Abstract
Overwhelming evidence supports the endosymbiosis theory that mitochondria originated once from the Alphaproteobacteria. However, its exact position in the tree of life remains highly debated. This is because systematic errors, including biased taxonomic sampling, high evolutionary rates and sequence composition bias have long plagued the mitochondrial phylogenetics. In this study, we address this issue by 1) increasing the taxonomic representation of alphaproteobacterial genomes by sequencing 18 phylogenetically novel species. They include 5 Rickettsiales and 4 Rhodospirillales, two orders that have shown close affiliations with mitochondria previously, 2) using a set of 29 slowly evolving mitochondria-derived nuclear genes that are less biased than mitochondria-encoded genes as the alternative “well behaved” markers for phylogenetic analysis, 3) applying site heterogeneous mixture models that account for the sequence composition bias. With the integrated phylogenomic approach, we are able to for the first time place mitochondria unequivocally within the Rickettsiales order, as a sister clade to the Rickettsiaceae and Anaplasmataceae families, all subtended by the Holosporaceae family. Our results suggest that mitochondria most likely originated from a Rickettsiales endosymbiont already residing in the host, but not from the distantly related free-living Pelagibacter and Rhodospirillales.
The origin of mitochondria was a seminal event in the history of life. It is now widely accepted that mitochondria evolved only once from bacteria living within their host cells, probably two billion years ago (known as the endosymbiosis theory). Specifically, phylogenetic analyses have indicated that mitochondria originated from Alphaproteobacteria, a subgroup of the purple non-sulfur bacteria1. However, exactly when it happened remains highly debated and this key piece of puzzle is still missing in our current assembly of the tree of life.
Defining precisely the alphaproteobacterial ancestry of the mitochondria has important implications. It is a prerequisite for elucidating the origin and early evolution of mitochondria and eukaryotic cells. Placing mitochondria firmly within the tree of life will allow us to use comparative methods to gain insights into the biology of the last common ancestor of mitochondria and Alphaproteobacteria – Was it a free-living bacterium or an endosymbiont? What was its genetic makeup2,3? Did the mitochondrion arise at the same time as, or subsequent to, the appearance of the eukaryotic nucleus4? Did it originate under initially anaerobic or aerobic conditions5? What was the driving force behind the initial symbiosis2,6?
Pinpointing the origin of mitochondria is inherently difficult, however, due to the compounding effects of at least three factors: 1) Weak phylogenetic signal. Most informative sites in the molecular sequence that allow us to resolve the deep evolutionary relationships have been erased by saturated mutations accumulated over a long period of time. As a result, individual genes such as the small subunit ribosomal RNA (SSU rRNA or 16S rRNA) usually do not contain sufficient phylogenetic signals to resolve this deep relationship. 2) Long-branch attraction (LBA). Mitochondria and the obligate intracellular Alphaproteobacteria have highly accelerated rates of evolution than the free-living bacteria. Therefore, molecular phylogenetic inference of the origin of the mitochondria is prone to the well-known LBA artifact, when fast-evolving but distantly related lineages are erroneously grouped together as sister nodes in the tree7,8. 3) Extreme sequence composition bias. Mitochondria and the obligate intracellular Alphaproteobacteria are in general extremely AT rich in their genome sequences. It is well established that sequence composition bias could adversely affect the phylogenetic reconstruction and lead to statistically robust but misleading conclusions9,10,11.
Due to these reasons, results from early studies based on the sequences of a few genes were often inconclusive. Mitochondria have been placed near the Rickettsiales order, a subgroup of Alphaproteobacteria that contains obligate intracellular bacterial parasites such as Rickettsia, Ehrlichia, and Anaplasma12,13. And often, the Rickettsia genus was asserted to be the closest modern relative of mitochondria14,15. Phylogenomic analysis using 32 genes shared by mitochondria and bacteria called into question the conjecture that Rickettsia genus is the closest relative of mitochondria16. Later it was suggested that Rhodospirillum rubrum within the Rhodospirillales order came as close to mitochondria as any Alphaproteobacteria investigated17. Recent genome-level phylogenetic analyses with increasingly more bacterial species showed an emerging trend that places mitochondria basal to the Rickettsiales order with very high statistical support18,19,20,21,22. However, who is the closest contemporary relative of mitochondria remains highly debated. Studies have suggested that a group of free-living bacteria known as the SAR11 group form the sister clade to mitochondria20,22. Members of SAR11 dominate in the ocean surface water and have the smallest cells and genomes of any free-living organisms. A sister-clade relationship with the SAR11 group would suggest that mitochondria originated from free-living marine bacteria and the endosymbiosis events of mitochondria and intracellular Rickettsiales were independent. However, this hypothesis has been convincingly refuted by more recent studies demonstrating that this sister-clade relationship is a tree reconstruction artifact resulted from sequence composition bias21,23,24.
Intriguingly, the conflicting sister-clade relationships of mitochondria all received high statistical support19,20,21,22. Obtaining a highly supported genome tree does not necessarily guarantee an accurate evolution reconstruction. It has been shown that highly supported branching patterns in a genome tree could be wrong because of unrealistic evolutionary models, composition biases in the sequence data, or the LBA25. Unlike the stochastic noise, systematic errors such as composition bias and LBA will not diminish but rather strengthen when the sequence alignment length is increased, ultimately leading the trees to converge toward the wrong tree with extremely high support (hence, be positively misleading)7. It has been demonstrated by many studies that genome trees with high bootstrap, jackknife or posterior probability support should be treated with greater caution than single-gene trees for possible misleading tree reconstruction artifacts9,26,27,28,29,30.
In this study, we first show that systematic errors in the current genome sequence dataset still present serious problems for precisely placing mitochondria in the tree of life. We then address the LBA and composition bias problems by 1) sequencing 18 strategically selected alphaproteobacterial isolates to substantially increase the taxonomic representation of the alphaproteobacterial genomes, 2) using a set of slowly evolving and less compositionally biased mitochondria-derived nuclear genes (compared to mitochondria-encoded genes) for phylogenetic reconstruction, 3) applying site heterogeneous mixture models that could account for composition bias. With the integrated phylogenomic approach, we are able to place mitochondria firmly within the Rickettsiales order, as a sister clade to the Rickettsiaceae/Anaplasmataceae families, all subtended by the free-living Alphaproteobacterium HIMB59 and the Holosporaceae family.
Results
Substantial systematic errors are present in the current genomic sequence dataset
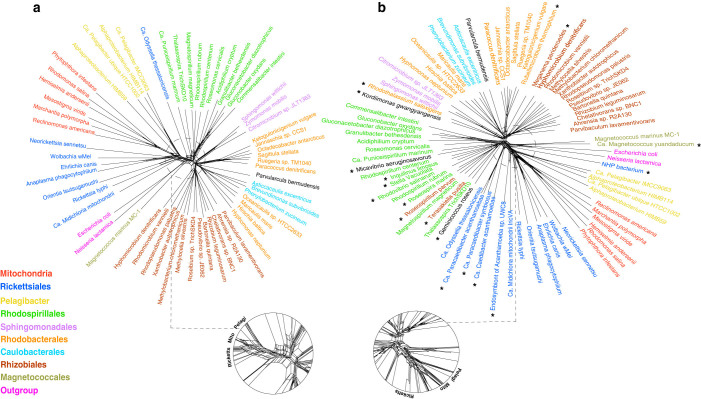
Because LBA and composition bias could produce conflicting signals competing against the true phylogenetic signal, they can be detected using split-based methods31,32,33,34. Split decomposition analysis produces a “neighbor net” where conflicting phylogenies are displayed as box-like structures. The more tree-like parts of the graph show where there is little conflict, and thus, little evidence of systematic errors. To determine whether there are significant systematic errors in the current genomic dataset, we performed a NeighborNet analysis on a concatenated protein sequence alignment of 26 mitochondria-encoded genes from genomes of 54 alphaproteobacterial and 6 mitochondrial representatives. Figure 1a shows that Alphaproteobacteria can be divided into at least 7 major groups (Rickettsiales, Rhodospirillales, Sphingomonadales, Rhodobacterales, Caulobacterales, Magnetococcales and Rhizobiales), by and large consistent with the taxonomic classification based on the SSU rRNA gene. Nevertheless, it also shows a large amount of networking or phylogenetic uncertainty around the base of mitochondria as observed previously17,18, indicating that the precise position of mitochondria within the Alphaproteobacteria is highly uncertain.
