Abstract
Salp15 is an Ixodes scapularis salivary protein that inhibits CD4+ T cell activation through the repression of TCR ligation-triggered calcium fluxes and IL-2 production. We show in this study that Salp15 binds specifically to the CD4 coreceptor on mammalian host T cells. Salp15 specifically associates through its C-terminal residues with the outermost two extracellular domains of CD4. Upon binding to CD4, Salp15 inhibits the subsequent TCR ligation-induced T cell signaling at the earliest steps including tyrosine phosphorylation of the Src kinase Lck, downstream effector proteins, and lipid raft reorganization. These results provide a molecular basis to understanding the immunosuppressive activity of Salp15 and its specificity for CD4+ T cells.
Ixodes scapularis ticks are the vectors for several pathogens including Borrelia burgdorferi and Anaplasma phagocytophilum (1, 2). Tick salivary proteins enter the host and exert pleiotropic immunosuppressive effects (3–9) that could also contribute to the efficient transmission of pathogens (10). I. scapularis salivary protein (Salp) 15 is a pleiotropic inducible Ag that may be responsible for the immunomodulatory action of tick saliva on acquired immune responses (3) and the protection of the causative agent of Lyme disease, B. burgdorferi, against Ab-mediated killing (11), which complements other immune evasion mechanisms by the spirochete (12). Inhibition of CD4+ T cell activation mediated by Salp15 results from the repression of IL-2 production upon recognition of cognate Ag (3). The interaction of Salp15 with CD4+ T cells results in decreased TCR-triggered calcium signals and the activation of the transcription factors, NF-κB and NF-AT (3). The characterization of Salp15 action in the mammalian host is important to understand the molecular basis of the immunosuppressive activities of I. scapularis saliva as well as vector-pathogen-host interactions.
We have now identified CD4 as the specific receptor for Salp15 on T cells. A direct association occurs between CD4 and the C-terminal amino acid residues of Salp15, which allows Salp15 to act at the very beginning of TCR ligation-induced signaling cascades. These results shed light on the mechanism of action of Salp15 during the activation of CD4+ T cells and provide the basis for its specificity.
Materials and Methods
Cell purification and activation
Murine CD4+ T cells were purified as described previously (3) and preincubated with His-tagged Salp15 (50 µg/ml) at 37°C for 20 min before stimulation with 10 µg/ml anti-CD3ε, 1 µg/ml anti-CD28, and 10 µg/ml anti-Armenian and Syrian hamster IgG (BD Biosciences). Jurkat (E6-1) cells were stimulated using 5 µg/ml OKT3 mAb (eBioscience) and 1 µg/ml anti-human CD28 cross-linked with 5 µg/ml anti-mouse IgG.
Antibodies
Anti-Lck, anti-pSrc family (Tyr416) to measure p-Lck364 (13), anti-pLck Tyr505, anti-pZap70 (Tyr319), and anti-Zap70 Abs were obtained from Cell Signaling Technologies. Anti-pTyr (pY99), anti-p phospholipase C (PLC)4 γ1 (Tyr783), anti-PLCγ1, polyclonal anti-CD4, anti-CD3, anti-CD28, anti-TCRβ Abs, and normal rabbit IgG were obtained from Santa Cruz Biotechnology. The polyclonal anti-His-HRP Ab was obtained from Novus Biologicals.
Purification of recombinant proteins
His-tagged Salp15 and His-tagged, thioredoxin (TR)-fused Salp13 (14) were purified from cultured Drosophila S2 cells, as described previously (3).
Confocal microscopy
For the visualization of lipid rafts, purified CD4+ and CD8+ cells were cytospun for 10 min at 1000 rpm onto positively charged microslides (Fisher Scientific), fixed with 3.7% paraformaldehyde (Sigma-Aldrich) for 15 min, and stained with CTB594 (Molecular Probes).
For colocalization experiments, CD4+ T cells were cytospun and fixed as described above, blocked with 5% normal serum and anti-CD16/CD32 (1: 100) in PBS, and stained with 1 µg of biotinylated anti-CD4 (L3T4; BD Biosciences), followed by streptavidin594 and His-tagged Salp15, labeled with Alexa Fluor-488 (His-tagged Salp15-488, Protein Labeling kit; Molecular Probes). For competition assays, cells were first blocked, incubated with either polyclonal anti-CD4 Ab, normal rabbit IgG, His-tagged Salp15 (100×), or lysozyme (100×) followed by staining with His-tagged Salp15-488. All samples were visualized with an Olympus Confocal microscope equipped with Fluorview 3.0 software.
