similar between Tnfaip3−/− single mutant and Tnfaip3−/−Ripk3−/− compound mutant cells (Figure S1C). Moreover, Tnfaip3−/−Ripk3−/− cells expressed similar amounts of pro-IL-1β and NLRP3 as Tnfaip3−/− single mutant cells (Figure S1C). Taken together, these observations suggest that A20 inhibits a TRIF- and RIPK3-dependent step in NLRP3 inflammasome activation that is distinct from TLR-triggered transcription.
A20 interacts with inflammasome proteins
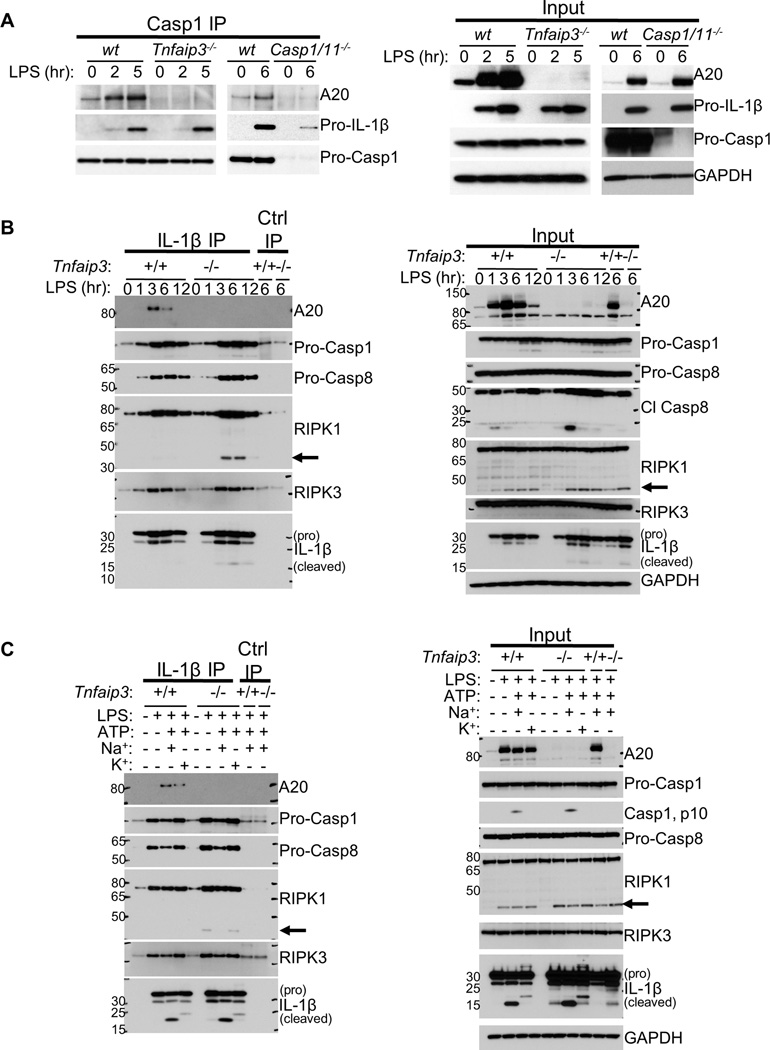
Our observations that A20 restricts LPS-induced spontaneous NLRP3 activation independently of its inhibitory role in NFκB signaling suggest that A20 may directly act on inflammasomes. To investigate whether A20 associates with inflammasome proteins, we IP’d Casp1 from BMDMs stimulated with LPS for 2–6 h and immunoblotted for A20. Inducible interaction between Casp1 and A20 was evident upon LPS stimulation alone that was not detected in LPS-stimulated Casp1/11−/− BMDMs (Figure 3A). To further investigate A20 interaction with inflammasome proteins, we IP’d pro-IL-1β from LPS-stimulated control and Tnfaip3−/− cells. Remarkably, A20 also co-IP’d with pro-IL-1β from Tnfaip3+/+ lysates, with maximal association occurring 3 h after LPS stimulation (Figure 3B). The presence of cleaved IL-1β was also evident in the Tnfaip3−/− IPs (Figure 3B). As we observed exaggerated NLRP3 activities in A20-deficient BMDMs to LPS plus ATP stimulation (Figures 2A, 2B), we asked whether ATP stimulation induced additional A20 recruitment to pro-IL-1β. We found that LPS-primed control cells recruited more A20 to pro-IL-1β within 15 minutes of ATP addition (Figure 3C). Blocking K+ efflux had minimal effect on the A20-pro-IL-1β association, suggesting that A20 is recruited to pro-IL-1β prior to P2X7-induced NLRP3 activation. Hence, A20 interacts with pro-IL-1β after LPS stimulation, and NLRP3 inflammasome activation by ATP further enhances these interactions.
Figure 3. A20 associates with pro-IL-1β complex consisting of Casp1, Casp8, RIPK1 and RIPK3.
(A) BMDMs cultured from wt, Tnfaip3−/−, or casp1/11−/− mice were stimulated with 500 ng/mL LPS for 0–6 h, lysed, and subjected to Casp1 IP, followed by IB analyses for A20 and pro-IL-1β interaction. Input for each IP is shown on the right. (B) Pro-IL-1β was IP’d from lysates of control and A20-deficient BMDMs stimulated with 500 ng/mL LPS for 0–12 h. Coassociation of each indicated protein with pro-IL-1β was analyzed by IB. Input IB analyses are shown on the right. (C) Control and A20-deficient cells were stimulated with LPS for 6 h, followed by 5 mM ATP for 30 minutes. Pro-IL-1β IPs were performed as in B to assay for changes in the interaction of each indicated protein with pro-IL-1β. To avoid potential complications by TNFα autocrine signaling to the assays in B and C, control and A20-deficient cells on Tnf−/− background were used. Similar results have been obtained using cells on Tnf+/+ background. Arrows indicate cleaved RIPK1.
Unexpectedly, the IP experiments above also revealed Casp1-pro-IL-1β pre-association in wt cells (Figures 3A, 3B, 3C), despite the absence of pro-IL-1β processing (Fig 1A). We also found Casp8, RIPK1, and RIPK3 to be present in this complex (Figures 3B, 3C). Since RIPK3 is necessary for the spontaneous secretion of IL-1β by A20-deficient cells (Figure 2F), these intriguing results suggest that (i) pro-IL-1β pre-associates with Casp1 prior to inflammasome activation and (ii) the absence of A20 allows for this complex to proceed with pro-IL-1β processing in a RIPK3-dependent manner. We thus determined the functional consequences of A20 deficiency on this complex. More Casp1, Casp8, and RIPK1 were associated with pro-IL-1β in A20-deficient cells (Figures 3B, 3C). A fraction of RIPK1 was cleaved in pro-IL-1β complexes from A20-deficient cells, suggesting the presence of active Casp8 (Figure 3B, arrows), since RIPK1 is a Casp8 substrate (Humphries et al., 2014). This observation is consistent with the increased active (cleaved) Casp8 in the lysates (Figure 3B). As Casp8 can cleave pro-IL-1β (Bossaller et al., 2012; Gringhuis et al., 2012; Gurung et al., 2014; Maelfait et al., 2008; Vince et al., 2012), the active Casp8 in this complex may be responsible for the residual IL-1β secretion by Tnfaip3−/−Casp1/11−/− cells (Figures 2C, 2E). This possibility was supported by our observation that this residual IL-1β secretion was inhibited by the Casp8 inhibitor, Z-IETD-FMK (not shown). Finally, ATP stimulation of either control or A20-deficient cells following LPS caused markedly more IL-1β cleavage, but did not augment Casp1 or Casp8 association with pro-IL-1β (Figure 3C). Hence, ATP activates—and A20 inhibits—the activities of Casp1 and/or Casp8 molecules that are pre-bound to pro-IL-1β.