Figure 1. Rooted genome trees of Alphaproteobacteria and mitochondria represented by NeighborNet graphs.
a) Original dataset. b) Original dataset + 18 newly sequenced genomes in this study (denoted with asterisks). Conflicting signal is represented by the network in the graph. The tree is rooted using Beta and Gammaproteobacteria as the outgroup.
To further investigate the source of the systematic errors, we carried out spectral analysis. Spectral analysis is an extremely useful tool that can be used to pinpoint and quantify the source of errors independently of any one particular tree35. If LBA is a problem, spectral analysis should indicate that there is support for two or more conflicting (i.e., mutually exclusive) splits, one of which grouping long-branch lineages together. Spectral analysis has been successfully applied to detect LBA in many datasets including mitochondrial genes36,37,38,39,40.
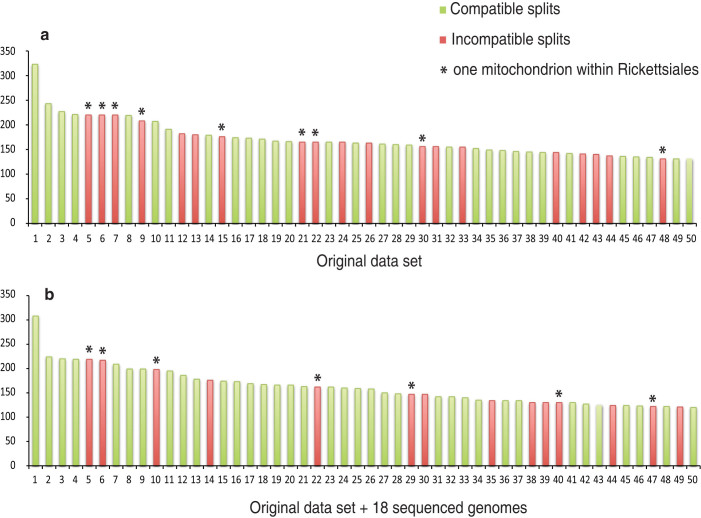
Figure 2 shows the split support spectrum of the same concatenated alignment used in the NeighborNet analysis. The strongest four splits are all compatible with the major groups shown in Figure 1, indicating that there is strong phylogenetic signal in the dataset. However, there are also substantial numbers of conflicting splits, many of them mutually incompatible. It is striking that incompatible splits in the top 50 splits are all associated with long-branch lineages (Supplementary Table 1). For example, most of these incompatible splits place a single mitochondrial species with long-branch lineages such as Rickettsiales, Pelagibacter and the outgroup (indicated by asterisks in Figure 2), but never with the “normal length” lineages. Conflicting splits placing a single species of Rickettsiales and Pelagibacter within other long-branch groups were also observed. The strong correlation between the conflicting splits and the long-branch lineages indicates that LBA presents a major problem in the current genomic dataset.
Figure 2. Split spectrum of the concatenated alignment of 26 mitochondria-encoded genes for a) the original dataset, b) the original dataset plus 18 genomes sequenced in this study.
Each bar represents a split and the height of bar (Y-axis) is the number of sites in the alignment supporting the split. The splits were ranked by their support and only the top 50 splits are shown. The splits were considered as compatible or incompatible by reconciling with well established phylogenetic relationships such as the monophyly of mitochondria or Rickettsiales. Compatible splits are in green and incompatible splits are in red. Asterisks indicate conflicting splits where a single mitochondrial species is placed within the Rickettsiales order.
Increasing the phylogenetic diversity of alphaproteobacterial genomes
Recent empirical phylogenomic studies have demonstrated that increasing taxon representation is very effective in mitigating LBA and improving the phylogenies26,28,41,42,43,44,45. At the beginning of this study, 425 alphaproteobacterial genomes had been sequenced according to the GenomeOnline database46. However, most of them were selected because of their economic and medical importance and did not take the phylogeny into consideration. As a result, many sequenced species were closely related and the taxonomic representation was extremely biased. For example, 220 or 52% of the sequenced alphaproteobacterial genomes came from one single order (Rhizobiales). 123 of them were actually from one single genus (Brucella). On the other hand, for the Rickettsiales order that has shown close phylogenetic relationship to mitochondria, two families (Holosporaceae and Incertae sedis 4) were completely missing. Consequently, many gaps remain in the alphaproteobacterial branch of the tree of life.
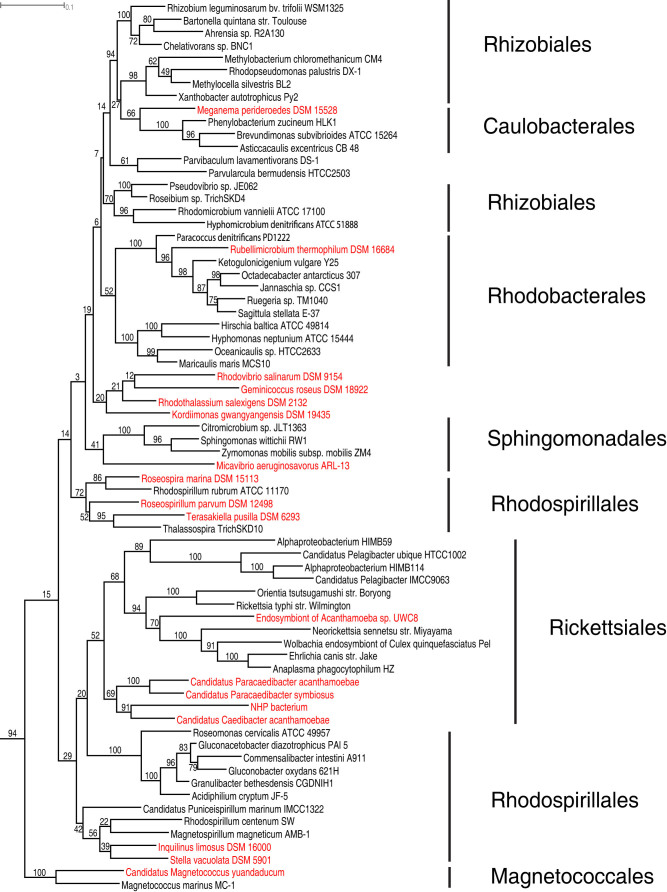
To fill the gaps in the tree, we selected alphaproteobacterial species for sequencing by maximizing the total amount of phylogenetic diversity they represented. We estimated the phylogenetic diversity based on the SSU rRNA tree. Although not perfect, SSU rRNA has been shown to be a sound predictor of an organism's position in the genome tree47. We downloaded the aligned SSU rRNA sequences of 9,817 alphaproteobacterial isolates from the Ribosomal Database Project48 and used them to construct a maximum likelihood tree. We then used a tree-based greedy algorithm described in49 to rank isolates by their phylogenetic novelty. Species that had been sequenced were removed from the list. The availability of an isolate's genomic DNA was also an important factor in our selection process. In total, 18 species from six orders (Rickettsiales, Rhodospirillales, Kordiimonadales, Magnetococcales, Rhizobiales and Rhodobacterales) were selected for sequencing (Table 1, also highlighted in Figure 3). Together, they represented 18.5% of the phylogenetic diversity of the Alphaproteobacteria in the tree (Figure 3) and increased the phylogenetic diversity significantly compared to a random set of 18 genomes (1.7–3.0 times, p = 7e-65). We note that 9 of 18 selected species belong to the Rickettsiales and Rhodospirillales orders, which have shown close affiliation with mitochondria previously.
