Abstract
Polymorphisms at Toll-like receptor 4 (TLR4) gene have been found to be associated with immune disorders. A murine macrophage cell line GG2EE derived from C3H/HeJ mice with a polymorphism site at TLR4 is hyposensitive to lipopolysacchride (LPS). To study the molecular base of diverse TLR4-mediated immune responses, the proteomic changes in both TLR4-deficient and –wild-type cell lines in response to the same LPS challenge were quantitatively compared by using multiplex amino acid coded mass-tagging (AACT)/SILAC-assisted mass spectrometry (MS). This strategy allows encoding of two distinct cell populations with different stable isotope-tagged lysine residues as the ‘in-spectra’ quantitative markers. In MS analysis of tryptic peptides derived from the equally mixed three cell populations, the lysine-containing peptides originated from two LPS stimulated cell populations can be clearly distinguished by their different mass shifts from the un-stimulated and unlabeled counterpart. The LPS-induced differential protein expression in TLR4 –deficient and –wild-type proteomes were obtained by comparing the intensities of isotopically encoded peptides. Among the more than 900 proteins identified, 35 were found to be deregulated at different levels in these two cell lines stimulated by LPS. This multiplex mass-tagging methodology can be readily extended to other comparative proteomic quantitation of different cell populations.
1 Introduction
Toll Like Receptors (TLRs) are critical sentinel surface receptors expressed on immune cells that recognize molecular motifs derived from pathogens including virus, bacteria, fungi, and protozoan.[1] Upon stimulation, TLRs typically elicit a signaling cascade including MyD88-IRAK-TRAF6-I-κB-NF-κB that ultimately leads to the production of proinflammatory cytokines and costimulatory molecules. Results from genetic knockout studies have firmly established the role of TLRs in initiating the innate responses toward various pathogens.[2]
TLR4 is the most studied receptor among the 13 reported TLRs (11 in human and 13 in mice). TLR4 is mainly expressed on antigen presenting cells, such as macrophages and dendritic cells. The natural ligand of TLR4 is the lipopolysacchride (LPS) derived from the cell wall of Gram-negative bacteria. Endogenous ligand, HSP70, has also been reported to activate TLR4 pathway.[3] Upon activation, TLR4 is able to utilize two different adaptor proteins, MyD88 and TRIF, to elicit immune responses that characterized as the production of proinflammatory cytokines or the induction of type I interferons.[4] In clinic cases, a severe bacterial infection may release a large dose of LPS, which activates TLR4 pathway to produce high amount of proinflammatory cytokines, such as TNFα. TNFα has been held responsible for the septic shock, an often-fatal consequence of widespread bacterial infection and kills about 20,000 people in USA and one million people worldwide each year. Meanwhile, polymorphisms of TLR4 gene in population have also been reported to be associated with disease pathogenesis.[5] Therefore, many studies target at characterizing the TLR4-mediated downstream protein expression corresponding to particular inflammatory responses for revealing critical factors in modulating the signaling, and then developing possible strategies for therapeutic intervention.
Despite the tremendous effort on the study of TLR4-mediated inflammatory responses using both molecular and cellular biology approaches, there is a lack of systems data describing the altered pattern of protein expression associated with the genetic variations in TLR molecules. Qian et al quantitatively analyzed the proteome of human plasma following in vivo LPS administration.[6] However, since it is reported that not all of the responses exerted by LPS is TLR4-dependent, it will be of significant interest to investigate the TLR4-dependent proteomic changes associated with clinically relevant TLR4 polymorphisms using more effective quantitative proteomic approaches.
To determine the proteome variations in different proteome states, a number of different isotope labeling techniques have been developed for improving the sensitivity, accuracy, and throughput of mass spectrometry (MS)-based comparative proteomics[7, 8], including isotope-coded affinity tagging (ICAT), 18O-labeling, and uniform or amino acid-specific metabolic labeling [9–13]. The basic scheme of these approaches is to utilize stable isotopes either enriched in chemical reagents or at particular amino acids as the ‘in-spectra’ quantitative markers to assist MS to analyze protein expression changes on large scale. Recently, an in vitro labeling technique named as iTRAQ, which tags the primary amine group in peptide digests, has recently been developed for quantitative analysis of proteomic changes in comparing up to 4 different cell populations.[14, 15] However, this peptide-based labeling strategy requests the preparation of individual samples involving protein separation and proteolysis, accurate equal mixing of proteolytic digests, etc. In addition, unsolved/co-eluting peptide signals with different sequences can lead to overlapping signals in low mass reporter region that the quantitative measurements are based on. Metabolic labeling strategy such as amino acid coded mass-tagging (AACT)[12] or SILAC naturally introduces the tags for quantitative measurements in MS spectra through cell culture. The protein extracts from different cell pools can be processed in a single experiment format by equal mixing of cell numbers or protein concentration, thus the procedure minimizes the experimental variations, contaminations, artifacts, etc originated from separate sample processing[16–18]. Meanwhile, by employing different stable isotopes or stable isotope-enriched amino acids, SILAC-based metabolic labeling can also quantify protein expression changes in 3 different cell populations.[19, 20] Using the multiplex technique, quantitative profiling of the proteome involved in S. Solfataricus,[19] and temporal quantitation of tyrosine phosphorylation after EGF stimulation in Hela cells [20] were accomplished.
Here we extended the multiplex AACT/SILAC-based quantitative method to simultaneously measure the relative proteomic changes in both TLR4-deficeint and –wild-type strains in response to same level LPS exposure. The LPS-induced differential changes in the proteome of TLR4-deficeint versus –wild-type macrophages were measured by comparing the MS intensities of the isotopically encoded peptides originated from the corresponding strains respectively, thus those proteins that showed the LPS-induced differential expression changes in both strains were identified. It is the first systems investigation of the proteomic immune responses in TLR4 –deficient and –wild-type macrophages.
2 Materials and Methods
2.1 Amino acid coded mass-tagging of macrophage cells
The C3H/HeN-derived macrophage cell line HeNC2 and the C3H/HeJ-derived macrophage cell line GG2EE were kindly provided by Dr. Steven B. Mizel (Wake Forest University, NC). The stable isotope-enriched amino acids were purchased from Cambridge Isotope Laboratories (Andover, MA). The murine macrophage GG2EE (TLR4-deficient) cell lines were grown in lysine-depleted D-MEM media supplemented with regular lysine and lysine-d4 respectively. The HeNC2 (TLR4-wild type) cell line was grown in lysine-depleted D-MEM media supplemented with lysine-13C615N2. After several cell doubling, the cells were harvested to examine the labeling efficiency by MS analysis as previously described [12].
2.2 Stimulation of macrophages
LPS (Sigma, St. Louis, MO) was added into the media with a final concentration of 500 ng/ml to stimulate the corresponding macrophage cell lines. Following 20 hrs stimulation, the cell were harvested and washed with PBS buffer twice. To examine the LPS-inducing time course-dependent expression of particular proteins of interest by Western blotting experiments, the LPS-stimulated macrophage cells for different period of time were collected and harvested at different time points.