Western blotting and coimmunoprecipitation
For tyrosine phosphorylation analysis, cells were lysed in 60 mM Tris (pH 7.4) buffer containing 1% Triton X-100 and protease inhibitors for 15 min on ice, sonicated 4 × 5 s and incubated at 37°C for 10 min. The lysate was cleared by centrifugation, subjected to SDS-PAGE, and immunoblotted with specific Abs.
For coimmunoprecipitation experiments, 6 × 106 murine CD4+ T, or 1–2 × 107 Jurkat, HeLa, or HeLa-CD4 cells were lysed in 20mMHEPES (pH 7.4) containing 1% Triton X-100 for 15 min on ice. The cell lysate was mixed with 50 µg/ml His-tagged-Salp15 or His-tagged TR-Salp13 as a control (14) and immunoprecipitated using an anti-polyhistidine Ab conjugated to agarose, anti-CD4, anti-CD3, anti-CD28, or normal rabbit IgG plus agarose-conjugated protein A (Sigma-Aldrich), overnight at 4°C.
Flow cytometry
Salp15 binding to NIH3T3-CD4 cells was determined by flow cytometry. The cells were blocked with 5% BSA in PBS, followed by staining with His-tagged Salp15-488 (15 µg/106 cells) in blocking solution for 30 min at 4°C. The cells were analyzed in a LSRII flow cytometer (BD Biosciences).
Microtiter binding assay
Purified soluble CD4 (sCD4) (D1D2 preparation) was coated overnight at 4°C at the indicated concentrations in 0.1 M sodium carbonate buffer (pH 9.5) in 96-well plates. The wells were blocked with 10% FCS/PBS, and incubated with His-tagged Salp15 (0.4 µM) and HRP-conjugated anti-His Ab followed by microwell peroxidase substrate (Kirkegaard & Perry Laboratories). Chemically synthesized overlapping peptides of Salp15 (3) were purchased from GenScript. To study the binding of Salp15 and peptides to sCD4, His-tagged Salp15 (5 µg) and the different peptides (0.5 µg) were coated as described above. The wells were then probed with sCD4-HRP (ImmunoDiagnostics). For competition assays, indicated concentrations of Salp15 or P11 were added to sCD4-HRP and incubated with plate-bound P11 (aa 95–114, GPNGQTCAE KNKCVGHIPGC, 0.1 µg). Kd were determined by nonlinear curve fit using GraphPad Prism, version 4.0 (GraphPad).
Analytical gel filtration
Purified sCD4 (D1–D4, 10 µg) and His-tagged Salp15 (20 µg) were eluted through two Superdex-200 HR 10/30 gel filtration columns (Amersham Pharmacia Biotech) connected in tandem using PBS as equilibration and elution buffer at a flow rate of 0.3 ml/min with a FPLC system (Amersham Pharmacia Biotech) equipped with a Liquid Chromatography Controller LCC-501 Plus and regulated using FPLC director version 1.10. Blue dextran was used to estimate the void volume, whereas BSA, OVA and lysozyme were used as standards with elution volumes of 14.78 ml, 26.45 ml, 28.45 ml, and 39.28 ml, respectively. To estimate the individual peak position of overlapping peaks, a Gaussian deconvolution algorithm (15) was used, using the software Origin version 5.0 (OriginLab).