Table 1. Overview of the 18 alphaproteobacterial genomes sequenced in this study.
| Genomes | Order | Draft genome size | No. of contigs | Coverage | GC content (%) | Protein coding genes | Mito markers | Nuclear markers | Phylum markers |
|---|---|---|---|---|---|---|---|---|---|
| Kordiimonas gwangyangensis DSM 19435 | Kordiimonadales | 4149991 | 272 | 320x | 57.6 | 3970 | 25 | 28 | 198 |
| Candidatus Magnetococcus yuandaducum | Magnetococcales | 2228395 | 649 | 23x | 58.9 | 2699 | 23 | 15 | 131 |
| Meganema perideroedes DSM 15528 | Rhizobiales | 3464569 | 324 | 209x | 67.1 | 3494 | 24 | 26 | 197 |
| Roseospirillum parvum DSM 12498 | Rhizobiales | 3436975 | 3024 | 323x | 69.6 | 4127 | 22 | 20 | 187 |
| Terasakiella pusilla DSM 6293 | Rhizobiales | 4067442 | 259 | 150x | 50.1 | 4098 | 24 | 27 | 200 |
| Rhodothalassium salexigens DSM 2132 | Rhodobacterales | 3156491 | 3163 | 294x | 68.0 | 4058 | 26 | 23 | 193 |
| Rubellimicrobium thermophilum DSM 16684 | Rhodobacterales | 3328337 | 361 | 99x | 69.2 | 3381 | 25 | 27 | 197 |
| Inquilinus limosus DSM 16000 | Rhodospirillales | 6772298 | 4283 | 83x | 69.3 | 8184 | 25 | 24 | 190 |
| Rhodovibrio salinarum DSM 9154 | Rhodospirillales | 4170570 | 258 | 117x | 65.9 | 4040 | 25 | 27 | 199 |
| Roseospira marina DSM 15113 | Rhodospirillales | 3635965 | 8906 | 91x | 67.0 | 6978 | 22 | 20 | 175 |
| Stella vacuolata DSM 5901 | Rhodospirillales | 4353044 | 1038 | 7x | 70.2 | 4337 | 20 | 22 | 145 |
| Candidatus Caedibacter acanthamoebae | Rickettsiales | 2175773 | 5 | 50x | 37.9 | 2332 | 26 | 26 | 193 |
| Candidatus Paracaedibacter acanthamoebae | Rickettsiales | 2454690 | 55 | 67x | 41.0 | 2535 | 26 | 26 | 197 |
| Candidatus Paracaedibacter symbiosus | Rickettsiales | 2668935 | 299 | 15x | 41.2 | 2967 | 23 | 26 | 195 |
| Endosymbiont of Acanthamoeba sp. UWC8 | Rickettsiales | 1615277 | 1 | 20x | 34.8 | 1608 | 24 | 26 | 196 |
| NHP bacterium | Rickettsiales | 1115609 | 15 | 927x | 49.8 | 1309 | 23 | 21 | 171 |
| Geminicoccus roseus DSM 18922 | unclassified | 5676036 | 1169 | 109x | 68.4 | 5909 | 24 | 27 | 191 |
| Micavibrio aeruginosavorus ARL-13 | unclassified | 2481983 | 1 | 60x | 54.7 | 2432 | 26 | 27 | 198 |
Figure 3. A rooted SSU rRNA maximum likelihood tree of alphaproteobacterial representatives using RAxML.
Highlighted in red are the 18 isolates selected for sequencing in this study. The tree was rooted using Beta and Gammaproteobacteria as the outgroup. Bootstrap values (out of 100 replicates) are shown.
The 18 alphaproteobacterial genomes were sequenced by whole-genome shotgun sequencing using a combination of 454 pyrosequencing and Illumina. The status and characteristics of the genomes are listed in Table 1.
Increasing the phylogenetic diversity reduced the systematic errors
We asked whether adding the 18 newly sequenced genomes reduced the systematic errors in the dataset. As shown in Figure 2, adding the 18 genomes visibly reduced the level of conflict in the split spectrum. Both the number of conflicting splits and their overall ranks decreased. Accordingly, the average systematic errors in the dataset, calculated as the proportion of incompatible split supporting values, decreased significantly from 0.02 to 0.014 (Mann-Whitney U-test, P = 0.008). The support for incompatible splits that grouped a single mitochondrial species within the Rickettsiales order also decreased. As a result, their ranks in the top 50 splits dropped. The improvement shows that the increased taxon sampling clearly has a positive effect on mitigating LBA.
Use of mitochondria-derived nuclear genes as alternative phylogenetic markers
As a consequence of their endosymbiotic lifestyle, mitochondria have gone through extensive genome reduction50. For example, the 16 Kbp human mitochondrial genome only encodes 13 proteins51. A large fraction of mitochondrial genes have simply been lost, while many others have been transferred into the nucleus at the early stage2. Once in the nucleus, these genes would be no longer subject to the same evolutionary forces that have driven mitochondria evolution to an extreme. Consequently, these nuclear genes will be less derived and will not have evolution rates and GC biases as extreme as the mitochondria-encoded genes. In theory, trees made from these nuclear genes will be more recalcitrant to the LBA and composition bias that have plagued the phylogenetic analysis of mitochondria. Because of their lower evolutionary rates and less composition biases, in some sense these genes could act as natural “time capsules” that when uncovered, will reveal cues about their distant past.
Mitochondria-to-nuclei gene transfers can be identified using a phylogenetic approach3,16,52,53. Unlike many other lateral gene transfer events, here we have the rare benefit of knowing the donor and the acceptor in advance. Therefore, mitochondria-derived nuclear genes can be identified by looking for a seemingly anomaly in the gene trees – the placement of eukaryotic nuclear genes within the Alphaproteobacteria. Here we leveraged the large number of bacterial, eukaryotic and mitochondrial genomes that are now available to systematically identify mitochondria-derived nuclear genes.
The mitochondria-to-nuclei gene transfer is an ongoing process54,55,56. Although there were parallel transfers, in general genes transferred at earlier stages should be found in a broader taxonomic range of eukaryotic nuclear genomes than these transferred at later stages. Therefore, genomes of phylogenetically diverse eukaryotes, especially those from deep-branching eukaryotes, would be very useful for identifying the early transferred genes. We limit our phylogenomic analyses to these early-transferred genes as they are expected to be less derived than those transferred at a later stage. It will also be much easier to distinguish them from the spurious transfers that happened more recently (e.g., direct transfers from Alphaproteobacteria to the nucleus57). From 2,527 eukaryotic protein families whose top BLAST hits included Alphaproteobacteria, our phylogenetic analysis identified 29 nuclear genes that were most likely transferred from the mitochondria early on (Table 2), as 28 of them were present in at least 4 eukaryotic phyla.
Table 2. Comparison between mitochondria-encoded genes and mitochondria-derived nuclear genes in terms of the evolutionary rate and composition bias.
| Mitochondria-encoded genes | Mitochondria-derived nuclear genes | ||
|---|---|---|---|
| Functional categories | Energy production and conversion | cob, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad9 | cox11, sdhB, sucD, petA, erpA, hesB, ybjS, nuoC, nuoD, nuoF, nuoG, nuoI |
| Translation and posttranslational modification | rpl2, rpl5, rpl6, rpl16, rps1, rps2, rps3, rps4, rps7, rps8, rps11, rps12, rps13, rps14, rps19 | rpl3, grpE, groEL, dnaK, clpB, clpP, hslV, engA, gidA, trmE | |
| Others | AFG1, apaG, bioC, hemN, ksgA, mraW, hypothetical | ||
| Mitochondrial/Nuclear average evolutionary rate (substitution/site) * | 1.713 (stdev 0.225) | 1.273 (stdev 0.088) | |
| Mitochondrial/Nuclear average aminoGC content ** | 0.152 (stdev 0.017) | 0.215 (stdev 0.004) | |
| Mitochondrial/Nuclear average compositional chi-square scores * | 662.4 (stdev 394.3) | 89.6 (stdev 41.4) | |
*T-test P < 0.01 ** T-test P < 0.001.
Evaluation of phylogenetic marker genes and tree reconstruction methods
We compared the mitochondria-derived nuclear genes and mitochondria-encoded genes in terms of their sequence composition biases and substitution rates (Supplementary Table 2). The mitochondrial proteins have significantly more extreme aminoGC than the nuclear proteins (p<0.001, Table 2), indicating that nuclear proteins are less biased than the mitochondrial proteins. We then measured the composition bias of the nuclear and mitochondrial sequences in the context of their alphaproteobacterial homologs using chi-square scores. The larger the chi-square score, the stronger the composition bias. Table 2 shows that the composition bias of the nuclear sequences is substantially smaller than that of the mitochondrial sequences (p<0.01).
Next we compared the substitution rates of the nuclear and mitochondrial genes. In the RAxML genome tree made with mitochondrial markers, the average branch length from the root to mitochondria is 1.713 substitutions/site (stdev 0.225). In comparison, the average branch length from the root to eukaryotes is 1.273 substitutions/site (stdev 0.088) in the genome tree made with nuclear markers. Therefore, the nuclear genes evolved significantly slower than the mitochondrial genes (p<0.01). A similar result was observed when comparing the PhyloBayes trees. Taken all these together, it suggests that mitochondria-derived nuclear genes could be used as a set of alternative “well-behaved” markers to improve the mitochondrial phylogeny.