2.3 Protein extraction, separation, and LC-MS analysis
The protocol for cell lysis and protein extraction was the similar one as previously described [12]. Briefly, similar amounts of 10 million cells from each of the stimulated and un-stimulated cells were suspended at 4×107 cells/ml in a lysis buffer (10 mM Hepes with pH at 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.34 M sucrose, 10% glycerol, and protease inhibitors). Following cell lysis and protein extraction, the concentration of the soluble proteins extracted from the control GG2EE cells, the lysine-d4 labeled LPS-treated GG2EE cells, and the lysine-13C615N2 labeled LPS-treated HeNC2 cells, were measured by Bio-Rad DCRC protein assay kit following the manufacture instruction. Then, the protein mixtures from each cell population were mixed at 1:1:1, loaded onto, and separated by a 14 × 14 cm 12% SDS PAGE. The separated proteins on a gel were cut into 62 bands and digested with trypsin (Promega, Madison, WI). The gel slices were destained with 50% acetonitrile and 50% 50 mM NH4HCO3 mixture. The destained gel slices were then dehydrated using acetonitrile and followed by speed-vac for 20 min. Trypsin was added in a final concentration of 25 ng/µL, and the mixture was incubated overnight 37 °C. The tryptic peptides were extracted from gel slices in two steps. First, the gel slices were suspended in 5% acetic acid solution and sonicated at 37 °C for 45 min. The supernatant was reserved in a fresh microcentrifuge tube on ice. The gel slices then continued to be extracted with 50% acetonitrle/5% acetic acid for an additional 45 min. Both supernatants were combined and dried by speed-vac. The samples were then re-suspended in a 0.1% TFA solution. The digested peptide mixtures were submitted to a nano-LC-MS/MS system (LC Packing Ultima, Dionex, Sunnyvale, CA) coupled with QSTAR XL mass spectrometer (AppliedBiosystems, Foster City, CA). 5 µl of samples was loaded into the LC column for separation. Mobile phase A is 0.1% formic acid and 5% acetonitrile. Mobile phase B is 0.1% formic acid and 95% acetonitrile. The gradient was kept at 5% B for 5 min, then ramped linearly from 5 to 50% B in 50 min, then jumped to 75% B and kept for 10 min. Then the gradient was set back to the start point and the column equilibrated for 10 min. The flow rate was 180 nL/min. The spray voltage was tuned to get stable background signals with the best signal-to-noise intensity. The two most intense ions with charge states between 2 to 4 in each survey scan were selected for the MS/MS experiment provided they passed the intensity threshold. The rolling collision energy feature was employed to fragment the peptide ions according to their charge states and m/z values.
2.4 Database searching and protein identification
The in-house licensed MASCOT (Version 2.0) server were used to interpret the LC-MS/MS data by searching against the Mus musculus protein sequence databases (2004.9.1) downloaded from the National Center for Biotechnology Information (NCBI) public ftp site. The lysine-d4 and lysine-13C615N2 modifications were added in the configuration file as static or variable modifications to constraint database searching. The parameters for database searching were setting as: (i) 0.2 Da mass error tolerance for both MS and MS/MS; (ii) variable modifications including phosphorylations on tyrosine/serine/threonine, oxidation of methionine and lysine-d4 and lysine-13C615N2; (iii) tryptic enzyme specificity and maximum 2 mis-cleavages; (iv) peaks with intensities less than 0.5% of the base peak in MS/MS were ignored for database searches; (v) no smoothing of spectra was applied. Proteins with 2 or more peptides with the matching score over 46 (p<0.05) were considered as positive identification. Only top rank peptide hits for given precursors were used for further protein identifications. Protein with 1 peptide match were manually validated by the following empirical rules: (i) both b- and y-series ions present in the spectrum; (ii) at least 4 adjacent fragment ions are observed in either b or y-series ions; (iii) major peaks in a spectrum must be explained by b- or y-series ions, or neutral loss from those ions, or a- ions; (iv) the mass error for matched fragment ions should be in an increasing or decreasing trend, but not a fluctuant trend. Further, a decoyed protein sequence database (reversed sequence of the mouse protein sequence database) was used to revalidate the empirical rules in the protein identification based on a single peptide. The decoyed database search gave few hits that could pass the default threshold at MASCOT as the identified peptides. The false-positive rate indicated from the decoyed database search was less than 5% by the search using MASCOT only. Combined with the manual verification for all hits of the identifications based on single peptide sequence, the false-positive rate was then reduced to less than 1%.
2.5 Protein quantitation
The spectra of those peptide precursors containing the labeled amino acids were further analyzed for protein quantification. This process also validates the database search results as the quantitative measurements were obtained. We first averaged all the TOF MS spectra of the unlabled, lysine-d4 labeled, and lysine-13C615N2 labeled forms of the peptides over their chromatographic elution profile. The quantitative results obtained by comparing their mono-isotopic peak intensities. Further, the background signals, including solvent cluster ions, chemical noise, can be subtracted in the Analyst QS, thus give rise of more reliable quantification results. To subtract background signals, two 10 sec-wide windows were selected before and after the chromatographic peak of the peptide of interest as the background reference, and the background was subtracted by Analyst QS. MSQuant (http://msquant.sourceforge.net) developed by Mann’s group at University of Southern Denmark were used to validate the manual quantification results. Whereas, MSQuant can only read the apparent isotopic intensity of a peptide in a single scan without considering the background signals, and sum the intensity across the chromatographic peak profile. Taking the quantitative measurement of carbonic anhydrase as an example, MSQuant reported a ratio of 3.0 of lysine-d4 labeled form with a standard deviation of 1.26, a ratio of 46.8 of lysine-13C615N2 labeled form with a standard deviation of 17.12. The large standard deviation was probably caused by poor signal-to-noise level. Although automatic quantification program sometime can not provide accurate information and need manual intervention, it still improved the throughput.
2.6 Immunoblotting analysis
3 million cells per well were seeded in a six-well plate the day before stimulation. Cells were lyzed in a lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40) supplemented with protease inhibitor cocktail (Sigma, St. Louis, MO). Soluble extracts were loaded onto 4–20% SDS-PAGE for separation. The proteins were transferred to a nitrocellulose membrane and incubated with antibodies against loading control--beta-actin (Abcam, Cambridge, MA), STAT1 (Stratagene, La Jolla, CA), and IL-1β (Abcam). The concentration and procedures of different antibodies were optimized according to the manufacture manual.
