Abstract
Emerging evidences indicate that blood platelets function in multiple biological processes including immune response, bone metastasis and liver regeneration in addition to their known roles in hemostasis and thrombosis. Global elucidation of platelet proteome will provide the molecular base of these platelet functions. Here, we set up a high throughput platform for maximum exploration of the rat/human platelet proteome using integrated proteomics technologies, and then applied to identify the largest number of the proteins expressed in both rat and human platelets. After stringent statistical filtration, a total of 837 unique proteins matched with at least two unique peptides were precisely identified, making it the first comprehensive protein database so far for rat platelets. Meanwhile, quantitative analyses of the thrombin-stimulated platelets offered great insights into the biological functions of platelet proteins and therefore confirmed our global profiling data. A comparative proteomic analysis between rat and human platelets was also conducted, which revealed not only a significant similarity, but also an across-species evolutionary link that the orthologous proteins representing ‘core proteome’, and the ‘evolutionary proteome’ is actually a relatively static proteome.
Keywords: Blood platelets, multidimensional separation, mass spectrometry, global profiling, cross-species comparison
1 Introduction
Platelets are anucleate cells that derive from megakaryocytes and circulate in the blood stream [1, 2]. Under normal conditions, platelets help maintain vascular integrity through their involvement in clot formation and repair of injury [3]. Meanwhile, disregulated platelets were found to contribute to pathological consequences such as many types of cardiovascular diseases which are the most common causes of mortality in modern society [1]. For example, arterial thrombosis appearing as the abnormal formation of clots is a major factor in developing heart attacks [4].
While many functional aspects of platelets have been investigated for centuries [5, 6], the growing line of evidences have been suggesting there are more functions of platelets yet to be explored [7-11]. For instance, platelets have recently been found to act as the immune and signaling cells in inflammation and innate immune responses [3, 12]. Under lipopolysaccharide (LPS) stimulation, platelets may express functional toll-like receptor 4 (TLR4) [7]. The interplay between platelets and neutrophils after TLR4 expression leads to the rapid formation of neutrophil extracellular traps (NETs) that can entrap and kill bacteria [8]. Blood platelets can also contribute to cancer metastasis [6, 13]. Platelets activated by the releasate from tumors may adhere to cancerous cells and form a shield that protects tumor cells from removal by the immune system [6]. Platelets may also facilitate metastasis by assisting circulating tumor cells to adhere to vascular tissues and move out of the vessel [6, 14, 15]. Furthermore, as platelets have no nuclei, they represent a simplified model for the study of signal-dependent protein transcription and programmed cell death [2, 9]. A recent study suggested that Bcl-xL triggers Bak mediated apoptosis in platelets [2]. Surprisingly, platelets have also been shown to play a remarkable role in liver regeneration [10].
Taken together of all these evidences, a global exploration of the platelet proteome, a yet still largely unknown treasure chest, represents a critical step to provide the molecular basis for understanding the multi-functional nature of platelets. Importantly, the characterization of those platelet proteins involved in discrete facets of physiological functions will reveal possible biomarkers for early diagnosis of the progression of certain diseases including cardiovascular diseases and cancers [16, 17].
Mass spectrometry (MS)-based proteomics technologies provide powerful approaches for profiling of particular proteomes including the expressed/secreted proteins in platelets [18, 19]. However, due to trade-offs involving sensitivity, accuracy, and throughput for each type of separation/identification scheme, it is difficult to obtain a relatively complete proteome by using a single separation/identification approach [20]. Remarkably, a prestigious group reported a global profiling of the membrane and cytosolic proteome of human red blood cells (RBCs) using combined protein identification technologies [21]. Furthermore, a direct comparison with mouse RBCs on the proteome level was also carried out, which indicated close concordance between the two proteomes [22]. Most recently, a cross-species comparison of Caenorhabditis elegans with fruitfly Drosophila melanogaster at the proteome level was performed, which illustrated their analogous proteins have similar relative amounts, even though the mRNA levels vary widely between the two species [23].
Here, we presented an integrated platform including multi-step protein extraction, multidimensional separation/identification approaches, and combined mass spectra interpretation algorithms to obtain the maximum coverage of both rat and human platelet proteome. To examine the functional relevance of our datasets, one of the popular labeling strategies, isobaric tags for relative and absolute quantitation (iTRAQ), was also used. In order to understand the mutual and diverse proteome between these closely related mammals, a comparative proteome analysis was conducted, which suggested the function diversity of platelet proteome for the first time.