RIPK1 catalytic activity promotes spontaneous pro-IL-1β processing in A20-deficient cells
RIPK3 activation usually requires the kinase activity of RIPK1 during programmed necrosis (Humphries et al., 2014). As we observed a requirement for RIPK3 in the spontaneous pro-IL-1β processing by A20-deficient cells (Figure 2F), as well as the presence of RIPK1 in the pro-IL-1β complex (Figures 3B, 3C, 4D), we asked whether RIPK1 activity was similarly required. Treatment of A20-deficient cells with LPS plus the RIPK1 inhibitor, necrostatin-1 (Nec1) (Degterev et al., 2008), reduced pro-IL-1β processing in a dose-dependent manner (Figure S2A). To determine whether the RIPK1 activity was accompanied by formation of RIPK1-RIPK3 complexes, or ripoptosomes (Feoktistova et al., 2011; Tenev et al., 2011), we IP’d RIPK1 from LPS-stimulated control and A20-deficient cells. More pro-IL-1β was co-IP’d with RIPK1 from A20-deficient cells (Figure S2B), confirming the RIPK1-pro-IL-1β association observed by pro-IL-1β IPs (Figures 3B, 3C). However, no increased RIPK1-RIPK3 interaction was observed regardless of LPS stimulation (Figure S2B). Taken together, our data suggest that A20 inhibits RIPK1 association with pro-IL-1β, RIPK1 catalytic activity and RIPK3 drive spontaneous pro-IL-1β processing in A20-deficient cells without ripoptosome formation.
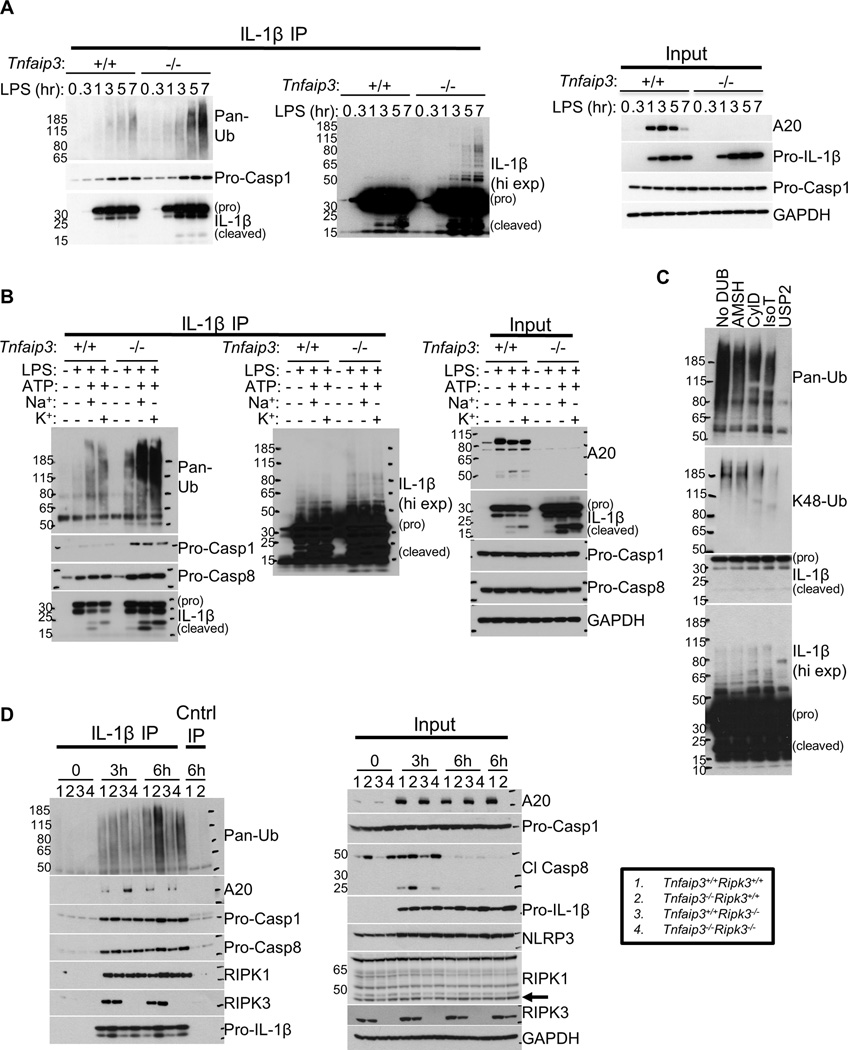
Figure 4. Pro-IL-1β in A20-deficient cells associate with RIPK3-dependent K63-linked and unanchored poly-Ub.
(A) wt and Tnfaip3−/− BMDMs were stimulated with 500 ng/mL LPS for 0–7 h. Pro-IL-1β IPs were performed as in Figure 3B and analyzed for Ub signals and Casp1 interaction. Input IB analyses are shown on the right. (B) Control and A20-deficient cells on Tnf−/− background were primed with 500 ng/mL LPS for 6 h and subsequently stimulated with 5 mM ATP for 15–20 min. Pro-IL-1β IPs, followed by IB analyses, were performed as in Figure 3C. IB analyses of each reaction input are shown on the right. (C) Pro-IL-1β IPs from A20-deficient BMDMs stimulated with 500 ng/mL LPS for 6 h were subjected to DUB proteolysis. Ub signals were subsequently analyzed by IB. (D) Control or A20-defcient BMDMs with or without RIPK3 deficiency were stimulated with 500 ng/mL LPS for 6 h. Subsequently, Pro-IL-1β IPs, followed by IB analyses, were performed to assay for Ub signals and the interaction of each indicated protein with pro-IL-1β. Arrow indicates cleaved RIPK1. Note the anti-IL-1β antibody used in IB analyses cannot recognize IL-1β that is modified by >10 Ub molecules, perhaps due to steric hindrance by the Ub modification.