3 Results
3.1 Overall experimental design
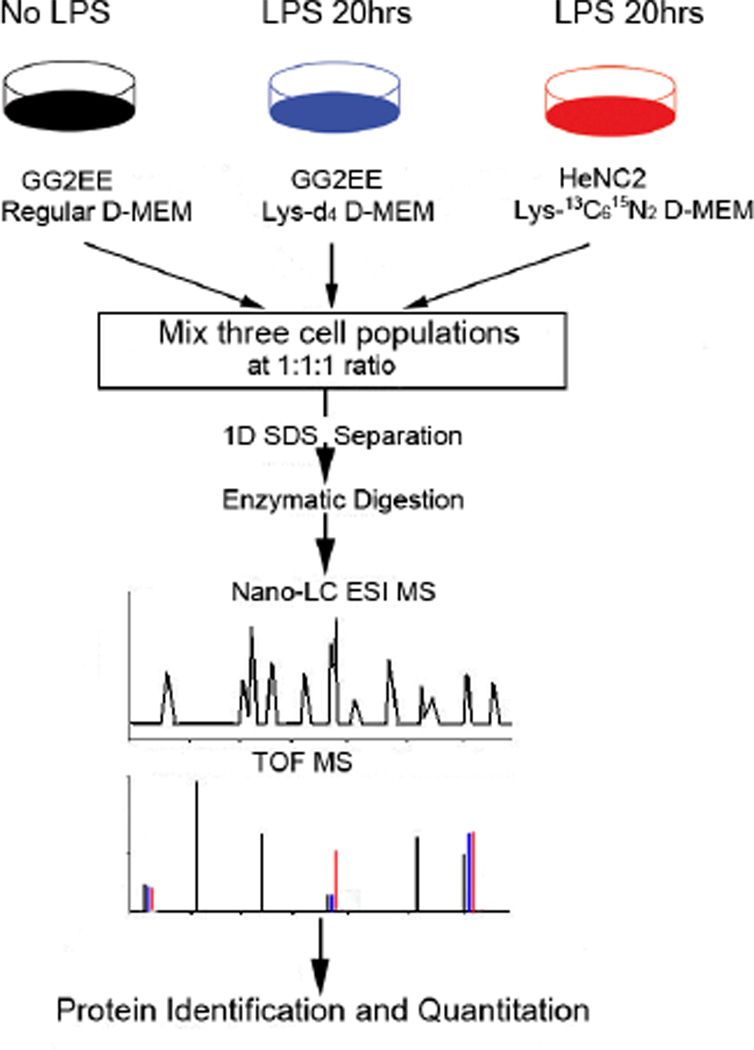
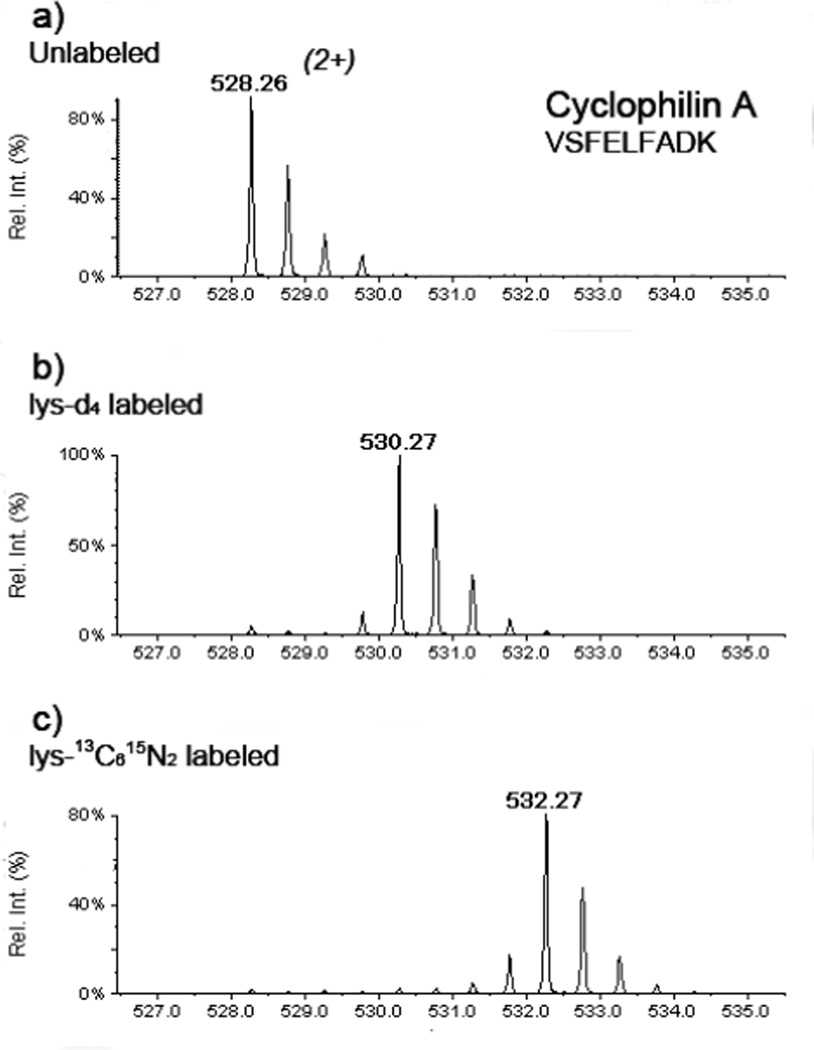
The workflow of the multiplex quantitative proteomic approach using multiple isotope-enriched amino acids for metabolic labeling is shown in Figure 1. The GG2EE cells were grown in a regular medium and the lysine-d4 supplemented medium respectively. The HeNC2 cells were grown in the lysine-13C615N2 supplemented medium. After 5–6 doubling (approximate 160–180 hrs), the labeling efficiency in cell culture using lysine-d4and lysine-13C615N2 were examined by MS analysis of the lysine-containing peptides. Briefly, small amount of protein extracts from unlabeled GG2EE, lysine-d4labeled CG2EE, and lysine-13C615N2 labeled HeNC2 cells respectively were separated on a mini SDS PAGE gel. Three randomly selected bands were excised from each lane for in-gel digestion. The extracted peptides were submitted to LC-MS/MS analysis. After been cultured in the “heavy” lysine-supplemented media for 5–6 doubling, more than 95% of individual proteins were successfully labeled. As shown in Figure 2, the lysine-d4and lysine-13C615N2 labeled peaks are predominant peaks in the mass spectra. The intensity of the unlabeled peptide is less then 5% of the labeled peptide. After achieving the desirable labeling efficiency, both lysine-d4- labeled CG2EE and lysine-13C615N2-labeled HeNC2 cells were stimulated by same dose LPS. All of the three cell populations were harvested 20 hrs after LPS stimulation. 80 µg soluble protein extracts from each of the three cell populations were mixed together. Totally, 240 µg protein were loaded and separated on a 14 × 14 cm SDS gel. The collected MS/MS data were searched against the NCBI mouse protein sequence database by MASCOT search engine. All identified proteins were quantified manually. Selected targets of differentially expressed proteins then were chosen for validation by Western blotting.
Figure 1.
Overall Experimental Design. Cells are grown in D-MEM medium supplemented with either regular lysine, lysine-d4, and lysine-13C615N2 for 160hrs to allow complete mass-tagging of the corresponding proteome. The lysine-d4 and lysine-13C615N2 labeled macrophages were stimulated respectively by LPS for 20 hrs. Soluble protein from the three different cell populations are mixed equally and separated on 1D-SDS PAGE, followed by in-gel tryptic digestion and LC-MS analysis.
Figure 2.
High specificity and efficiency of multiplex amino acid coded mass-tagging. The mass spectra of cyclophilin A peptide from the cells grown in (a) regular, (b) lysine-d4 supplemented, and (c) lysine-13C615N2 supplemented media showed the labeling specificity is more than 95%.