Results and Discussion
CD4 is the receptor for Salp15
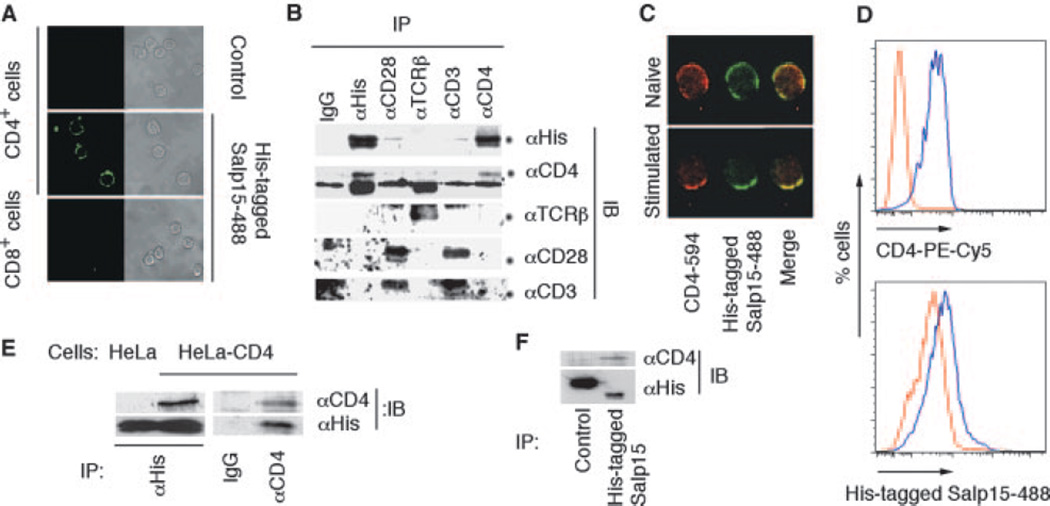
Salp15 binds to a protein component on the surface of CD4+ T cells (3). We first examined the specificity of Salp15 binding on T lymphocytes. Alexa-Fluor-labeled Salp15 (His-tagged Salp15-488) bound specifically to CD4+ T cells, whereas no staining of CD8+ cells was detected by confocal microscopy (Fig. 1A). Because Salp15 inhibits TCR ligation-induced T cell signaling, which involves a number of key receptors including CD4, CD3, and CD28, the interaction of Salp15 with these receptors was studied. His-tagged Salp15 coimmunoprecipitated CD4, but not other key components of the TCR complex, particularly CD3ε, CD28, and TCRβ (Fig. 1B). The simultaneous visualization of CD4 and His-tagged Salp15-488 binding on unstimulated and activated CD4+ T cells by confocal microscopy revealed the colocalization of both proteins (Fig. 1C).
FIGURE 1.
Salp15 binds to CD4. A, Left panels, Analysis of His-tagged Salp15-488 binding to CD4+ and CD8+ T cells by confocal microscopy compared with unstained CD4+ T cells (control). The panels on the right show the same field under brightfield microscopy. B, Murine primary CD4+T cells lysates containing His-tagged Salp15 were immunoprecipitated (IP) using anti-His, anti-CD3ε, anti-CD28, anti-CD4, anti-TCRβ, or IgG (control). The immunoprecipitate was immunoblotted (IB) using anti-His, anti-CD4, anti-CD3ε, anti-TCRβ, and anti-CD28 Abs. C, Colocalization of anti-CD4 staining and His-tagged Salp15-488 binding on naive and stimulated CD4+ T cells is indicated by the yellow color in the merged confocal micrograph. D, Flow cytometric analysis of CD4 expression using PE-Cy5-labeled anti-CD4 (CD4PE-Cy5) (upper panel) and His-tagged Salp15-488 binding (lower panel) in 3T3 (red) and 3T3-CD4 (blue) cells. E, HeLa and HeLa-CD4 cell lysates containing His-tagged Salp15 were immunoprecipitated using anti-His Ab. The immunoprecipitates were subjected to immunoblotting using anti-CD4 and anti-His Abs. The reciprocal immunoprecipitation from a HeLa-CD4 cell lysate was performed using anti-CD4 or IgG followed by immunoblotting with anti-CD4 or anti-His Abs. F, Immunoprecipitation from HeLa-CD4 cell lysate containing either His-tagged TR-Salp13 (control) or His-tagged Salp15 using an anti-His Ab followed by immunoblotting with anti-CD4 or anti-His Abs.
To confirm a direct association between CD4 and Salp15, nonlymphocyte cell lines expressing CD4 were used. Flow cytometry analysis showed an increase in His-tagged Salp15-488 binding to 3T3-CD4 cells compared with nontransfected controls (Fig. 1D). Coimmunoprecipitation experiments using a HeLa-CD4 cell lysate further supported the direct interaction of Salp15 with CD4 (Fig. 1E). Immunoprecipitation of a HeLa-CD4 cell lysate with His-tagged Salp15 or a control His-tagged-TR-fused tick saliva protein (His-tagged TR-Salp13) (14) using an anti-His Ab confirmed the specificity of the interaction (Fig. 1F).