Pro-IL-1β complex contains K63-linked and unanchored poly-Ub
As A20 restricts NFκB signaling by modifying Ub chains on key signaling proteins, we investigated whether inflammasome activation involves ubiquitination. We IP’d pro-IL-1β from LPS-stimulated wt or Tnfaip3−/− cells and probed for Ub. A high molecular weight complex containing Ub was evident in pro-IL-1β IPs from both wt and Tnfaip3−/− BMDMs (Figure 4A). Longer exposures of IL-1β immunoblot following pro-IL-1β IPs also revealed that pro-IL-1β itself is modified by high molecular weight moieties with laddering patterns suggestive of ubiquitination (Figure 4A, middle panel). Strikingly, ATP robustly augmented the formation of this Ub-containing complex in both control and A20-deficient cells (Figure 4B, left panel, 3rd and 7th lanes, suggesting that NLRP3 inflammasome are ubiquitinated. Moreover, the pro-IL-1β complex ubiquitination was markedly increased in A20-deficient cells (Figure 4A), and the timing of pro-IL-1β-associated Ub signals in A20-deficient cells correlated with the temporal increase in IL-1β secretion (Figure 1D). Preventing K+ efflux had no effects on the ATP-induced ubiquitination (Figure 4B, left panel, 4th and 8th lanes), indicating that the poly-Ub chains are formed prior to NLRP3 oligomerization. Thus, these observations collectively suggest that A20 restricts pro-IL1β complex ubiquitination prior to NLRP3-ASC inflammasome assembly.
To better characterize the pro-IL-1β complex ubiquitination in LPS-stimulated A20-deficient cells, we utilized linkage-specific de-ubiquitinases (DUBs) to determine the types of Ub chains present in the pro-IL-1β complex (Mevissen et al., 2013). We first treated pro-IL-1β IPs from these cells with the non-specific DUB USP2 (Ubiquitin carboxyl-terminal hydrolase 2). USP2 eliminated all immunoreactive Ub from the pro-IL-1β IP, confirming that our anti-Ub immunoblots detected ubiquitination (Figure 4C, last lane). Much of the covalent, high molecular weight modifications on pro-IL-1β were also eliminated by USP2 treatment, reinforcing the notion that pro-IL-1β itself is modified by poly-Ub (Figure 4C). To determine whether the pro-IL-1β-associated Ub chains contained non-degradative Ub linkages, we treated the pro-IL-1β IPs with the K63-specific DUB AMSH (associated molecule with a Src homology 3 domain of signal transducing adaptor molecule) and CylD (Cylindromatosis), which cleaves both linear and K63-linked chains (Komander et al., 2008; 2009). Both AMSH and CylD reduced the pro-IL-1β-associated poly-Ub (Figure 4C, 2nd and 3rd lanes), indicating the presence of K63-linked Ub chains in the pro-IL-1β complex. To determine whether the ubiquitin chains in this complex were anchored, we treated the pro-IL-1β IP with isopeptidase T (IsoT), a DUB specific for unanchored chains (Wilkinson et al., 1995). IsoT also reduced the pro-IL-1β-associated ubiquitin (Figure 4C, 4th lane). Thus, the ubiquitin chains associated with pro-IL-1β include unanchored as well as K63-linked poly-Ub, both types known to support signal propagation by encouraging non-degradative protein-protein interactions (Corn and Vucic, 2014; Xia et al., 2009).
We next determined whether K48 Ub linkages existed in the complex by probing the DUB-treated pro-IL-1β IPs with an anti-K48-linked poly-Ub antibody. Without DUB treatment, the presence of K48-linked poly-Ub in the pro-IL-1β complex was clearly evident (Figure 4C, 1st lane). AMSH treatment had no effects on the K48-linked Ub signals, confirming its specificity for K63 linkages (Figure 4C, 2nd lane). Interestingly, IsoT treatment removed much of the K48-linked Ub signals, indicating that the majority of K48 linkages were present in unanchored chains (Figure 4C, 4th lane). It has been reported that K48 linkages in unanchored poly-Ub or mixed chains can also support positive roles in signaling independently of proteasomal degradation (Jiang et al., 2012; Rajsbaum et al., 2014). Hence, the pro-IL-1β complex is ubiquitinated with chains that can support non-proteasomal signal propagation.
Pro-IL-1β-associated ubiquitination depends on RIPK3, but not NLRP3, ASC, or Casp1
Our data above suggest that ubiquitination of pro-IL-1β complex correlates with inflammasome activity. As RIPK3 deficiency abolished most of IL-1β secretion by A20-deficient cells, we next determined whether rendering Tnfaip3−/− cells RIPK3 deficient would similarly affect the pro-IL-1β-associated Ub signals. Strikingly, Tnfaip3−/−Ripk3−/− cells displayed markedly less ubiquitination of pro-IL-1β complex than Tnfaip3−/−Ripk3+/+ cells after LPS stimulation (Figure 4D). Of note, Tnfaip3−/−Ripk3−/− cells also displayed reduced Casp8 activity, as indicated by the decreased presence of cleaved Casp8 as well as its putative substrate, RIPK1 (Figure 4D, right panel, arrow). To determine whether formation of this ubiquitinated complex required NLRP3, ASC, or Casp1, we tested Tnfaip3−/−Nlrp3−/−, Tnfaip3−/−Asc−/−, and Tnfaip3−/−Casp1/11−/− compound mutant cells in a similar fashion. In contrast to Tnfaip3−/−Ripk3−/− cells, ubiquitination of pro-IL-1β complex in these compound mutant cells resembled those seen in single mutant Tnfaip3−/− cells (Figures S3A, S3C). Furthermore, ATP stimulation of these compound mutant cells induced similar amounts of Ub modification on pro-IL-1β complexes (Figures S3A, S3B). Taken together, these data suggest that exaggerated ubiquitination of pro-IL-1β complexes in A20-deficient cells occurs downstream of RIPK3 but upstream of NLRP3, ASC, and Casp1/Casp11 inflammasome assembly.