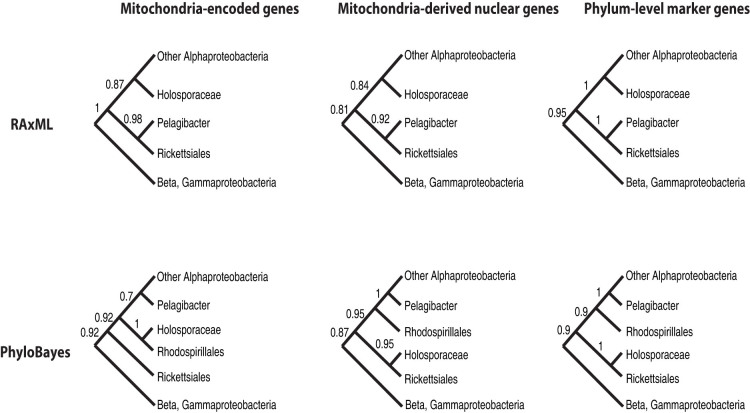
We carried out phylogenetic analyses using the concatenated protein sequences of the nuclear and mitochondrial marker genes respectively. As a reference, the analyses also included the phylum-level markers, a set of 200 single-copy marker genes that were shared by the Alphaproteobacteria58. We used both maximum likelihood and Bayesian methods to infer the phylogeny. To evaluate the effect of composition bias on the phylogeny, we applied the CAT mixture model in PhyloBayes to account for compositional heterogeneity. In contrast, the evolutionary models used to make RAxML maximum likelihood trees did not take compositional heterogeneity into account. Six unique combinations of datasets and methods yielded three different topologies (Figure 4 and Figures S1-6). They differ primarily in the positions of the Pelagibacter and the Holosporaceae family, a group of mostly obligate endosymbionts in the protist acanthamoeba.
Figure 4. Schematic phylogenetic trees based on the mitochondrial, nuclear and phylum-level marker datasets and reconstructed using RAxML and PhyloBayes.
Bootstrap values (for RAxML trees) and posterior probability values (for PhyloBayes trees) for internal nodes are shown beside them.
In all the RAxML trees, Pelagibacter forms a sister clade relationship with the Rickettsiales. It has been well demonstrated that this is a tree reconstruction artifact caused by sequence composition bias20,24,59. Accordingly, the PhyloBayes trees of different markers are in agreement with each other in that they all group Pelagibacter with the free-living Alphaproteobacteria. However, they differ in terms of the position of the Holosporaceae. Trees based on the 200 phylum-level markers and the nuclear markers are congruent and both place Holosporaceae within the Rickettsiales. The tree based on the mitochondrial markers, on the other hand, places Holosporaceae next to the free-living Rhodospirillales.
We then used gene order to evaluate the conflicting evolutionary relationships between Holosporaceae, Pelagibacter and Rickettsiales. In particular, we identified unique genome rearrangement events shared by Holosporaceae and other Rickettsiales in a number of gene clusters, which are otherwise highly syntenic between Pelagibacter and free-living Alphaproteobacteria. Figure 5 shows one such gene cluster encoding 12 proteins, most of which are involved in the TCA cycle and ATP synthesis. The 12 genes form a highly conserved cluster in Pelagibacter and free-living Alphaproteobacteria, with one deletion event occurred in Pelagibacter between genes priA and pdhD. However in Holosporaceae and Rickettsia, the gene cluster has been broken apart at several “hot spots”. For example, the cluster was split on both sides of the priA gene in Holosporaceae and Rickettsia, and it was further split on both sides of the sucCD genes in Rickettsia. The similar gene order patterns in Holosporaceae and Rickettsia suggest that they are closely related and the genome rearrangement events likely occurred in their last common ancestor. Therefore, the independent gene order information is consistent with placing the Holosporaceae with Rickettsiales, and Pelagibacter with the free-living Alphaproteobacteria. Based on the additional gene order information, we believe that the PhyloBayes trees of the phylum-level markers and the nuclear markers make more sense than the tree of the mitochondrial markers.
Figure 5. The gene orders of a gene cluster of 12 protein-coding genes in Rickettsiales (red), Holosporaceae (green), the SAR11 group (purple) and the free-living Rhodospirillum rubrum (black).
Each arrow represents a gene in the cluster. Arrows with dotted lines represent a missing gene. Genome rearrangements are shown as dotted lines between two genes, with the distance between them shown above the lines. Because of the incomplete nature of some genome assemblies, the exact distance between two genes could not be determined. In this case, a minimum distance was estimated as the sum of distances of each gene to the end of the contig it was located on. For the same reason, the orientation of some genes could not be determined (indicated by asterisks below the genes).
Assembly of the alphaproteobacterial and mitochondrial branch of tree of life
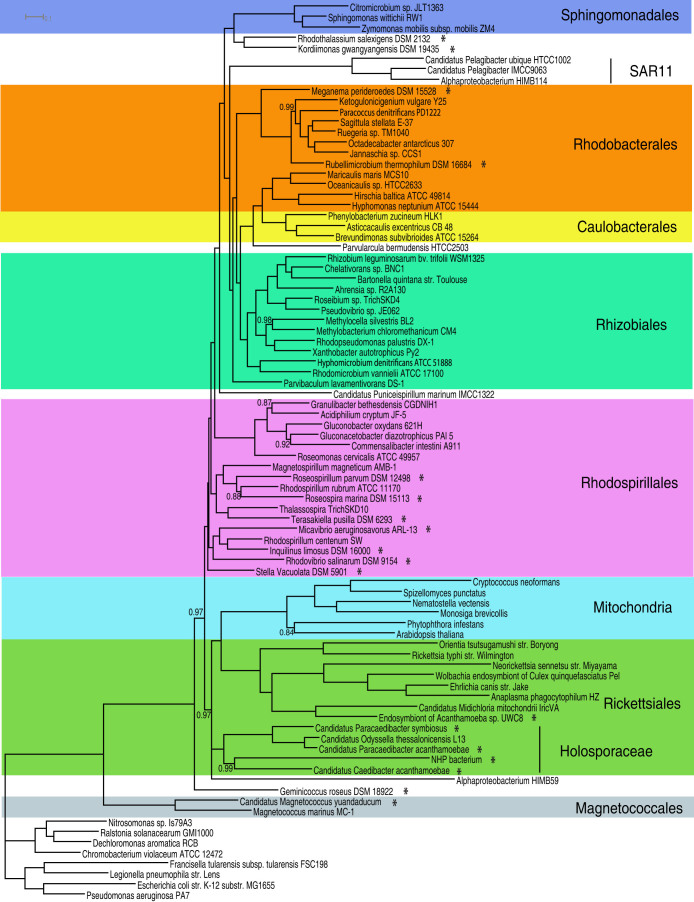
Since mitochondria-derived nuclear genes have less composition bias, lower substitution rates and produce a phylogenetic tree that is consistent with the gene order patterns, we chose to use mitochondria-derived nuclear genes as the marker genes in our final phylogenomic analysis to infer the origin of mitochondria. The final concatenated protein sequence alignment consisted of 6,201 amino acids after the ambiguous alignment regions were removed using the program ZORRO60. Our genome tree using the CAT + GTR model divides the Alphaproteobacteria into at least 7 major groups, corresponding to 7 orders. It places mitochondria within Rickettsiales as a sister clade to the Anaplasmataceae/Rickettsiaceae families, all subtended by the free-living Alphaproteobacterium HIMB59 and the Holosporaceae family (Figure 6). We also used the CAT + BP model to account for both across-site and across-branch compositional heterogeneity. CAT + BP models are extremely computationally expensive for large-datasets like ours24,61. Although the chains have not converged, it is reassuring that preliminary analysis indicates that the consensus tree is congruent with the tree in Figure 6 and places mitochondria at exactly the same position within Rickettsiales.
Figure 6. A rooted Bayesian consensus tree made with the nuclear dataset of 72 Alphaproteobacteria and 6 eukaryotes.
Asterisks indicate the 18 genomes sequenced in this study. The tree was rooted using Beta and Gammaproteobacteria as the outgroup. The posterior probability support values of the internal nodes are 1.0 unless as indicated in the tree.
As a comparison, we also reconstructed genome trees with different combinations of datasets (the nuclear or mitochondrial markers), tree methods (RAxML or PhyloBayes) and data types (original or recoded). The trees are shown in Figures S7–13. Like their counterpart trees with only alphaproteobacterial lineages (Figures S1–S4), these trees either show artifacts of sequence composition bias or topologies inconsistent with the gene order patterns, except for the tree in Figure S11. Figures S11 shows a Bayesian tree made with recoded nuclear marker genes that is congruent with the tree in Figure 6. PhyloBayes tree made using a broader range of 22 eukaryotic genomes (Figure S14) is congruent with the tree in Figure 6, indicating that removing long-branch lineages such as Alveolata and Amoebozoa does not affect the placement of mitochondria in the tree. 8 eukaryotic genomes were not included in the tree in Figure S14 because they were missing most of the 29 nuclear marker genes (Supplementary file 1).