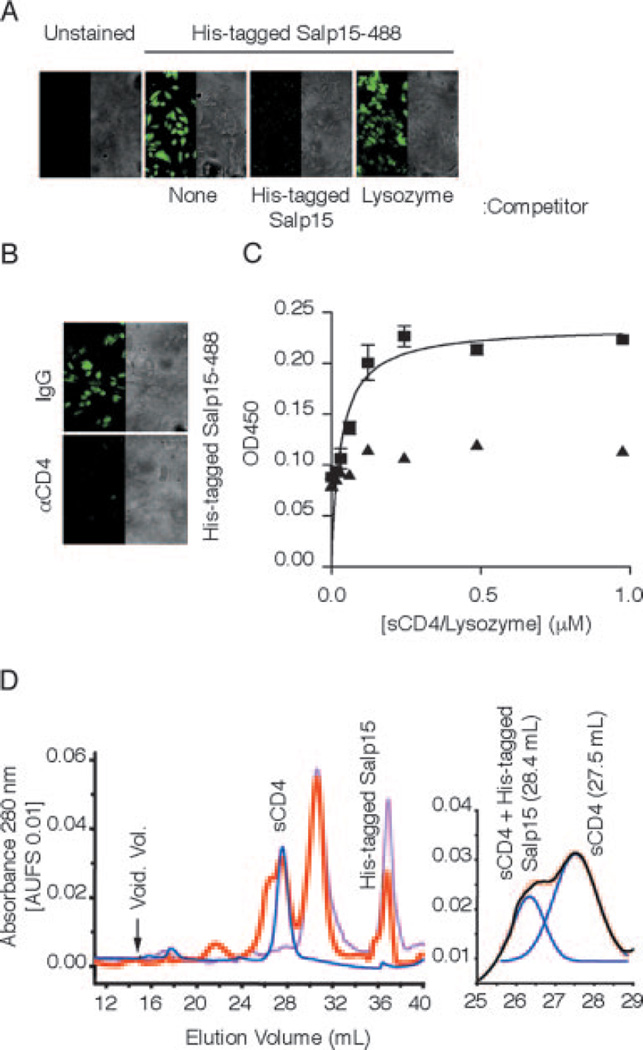
Binding of His-tagged Salp15-488 to HeLa-CD4 cells was competed by preincubating the cells with unlabeled His-tagged Salp15 or a polyclonal anti-CD4 Ab (Fig. 2, A and B), confirming the specific interaction between Salp15 and CD4. His-tagged Salp15 showed a saturable binding to sCD4 (D1–D2, containing the extracellular outer two domains) in a microtiter assay compared with a nonspecific control (lysozyme; Fig. 2C) with a Kd of 47 nM. His-tagged Salp15 also showed a saturable binding to a sCD4 preparation that contains all four extracellular domains (D1–D4) with similar kinetics (data not shown). To further verify the interaction between Salp15 and sCD4, the gel filtration profile of a His-tagged Salp15-CD4 solution was studied. We first studied the elution profiles of His-tagged Salp15 and sCD4 (D1–D4). His-tagged Salp15 preparations consisted primarily of monomer and dimer populations, whereas the gel filtration profile of sCD4 contained a single peak corresponding to monomeric sCD4 (Fig. 2D). An additional peak overlapping with the sCD4 peak was observed in a His-tagged Salp15-CD4 solution compared with the profiles of His-tagged Salp15 and sCD4 alone (Fig. 2D). The overlapping peaks were resolved by Gaussian fit analysis (15), which showed the elution volume of the complex to be 26.38 ml, compared with 27.53 ml for the sCD4 peak (Fig. 2D). The elution volumes of the His-tagged Salp15-sCD4 complex and standards were used to generate a linear fit in the natural log(molecular mass) vs elution volume plot, which estimated a molecular mass of 67 ± 8 kDa, suggesting that the molar ratio of Salp15-CD4 interaction is 1:1. The peak height of His-tagged Salp15-CD4 was directly related to monomer content of His-tagged Salp15 in different experiments.
FIGURE 2.
Salp15 binds to the extracellular domains of CD4. A, Unlabeled Salp15 but not lysozyme pretreatment blocks His-tagged Salp15-488 binding to HeLa-CD4 cells. Control represents HeLa-CD4 cells in the absence of His-tagged Salp15-488. Right panel, The brightfield of the image on the left. B, Preincubation of HeLa-CD4 cells with a polyclonal anti-CD4 Ab abolishes His-tagged Salp15-488 binding compared with control IgG pretreatment. C, His-tagged Salp15 (0.4 µM) was incubated with increasing amounts of immobilized sCD4 (D1D2, ■) or lysozyme (▲) in a microtiter assay showing saturable binding. The results are expressed as mean ± SE of three independent experiments. D, Elution profiles of sCD4 (D1–D4) (blue), His-tagged Salp15 (pink), and His-tagged Salp15-sCD4 (red) from Superdex-200 gel filtration columns (left panel). The Gaussian deconvolution of His-tagged Salp15-sCD4 and sCD4 peaks is shown on the right panel. The results shown are representative of three to five individual experiments performed.