3.2 Protein Identification
In this work, the proteins with 2 peptide matches with better than 95% probability were considered as validated identifications. Proteins with single peptide match at > 95% probability were validated by visual inspection of the MS/MS spectra. As a result, more than 900 proteins were identified by the 1D SDS-PAGE LC-MS/MS platform which has been demonstrated as a generic and powerful tool in proteomic studies [20, 21]. Searching the collected MS/MS data with the decoyed protein sequence database showed the empirical rules we used in single peptide-based protein identification is highly strict. Although there are few single-peptide matches passed the threshold (p<0.05) determined by MASCOT when searching against the decoyed database, none of these matches passed the empirical rules described in the Materials and Methods section. Therefore, manual inspection of those single peptide matches with stringent criteria largely removed the false-positive identifications in the database search.
3.3 Protein quantification
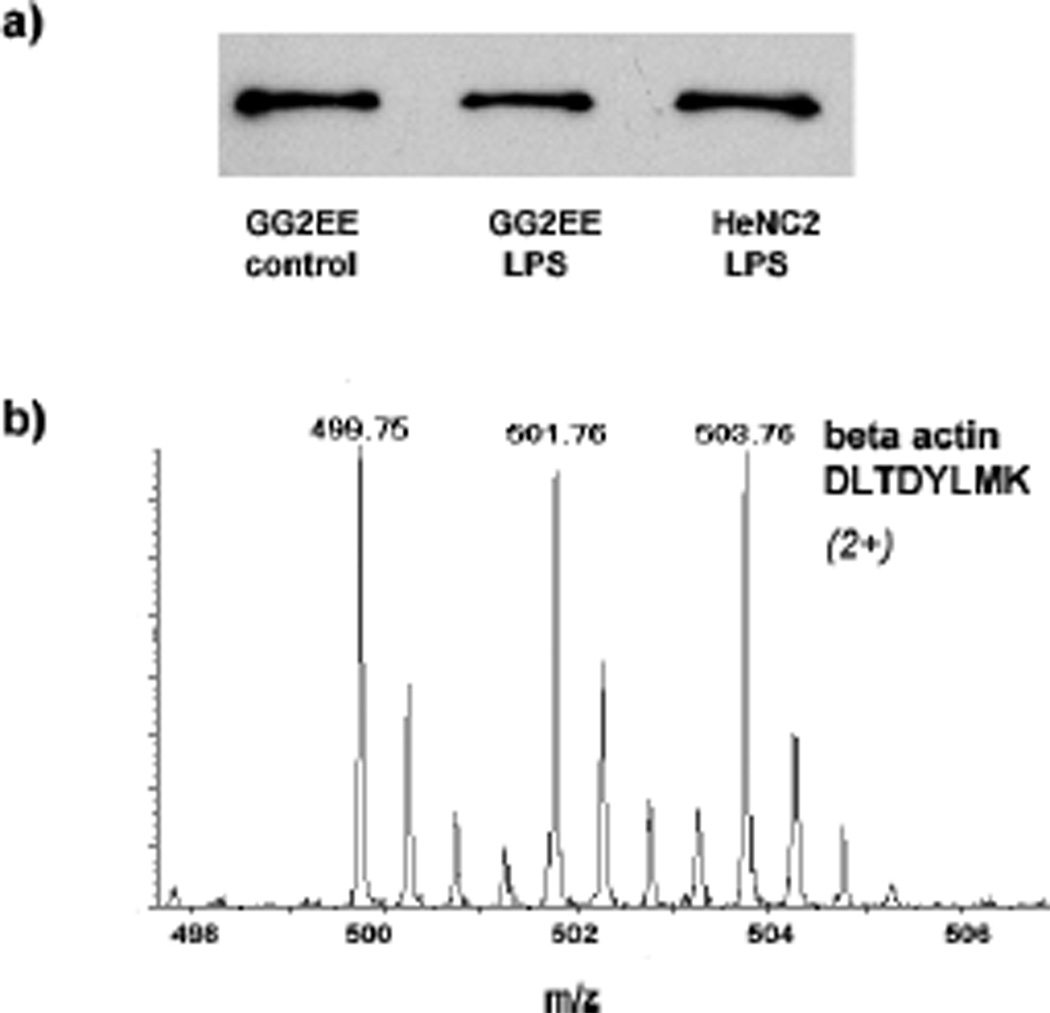
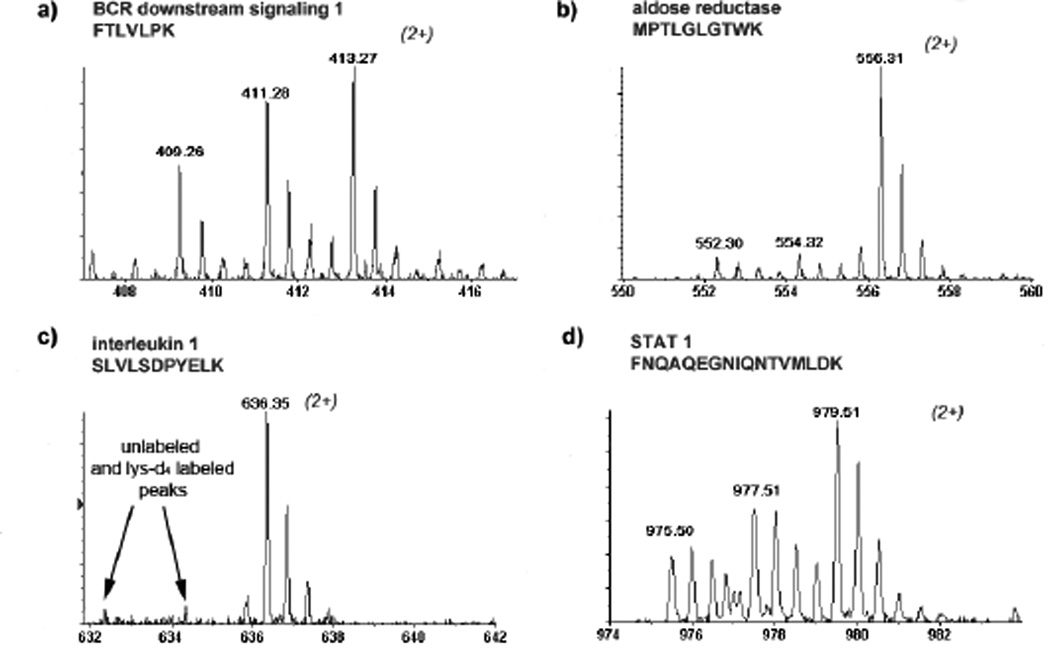
Further, more than 500 proteins that contain specific AACT signatures, i.e., light or heavy lysine residues, were quantified. The expression levels of actin in these three different cell populations were estimated by a western blot (Figure 3a), which shows the expression of actin is not affected by LPS stimulation. Meanwhile, Figure 3b shows that the intensities of the isotope-encoded signals of actin peptide were the same derived from the equally mixed cell populations. On the other hand, interleukin 1β (IL-1β) showed a 25-fold increase in its expression in LPS-stimulated HeNC2 cells (Table 1 and Figure 4a). It is known that IL-1β is a marker of macrophage immune response under LPS stimulation and was identified in the protein band around 35 kDa. The intracellular form of IL-1β has been reported to migrate around 33 kDa, whereas the molecular mass of matured IL-1β is 17 kDa.[22] All of these results validated the accuracy of our experimental proteomics design. Twenty proteins were found largely up-regulated in the LPS-stimulated HeNC2 cells, whereas 5 of them were also significantly up-regulated in the LPS-stimulated GG2EE cells. Importantly, for those up-regulated proteins found in both strains, the differences in their expression in response to the LPS challenge were accurately measured simultaneously. The list of up-regulated proteins and the quantitative measurement of the level of LPS-induced expression changes are given in Table 1. In our results, several proteins known to be involved in LPS-induced immune response were found up-regulated. For example, BCR downstream signaling protein 1 was found up-regulated in both LPS-stimulated GG2EE and HeNC2 cells. Some proteins, such as aldo-keto reductase, RAS-related C3 botulinum substrate 2, heat shock protein 105, and interleukin 1 receptor antagonist, were specifically up-regulated in LPS-stimulated HeNC2 cells. Interestingly, the multi-functional carbonic anhydrase was also found significantly up-regulated, i.e., more than 3 folds in LPS-stimulated GG2EE and more than 50 folds in LPS-stimulated HeNC2 cells.