A20 suppresses Casp1 activity
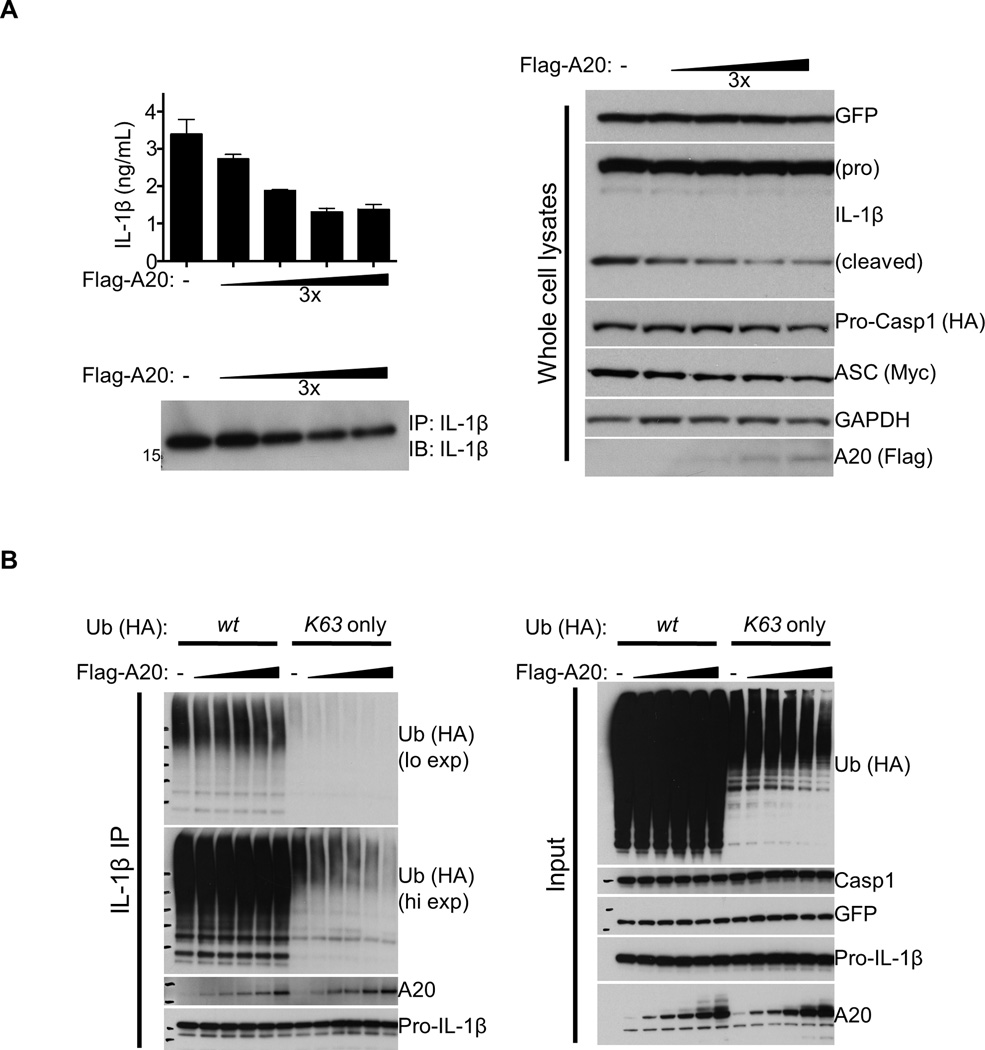
Our data above suggested that A20 may act directly on inflammasome complexes to prevent their spontaneous activity. To test whether A20 can inhibit Casp1 activity independently of TLR signaling, 293T cells were transiently transfected with expression plasmids of ASC, Casp1, and pro-IL-1β coupled to GFP by an IRES sequence, with or without A20. As previously reported (Srinivasula et al., 2002), overexpression of ASC and Casp1 led to their self assembly, and the resultant Casp1 activity was evident through the secretion of mature IL-1β (Figure 5A). Importantly, co-expression of A20 dampened this process in a dose-dependent manner (Figure 5A, left panels) without affecting GFP or pro-IL-1β expression (Figure 5A, right panels). These results reinforce the point that A20 can suppress pro-IL-1β processing independently of the amount of pro-IL-1β expressed. We next determined whether this Casp1 activity involved ubiquitination by including constructs for the expression of HA-tagged wild-type Ub as well as a mutant that can undergo only K63-linked conjugation. As observed with BMDMs, K63-linked Ub signals were detected in pro-IL-1β IPs, and overexpression of A20 dampened this signal in a dose-dependent manner (Figure 5B). These results reinforce our data above that A20 restricts K63-linked ubiquitination of the pro-IL-1β complex (Figure 4C).
Figure 5. Over-expression of A20 suppresses Casp1 activity.
(A) 293T cells seeded in 6-well plates were transiently transfected with 150 ng/well of each plasmid encoding HA-Casp1, Myc-ASC, or pro-IL-1β linked to GFP via an IRES sequence, plus 0–50 ng/well of plasmid encoding Flag-A20. 24 h later, IL-1β secretions in the supernatant were quantified by ELISA (upper left panel) and analyzed by IB following IL-1β IP (lower left panel). IB analyses of the transfected cell lysates are shown on the right. (B) 293T cells seeded in 6-cm dishes were transfected with 500 ng/well of each construct for Streptag-Casp1, Myc-ASC, and pro-IL-1β linked to GFP via an IRES sequence. 6 µg of plasmid encoding HA-tagged wt or K63-only Ub was also included along with 0–50 ng/well of Flag-A20 construct. 24 h later, cells were lysed, subjected to pro-IL-1β IP, and analyzed by IB for the indicated proteins. Input immunoblots are shown on the right.
Ubiquitination of pro-IL-1β supports Casp1 proteolytic processing
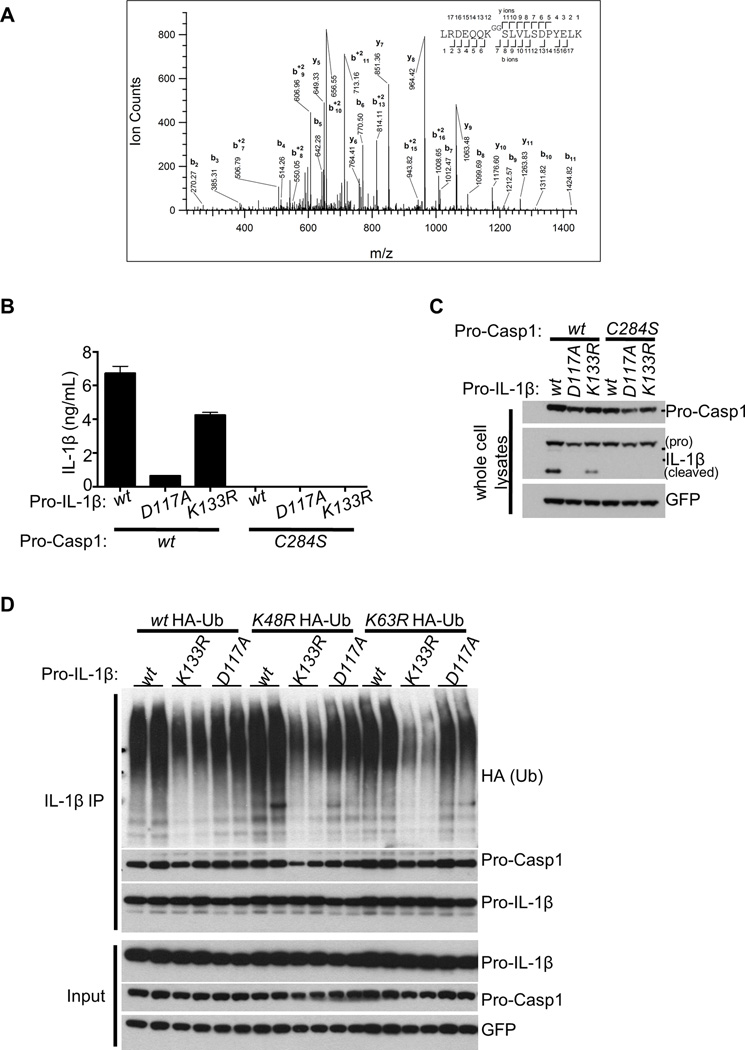
Our finding that pro-IL-1β-associated ubiquitination in Tnfaip3−/− cells was not eliminated by NLRP3, ASC, or Casp1/11 deficiency (Figures S3A, S3B, S3C) suggested that pro-IL-1β itself might undergo ubiquitination. This notion is also supported by our observation that pro-IL-1β in activated macrophages is modified by high molecular weight moieties, which were removed by USP2 (Figures 4A, 4B, 4C). We thus utilized anti-G-G antibody assisted mass spectrometry to find potential ubiquitinated IL-1β peptides from LPS-stimulated Tnfaip3−/− BMDMs. These analyses revealed that an evolutionarily conserved K133 of mouse pro-IL-1β undergoes ubiquitination (Figure 6A). As these studies were performed on endogenous proteins in primary cells undergoing LPS stimulation, and as K133 ubiquitination was detected repeatedly, the ubiquitination at this site is likely to be a dominant and physiological event. To investigate the functional impact of pro-ILβ ubiquitination, we generated a K133R mutant of pro-IL-1β and tested its processing in 293T cells. Mutant K133R pro-IL-1β was cleaved less efficiently than wild type protein, leading to less secretion of mature IL-1β (Figures 6B, 6C). As expected, mutating the cleavage site of pro-IL-1β at D117 as well as the catalytic site of Casp1 at C284 abolished pro-IL-1β processing (Figures 6B, 6C). K133R pro-IL-1β protein also exhibited less total, non-K48, and non-K63 ubiquitination than wild-type pro-IL-1β (Figure 6D). By contrast, the non-cleavable D117A mutant form of pro-IL-1β exhibited no obvious defects in ubiquitination (Figure 6D). Thus, these data support the notion that pro-IL-1β ubiquitination at K133 promotes its proteolytic cleavage.