2 Materials and methods
2.1 Vital platelet isolation from rat and human blood
Fresh blood obtained from healthy rats was placed into plastic tubes containing acid/citrate/dextrose (ACD) antigoagulant (70 mM citric acid, 85 mM sodium citrate, 110 mM glucose) [24]. The subsequent isolation was carried out as previously reported [25]. Minor modifications included altering the ratio between ACD buffer and blood volume, multiple steps but higher centrifugation speed to avoid any potential contaminants from red blood cells (RBCs), and different but compatible lysis buffer for subsequent MS analysis. Briefly, 5 mL ACD buffer were added into 10mL fresh blood, and this increased ratio could successfully prevent potential precipitation [24]. After centrifugation at 200×g for 20 minutes, the upper two-thirds of the platelet-rich plasma (PRP) was carefully collected followed by 12 minutes centrifugation at 1000×g [26]. The platelet pellets on the bottom of the tube were gently collected, then resuspended in Tyrodes buffer and washing buffer described previously [24]. After 1 hour incubating at room temperature, the platelets were spun down again at 1000×g for 12 minutes to eliminate some fragments of RBCs. The platelet pellets were then subjected to protein extraction and drawn into ACD buffer to prevent precipitation. Similar procedures as described above were then applied to isolate blood platelets. Based on the procedures described above, human platelets were prepared from fresh blood of healthy volunteers who had not been on medication for the previous ten days. Use of human blood was approved by the ethics committee of the Fudan University and informed consent was granted by the donors.
2.2 Thrombin stimulation of vital platelets
The platelets freshly isolated from rat blood following the above procedures were re-suspended in 2mL Tyrodes buffer to the final concentration of 1×109 cells/mL. 100μL stimulation solution (1 unit thrombin, 20 mM CaCl2, 20 mM MgCl2 in Tyrodes buffer) was added to stimulate the platelets for 3 min at 37°C. The platelets were washed and pelleted at 5000×g for 3 min, and the supernatant was removed.
2.3 Protein extraction, SDS PAGE separation, and in-solution/in-gel digestion
An optimized two-step strategy was carried out to extract platelet proteins. Firstly, the platelet pellets obtained from rat and human blood were added into 8M urea to extract water-soluble proteins (Extraction I). After 30 minutes incubation on ice, the lysate was centrifuged at 15,000 rpm for 15 minutes. Half of the Extraction I was subjected to in-solution digestion after diluting the urea concentration to lower than 2M. The remaining proteins were subjected to IEF separation. The pellet debris was incubated for 30 minutes on ice in a second lysis buffer (7M urea, 2M thiourea, 2% CHAPS, 15 μL protease inhibitor, 100mM fresh-made DTT) to obtain Extraction II. Then no visible insoluble debris was left in the tube. Extracted proteins were then separated by SDS PAGE under a standard protocol [27]. About 200 μg Extraction II were loaded into 2 parallel lanes of a homemade 12% SDS-PAGE gel. After coomassie blue staining, the gel was cut into 14 continuous sections (Supplement Figure 1), diced into small pieces and placed in 1-mL Eppendorf tubes. In-gel digestion and peptide extraction were performed using common protocol [27].
2.4 Isoelectric focusing (IEF) separation
Urea-extracted proteins were resuspended in a modified IPG buffer that contained only 3-10 pH nonlinear IPG buffers (Amersham Bioscience), so harmful contaminations of the CHAPS detergent and bromophenol blue dye were intentionally avoided [28]. The protein solution was then applied to an 18 cm 3-10 nonlinear Immobiline DrySrip (Amersham Bioscience) according to the manufacture's procedure.
Similarly, IEF separation of peptides was carried out on the in-solution digested peptides using similar IPG strip. The focusing protocol was slightly different from the IEF of proteins. Excess cover oil was carefully removed, and the strip was cut into 24 equal sections within 3 minutes to avoid potential diffusion [29]. As for proteins on the strip, in-gel digestion was carried out as described previously [27]. For peptides on the strip, a series of sequential extractions was performed as described elsewhere [28].