Discussion
Placing mitochondria precisely in the tree of life has been problematic. Sparse taxonomic sampling, sequence composition biases, high evolutionary rates have all plagued the molecular phylogenetic inference of the origin of mitochondria. Here we address this issue with an integrated phylogenomic approach by using a broad taxonomic sampling, better-behaved marker genes and sophisticated models of sequence evolution.
Using NeighborNet and spectral analyses, we first demonstrated that there were significant systematic errors in the current genomic dataset. Of particular concern was the potential LBA problem. We alleviated this problem by filling the gaps in the tree with 18 genomes of novel phylogenetic lineages that had not been sequenced before. In particular, we sequenced five Rickettsiales and four Rhodospirillales, two orders that had shown close affiliations with mitochondria previously. We showed that with the broad taxonomic sampling we were able to reduce the systematic errors, evident by the less prominent incompatible splits observed in the spectral analysis after adding the novel lineages.
One big hurdle in mitochondrial phylogenetic analysis is the extreme composition biases and high evolutionary rates of the mitochondria-encoded genes. To address this issue, we resorted to well-behaved nuclear genes. We showed that mitochondria-derived nuclear genes have significantly less composition biases and lower rates of evolution than mitochondria-encoded genes. As expected, the tree topologies were sensitive to both the marker datasets and methods used to infer the phylogeny. Because the tree made from the nuclear dataset with the CAT site heterogeneous mixture model was congruent with the tree based on the 200 phylum-level marker genes and was most consistent with the gene order patterns, we chose to make the final tree using this setting.
Placing mitochondria firmly within Alphaproteobacteria depends on a robust alphaproteobacterial phylogeny. Overall our final tree using the nuclear dataset is similar to the previously published alphaproteobacterial species trees based on either mitochondrial or phylum-level marker genes18,19,20,21,22,24 in that they all recover the major alphaproteobacterial groups. However, our genome tree does present novel and interesting branching patterns of alphaproteobacterial species that are particularly relevant to the placement of mitochondria. We discuss these new patterns first.
The Holosporaceae family consists of mostly obligate endosymbionts from acanthamoeba. Traditionally it has been assigned to the Rickettsiales order based on the SSU rRNA phylogeny62. With only one draft genome (Odyssella thessalonicensis) sequenced recently, this family was either absent or very poorly represented in all the previous published genome trees16,17,18,19,20,21,22,24,47,59,63. In a recent study with O. thessalonicensis as the sole representative, Holosporaceae was placed outside of the Rickettsiales order and close to the Rhodospirillales20. With a much broader taxonomic representation of this family, we placed Holosporaceae as a deep lineage within Rickettsiales, which is consistent with the traditional taxonomy (Figure 6). We think the topology of Georgiades' study is most likely an artifact of sequence composition bias in the data because when we used mitochondria-encoded genes or did not apply the CAT mixture model to account for compositional heterogeneity, we observed topologies similar to that of Georgiades' study as well (Figure S1–3, S5). In addition, our topology is consistent with the gene order patterns and is congruent with the SSU rRNA tree and the genome tree based on 200 phylum-level marker genes.
While traditionally SAR11 has been placed within the Rickettsiales clade19,64, and as a sister clade to mitochondria20,22, recent studies have conclusively shown that this placement is a tree artifact caused by composition bias, as mitochondria, Rickettsiales and SAR11 all have AT rich genomes21,23,24. Indeed, when we used models that did not account for composition bias, we observed the traditional topology (Figure S1, S3, S5). However, when we applied models that accounted for compositional heterogeneity, only HIMB59 was mostly placed within the Rickettsiales, while all the other SAR11 members clustered with the free-living bacteria (Figure S2, S4, S6). The paraphyletic nature of the SAR11 group has been well documented previously21,59, but there is still uncertainty about the exact position of HIMB5959. In the Viklund study, HIMB59 has been positioned either within the Rickettsiales or the Rhodospirillales order depending on the marker datasets used. In our analyses, HIMB59 is almost always positioned within the Rickettsiales regardless of the markers (mitochondrial, nuclear or phylum-level markers) or the methods used (RAxML or PhyloBayes). The only exception is in the PhyloBayes tree of the mitochondrial dataset, where HIMB59 and other SAR11 species together group with free-living bacteria (Figures S1–6). The placement of HIMB59 within Rickettsiales is unlikely caused by the composition bias because the other SAR11 members with more biased AT rich genomes have been separated from the Rickettsiales. We note however that the branch leading to HIMB59 is not completely resolved from other Rickettsiales (Figure 6), indicating that the position of HIMB59 is unstable. Therefore, we consider the position of HIMB59 tentative and sampling of additional taxa close to HIMB59 should help resolve this issue.
Recent phylogenomic studies have supported two alternative topologies regarding the position of mitochondria: 1) grouping with the free-living Rhodospirillales order17, 2) grouping with the Rickettsiales order16,18,19,20,21,22. Resolving this conflict has clear bearing on our understanding of the driving force behind the initial endosymbiosis event. For example, the “hydrogen hypothesis” proposes the metabolic syntrophy between a H2-producing alphaproteobacterial symbiont and a H2-dependant archaeon as the driving force behind the endosymbiosis6. The “oxygen scavenger” hypothesis, on the other hand, proposes that the removal of the toxic oxygen by the Alphaproteobacterium from the anaerobic host has driven the initial symbiosis65. A key piece of support for the “hydrogen hypothesis” necessitates that the alphaproteobacterial ancestor of mitochondria possessed a H2-producing machinery. Members of the Rhodospirillales order are capable of producing H2 by fermentation while Rickettsiales species are not. Grouping mitochondria with Rhodospirillales certainly lends stronger support to the “hydrogen hypothesis”. With a much broader taxon sampling of both Rickettsiales and Rhodospirillales, our phylogenomic analyses have almost always placed mitochondria with Rickettsiales and never with Rhodospirillales, regardless of the marker datasets and phylogenetic methods used (Figures 6, S7–13). Using the same dataset in Esser et al. study but a more sophisticated trimming method to remove fast-evolving sites, Fizpatrick et al. have shown that mitochondria are grouped with Rickettsiales and not with Rhodospirillales18. Taking our and Fizpatrick et al. 's results together, we suspect the topology observed by Esser et al. might be a phylogenetic tree reconstruction artifact caused by either inadequate taxonomic sampling or sequence alignment trimming.
Our genome tree shows that the Rickettsiaceae/Anaplasmataceae families are the closest relatives of mitochondria (posterior probability 1.0, Figure 6). This suggests that the ancestor of mitochondria was most likely a Rickettsiales endosymbiont that had been already living inside the host cells. We note, however, that the endosymbiont did not have to be an obligate intracellular bacterium at the time of the initial endosymbiosis event. As a result, it could have escaped the host later on and given rise to obligate intracellular Rickettsiales lineages as we see today. For the first time, we are able to place mitochondria firmly within the Rickettsiales order. Previous studies have all placed mitochondria as a sister clade to Rickettsiales but never unequivocally within Rickettsiales (if we discount the sister clade relationship of Pelagibacter and mitochondria). In our genome tree, Holosporaceae forms the deepest branch within the Rickettsiales. Mitochondria originated sometime after the divergence of Holosporaceae from the rest of the Rickettsiales. The Rickettsiales/mitochondria clade has a very strong posterior probability support value of 0.97. Therefore, we conclude that mitochondria evolved as a derived lineage from within the Rickettsiales order.
The multiple novel Holosporaceae genomes will be extremely valuable in providing insights into the genetic complement of mitochondrial ancestor. Because they are the immediate outgroup of the mitochondria/Rickettsiaceae/Anaplasmataceae clade, they have great potentials to improve the accuracy of the mitochondrial ancestral reconstruction. For example, based on the genome sequence of Candidatus Midichloria mitochondrii, a novel phylogenetic lineage within Rickettsiales, it has been recently predicted that mitochondrial ancestor possessed flagella and could undergo oxidative phosphorylation under both aerobic and microoxic conditions66.
In conclusion, using an integrated phylogenomic approach, we placed mitochondria firmly within the tree of life and moved a step closer toward pinpointing the origin of mitochondria. Our results suggest that mitochondria most likely originated from the Rickettsiales lineage, but not from the distantly related free-living Pelagibacter and Rhodospirillales.