To map the residues of Salp15 that interact with sCD4, we generated synthetic 20-aa-long peptides with 10-aa overlaps spanning the entire Salp15 sequence (3). Only the C-terminal peptide of Salp15 (P11) showed binding to sCD4 (Fig. 3A). The binding was saturable, with a Kd of 50 nM (Fig. 3B). We could compete P11 binding to sCD4 by adding increasing concentrations of the free peptide (Fig. 3C), suggesting a specific interaction. The binding of P11 to sCD4 could also be competed by adding increasing concentrations of His-tagged Salp15 (Fig. 3D). These data demonstrated that Salp15 binds to CD4 through its C-terminal residues.
FIGURE 3.
The C-terminal peptide of Salp15 binds CD4. A, Binding of His-tagged Salp15 (5 µg) and overlapping synthetic peptides of Salp15 (0.5 µg) to sCD4 (D1–D2). B, Increasing concentrations of P11 (■) but not P8 (▲) show saturable binding to sCD4 (D1–D2). C, Competition of increasing concentrations of free P11 with immobilized P11 (50 nmol) for binding to sCD4 (D1–D2). D, Competition of P11 (50 nmol) binding to sCD4 (D1–D2) by increasing concentrations of His-tagged Salp15. Control represents P11 binding in the absence of Salp15. The results are expressed as mean ± SE of at least three independent experiments.
CD4 binds to several physiological ligands that result in varied signals to the T cell. The coreceptor interacts with nonpolymorphic regions of MHC class II molecules (16) and also binds an array of distinct ligands, including the chemotactic cytokine IL-16 (17), human plasma glycoprotein gp17 (18), and the gross cystic fluid protein of 15 kDa (GCDFP-15) (19). In addition, human CD4 also acts as a receptor for the HIV-1 retrovirus envelope protein (20). Our data show that Salp15 also acts as a specific ligand for the CD4 coreceptor.
Salp15 inhibits early TCR signaling
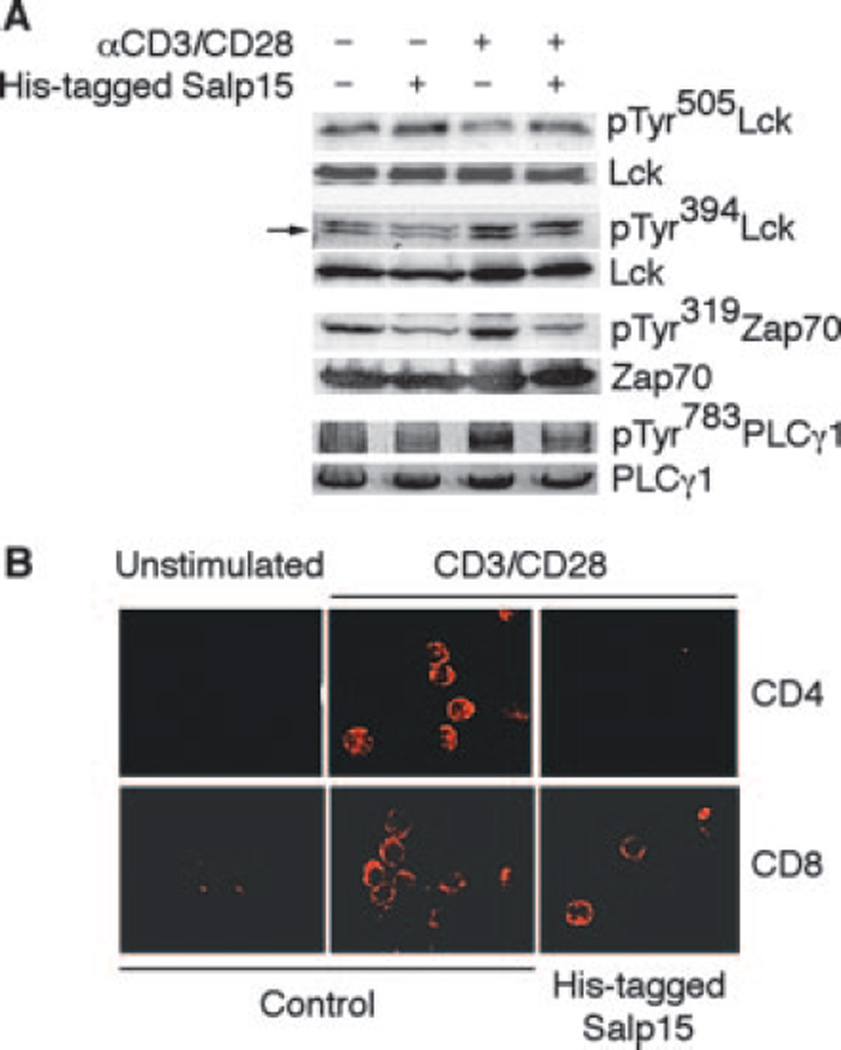
Salp15 inhibits the activation of CD4+ T cells stimulated with anti-CD3/CD28, as well as through MHC II-CD4-dependent signals (3), suggesting that its effect does not occur through interference with the interaction between MHC II and CD4, but rather with the interaction and proper alignment of CD4 and the associated Src family protein Lck with the TCR complex, which is critical for T cell activation. Lck is activated through a sequential series of events that involve its dephosphorylation at Tyr505, allowing its autophosphorylation at Tyr394 and its colocalization with downstream targets, including elements of the TCR complex and other intermediate signaling molecules, such as Zap70 (21, 22). We thus hypothesized that Salp15 would induce the disruption of early signaling events during CD4+ T cell activation. We examined tyrosine phosphorylation at residues 505 and 394 of Lck, upon TCR ligation in the presence or absence of Salp15. His-tagged Salp15 prevented the dephosphorylation of Lck at Tyr505, compared with control-activated cells, accompanied by a reduction in the phosphorylation levels of the protein at Tyr394 (Fig. 4A). Preincubation of CD4+ T cells with His-tagged Salp15 also prevented the phosphorylation of Zap70 (Fig. 4A), indicating that Salp15 hindered the activation of Lck. CD4+ T cell immunosuppression caused by Salp15 shared similarities with previously described interactions of CD4 with