2.5 Liquid chromatography and mass spectrometry
Two dimensional liquid chromatography tandem mass spectrometry (2D LC MS/MS) approach involved an LTQ Orbitrap mass spectrometer (Thermo Electron) in connection with an LC Packings Ultimate chromatography system (LC Packings Ultimate System, Dionex, CA). Peptides were bounded to a strong cation exchange (SCX) column (15 cm×4.6 mm, 5 μm, 300Ǻ, PolyLC) and eluted with eight sequential injections of ammonium acetate solutions with various concentrations (0, 50, 100, 200, 400, 800, 1000, 2000 mM), which was followed by a 2-hour reversed phase gradient (15 cm×300 μm column, 5μm C18, LC Packings) to further separate the peptides containing in each elution step. Mobile phase A was 0.1% formic acid (FA) in 5% acetonitrile (ACN). Mobile phase B was 0.1% FA in 95% ACN. Gradient parameters: 0-10 minutes, 10%B; 10-90 minutes, 40%B; 90-110 minutes, 90%B; 110-120 minutes, 0%B. The 1D LC separations was performed on the same LC system mentioned above or a Micromass CapLC Pump (Waters, UK). The column and gradient settings were almost the same as described above except some minor changers in the column equilibration time in the Micromass CapLC.
Tandem MS (MS/MS) analysis was performed on the LTQ Orbitrap or QTOF (Micromass, QTOF API-US, Waters, UK) mass analyzer. The five most intense ions fully scanned by Orbitrap (resolution is 60000 at m/z 400, scan range is from m/z 300 to 2000) were sequenced in the LTQ. Ion charge state screening was enabled and all the singly charged species were rejected. For QTOF, mass spectra were acquired in positive mode over the range m/z 400-1900. The top four most-abundant precursors were selected for MS/MS analysis using 38V cone voltage.
2.6 Database searching and protein identification
All raw MS data was searched against the rat International Protein Index (IPI) protein database (version 3.07) using the Mascot (version 2.0; Matrix Science, London, UK) and Sequest search engines (ThermoFisher Scientific, CA), respectively. All the “.raw” files acquired from LTQ Orbitrap were converted to “.mgf” (Mascot Generic Format) files using the Trans Proteomic Pipeline (TPP) platform[30] to make them compatible with Mascot. The merged “.mgf” files from different salt fractions of 2D LC experiments were then submitted to a 64 bite dualprocessor server in the UNC-Duke Michael Hooker Proteomics Center (http://proteomics.unc.edu). The Sequest search engine is an in-house cluster with 72.2-Ghz dual processor nodes.
For protein assignment, positive identifications were made based on stringent filters. In the case of Sequest engine, the ΔCn values for all matched peptides were set higher than 0.1, and the cross-correlation scoring (XCorr) thresholds were at least 1.90 (z=2), 2.70 (z=3), and 3.11 (z=4) according to the reversed database search evaluation and the ensured false discovery rate (FDR) was less than 0.05. Additionally, the computed probability through TPP tools[30] must be higher than 0.95 for each matched peptide. Protein assignments were only made if the protein had at least two unique peptides passing the criteria mentioned above. As for the Mascot search engine, all assigned peptides should be in the first rank and then filtered by the peptide score originating from reversed database searching (34 for the QTOF-based analyses, 41 for the others). To further eliminate redundant proteins matched with the same set of peptides (group proteins), a stringent interpretation method was employed, thus leading to a minimized dataset. In brief, proteins that span the same set of peptides, or a subset, were collapsed into one representative protein, which has the max hits or shortest sequence length [31, 32].
2.7 iTRAQ labeling of platelet peptides, and iTRAQ-based quantitative analysis of thrombin-induced proteome changes in vital platelets
The proteins extracted from both non-stimulated and thrombin-stimulated platelets were treated with tris-(2-carboxyethyl) phosphine (TCEP) and iodoacetamide, in-solution digested with trypsin and then labeled with an isobaric tag reagent according to the manufacturer's protocol (Applied Biosystems, Foster City, CA). The above two samples were equally mixed and separated using an off-line 2D SCX-RPLC approach. The LC conditions were similar to those used in on-line 2DLC separation described above, except that the samples were washed 30 min using loading buffer (10 mM KH2PO4 in 25% ACN at pH 3.0) before the salt elution in order to remove detergents and excess reagents.[33] Eight salt plugs (25, 37.5, 50, 75, 100, 125, 150, 175, 200, and 350 mM KCl at pH 3.0) were injected sequentially and peptide fractions were collected, lyoplized and then cleaned using PepClean C18 spin column (Pierce, Rockford, IL).