Methods
NeighborNet and spectral analyses
Mitochondria-encoded genes from16 were used as the marker genes for NeighborNet and spectral analyses. Six genes (atp6, atp9, atpA, cox1, yejR, yejU) were excluded from the original list because of their potential involvement in lateral gene transfer, resulting in a total of 26 genes (Table 2). These genes from 54 alphaproteobacterial genomes and 6 mitochondria representatives (Figure 1) were identified, aligned, trimmed using AMPHORA267. The 54 alphaproteobacterial genomes were selected using a tree-based greedy algorithm49 to maximize the phylogenetic diversity. The 6 mitochondrial representatives (Reclinomonas americana, Marchantia polymorpha, Hemiselmis andersenii, Mesostigma viride, Rhodomonas salina and Phytophthora infestans) were selected because they were primitive, gene rich and represented a broad range of phylogenetic diversities. NeighborNet analysis was performed using the SplitsTree program68 on the concatenated alignment of the 26 mitochondria-encoded proteins with the default parameters. The spectral analysis was performed using the Split Analyses Methods (SAMS)38 with the same dataset after recoding amino acids into 4 categories according to their physicochemical properties (AVFPMILW, DE, RK and STYHCNGQ). In the spectral analysis, the support for each split was calculated as the number of sites in the alignment supporting that split. The splits were then ranked by their supporting values. To evaluate the systematic errors in the dataset, each of the 50 top-ranked splits was manually evaluated to determine whether it was compatible with well established phylogenetic relationships such as the monophyly of mitochondria or Rickettsiales. The systematic errors in the dataset were quantified as Sinc/Stotal, where Sinc is the support value of an incompatible split and Stotal is the total split support values in the 50 top-ranked splits. The statistical significance of the difference in systematic errors with and without 18 new genomes was tested using the Mann-Whitney U-test.
Selection of novel alphaproteobacterial species for sequencing
The aligned SSU rRNA gene sequences of 9,817 alphaproteobacterial isolates were retrieved from the Ribosomal Database Project48 and were used to construct a maximum likelihood tree using FastTree69. A tree-based greedy algorithm was then used to rank isolates by their phylogenetic novelty49, taking into consideration at the same time whether genome sequences of closely related species were available. The availability of an isolate's genomic DNA was also considered in the selection process. In total, 18 isolates were selected for genome sequencing. A SSU rRNA maximum likelihood tree of 70 alphaproteobacterial representatives including the 18 targeted species was then made by RAxML70 using the GTR + Gamma model.
Genome sequencing, assembly and annotation
Genomes of the 18 bacterial strains were sequenced by 454 and Illumina sequencing. 7 bacterial strains (Micavibrio aeruginosavorus, endosymbiont of acanthamoeba UWC8, Candidatus Caedibacter acanthamoebae, Candidatus Paracaedibacter acanthamoebae, Candidatus Paracaedibacter symbiosus, Stella vacuolata, Candidatus Magnetococcus yuandaducum) were sequenced by 454 using a combination of indexed shotgun and 3 kb paired-end libraries, and assembled using Newbler 2.5.3. The rest 11 strains were sequenced by the Illumina paired-end sequencing using HiSeq 2000, and assembled using the CLCGenomicWorkbench 6.0.1. PCR and Sanger sequencing were used to close the gaps between contigs when necessary. Protein-coding genes of all 18 genomes were predicted using the GLIMMER software package71. The genome sequence of M. aeruginosavorus has already been reported previously72.
Systematic identification of mitochondria-derived nuclear genes
The phylogenetic distribution of all sequenced eukaryotic genomes was retrieved from the GenomeOnline database46. A total of 30 eukaryotic genomes, representing a broad range of phylogenetic diversity, were selected for identifying the mitochondria-derived nuclear genes (Supplementary Table 3). For every single protein in the 30 eukaryotic nuclear genomes, an initial BLASTP search was performed against a local database containing all complete bacterial, archaeal and mitochondrial genomes. A eukaryotic gene was retained for further analysis if its top 5 hits contained an alphaproteobacterial or mitochondrial sequence (e-value cutoff 1e-4). The eukaryotic genes passing the initial BLASTP screening were clustered into protein families using the Markov Cluster Algorithm73 and only families that were present in at least 8 eukaryotic species were selected for phylogenetic analysis. For each of retained protein families, its homologs from all complete bacterial genomes were retrieved by BLASTP search (e-value cutoff 1e-15). Protein sequences of each family were aligned by MAFFT74 and trimmed by ZORRO60. Phylogenetic trees constructed using FastTree were subject to manual inspection. Paralogs, if existed in a family, were separated and each was treated as a new family so that only orthologous genes were used for inferring phylogeny. We looked for a specific branching pattern in the trees where eukaryotic sequences clustered with Alphaproteobacteria and/or mitochondria. Families with less than 8 eukaryotic species, or few alphaproteobacterial species, or a complex evolutionary history (e.g., alpha, beta and gammaproteobacterial lineages were not clustered together) were removed. In the end, 29 mitochondria-derived nuclear genes were identified as the marker genes for phylogenomic analysis (Table 2). All genes are present in at least 4 phyla except for apaG, which is present in 2 eukaryotic phyla (Supplementary file 1).
Assembly of mitochondrial, nuclear and phylum-level marker datasets
For each of 26 mitochondrial and 29 nuclear marker genes, its homologs in 200 alphaproteobacterial genomes (Supplementary Table 4) and mitochondrial/eukaryotic representatives were identified, aligned and trimmed using the program AMPHORA267. With very few exceptions, the nuclear marker genes were single-copy genes in all of the bacterial and nuclear genomes analyzed. More duplications were present in mitochondrial marker genes (Supplementary file 2). In those cases in which two or more homologs were identified within a single genome, a tree-guided approach was used to resolve the redundancy as described in63. If the redundancy was caused by a species-specific duplication event, then one homolog was randomly chosen as the representative. Otherwise, to avoid potential complications in interpreting the phylogeny, we treated the marker as ‘missing’ in that particular genome. We also identified 200 single-copy marker genes that were present in all the alphaproteobacterial genomes using Phyla-AMPHORA58 and we called them the phylum-level marker dataset. Aligned and trimmed protein sequences within each dataset were concatenated by species and were used as the master datasets for the downstream analyses. The final mitochondrial, nuclear and phylum-level marker alignments contain 5,790, 6,201 and 54,006 amino acids respectively (Supplementary file 3, 4).
Evaluation of marker datasets and phylogenetic methods
We selected 47 representatives of alphaproteobacterial genomes using the tree-based greedy algorithm described above49 and used this set of taxa as a benchmark to evaluate the different datasets (mitochondrial, nuclear and phylum-level markers) and tree construction methods (RAxML and PhyloBayes). We limited this analysis to 47 alphaproteobacterial genomes to reduce the computational cost associated with reconstructing the PhyloBayes tree from the phylum-level marker alignment, which contained 54,006 amino acids. For each concatenated dataset, a maximum likelihood (ML) tree and a Bayesian tree were made. ML trees were reconstructed using RAxML70 with the best model selected by the program, and was bootstrapped with 100 replicates. Bayesian consensus trees were reconstructed using PhyloBayes75 with the CAT + GTR options, as recommended in the manual. Two independent MCMC chains were run and the chains were considered converged when the maxdiff dropped below 0.3, as suggested in the manual. The trees were sampled every 10 cycles and the beginning one fifth of the trees from each chain were discarded as burn-in.
Estimation of the composition biases and evolutionary rates of the mitochondrial and nuclear marker genes
To estimate the composition biases and evolutionary rates of the mitochondrial and nuclear marker genes, we selected a larger set of 72 Alphaproteobacteria representatives (including 18 genomes sequenced in this study). For the mitochondrial marker dataset, we added 6 mitochondrial representatives described above. For the nuclear marker dataset, we added 6 eukaryotic representatives (Cryptococcus neoformans, Arabidopsis thaliana, Nematostella vectensis, Spizellomyces punctatus, Monosiga brevicollis, Phytophthora infestans) that represented the major eukaryotic phyla that have sequenced genomes. Alveolata, Amoebozoa, Euglenozoa and Diplomonadida were not included because they had extremely long branches and therefore were prone to LBA. To quantify the GC bias in the data, first we calculated aminoGC, the frequencies of amino acids (Gly, Ala and Pro) that are encoded by GC rich codons24. AminoGC essentially measures the effect of GC bias on the protein sequences. The composition bias of each taxon was also measured as a chi-square score using a scheme described in24. To better account for the missing data in the alignment, we modified the scheme and used the normalized frequency of each amino acid instead of the absolute count. RAxML and PhyloBayes trees were reconstructed using the mitochondrial and nuclear marker alignments. The overall mitochondria/eukaryotes evolutionary rate was estimated as the average branch length from the root of the tree to all the mitochondrial/eukaryotic lineages.