Abs specific for CD4 epitopes or HIV gp120 (3). However, Salp15 did not induce Lck phosphorylation in the absence of T cell stimulation, in contrast to the described effect of gp120 (23). Moreover, Salp15 does not affect AP-1 DNA-binding activity (3) or TCR-mediated ERK activation (data not shown) compared with the ERK-mediated inhibition of AP-1 DNA-binding activity observed in cells treated with gp120 (24), further distinguishing the effect of Salp15 to those of HIV gp120 and anti-CD4 cross-linking.
FIGURE 4.
Salp15 inhibits early steps during T cell activation. A, Western blots showing the increase in tyrosine phosphorylation of Lck at the residue 505, and decreased phosphorylation of Lck at Tyr394, Zap70 at Tyr319, and PLCγ1 at Tyr783 in anti-CD3/CD28-stimulated mouse CD4+ T cells in the presence or absence of His-tagged Salp15. B, Representative immunofluorescence micrographs showing CTB594 staining in CD4+ and CD8+ T cells stimulated with anti-CD3/CD28 in the presence or absence of His-tagged Salp15.
We also analyzed downstream events associated with CD4+ T cell activation that could be affected by the interaction of Salp15 with CD4. Stimulated CD4+ T lymphocytes that were pretreated with His-tagged Salp15 presented a reduction in the phosphorylation of PLCγ1 at Tyr783 (Fig. 4A). During TCR signaling, PLCγ1 hydrolyzes phosphatidyl-inositol (4,5)-bisphosphate to inositol (1,4,5)-trisphosphate and diacylglycerol, leading to calcium mobilization and protein kinase-C activation, respectively (25). The inhibition of PLCγ1 phosphorylation thus explains Salp15-mediated repression of calcium fluxes during T cell activation (3).
We next analyzed the stimulation-induced redistribution of lipid rafts on CD4+ T cells using Alexa-Fluor594-labeled cholera-toxin B (CTB)594. CTB594 staining of the stimulated CD4+ T cell membrane was confined in a dense cap structure, as described (26) in contrast to unstimulated T cells (Fig. 4B), indicating that rafts had aggregated during the activation process. Lipid raft clustering, however, was reduced by His-tagged Salp15 treatment before the stimulation with anti-CD3/CD28 (Fig. 4B), whereas lipid raft reorganization during CD8+ T cell activation in response to anti-CD3/CD28 signals was unaffected by His-tagged Salp15 pretreatment (Fig. 4B). These data indicate that Salp15 does not affect lipid raft reorganization per se, and confirms that the interaction of Salp15 with CD4 affects specific upstream pathways leading to lipid raft clustering and inhibiting the amplification of primary signaling events induced by TCR ligation.
In light of our results, we propose that upon contact with CD4, Salp15 blocks TCR-mediated Lck activation, leading to the improper activation of downstream signaling proteins. This results in a reduction in the clustering of signaling proteins in lipid rafts, which are essential for the amplification of T cell signals that result in IL-2 production. I. scapularis ticks modulate the host immune response in many different ways, resulting in efficient transmission of several pathogens (10). In this study, we report the first example of an interaction between an arthropod protein and a mammalian T cell coreceptor that clarifies the mechanism by which ticks withstand host immune responses.