Figure 3.
Expression of actin is not affected by LPS treatment. (a) Mass spectrum of actin peptide –DLTDYLMK from equally mixed sample. (b) Western blot of actin in three different cell populations before the mix.
Table 1.
LPS-induced differentially regulated proteins in the macrophage pair.
| Accession Number | Protein Description | Molecular Function / Biological Processa |
Subcellular Location b |
Number of Peptides for Quantification |
GG2EE (TLR4 deficient) +LPS |
HeNC2 (TLR4 functional) +LPS |
||
|---|---|---|---|---|---|---|---|---|
| lysine-d4 unlabeled |
S.D. c | lysine- 13C615N2 : unlabeled |
S.D. c | |||||
| gi|31981657 | carbonic anhydrase 2 | reversible hydration of carbon dioxide; zinc ion binding |
cytoplasmic | 5 | 3.18 | 0.65 | 54.73 | 8.01 |
| gi|6680415 | interleukin 1 beta | cytokine activity; cell proliferation; Inflammatory response |
extracellular | 2 | 1.01 | 0.11 | 24.58 | 2.66 |
| gi|31981909 | aldo-keto reductase family 1, member B3 |
aldehyde reductase activity; Oxidoreductase activity |
cytoplasmic | 2 | 1.01 | 0.10 | 7.84 | 0.63 |
| gi|6754450 | fatty acid binding protein 5 |
glucose/lipid metabolism; phosphatidylcholine biosynthesis |
cytoplasmic mitochondrial |
2 | 1.18 | 0.03 | 2.05 | 0.12 |
| gi|21644585 | 2'-5' oligoadenylate synthetase 3 |
nucleic acid binding | cytoplasmic nuclear |
1 | 1.45 | / | 2.00 | / |
| gi|6671666 | CAP, adenylate cyclase-associated protein 1 |
cellular morphogenesis; receptor mediated endocytosis; Actin cytoskeleton organization and biogenesis |
membrane | 4 | 1.19 | 0.09 | 1.68 | 0.16 |
| gi|6680457 | Inositol polyphosphate-1- phosphatase |
Phosphatidylinositol signaling; magnesium ion binding |
cytoplasmic nuclear |
2 | 1.08 | 0.02 | 1.86 | 0.12 |
| gi|6679601 | RAS-related C3 botulinum substrate 2 |
small GTPase mediated signal transduction |
cytoplasmic; membrane- associated when activated |
2 | 0.92 | 0.17 | 1.73 | 0.15 |
| gi|31543778 | signal transducer and activator of transcription 1 |
intracellular signaling cascade; Regulation of transcription, DNA- dependent |
cytoplasmic; translocated into the nucleus in response to phosphorylatio n |
2 | 1.68 | 0.08 | 2.54 | 0.06 |
| gi|7305169 | heat shock protein 105 | chaperone cofactor dependent protein folding; ATP binding |
cytoplasmic | 2 | 1.07 | 0.11 | 1.70 | 0.08 |
| gi|9910568 | BCR downstream signaling 1 |
protein tyrosine kinase signaling pathway; intracellular signaling cascade; myeloid cell differentiation |
mitochondrial cytoplasmic |
1 | 1.54 | / | 1.76 | / |
| gi|38090997 | methionine-tRNA synthetase |
protein biosynthesis; TRNA aminoacylation for protein translation |
mitochondrial | 2 | 1.71 | 0.17 | 1.48 | 0.11 |
| gi|47894398 | tropomyosin 4 | actin binding; muscle development |
mitochondrial nucelar |
3 | 0.92 | 0.09 | 1.63 | 0.11 |
| gi|12963663 | magnesium-dependent phosphatase-1 |
/ | cytoplasmic | 1 | 0.73 | / | 1.36 | / |
| gi|40254244 | loss of heterozygosity 11 |
DNA repair; pathogenesis; cell cycle |
nuclear | 1 | 0.82 | / | 1.75 | / |
| gi|13624317 | interleukin 1 receptor antagonist |
immune response; cell surface receptor linked signal transduction; lipid metabolism |
extracellular integral to plasma membrane |
1 | 0.49 | / | 1.48 | / |
| gi|34328230 | adenylate kinase 2 | nucleobase, nucleoside, nucleotide and nucleic acid metabolism |
Mitochondrial Intermembrane space |
0.89 | 0.05 | 1.48 | 0.06 | |
| gi|7949047 | hydroxyacyl- Coenzyme A dehydrogenase type II |
oxidoreductase activity | mitochondrial cytoplasmic |
1 | 0.95 | / | 1.34 | / |
| gi|6678483 | ubiquitin-activating enzyme E1 |
ubiquitin-dependent protein catabolism |
cytoplasmic vesicles of secretory system |
1 | 1.02 | / | 1.33 | / |
| gi|6680187 | hematopoietic cell specific Lyn substrate |
substrate of the antigen receptor-coupled tyrosine kinase |
mitochondrial | 2 | 0.71 | 0.05 | 1.39 | 0.01 |
| gi|6671509 | actin beta | cytoplasmic | 4 | 0.99 | 0.05 | 1.00 | 0.09 | |
| gi|6671549 | peroxiredoxin 6 | redox regulation of the cell; response to reactive oxygen species |
cytoplasmic | 7 | 1.02 | 0.05 | 0.73 | 0.06 |
| gi|30519911 | transgelin 2 | muscle development | nuclear | 3 | 0.97 | 0.17 | 0.54 | 0.08 |
| gi|6755114 | peroxiredoxin 5 | antioxidant protein with alkyl hydroperoxidase activity |
cytoplasmic | 7 | 0.52 | 0.03 | 0.51 | 0.11 |
| gi|7106301 | microtubule- associated protein |
mitosis; cell cycle | microtubule Golgi apparatus |
2 | 1.04 | 0.10 | 0.50 | 0.02 |