Figure 6. Lys133 of pro-IL-1β supports ubiquitination and processing.
(A) Tnfaip3−/− BMDMs stimulated with 500 ng/mL LPS for 6 h were subjected to mass spectrometric analyses. CID tandem mass spectrum was obtained from a precursor ion with m/z value 759.0690+3, present in the pull-downs of K-ε-GG peptides from the trypsin-digested lysates. Database search identifies it as the peptide expanding amino acids 127 to 144 of pro-IL-1β, carrying a diglycine remnant on K133 (theoretical monoisotopic m/z= 759.0691+3). Expectation value for the sequence assignment is 8.1 × 10−5. Sequence ions are labeled in the figure and indicated with marks over (C-terminal ions) and below (N-terminal ions) the sequence of the peptide. (B–C) 293T cells seeded in 6-well plates were transiently transfected with 500 ng/well of Myc-ASC expression plasmid, plus 500 ng/well of construct encoding either wt or mutant pro-IL-1β linked to GFP via an IRES sequence. IL-1β secretion in each condition was subsequently measured by ELISA (B). C284S mutant form of Casp1 was included as a negative control for Casp1 activity. The noncleavable D117A pro-IL-1β mutant was additionally included as a negative control. (C) IB analyses of the cells transfected in B were performed to illustrate equal transfection in the conditions. (D) 293T cells seeded in 6-well plates were transfected with 500 ng/well of each construct for Myc-ASC, Streptag-Casp1, and wild-type or K133R pro-IL-1β linked to GFP via IRES. 3 µg of expression plasmid encoding HA-tagged wild-type, K48R, or K63R Ub was also included. 24 h later, cells were lysed and subjected to pro-IL-1β IP followed by IB analyses for Ub signals and Casp1 association. Input immunoblots are shown below.
DISCUSSION
In this study, we have uncovered a role for A20 in restricting NLRP3 inflammasome functions. A20-deficient macrophages exhibit spontaneous NLRP3 activity to various stimuli that are otherwise insufficient to induce pro-IL-1β processing in normal cells. Interestingly, TLRs that only utilize MyD88 to signal did not trigger this response. AIM2 inflammasome response to poly(dA:dT) in Tnfaip3−/− cells were also normal following TLR2 or TLR7 priming (Figures S4A and S4B). Hence, A20 may be particularly important for preventing spontaneous NLRP3 activation by receptors that share common components with TLR3 signaling. The absence of DAMPs in the supernatants of stimulated Tnfaip3−/− cells suggests an intrinsic mechanism of NLRP3 activation.
We previously described an important role for A20 in restricting TLR-induced NFκB signaling (Boone et al., 2004; Turer et al., 2008), and a recent study showed that exaggerated TLR-induced expression of inflammasome proteins causes arthritis in mice lacking A20 in myeloid cells (Vande Walle, L et al., 2014). Our current study, however, reveals that the spontaneous NLRP3 activity in A20-deficient cells does not correlate entirely with the amount of NFκB-induced pro-IL-1β or NLRP3 protein. Hence, our multiple lines of evidence indicate that A20 restricts NLRP3 inflammasome activity by additional mechanisms.
Genetic defects that lead to spontaneous IL-1β secretion by innate immune cells have been reported in three other instances. In one case, ablation of the Atg16L1 gene involved in autophagy resulted in spontaneous NLRP3 activation by LPS alone (Saitoh et al., 2008). In another, BMDMs from mice rendered deficient in all three IAPs similarly exhibited spontaneous NLRP3 inflammasome activity (Vince et al., 2012). Most recently, loss of second signal requirement for pro-IL-1β processing was reported for Casp8-deficient DCs (Kang et al., 2013). The molecular mechanisms by which Tnfaip3−/− cells acquire spontaneous NLRP3 activity are likely to be distinct from these reported studies. First, A20 deficiency exaggerates, not blunts, autophagic response to LPS (Shi and Kehrl, 2010a; 2010b). Second, as observed here and previously reported (Jin et al., 2009), Tnfaip3−/− cells exhibit increased Casp8 activity. While the spontaneous NLRP3 activity of Tnfaip3−/− cells resembles that of the IAP triple mutants (Vince et al., 2012), marked differences in their responses are also noted. As in IAP triple deficient cells, RIPK3 may drive NLRP3 inflammasome activation in Tnfaip3−/− cells via ROS generation (not shown). However, unlike the IAP triple mutants, which exhibit spontaneous pro-IL-1β processing in response to all TLR ligands, A20-deficient cells selectively display this response after TLR signals that utilize TRIF. Moreover, we found no evidence for increased formation of RIPK1-RIPK3 complexes in Tnfaip3−/− cells after LPS stimulation. Whereas RIPK1 catalytic activity is dispensable in IAP triple mutants, it appears necessary for the spontaneous NLRP3 activity in Tnfaip3−/− cells. Thus, our studies build on the concept that RIPK3 supports NLRP3 inflammasome activation, but identify a novel pathway toward this outcome that is inhibited by A20.