Acknowledgments
HeLa-CD4, National Institutes of Health/3T3-CD4 cells, and purified sCD4 (D1D2) were obtained through the National Institutes of Health AIDS Research and Reference Reagent Program, Division of AIDS, National Institute of Allergy and Infectious Diseases, National Institutes of Health. We thank Nataliya Kubalik for technical support.
Footnotes
This work was supported by grants from the National Institutes of Health (to V.T., E.F., M.R., and J.A.) and American Health Association (to N.R. and C.M.Y.). E.F. is the recipient of a Burroughs Wellcome clinical scientist award in translational research. J.K.K. is the recipient of a University of North Carolina at Charlotte faculty research award.
Abbreviations used in this paper: PLC, phospholipase C; TR, thioredoxin; sCD4, soluble CD4; CTB, cholera-toxin B.
Disclosures
The authors have no financial conflict of interest.
References
- 1.Burgdorfer W, Barbour AG, Hayes SF, Benach JL, Grunwaldt E, Davis JP. Lyme disease-a tick-borne spirochetosis? Science. 1982;216:1317–1319. doi: 10.1126/science.7043737. [DOI] [PubMed] [Google Scholar]
- 2.Chen SM, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 1994;32:589–595. doi: 10.1128/jcm.32.3.589-595.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anguita J, Ramamoorthi N, Hovius JW, Das S, Thomas V, Persinski R, Conze D, Askenase PW, Rincon M, Kantor FS, Fikrig E. Salp15, an Ixodes scapularis salivary protein, inhibits CD4+ T cell activation. Immunity. 2002;16:849–859. doi: 10.1016/s1074-7613(02)00325-4. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira BR, Silva JS. Saliva of Rhipicephalus sanguineus tick impairs T cell proliferation and IFN-γ-induced macrophage microbicidal activity. Vet. Immunol. Immunopathol. 1998;64:279–293. doi: 10.1016/s0165-2427(98)00135-4. [DOI] [PubMed] [Google Scholar]
- 5.Kopecky J, Kuthejlova M. Suppressive effect of Ixodes ricinus salivary gland extract on mechanisms of natural immunity in vitro. Parasite Immunol. 1998;20:169–174. [PubMed] [Google Scholar]
- 6.Ribeiro JM, Schneider M, Guimaraes JA. Purification and characterization of prolixin S (nitrophorin 2), the salivary anticoagulant of the blood-sucking bug. Rhodnius prolixus Biochem. J. 1995;308(Pt. 1):243–249. doi: 10.1042/bj3080243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schoeler GB, Manweiler SA, Wikel SK. Ixodes scapularis: effects of repeated infestations with pathogen-free nymphs on macrophage and T lymphocyte cytokine responses of BALB/c and C3H/HeN mice. Exp. Parasitol. 1999;92:239–248. doi: 10.1006/expr.1999.4426. [DOI] [PubMed] [Google Scholar]
- 8.Urioste S, Hall LR, Telford SR, 3rd, Titus RG. Saliva of the Lyme disease vector, Ixodes dammini, blocks cell activation by a nonprostaglandin E2-dependent mechanism. J. Exp. Med. 1994;180:1077–1085. doi: 10.1084/jem.180.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wikel SK, Bergman D. Tick-host immunology: Significant advances and challenging opportunities. Parasitol. Today. 1997;13:383–389. doi: 10.1016/s0169-4758(97)01126-5. [DOI] [PubMed] [Google Scholar]
- 10.Wikel SK. Tick modulation of host immunity: an important factor in pathogen transmission. Int. J. Parasitol. 1999;29:851–859. doi: 10.1016/s0020-7519(99)00042-9. [DOI] [PubMed] [Google Scholar]
- 11.Ramamoorthi N, Narasimhan S, Pal U, Bao F, Yang XF, Fish D, Anguita J, Norgard MV, Kantor FS, Anderson JF, et al. The Lyme disease agent exploits a tick protein to infect the mammalian host. Nature. 2005;436:573–577. doi: 10.1038/nature03812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anguita J, Hedrick MN, Fikrig E. Adaptation of Borrelia burgdorferi in the tick and the mammalian host. FEMS Microbiol. Rev. 2003;27:493–504. doi: 10.1016/S0168-6445(03)00036-6. [DOI] [PubMed] [Google Scholar]