| gi|24762230 | ribosomal protein S15a | protein translation | cytoplasmic | 2 | 0.93 | 0.07 | 0.49 | 0.10 |
| gi|6755682 | serine/threonine kinase receptor associated protein (Strap) |
negative regulation of transcription from RNA polymerase II promoter; negative regulation of transforming growth factor beta receptor signaling pathway |
cytoplasmic mitochondrial |
3 | 1.00 | 0.04 | 0.48 | 0.04 |
| gi|6680047 | guanine nucleotide binding protein beta 2 |
protein kinase C binding; protein localization; intracellular signaling cascade |
cytoplasmic | 2 | 1.00 | 0.08 | 0.46 | 0.06 |
| gi|42476345 | ribosomal protein large P2 |
tanslational elongation | extracellular | 3 | 1.04 | 0.07 | 0.44 | 0.05 |
| gi|31981458 | glutaredoxin 1 | cell redox homeostasis; electron transport |
cytoplasmic | 0.89 | 0.05 | 0.40 | 0.06 | |
| gi|51829008 | protein phosphatase 2A inhibitor-2 |
DNA replication; Cellular physiological process |
nuclear | 2 | 1.05 | 0.06 | 0.36 | 0.01 |
| gi|6754570 | annexin A1 | calcium ion binding; regulation of cell proliferation |
cytoplasmic | 4 | 0.31 | 0.15 | 0.33 | 0.08 |
| gi|9789995 | stathmin 1 | mitotic spindle organization and biogenesis; microtubule depolymerization; intracellular signaling cascade |
cytoplasmic | 3 | 0.95 | 0.03 | 0.22 | 0.04 |
| gi|6753060 | annexin A5 | calcium ion binding; calcium-dependent phospholipid binding |
cytoplasmic | 2 | 0.31 | 0.14 | 0.21 | 0.08 |
| gi|25023967 | endothelial monocyte- activating polypeptide |
calcium ion binding; cytokine activity |
nuclear | 2 | 0.50 | 0.01 | 0.21 | 0.05 |
Functional annotation was extracted from Swissprot database;
Subcellular location was obtained from swissprot database if such information is available. PSORT II (http://psort.nibb.ac.jp) was used to predict the subcellular location if no information exists in Swissprot.
Standard Deviation was calculated from the quantification ratio of several peptides. “/” indicates the quantification ratio is obtained from one lysine containing peptide.
Figure 4.
Mass spectra of several peptides derived from (a) BCR downstream signaling, (b) aldose reductase, (c) interleukin 1β, and (d) STAT 1 were shown to represent the signal class from those up-regulated proteins. The mass spectra of the peptide derived from those up-regulated proteins show higher isotopic intensity of the lysine-d4 or lysine-13C615N2 labeled peptides.
3.4 Immunoblotting analysis
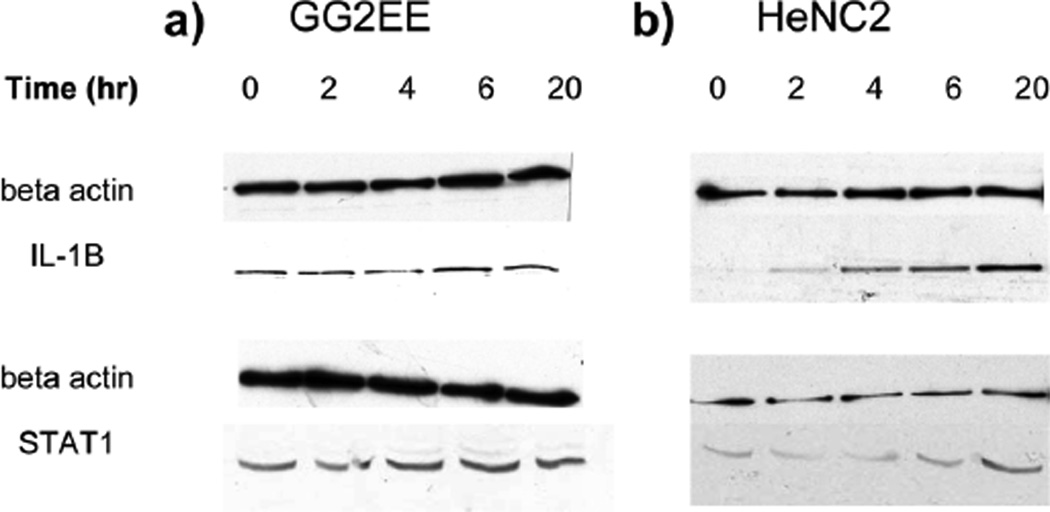
IL-1β and signal transducer and activator of transcription 1 (STAT1) were chosen for validating our quantitative proteomic profile of the LPS-induced differentially expressed proteins by Western blotting. As shown in Figure 5, the expression level of actin in both cell lines was not affected by LPS stimulation up to 20 hrs, and it was used as loading control for western blot. The basal level of IL-1β in GG2EE cells was not affected by LPS stimulation, whereas, the expression IL-1β in HeNC2 cells was obviously increased along with the LPS stimulating time. We also observed the differentially elevated STAT1 expression in both LPS stimulated cell lines as it is more obvious in HeNC2 cells as shown in Figure 5b. Our western blot results were consistent with the quantitative proteomic results obtained from our multiplex AACT/SILAC quantitative approach.
Figure 5.
The time-course study of IL-1β and STAT1 expression at protein level after LPS stimulation. (a) TLR4-deficient macrophage cell line GG2EE. (b) TLR4 sufficient or wild-type macrophage cell line HeNC2. Note that the expression level of beta-actin did not change during stimulation of both cell lines, whereas IL-1β was induced only in HeNC2 cells. Upon stimulation, at protein level STAT1 was induced by 2 folds in HeNC2 cells and was less elevated in GG2EE cells.
4 Discussion
4.1. Multiplex AACT/SILAC-based quantitative analysis
In the past two years, AACT (synonym- SILAC) has been widely used in quantitative proteomics and proven to be an efficient and precise method. In most of the quantitative proteomic studies, only two states of cell/tissue populations could be characterized. Recently, the emerging iTRAQ technology provided the possibility to quantify up to 4 different cell populations.[14, 15] Further, triple amino acid-specific metabolic labeling to study temporal phosphotyrosine signaling[20] and universal 15N/13C triple labeling were applied in quantitative proteomics and peptide de novo sequencing[19].