Using pro-IL-1β IPs, we have discovered that A20 exists in a complex that contains Casp1, Casp8, RIPK1, and RIPK3 in LPS-stimulated BMDMs. These protein interactions appear specific because (i) pull-downs using the isotype control antibody failed to co-IP these proteins and (ii) pro-IL-1β IPs from lysates of unstimulated cells, which do not express any pro-IL-1β, yielded little, if any, co-associated proteins. These results suggest two potential mechanisms by which A20 suppresses NLRP3 inflammasomes. First, the increased amounts of Casp1, Casp8 and RIPK1 associating with pro-IL-1β in A20-deficient cells indicate that A20 regulates the composition of this complex. Second, as ubiquitination of pro-IL-1β complex is increased in A20-deficient cells in response to LPS alone or LPS plus ATP, these Ub chains might support pro-IL-1β processing and subsequent secretion. Neither ASC deficiency (not shown) nor preventing K+ efflux affected the composition or ubiquitination of the pro-IL-1β complex. Thus, both ubiquitination and formation of this complex occur prior to NLRP3-ASC inflammasome assembly.
We have observed increased Casp8 activity in A20-deficient cells that tightly correlates with spontaneous IL-1β secretion. Exaggerated Casp8 activity in LPS-stimulated A20-deficient BMDMs might reflect A20’s ability to restrict Casp8 (Jin et al., 2009). As Casp8 can cleave pro-IL-1β in the absence of Casp1 (Bossaller et al., 2012; Gringhuis et al., 2012; Gurung et al., 2014; Maelfait et al., 2008; Vince et al., 2012), the observed residual IL-1β secretions by Tnfaip3−/− Casp1/11−/− cells may reflect pro-IL-1β processing by Casp8. Additionally, abolishing RIPK3 in A20-deficient cells reduced Casp8 activity as well as pro-IL-1β processing. A recent finding suggests that Casp8 activity can prime and activate NLRP3 inflammasomes (Gurung et al., 2014). It is thus tempting to speculate that Casp8 activity initiates NLRP3 and Casp1 activation in A20-deficient cells.
We have discovered the presence of K63-linked and unanchored poly-Ub in the pro-IL1β complex, both chain types known to play critical roles in signal propagation (Corn and Vucic, 2014; Xia et al., 2009). While K48-linked poly-Ub is also present, the majority of K48 linkages appeared to be associated with unanchored chains. Recently, K48 linkages in unanchored or mixed Ub chains have been reported to propagate signaling independently of the proteasomes (Jiang et al., 2012; Rajsbaum et al., 2014). Thus, the presence of K63 linkages (and potentially, K48 linkages in unanchored chains) in pro-IL1β complex suggests that NLRP3 inflammasome activation involves ubiquitination, a step that A20 inhibits.
NLRP3, ASC, and Casp1 deficiency had no effects on the pro-IL-1β-associated Ub signals in LPS-stimulated A20-deficient cells. These observations indicate that this ubiquitination event is distinct from those previously reported for Casp1 and ASC during NLRP3 inflammasome activation (Labbé et al., 2011; Rodgers et al., 2014). Our IP/IB, mass-spectrometry and mutational analyses revealed that K133 of mouse pro-IL-1β is ubiquitinated and that ubiquitination of this site supports pro-IL-1β proteolytic cleavage. IL-1β ubiquitination may support pro-IL-1β cleavage by encouraging higher order oligomerization of inflammasome components; future studies will be needed to define this mechanism more precisely. Given the known roles of ubiquitination in autophagy (Shaid et al., 2013), IL-1β ubiquitination could also potentially support IL-1β secretion through an autophagy-based unconventional secretory pathway (Dupont et al., 2011).
In summary, we have discovered that A20 plays a critical role in preventing spontaneous NLRP3 inflammasome activation and in limiting its activity by exogenous stimuli. Given the genetic links of A20 to multiple human inflammatory diseases, our findings establish links between A20, ubiquitination, inflammasomes, and human diseases.
EXPERIMENTAL PROCEDURES
Mice
Tnfaip3−/−, Nlrp3−/−, Asc−/−, Casp1/11−/−, Tnf−/−, and Ripk3−/− mutant mice have been previously described. All mice used in this study were housed and bred in a specific pathogen-free facility according to the IACUC guidelines of UCSF.
BMDM cultures and treatments
Primary BMDMs were cultured from mouse femurs and tibia using standard protocols described in Supplemental Experimental Procedures. For ELISAs, cells were plated at 1–2 million cells in 0.5 mL per well in 24-well format. Innate immune receptor ligands were purchased from InvivoGen. Anti-CD40 antibody, clone FGK4.5, was produced by the UCSF antibody core facility. Recombinant murine TNFα and Necrostatin-1 were purchased from R&D Systems and Enzo Life Sciences, respectively. Supernatants were typically harvested between 2–18 h poststimulation. To induce NLRP3 activation by ATP, BMDMs were primed with LPS at 50–500 ng/mL for 4–16 h and subsequently treated with PBS containing 5 mM ATP for 15–30 min as described in Supplemental Experimental Procedures.
Protein overexpression analyses
293T cells were grown to ~70–80% confluence in DMEM containing 10% FBS, Pen-Strep, and L-Glut before transfection using Lipofectamine LTX without Plus Reagent (Invitrogen). The following full-length cDNA expression plasmids were either generated using commercially available vectors or purchased from the indicated vendors: Myc-DDK tagged ASC in pCMV6-Entry (Origene); N-terminally HA-tagged Casp1 in pCMV-HA (Clonetech); N-terminally-StrepTag mouse Casp1 in pCMV-Script (Stratagene); Full-length IL-1β in pIRES2-AcGFP (Clonetech); N-terminally Flag-tagged A20 in pCMV-Tag2 (Stratagene). 3x HA-tagged wildtype Ub, K63-only Ub, K48R Ub, and K63R Ub pcDNA3.1 constructs were kind gifts from Dr. Vishva Dixit (Genentech). Cells were typically transfected in 6-well format in 2 mL media with 150 ng/well of each vector, except for Ub constructs (which were used at 3 µg/well) and A20 (which varied from 0–50 ng/well).
ELISAs and LDH release assays
IL-1β (BD Biosciences) and IL-18 (eBioscience) ELISAs and LDH release assays (Cytotoxicity Detection Kit [Roche Life Science]) were performed per manufacturer’s instructions.
Immunoblotting and immunoprecipitation
IB and IP analyses were performed using standard protocols described in Supplemental Experimental Procedures.
DUB assays
Five 10-cm plates of Tnfaip3−/− cells (corresponding to 2 × 108 cells) were stimulated with 500 ng/mL LPS for 6 h. Cells were lysed, and IL-1β IPs were performed in 5 separate tubes as described in Supplemental Experimental Procedures. During the last wash, the beads containing IL-1β-associated Ub complex were combined, split into several equal aliquots, and subjected to DUB treatments using protocols similar to those included in the UbiCREST kit (Boston Biochem) as detailed in Supplemental Experimental Procedures.