- 13.Alonso A, Bottini N, Bruckner S, Rahmouni S, Williams S, Schoenberger SP, Mustelin T. Lck dephosphorylation at Tyr-394 and inhibition of T cell antigen receptor signaling by Yersinia phosphatase YopH. J. Biol. Chem. 2004;279:4922–4928. doi: 10.1074/jbc.M308978200. [DOI] [PubMed] [Google Scholar]
- 14.Das S, Banerjee G, DePonte K, Marcantonio N, Kantor FS, Fikrig E. Salp25D, an Ixodes scapularis antioxidant, is 1 of 14 immunodominant antigens in engorged tick salivary glands. J. Infect. Dis. 2001;184:1056–1064. doi: 10.1086/323351. [DOI] [PubMed] [Google Scholar]
- 15.Lofvenberg L, Backman L. High-performance liquid chromatography analysis of spectrin oligomerization. Anal. Biochem. 2001;292:222–227. doi: 10.1006/abio.2001.5036. [DOI] [PubMed] [Google Scholar]
- 16.Konig R, Huang LY, Germain RN. MHC class II interaction with CD4 mediated by a region analogous to the MHC class I binding site for CD8. Nature. 1992;356:796–798. doi: 10.1038/356796a0. [DOI] [PubMed] [Google Scholar]
- 17.Cruikshank WW, Lim K, Theodore AC, Cook J, Fine G, Weller PF, Center DM. IL-16 inhibition of CD3-dependent lymphocyte activation and proliferation. J. Immunol. 1996;157:5240–5248. [PubMed] [Google Scholar]
- 18.Gaubin M, Autiero M, Basmaciogullari S, Metivier D, Misëhal Z, Culerrier R, Oudin A, Guardiola J, Piatier-Tonneau D. Potent inhibition of CD4/TCR-mediated T cell apoptosis by a CD4-binding glycoprotein secreted from breast tumor and seminal vesicle cells. J. Immunol. 1999;162:2631–2638. [PubMed] [Google Scholar]
- 19.Haagensen DE, Jr, Dilley WG, Mazoujian G, Wells SA., Jr Review of GCDFP-15. An apocrine marker protein. Ann. NY Acad. Sci. 1990;586:161–173. doi: 10.1111/j.1749-6632.1990.tb17804.x. [DOI] [PubMed] [Google Scholar]
- 20.Diamond DC, Sleckman BP, Gregory T, Lasky LA, Greenstein JL, Burakoff SJ. Inhibition of CD4+ T cell function by the HIV envelope protein, gp120. J. Immunol. 1988;141:3715–3717. [PubMed] [Google Scholar]
- 21.Huppa JB, Davis MM. T-cell-antigen recognition and the immunological synapse. Nat. Rev. Immunol. 2003;3:973–983. doi: 10.1038/nri1245. [DOI] [PubMed] [Google Scholar]
- 22.Li QJ, Dinner AR, Qi S, Irvine DJ, Huppa JB, Davis MM, Chakraborty AK. CD4 enhances T cell sensitivity to antigen by coordinating Lck accumulation at the immunological synapse. Nat. Immunol. 2004;5:791–799. doi: 10.1038/ni1095. [DOI] [PubMed] [Google Scholar]
- 23.Briand G, Barbeau B, Tremblay M. Binding of HIV-1 to its receptor induces tyrosine phosphorylation of several CD4-associated proteins, including the phosphatidylinositol 3-kinase. Virology. 1997;228:171–179. doi: 10.1006/viro.1996.8399. [DOI] [PubMed] [Google Scholar]
- 24.Jabado N, Pallier A, Jauliac S, Fischer A, Hivroz C. gp160 of HIV or anti-CD4 monoclonal antibody ligation of CD4 induces inhibition of JNK and ERK-2 activities in human peripheral CD4+ T lymphocytes. Eur. J. Immunol. 1997;27:397–404. doi: 10.1002/eji.1830270209. [DOI] [PubMed] [Google Scholar]
- 25.Rhee SG, Bae YS. Regulation of phosphoinositide-specific phospholipase C isozymes. J. Biol. Chem. 1997;272:15045–15048. doi: 10.1074/jbc.272.24.15045. [DOI] [PubMed] [Google Scholar]
- 26.Janes PW, Ley SC, Magee AI, Kabouridis PS. The role of lipid rafts in T cell antigen receptor (TCR) signalling. Semin. Immunol. 2000;12:23–34. doi: 10.1006/smim.2000.0204. [DOI] [PubMed] [Google Scholar]