In this work, we applied multiplex AACT to study the TLR4-dependent and –independent proteomic changes in macrophages upon LPS stimulation. The labeling efficiency and specificity of AACT with different isotopically enriched lysine reached more than 95% (as shown in Figure 2), therefore, the unlabeled portion in the labeled samples causes little interference in quantitation. Multiplex labeling enables the simultaneous comparison of 3 different cell populations in a single experiment, thus greatly improves the accuracy, efficiency, and throughput in quantitative proteomics as multiplex labeling avoids the possible variations involved in multiple experiments. However, the increased number of labeling precursors also increases the complexity of the mass spectra. We observed many labeled peptides overlapped with other different peptides in the spectra (data not shown), especially when analyzed large gel bands. The overlapped isobaric peptides may cause large errors in automatic quantification using MSQuant or other similar programs. In manual quantification, the error caused by overlapped isobaric peptides can be reduced by manually subtracting the spectral background. In addition, when the overlapped peptides co-eluted, it becomes impossible to subtract all the background. Therefore, the quantification results obtained from co-eluted isobaric peptides were not included in this report. Compared to the standard deviations (S.D.s) of manual quantification, the S.D.s of quantification results obtained from automated software, MSQuant, is relatively significant. The possible reason is that the deuterium-labeled lysine creates minor chromatographic shift on reverse phase LC separation, and the computer software cannot take this factor into calculation. However, the manual quantification can subtract background and reduce the error caused by the chromatographic shift, therefore, provides more accurate quantification results.
We chose to use strict trypsin specificity in protein identification by MS/MS searching using MASCOT. Some previous studies showed that proteins in a complex protein mixture were often cleaved at non-specific site,[23] therefore, some researchers suggested using non-enzyme specificity during database search to improve sensitivity. [24] Recent studies also suggested trypsin cleaves proteins exclusively at C-terminal of arginine and lysine.[25] We found that by using strict trypsin specificity in database search could significantly reduce the searching time (from more than 10 hours to several minutes for each search). More interestingly, we found that using strict trypsin specificity can actually identify more proteins than using non-tryptic specificity (data not shown). The sensitivity of protein identification by database search is not improved by using non-tryptic specificity. Without the support of a high computer power, it is impractical to perform database search without enzyme specificity. By using strict trypsin specificity in database search, it ensured the database searching results would have less false-positive rate. Therefore, it requires less manual intervention in the result revalidation.
Manual inspection of the automatic protein database search results is an effective way to remove false-positives in protein identification,[26] although it is not as fast as using computational approaches. Combining decoyed database (i.e., sequence reversed database) search and manual inspection, it can greatly reduce the false-positive rate in protein identification by database search.
Some researchers had found that semi-essential amino acid arginine could be converted to proline partly through the urea metabolic pathway. Thus, the 13C on the backbone of arginine could be incorporated into proline, and some proline containing peptides will then show the part of specific isotopic pattern (5 Da split).[27] In this work, we did not find isotope scrambling caused by lysine-specific mass tagging.
4.2. Biological implication of our proteomic dataset
Previously TLR4 has been established as the LPS receptor by genetic approaches. However, several reports indicated that not all cellular responses toward LPS stimulation are via TLR4.[1] In the present study we applied a multiplex labeling AACT/SILAC to distinguish those proteins whose expression changes depend on TLR4 pathway from those that are not TLR4-dependent. Specifically, both HeNC2 and GG2EE are macrophage cell lines derived from congenic mice strains, C3H/HeN and C3H/HeJ. These two strains are genetically identical except that C3H/HeJ harbors a single point mutation on its TLR4 intracellular domain that abolishes TLR4-mediated response [28]. As indicated in Table 1, 20 proteins were significantly up-regulated in HeNC2 cells, of which only 5 were induced in GG2EE cells. Interleukine 1 (IL-1) is the well-known proinflammatory cytokine that is induced in macrophage by LPS through TLR4 pathway. IL-1 receptor antagonist, etc, is also reported as LPS-inducible genes elsewhere.[29] The requirement of TLR4 for the induction of these genes was validated by western blot as in Figure 5. We also identified cytokines such as TNF-α associate protein 1 (TRAP 1) and IFN-α response protein (IFNG15) in the study, but did not detect their lysine-containing peptides for the quantification purpose (Suppl. Figure 1). If we increase the sample amount or using alternative amino acid precursors for labeling, it is likely to quantify these proteins too. Nevertheless, using a systems investigation, our results support the previous notion that not all LPS-inducible protein expression is dependent on TLR4. One example of these proteins is called BCR downstream signaling 1 (or Signal-transducing adaptor protein 1, STAP-1) that contains a SH2 domain that normally functions as an adaptor for tyrosine kinase pathways.[30] Its close homologue STAP-2 was shown to mediate the IL-6 induced STAT3 activation.[30] One may speculate that STAP-1 may function as an adaptor to mediate some of the LPS-mediated cellular responses.
Previously Hirotani et.al. performed a comprehensive microarray analysis of LPS-inducible genes in mouse macrophages from wild-type, MyD88−/−, TRIF−/−, and MyD88−/−TRIF−/− mice. 148 genes in wild-type macrophages were found to be induced in response to LPS.[31] Compared to genomic approach, our proteomic approach did not detect as many changes at protein level, which is probably due to the higher complexity of proteome. However, proteomic approach can provide supplementary information to what genomic approach can do. From example, among these identified proteins, carbonic anhydrase 2 was induced to the most. Its induction is mainly TLR4-dependent although in GG2EE cells there was a three-fold induction upon LPS stimulation. Interestingly, in a report Carbonic Anhydrase 2 was indicated as transcriptionally uninduced by LPS.[32, 33] This serves as solid evidence why a proteome analysis is important for a full understanding of the cellular responses. In fact, carbonic anhydrase is important in inflammatory processes because it maintains the intracellular pH by controlling the concentration of pCO2. Furthermore, the ambient pCO2 modulates intracellular pH, oxidant generation and IL-8 secretion in human neutrophils.[32]
We also noticed that there are several down-regulated proteins in the study. Protein phosphatase 2A inhibitor-2 (PP2Ai-2) was specifically down-regulated in the TLR4 wild type cell but not in the TLR4-deficienct cell line. Protein phosphatase 2A (PP2A) has been shown to down-regulate mitogen-activated protein (MAP) kinase pathway by decreasing phosporylated MAP kinase[34]. Because PP2Ai-2 functions as an inhibitor of PP2A, we therefore suspect that TLR4 signaling activation suppresses PP2Ai-2 expression so that PP2A can deactivate the MAPK signaling events following TLR4 activation. This might serve as a negative regulatory mechanism to shut down the TLR4 signaling. On the other hand, proteins such as annexin A1 and A5 were equally down-regulated in both TLR4 WT and deficient lines, suggesting that these proteins are probably involved in a more global response to a cellular stress exerted by LPS stimulation.
In summary, we have designed a multiplex AACT quantitative approach to comparatively study the TLR4-dependent and -independent cellular protein changes in macrophage upon LPS stimulation. Our study provided first proteomic view supporting the notion that not all LPS-inducible events are TLR4-dependent, which will shed light to our understanding of how immune cells respond to LPS challenge.
Supplementary Material
Acknowledgements
This research was supported by NIH 1R01AI064806-01A2, and the Office of Science (BER), U.S. Department of Energy, Grant No. DE-FG02-07ER64422.