Pro-IL-1β ubiquitination site determination
Day-6 Tnfaip3−/− BMDMs were seeded in twenty 15-cm plates and stimulated with 500 ng/mL LPS for 6 h as described above. Subsequently, the cells were lifted using PBS containing 5 mM EDTA, pooled, centrifuged, and snap-frozen upon removal of PBS. The frozen cells were then subjected to lysis, followed by anti-Gly-Gly immunoprecipitation and UbiScan-assisted mass spectrometric analyses as detailed in Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Drs. V. Dixit, S. Akira, R. Medzhitov, X. Wang, R. Vance, and A. Winoto for generously sharing mice. This work was supported by the NIH and the UCSF Liver Center. BD was partially supported by T32 AI007334, T32 DK007007, and a CCFA Research Fellowship Award. The Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A. Burlingame, Director) is supported by the Howard Hughes Medical Institute and the Biomedical Technology Research Resources Program (8P41GM103481) at the National Institute of General Medical Sciences.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bauernfeind FG, Horvath G, Stutz A, Alnemri ES, MacDonald K, Speert D, Fernandes-Alnemri T, Wu J, Monks BG, Fitzgerald KA, et al. Cutting edge: NF-kappaB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. Journal of Immunology (Baltimore, Md. : 1950) 2009;183:787–791. doi: 10.4049/jimmunol.0901363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad R-C, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, et al. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nature Immunology. 2004;5:1052–1060. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- Bossaller L, Chiang P-I, Schmidt-Lauber C, Ganesan S, Kaiser WJ, Rathinam VAK, Mocarski ES, Subramanian D, Green DR, Silverman N, et al. Cutting edge: FAS (CD95) mediates noncanonical IL-1β and IL-18 maturation via caspase-8 in an RIP3-independent manner. Journal of Immunology (Baltimore, Md. : 1950) 2012;189:5508–5512. doi: 10.4049/jimmunol.1202121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown GD, Taylor PR, Reid DM, Willment JA, Williams DL, Martinez-Pomares L, Wong SY, Gordon S. Dectin-1 is a major beta-glucan receptor on macrophages. The Journal of Experimental Medicine. 2002;196:407–412. doi: 10.1084/jem.20020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catrysse L, Vereecke L, Beyaert R, van Loo G. A20 in inflammation and autoimmunity. Trends in Immunology. 2014;35:22–31. doi: 10.1016/j.it.2013.10.005. [DOI] [PubMed] [Google Scholar]
- Chu Y, Vahl JC, Kumar D, Heger K, Bertossi A, Wójtowicz E, Soberon V, Schenten D, Mack B, Reutelshöfer M, et al. B cells lacking the tumor suppressor TNFAIP3/A20 display impaired differentiation and hyperactivation and cause inflammation and autoimmunity in aged mice. Blood. 2011;117:2227–2236. doi: 10.1182/blood-2010-09-306019. [DOI] [PubMed] [Google Scholar]
- Corn JE, Vucic D. Ubiquitin in inflammation: the right linkage makes all the difference. Nature Structural & Molecular Biology. 2014;21:297–300. doi: 10.1038/nsmb.2808. [DOI] [PubMed] [Google Scholar]
- Davis BK, Wen H, Ting JP-Y. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annual Review of Immunology. 2011;29:707–735. doi: 10.1146/annurev-immunol-031210-101405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degterev A, Hitomi J, Germscheid M, Ch'en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G, et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nature Chemical Biology. 2008;4:313–321. doi: 10.1038/nchembio.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. Embo J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Häcker G, Leverkus M. cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Molecular Cell. 2011;43:449–463. doi: 10.1016/j.molcel.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gringhuis SI, Kaptein TM, Wevers BA, Theelen B, van der Vlist M, Boekhout T, Geijtenbeek TBH. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nature Immunology. 2012;13:246–254. doi: 10.1038/ni.2222. [DOI] [PubMed] [Google Scholar]
- Gurung P, Anand PK, Malireddi RKS, Vande Walle L, Van Opdenbosch N, Dillon CP, Weinlich R, Green DR, Lamkanfi M, Kanneganti T-D. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. Journal of Immunology (Baltimore, Md. : 1950) 2014;192:1835–1846. doi: 10.4049/jimmunol.1302839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer GE, Turer EE, Taylor KE, Fang CJ, Advincula R, Oshima S, Barrera J, Huang EJ, Hou B, Malynn BA, et al. Expression of A20 by dendritic cells preserves immune homeostasis and prevents colitis and spondyloarthritis. Nature Immunology. 2011;12:1184–1193. doi: 10.1038/ni.2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Liang Y, Shao F, Wang X. Toll-like receptors activate programmed necrosis in macrophages through a receptor-interacting kinase-3-mediated pathway. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:20054–20059. doi: 10.1073/pnas.1116302108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger K, Fierens K, Vahl JC, Aszodi A, Peschke K, Schenten D, Hammad H, Beyaert R, Saur D, van Loo G, et al. A20-deficient mast cells exacerbate inflammatory responses in vivo. PLoS Biology. 2014;12:e1001762. doi: 10.1371/journal.pbio.1001762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe K, Du X, Georgel P, Janssen E, Tabeta K, Kim SO, Goode J, Lin P, Mann N, Mudd S, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424:743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- Humphries F, Yang S, Wang B, Moynagh PN. RIP kinases: key decision makers in cell death and innate immunity. Cell Death Differ. 2014 doi: 10.1038/cdd.2014.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ. Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity. 2012;36:959–973. doi: 10.1016/j.immuni.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- Kaiser WJ, Sridharan H, Huang C, Mandal P, Upton JW, Gough PJ, Sehon CA, Marquis RW, Bertin J, Mocarski ES. Toll-like receptor 3-mediated necrosis via TRIF, RIP3, and MLKL. The Journal of Biological Chemistry. 2013;288:31268–31279. doi: 10.1074/jbc.M113.462341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T-B, Yang S-H, Toth B, Kovalenko A, Wallach D. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity. 2013;38:27–40. doi: 10.1016/j.immuni.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Kayagaki N, Warming S, Lamkanfi M, Vande Walle L, Louie S, Dong J, Newton K, Qu Y, Liu J, Heldens S, et al. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- Komander D, Lord CJ, Scheel H, Swift S, Hofmann K, Ashworth A, Barford D. The structure of the CYLD USP domain explains its specificity for Lys63-linked polyubiquitin and reveals a B box module. Molecular Cell. 2008;29:451–464. doi: 10.1016/j.molcel.2007.12.018. [DOI] [PubMed] [Google Scholar]
- Komander D, Reyes-Turcu F, Licchesi JDF, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Reports. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, van Loo G, Waelput W, De Prijck S, Muskens F, Sze M, van Praet J, Branco-Madeira F, Janssens S, Reizis B, et al. The ubiquitin-editing protein A20 prevents dendritic cell activation, recognition of apoptotic cells, and systemic autoimmunity. Immunity. 2011;35:82–96. doi: 10.1016/j.immuni.2011.05.013. [DOI] [PubMed] [Google Scholar]