Reconstruction of final genome tree
For the final genome tree reconstruction, we used the nuclear dataset of 72 Alphaproteobacteria representatives, 6 eukaryotic representatives described above and 8 outgroups (Nitrosomonas sp. Is79A3, Ralstonia solanacearum GMI1000, Dechloromonas aromatica RCB, Chromobacterium violaceum ATCC 12472, Francisella tularensis subsp. tularensis FSC198, Legionella pneumophila str. Lens, Escherichia coli str. K-12 substr. MG1655 and Pseudomonas aeruginosa PA7). The outgroup species were taken from a previous study21. We excluded Buchnera because it had a long branch and might cause the LBA artifact. A Bayesian consensus tree was made using PhyloBayes as described above. A Bayesian analysis was also performed with the CAT + BP model as implemented in nh_phylobayes76, with two separate chains running for 6,000 generations each. The chains have not converged after 3 months. As noted by24,61, nh_phylobayes is computationally expensive.
As a comparison, we also reconstructed both RAxML and PhyloBayes trees from mitochondrial and nuclear markers with and without amino acid recoding. For the Bayesian analysis, amino acids were recoded to 6 Dayhoff categories. Bayesian consensus trees were made using PhyloBayes as described above plus the ‘–recode dayhoff6’ option. For the RAxML analysis, amino acids were recoded to 4 Dayhoff categories. ML trees were made using the GTR + Gamma model.
Author Contributions
Conceived and designed the experiments: M.W. Performed the experiments: Z.W. Analyzed the data: Z.W. M.W. Contributed to the writing of the manuscript: Z.W. M.W.
Supplementary Material
Supplementary information
Dataset 1
Dataset 2
Dataset 3
Dataset 4
Dataset 5
Dataset 6
Dataset 7
Dataset 8
Acknowledgments
This research was funded by an award from Thomas F. & Kate Miller Jeffress Memorial Trust to MW. We would like to thank John Dustin Loy of University of Nebraska-Lincoln for providing shrimp tissues that contained the NHP bacterium, and Wei Lin and Yongxin Pan of Institute of Geology and Geophysics, Chinese Academy of Sciences for providing DNA of Candidatus Magnetococcus yuandaducum.
References
- Lang B. F., Seif E., Gray M. W., O'Kelly C. J. & Burger G. A comparative genomics approach to the evolution of eukaryotes and their mitochondria. J. Eukaryot. Microbiol. 46, 320–326 (1999). [DOI] [PubMed] [Google Scholar]
- Kurland C. G. & Andersson S. G. Origin and evolution of the mitochondrial proteome. Microbiol. Mol. Biol. Rev. 64, 786–820 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T. & Huynen M. A. Reconstruction of the proto-mitochondrial metabolism. Science 301, 609 (2003). [DOI] [PubMed] [Google Scholar]
- Gray M. W., Burger G. & Lang B. F. Mitochondrial evolution. Science 283, 1476–1481 (1999). [DOI] [PubMed] [Google Scholar]
- Gray M. W., Burger G. & Lang B. F. The origin and early evolution of mitochondria. Genome Biol. 2, REVIEWS1018 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W. & Muller M. The hydrogen hypothesis for the first eukaryote. Nature 392, 37–41 (1998). [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst. Zool. 27, 401–410 (1978). [Google Scholar]
- Hillis D. M., Huelsenbeck J. P. & Swofford D. L. Hobgoblin of phylogenetics? Nature 369, 363–364 (1994). [DOI] [PubMed] [Google Scholar]
- Foster P. G. & Hickey D. A. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J. Mol. Evol. 48, 284–290 (1999). [DOI] [PubMed] [Google Scholar]
- Woese C. R., Achenbach L., Rouviere P. & Mandelco L. Archaeal phylogeny: reexamination of the phylogenetic position of Archaeoglobus fulgidus in light of certain composition-induced artifacts. Syst. Appl. Microbiol. 14, 364–371 (1991). [DOI] [PubMed] [Google Scholar]
- Hasegawa M. & Hashimoto T. Ribosomal RNA trees misleading? Nature 361, 23 (1993). [DOI] [PubMed] [Google Scholar]
- Viale A. M. & Arakaki A. K. The chaperone connection to the origins of the eukaryotic organelles. FEBS Lett. 341, 146–151 (1994). [DOI] [PubMed] [Google Scholar]
- Gupta R. S. Evolution of the chaperonin families (Hsp60, Hsp10 and Tcp-1) of proteins and the origin of eukaryotic cells. Mol. Microbiol. 15, 1–11 (1995). [DOI] [PubMed] [Google Scholar]
- Karlin S. & Brocchieri L. Heat shock protein 60 sequence comparisons: duplications, lateral transfer, and mitochondrial evolution. Proc. Natl. Acad. Sci. U. S. A. 97, 11348–11353 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov V. V. Mitochondrial connection to the origin of the eukaryotic cell. Eur. J. Biochem. 270, 1599–1618 (2003). [DOI] [PubMed] [Google Scholar]
- Wu M. et al. Phylogenomics of the Reproductive Parasite Wolbachia pipientis wMel: A Streamlined Genome Overrun by Mobile Genetic Elements. PLoS Biol. 2, E69 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esser C. et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol. Biol. Evol. 21, 1643–1660 (2004). [DOI] [PubMed] [Google Scholar]
- Fitzpatrick D. A., Creevey C. J. & McInerney J. O. Genome Phylogenies Indicate a Meaningful Alphaproteobacterial Phylogeny and Support a Grouping of the Mitochondria with the Rickettsiales. Mol. Biol. Evol. 23, 74–85 (2006). [DOI] [PubMed] [Google Scholar]
- Williams K. P., Sobral B. W. & Dickerman A. W. A robust species tree for the alphaproteobacteria. J. Bacteriol. 189, 4578–4586, 10.1128/JB.00269-07 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgiades K., Madoui M. A., Le P., Robert C. & Raoult D. Phylogenomic analysis of Odyssella thessalonicensis fortifies the common origin of Rickettsiales, Pelagibacter ubique and Reclimonas americana mitochondrion. PLoS ONE 6, e24857, 10.1371/journal.pone.0024857 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N. & Embley T. M. The SAR11 group of alpha-proteobacteria is not related to the origin of mitochondria. PLoS ONE 7, e30520, 10.1371/journal.pone.0030520 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash J. C. et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep 1, 13, 10.1038/srep00013 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brindefalk B., Ettema T. J., Viklund J., Thollesson M. & Andersson S. G. A phylometagenomic exploration of oceanic alphaproteobacteria reveals mitochondrial relatives unrelated to the SAR11 clade. PLoS ONE 6, e24457, 10.1371/journal.pone.0024457 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund J., Ettema T. J. & Andersson S. G. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol. Biol. Evol. 29, 599–615, 10.1093/molbev/msr203 (2012). [DOI] [PubMed] [Google Scholar]
- Jeffroy O., Brinkmann H., Delsuc F. & Philippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 22, 225–231 (2006). [DOI] [PubMed] [Google Scholar]
- Stefanovic S., Rice D. W. & Palmer J. D. Long branch attraction, taxon sampling, and the earliest angiosperms: Amborella or monocots? BMC Evol. Biol. 4, 35 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips M. J., Delsuc F. & Penny D. Genome-scale phylogeny and the detection of systematic biases. Mol. Biol. Evol. 21, 1455–1458 (2004). [DOI] [PubMed] [Google Scholar]
- Delsuc F., Brinkmann H. & Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat. Rev. Genet. 6, 361–375 (2005). [DOI] [PubMed] [Google Scholar]
- Soltis D. E. et al. Genome-scale data, angiosperm relationships, and “ending incongruence”: a cautionary tale in phylogenetics. Trends Plant Sci. 9, 477–483 (2004). [DOI] [PubMed] [Google Scholar]
- Lockhart P. J. & Penny D. The place of Amborella within the radiation of angiosperms. Trends Plant Sci. 10, 201–202 (2005). [DOI] [PubMed] [Google Scholar]
- Bandelt H. J. & Dress A. W. M. A canonical decomposition theory for metrics on a finite set. Adv. Math. 92, 47–105 (1992). [Google Scholar]
- Lockhart P. J. & Cameron S. A. Trees for bees. Trends Ecol Evol 16, 84–88 (2001). [DOI] [PubMed] [Google Scholar]
- Waddell P. J., Cao Y., Hauf J. & Hasegawa M. Using novel phylogenetic methods to evaluate mammalian mtDNA, including amino acid-invariant sites-LogDet plus site stripping, to detect internal conflicts in the data, with special reference to the positions of hedgehog, armadillo, and elephant. Syst. Biol. 48, 31–53 (1999). [DOI] [PubMed] [Google Scholar]