References
- 1.Beutler B. Inferences, questions and possibilities in Toll-like receptor signalling. Nature. 2004;430:257–263. doi: 10.1038/nature02761. [DOI] [PubMed] [Google Scholar]
- 2.Akira S. Mammalian Toll-like receptors. Curr Opin Immunol. 2003;15:5–11. doi: 10.1016/s0952-7915(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 3.Vabulas RM, Ahmad-Nejad P, Ghose S, Kirschning CJ, et al. HSP70 as endogenous stimulus of the Toll/interleukin-1 receptor signal pathway. J Biol Chem. 2002;277:15107–15112. doi: 10.1074/jbc.M111204200. [DOI] [PubMed] [Google Scholar]
- 4.Oshiumi H, Sasai M, Shida K, Fujita T, et al. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to toll-like receptor 4 TICAM-1 that induces interferon-beta. J Biol Chem. 2003;278:49751–49762. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- 5.Smirnova I, Poltorak A, Chan EK, McBride C, Beutler B. Phylogenetic variation and polymorphism at the toll-like receptor 4 locus (TLR4) Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-research002. RESEARCH002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian WJ, Monroe ME, Liu T, Jacobs JM, et al. Quantitative proteome analysis of human plasma following in vivo lipopolysaccharide administration using 16O/18O labeling and the accurate mass and time tag approach. Mol Cell Proteomics. 2005;4:700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Righetti PG, Campostrini N, Pascali J, Hamdan M, Astner H. Quantitative proteomics: a review of different methodologies. Eur J Mass Spectrom (Chichester, Eng) 2004;10:335–348. doi: 10.1255/ejms.600. [DOI] [PubMed] [Google Scholar]
- 8.Tao WA, Aebersold R. Advances in quantitative proteomics via stable isotope tagging and mass spectrometry. Curr Opin Biotechnol. 2003;14:110–118. doi: 10.1016/s0958-1669(02)00018-6. [DOI] [PubMed] [Google Scholar]
- 9.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. Proc Natl Acad Sci U S A. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gygi SP, Rist B, Gerber SA, Turecek F, et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 11.Yao X, Freas A, Ramirez J, Demirev PA, Fenselau C. Proteolytic 18O labeling for comparative proteomics: model studies with two serotypes of adenovirus. Anal Chem. 2001;73:2836–2842. doi: 10.1021/ac001404c. [DOI] [PubMed] [Google Scholar]
- 12.Zhu H, Pan S, Gu S, Bradbury EM, Chen X. Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid Commun Mass Spectrom. 2002;16:2115–2123. doi: 10.1002/rcm.831. [DOI] [PubMed] [Google Scholar]
- 13.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 14.Ross PL, Huang YN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.DeSouza L, Diehl G, Rodrigues MJ, Guo J, et al. Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J Proteome Res. 2005;4:377–386. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 16.Zhu H, Hunter TC, Pan S, Yau PM, et al. Residue-specific mass signatures for the efficient detection of protein modifications by mass spectrometry. Anal Chem. 2002;74:1687–1694. doi: 10.1021/ac010853p. [DOI] [PubMed] [Google Scholar]
- 17.Ibarrola N, Kalume DE, Gronborg M, Iwahori A, Pandey A. A proteomic approach for quantitation of phosphorylation using stable isotope labeling in cell culture. Anal Chem. 2003;75:6043–6049. doi: 10.1021/ac034931f. [DOI] [PubMed] [Google Scholar]
- 18.Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004;1:119–126. doi: 10.1038/nmeth715. [DOI] [PubMed] [Google Scholar]
- 19.Snijders AP, de Vos MG, Wright PC. Novel approach for peptide quantitation and sequencing based on 15N and 13C metabolic labeling. J Proteome Res. 2005;4:578–585. doi: 10.1021/pr0497733. [DOI] [PubMed] [Google Scholar]
- 20.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;22:1139–1145. doi: 10.1038/nbt1005. [DOI] [PubMed] [Google Scholar]
- 21.Gu S, Chen J, Dobos KM, Bradbury EM, et al. Comprehensive Proteomic Profiling of the Membrane Constituents of a Mycobacterium tuberculosis Strain. Mol Cell Proteomics. 2003;2:1284–1296. doi: 10.1074/mcp.M300060-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Schonbeck U, Herzberg M, Petersen A, Wohlenberg C, et al. Human vascular smooth muscle cells express interleukin-1beta-converting enzyme (ICE), but inhibit processing of the interleukin-1beta precursor by ICE. J Exp Med. 1997;185:1287–1294. doi: 10.1084/jem.185.7.1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alberghina G, Cozzolino R, Fisichella S, Garozzo D, Savarino A. Proteomics of gluten: mapping of the 1Bx7 glutenin subunit in Chinese Spring cultivar by matrix-assisted laser desorption/ionization. Rapid Commun Mass Spectrom. 2005;19:2069–2074. doi: 10.1002/rcm.2032. [DOI] [PubMed] [Google Scholar]
- 24.Griffin TJ, Gygi SP, Ideker T, Rist B, et al. Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics. 2002;1:323–333. doi: 10.1074/mcp.m200001-mcp200. [DOI] [PubMed] [Google Scholar]
- 25.Olsen JV, Ong SE, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Kwon SW, Kim SC, Zhao Y. Integrated approach for manual evaluation of peptides identified by searching protein sequence databases with tandem mass spectra. J Proteome Res. 2005;4:998–1005. doi: 10.1021/pr049754t. [DOI] [PubMed] [Google Scholar]
- 27.Liang XQ, Hajivandi MR, Pope M. 53rd ASMS Conference on Mass Spectrometry and Apllied Topics. San Antonio, TX: 2005. [Google Scholar]
- 28.Poltorak A, He X, Smirnova I, Liu MY, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 29.Krzesicki RF, Hatfield CA, Bienkowski MJ, McGuire JC, et al. Regulation of expression of IL-1 receptor antagonist protein in human synovial and dermal fibroblasts. J Immunol. 1993;150:4008–4018. [PubMed] [Google Scholar]
- 30.Minoguchi M, Minoguchi S, Aki D, Joo A, et al. STAP-2/BKS, an adaptor/docking protein, modulates STAT3 activation in acute-phase response through its YXXQ motif. J Biol Chem. 2003;278:11182–11189. doi: 10.1074/jbc.M211230200. [DOI] [PubMed] [Google Scholar]
- 31.Hirotani T, Yamamoto M, Kumagai Y, Uematsu S, et al. Regulation of lipopolysaccharide-inducible genes by MyD88 and Toll/IL-1 domain containing adaptor inducing IFN-beta. Biochem Biophys Res Commun. 2005;328:383–392. doi: 10.1016/j.bbrc.2004.12.184. [DOI] [PubMed] [Google Scholar]
- 32.Coakley RJ, Taggart C, Greene C, McElvaney NG, O'Neill SJ. Ambient pCO2 modulates intracellular pH, intracellular oxidant generation, and interleukin-8 secretion in human neutrophils. J Leukoc Biol. 2002;71:603–610. [PubMed] [Google Scholar]
- 33.Frevel MA, Bakheet T, Silva AM, Hissong JG, et al. p38 Mitogen-activated protein kinase-dependent and -independent signaling of mRNA stability of AU-rich element-containing transcripts. Mol Cell Biol. 2003;23:425–436. doi: 10.1128/MCB.23.2.425-436.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Santoro MF, Annand RR, Robertson MM, Peng YW, et al. Regulation of protein phosphatase 2A activity by caspase-3 during apoptosis. J Biol Chem. 1998;273:13119–13128. doi: 10.1074/jbc.273.21.13119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.