- Labbé K, McIntire CR, Doiron K, Leblanc PM, Saleh M. Cellular inhibitors of apoptosis proteins cIAP1 and cIAP2 are required for efficient caspase-1 activation by the inflammasome. Immunity. 2011;35:897–907. doi: 10.1016/j.immuni.2011.10.016. [DOI] [PubMed] [Google Scholar]
- Lamkanfi M, Dixit VM. Inflammasomes and their roles in health and disease. Annual Review of Cell and Developmental Biology. 2012;28:137–161. doi: 10.1146/annurev-cellbio-101011-155745. [DOI] [PubMed] [Google Scholar]
- Lee EG, Boone DL, Chai S, Libby SL, Chien M, Lodolce JP, Ma A. Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science (New York, N.Y.) 2000;289:2350–2354. doi: 10.1126/science.289.5488.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans JC, Cassel SL, Sutterwala FS. Sensing damage by the NLRP3 inflammasome. Immunological Reviews. 2011;243:152–162. doi: 10.1111/j.1600-065X.2011.01043.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nature Reviews. Immunology. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maelfait J, Vercammen E, Janssens S, Schotte P, Haegman M, Magez S, Beyaert R. Stimulation of Toll-like receptor 3 and 4 induces interleukin-1beta maturation by caspase-8. The Journal of Experimental Medicine. 2008;205:1967–1973. doi: 10.1084/jem.20071632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Weiss DS, Newton K, McBride J, O'Rourke K, Roose-Girma M, Lee WP, Weinrauch Y, Monack DM, Dixit VM. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature. 2006;440:228–232. doi: 10.1038/nature04515. [DOI] [PubMed] [Google Scholar]
- Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Molecular Cell. 2002;10:417–426. doi: 10.1016/s1097-2765(02)00599-3. [DOI] [PubMed] [Google Scholar]
- Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annual Review of Immunology. 2009;27:229–265. doi: 10.1146/annurev.immunol.021908.132715. [DOI] [PubMed] [Google Scholar]
- Matmati M, Jacques P, Maelfait J, Verheugen E, Kool M, Sze M, Geboes L, Louagie E, Mc Guire C, Vereecke L, et al. A20 (TNFAIP3) deficiency in myeloid cells triggers erosive polyarthritis resembling rheumatoid arthritis. Nature Genetics. 2011;43:908–912. doi: 10.1038/ng.874. [DOI] [PubMed] [Google Scholar]
- Mevissen TET, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, Oualid, El F, et al. OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell. 2013;154:169–184. doi: 10.1016/j.cell.2013.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Planillo R, Kuffa P, Martínez-Colón G, Smith BL, Rajendiran TM, Núñez G. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Sun X, Dixit VM. Kinase RIP3 is dispensable for normal NF-kappa Bs, signaling by the B-cell and T-cell receptors, tumor necrosis factor receptor 1, and Toll-like receptors 2 and 4. Molecular and Cellular Biology. 2004;24:1464–1469. doi: 10.1128/MCB.24.4.1464-1469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, Versteeg GA, Schmid S, Maestre AM, Belicha-Villanueva A, Martínez-Romero C, Patel JR, Morrison J, Pisanelli G, Miorin L, et al. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKε kinase-mediated antiviral response. Immunity. 2014;40:880–895. doi: 10.1016/j.immuni.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VAK, Vanaja SK, Waggoner L, Sokolovska A, Becker C, Stuart LM, Leong JM, Fitzgerald KA. TRIF licenses caspase-11-dependent NLRP3 inflammasome activation by gram-negative bacteria. Cell. 2012;150:606–619. doi: 10.1016/j.cell.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers MA, Bowman JW, Fujita H, Orazio N, Shi M, Liang Q, Amatya R, Kelly TJ, Iwai K, Ting J, et al. The linear ubiquitin assembly complex (LUBAC) is essential for NLRP3 inflammasome activation. The Journal of Experimental Medicine. 2014;211:1333–1347. doi: 10.1084/jem.20132486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Fujita N, Jang MH, Uematsu S, Yang B-G, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:21–30. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shembade N, Harhaj EW. Regulation of NF-κB signaling by the A20 deubiquitinase. Cellular & Molecular Immunology. 2012;9:123–130. doi: 10.1038/cmi.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C-S, Kehrl JH. Traf6 and A20 differentially regulate TLR4-induced autophagy by affecting the ubiquitination of Beclin 1. Autophagy. 2010a;6:986–987. doi: 10.4161/auto.6.7.13288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C-S, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Science Signaling. 2010b;3:ra42. doi: 10.1126/scisignal.2000751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasula SM, Poyet J-L, Razmara M, Datta P, Zhang Z, Alnemri ES. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. The Journal of Biological Chemistry. 2002;277:21119–21122. doi: 10.1074/jbc.C200179200. [DOI] [PubMed] [Google Scholar]
- Strowig T, Henao-Mejia J, Elinav E, Flavell R. Inflammasomes in health and disease. Nature. 2012;481:278–286. doi: 10.1038/nature10759. [DOI] [PubMed] [Google Scholar]
- Tavares RM, Turer EE, Liu CL, Advincula R, Scapini P, Rhee L, Barrera J, Lowell CA, Utz PJ, Malynn BA, et al. The ubiquitin modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity. 2010;33:181–191. doi: 10.1016/j.immuni.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K, et al. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Molecular Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, Lee B, Shifrin N, Malynn BA, Ma A. Homeostatic MyD88-dependent signals cause lethal inflamMation in the absence of A20. The Journal of Experimental Medicine. 2008;205:451–464. doi: 10.1084/jem.20071108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–815. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- Vande Walle L, Van Opdenbosch N, Jacques P, Fossoul A, Verheugen E, Vogel P, Beyaert R, Elewaut D, Kanneganti TD, van Loo G, et al. Negative regulation of the NLRP3 inflammasome by A20 protects against arthritis. Nature. 2014;512:69–73. doi: 10.1038/nature13322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereecke L, Sze M, Mc Guire C, Rogiers B, Chu Y, Schmidt-Supprian M, Pasparakis M, Beyaert R, van Loo G. Enterocyte-specific A20 deficiency sensitizes to tumor necrosis factor-induced toxicity and experimental colitis. The Journal of Experimental Medicine. 2010;207:1513–1523. doi: 10.1084/jem.20092474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen L, Verhelst K, van Loo G, Carpentier I, Ley SC, Beyaert R. Expression, biological activities and mechanisms of action of A20 (TNFAIP3) Biochemical Pharmacology. 2010;80:2009–2020. doi: 10.1016/j.bcp.2010.06.044. [DOI] [PubMed] [Google Scholar]
- Vince JE, Wong WW-L, Gentle I, Lawlor KE, Allam R, O'Reilly L, Mason K, Gross O, Ma S, Guarda G, et al. Inhibitor of apoptosis proteins limit RIP3 kinase-dependent interleukin-1 activation. Immunity. 2012;36:215–227. doi: 10.1016/j.immuni.2012.01.012. [DOI] [PubMed] [Google Scholar]
- Wilkinson KD, Tashayev VL, O'Connor LB, Larsen CN, Kasperek E, Pickart CM. Metabolism of the polyubiquitin degradation signal: structure, mechanism, and role of isopeptidase T. Biochemistry. 1995;34:14535–14546. doi: 10.1021/bi00044a032. [DOI] [PubMed] [Google Scholar]
- Xia Z-P, Sun L, Chen X, Pineda G, Jiang X, Adhikari A, Zeng W, Chen ZJ. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461:114–119. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.