- Clements K. D., Gray R. D. & Howard Choat J. Rapid evolutionary divergences in reef fishes of the family Acanthuridae (Perciformes: Teleostei). Mol. Phylogenet. Evol. 26, 190–201 (2003). [DOI] [PubMed] [Google Scholar]
- Hendy M. D. & Penny D. Spectral analysis of phylogenetic data. J Classif 10, 5–24 (1993). [Google Scholar]
- Lento G. M., Hickson R. E., Chambers G. K. & Penny D. Use of spectral analysis to test hypotheses on the origin of pinnipeds. Mol. Biol. Evol. 12, 28–52 (1995). [DOI] [PubMed] [Google Scholar]
- Kennedy M., Holland B. R., Gray R. D. & Spencer H. G. Untangling long branches: identifying conflicting phylogenetic signals using spectral analysis, neighbor-net, and consensus networks. Syst. Biol. 54, 620–633 (2005). [DOI] [PubMed] [Google Scholar]
- Wagele J. W. & Mayer C. Visualizing differences in phylogenetic information content of alignments and distinction of three classes of long-branch effects. BMC Evol. Biol. 7, 147 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy M. et al. The long and short of it: branch lengths and the problem of placing the New Zealand short-tailed bat, Mystacina. Mol. Phylogenet. Evol. 13, 405–416 (1999). [DOI] [PubMed] [Google Scholar]
- Mallatt J. & Winchell C. J. Testing the new animal phylogeny: first use of combined large-subunit and small-subunit rRNA gene sequences to classify the protostomes. Mol. Biol. Evol. 19, 289–301 (2002). [DOI] [PubMed] [Google Scholar]
- Brinkmann H., van der Giezen M., Zhou Y., Poncelin de Raucourt G. & Philippe H. An empirical assessment of long-branch attraction artefacts in deep eukaryotic phylogenomics. Syst. Biol. 54, 743–757 (2005). [DOI] [PubMed] [Google Scholar]
- Philippe H. Rodent monophyly: pitfalls of molecular phylogenies. J. Mol. Evol. 45, 712–715 (1997). [PubMed] [Google Scholar]
- Philippe H., Lartillot N. & Brinkmann H. Multigene analyses of bilaterian animals corroborate the monophyly of Ecdysozoa, Lophotrochozoa, and Protostomia. Mol. Biol. Evol. 22, 1246–1253 (2005). [DOI] [PubMed] [Google Scholar]
- Leebens-Mack J. et al. Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one's way out of the Felsenstein zone. Mol. Biol. Evol. 22, 1948–1963 (2005). [DOI] [PubMed] [Google Scholar]
- Yoon H. S. et al. Broadly sampled multigene trees of eukaryotes. BMC Evol. Biol. 8, 14 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani I. et al. The Genomes OnLine Database (GOLD) v.4: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 40, D571–579, 10.1093/nar/gkr1100 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D. et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462, 1056–1060, 10.1038/nature08656 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. R. et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37, D141–145, 10.1093/nar/gkn879 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel M. Phylogenetic diversity and the greedy algorithm. Syst. Biol. 54, 527–529 (2005). [DOI] [PubMed] [Google Scholar]
- Burger G., Gray M. W. & Lang B. F. Mitochondrial genomes: anything goes. Trends Genet 19, 709–716 (2003). [DOI] [PubMed] [Google Scholar]
- Anderson S. et al. Sequence and organization of the human mitochondrial genome. Nature 290, 457–465 (1981). [DOI] [PubMed] [Google Scholar]
- Karlberg O., Canback B., Kurland C. G. & Andersson S. G. The dual origin of the yeast mitochondrial proteome. Yeast 17, 170–187 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabaldon T. & Huynen M. A. From Endosymbiont to Host-Controlled Organelle: The Hijacking of Mitochondrial Protein Synthesis and Metabolism. PLoS Comput. Biol. 3, e219 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent J. M. & Palmer J. D. RNA-mediated transfer of the gene coxII from the mitochondrion to the nucleus during flowering plant evolution. Cell 66, 473–481 (1991). [DOI] [PubMed] [Google Scholar]
- Adams K. L. et al. Intracellular gene transfer in action: dual transcription and multiple silencings of nuclear and mitochondrial cox2 genes in legumes. Proc. Natl. Acad. Sci. U. S. A. 96, 13863–13868 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello P. S. & Gray M. W. Silent mitochondrial and active nuclear genes for subunit 2 of cytochrome c oxidase (cox2) in soybean: evidence for RNA-mediated gene transfer. EMBO J. 11, 3815–3820 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp J. C. et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 (2007). [DOI] [PubMed] [Google Scholar]
- Wang Z. & Wu M. A phylum-level bacterial phylogenetic marker database. Mol Biol Evol 30, 1258–1262, 10.1093/molbev/mst059 (2013). [DOI] [PubMed] [Google Scholar]
- Viklund J., Martijn J., Ettema T. J. & Andersson S. G. Comparative and Phylogenomic Evidence That the Alphaproteobacterium HIMB59 Is Not a Member of the Oceanic SAR11 Clade. PLoS ONE 8, e78858, 10.1371/journal.pone.0078858 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M., Chatterji S. & Eisen J. A. Accounting for alignment uncertainty in phylogenomics. PLoS One 7, e30288, 10.1371/journal.pone.0030288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesnidal M. P., Helmkampf M., Bruchhaus I. & Hausdorf B. Compositional heterogeneity and phylogenomic inference of metazoan relationships. Mol. Biol. Evol. 27, 2095–2104, 10.1093/molbev/msq097 (2010). [DOI] [PubMed] [Google Scholar]
- Garrity G. M., Bell J. A. & Lilburn T. G. Taxonomic outline of the prokaryotes. Bergey's manual of systematic bacteriology, Second Edition., (Springer-Verlag, New York., 2004). [Google Scholar]
- Wu M. & Eisen J. A. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 9, R151 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferla M. P., Thrash J. C., Giovannoni S. J. & Patrick W. M. New rRNA gene-based phylogenies of the Alphaproteobacteria provide perspective on major groups, mitochondrial ancestry and phylogenetic instability. PLoS ONE 8, e83383, 10.1371/journal.pone.0083383 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson S. G., Karlberg O., Canback B. & Kurland C. G. On the origin of mitochondria: a genomics perspective. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 358, 165–177; discussion 177–169 10.1098/rstb.2002.1193 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassera D. et al. Phylogenomic evidence for the presence of a flagellum and cbb(3) oxidase in the free-living mitochondrial ancestor. Mol. Biol. Evol. 28, 3285–3296, 10.1093/molbev/msr159 (2011). [DOI] [PubMed] [Google Scholar]
- Wu M. & Scott A. J. Phylogenomic analysis of bacterial and archaeal sequences with AMPHORA2. Bioinformatics 28, 1033–1034, 10.1093/bioinformatics/bts079 (2012). [DOI] [PubMed] [Google Scholar]
- Huson D. H. & Bryant D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267 (2006). [DOI] [PubMed] [Google Scholar]
- Price M. N., Dehal P. S. & Arkin A. P. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490, 10.1371/journal.pone.0009490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- Delcher A. L., Bratke K. A., Powers E. C. & Salzberg S. L. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23, 673–679 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Kadouri D. E. & Wu M. Genomic Insights into An Obligate Epibiotic Bacterial Predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics 12, 453, 10.1186/1471-2164-12-453 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A. J., Van Dongen S. & Ouzounis C. A. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 30, 1575–1584 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K. & Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N. & Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol Biol Evol 21, 1095–1109, 10.1093/molbev/msh112 (2004). [DOI] [PubMed] [Google Scholar]
- Blanquart S. & Lartillot N. A site- and time-heterogeneous model of amino acid replacement. Mol. Biol. Evol. 25, 842–858 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Dataset 1
Dataset 2
Dataset 3
Dataset 4
Dataset 5
Dataset 6
Dataset 7
Dataset 8