LC-MS/MS analyses were performed on a nanoflow LC Packings System (Dionex, CA) interfaced to a QSTAR Elite Hybrid mass spectrometer (Applied Biosystems). Peptides were separated at a flow rate of 200 nL/min over a C18 column (15 cm×75 μm column, 5μm particle size, LC Packings) using an identical 2-hour gradient described above. MS scans were acquired in a data-dependant mode from m/z 300 to 2000 and up to three most intense parent ions were selected for MS/MS scan.
ProteinPilot (version 2.0.1, Revision 67476, Applied Biosystems) with the Paragon Algorithm was used for the identification and quantification of relative abundance of platelet proteins expressed in different pathological conditions. All the MS/MS spectra were searched against the NCBI protein database using the following criteria: 95% confidence for protein identification, trypsin cleavage specificity, methyl methanethiosulfonate (MMTS) as the defined modification.
3 Results and Discussion
3.1 Integrated platform maximizes the coverage of MS-detectable platelet proteome
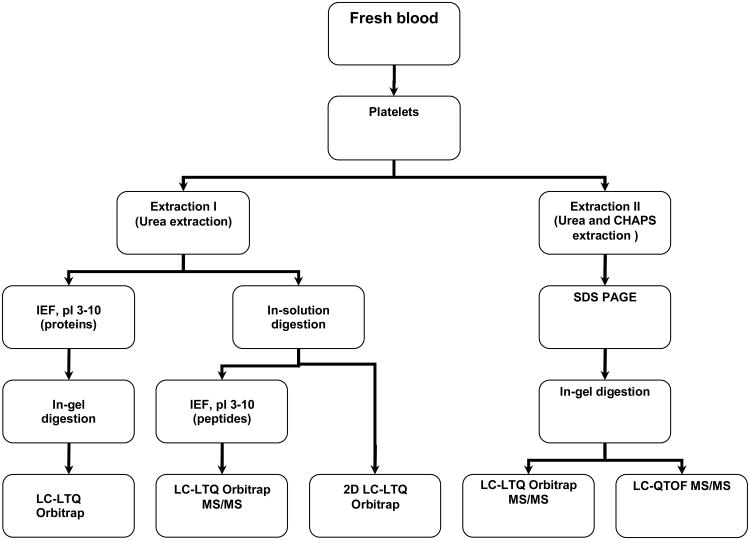
When a single MS-based proteomic approach is taken trade-offs involving the sensitivity, accuracy, and throughput often made in profiling proteins with diverse physical properties. To identify maximum numbers of the platelet proteins with different pI and hydrophobicity, variable expressions in a wide dynamic range, etc, our current platform integrates multiple components including a two-step protein extraction approach, multiple gel-based and gel-free separation methods in a complimentary manner, and different MS analysis strategies (Figure 1).
Figure 1. Schematic illustration of rat and human platelet proteome analysis.
A two-step extraction method with 8M urea and the lysis buffer was applied to extract platelet proteins for either SDS gel-based separation and LC-MS/MS or in-solution digestion respectively.
First, the two-step extraction protocol was used to recover most of the proteins from the purified platelets. In contrast to the common extraction methods, such as repeated freeze-to-thaw cycling and acetone precipitation, which collect soluble proteins and normally discard the undissolved parts, our method could recover most of the platelet proteins which ensured a comprehensive starting material [22, 34, 35]. Secondly, the subsequent gel-based and gel-free separation methods were chosen for the proteins extracted from different lysis buffers accordingly. The proteins in surfactant-based lysis buffer were loaded onto the SDS gel. Then the CHAPS detergents, which are notoriously incompatible with RPLC-MS/MS, were removed. Meanwhile, the proteins extracted from surfactant-free lysis buffer were introduced to IPG strip and digested in solution, respectively. Thus the resulted peptides were compatible with the following RPLC-MS/MS approach because urea was readily removed by reversed phase column without pretreatment [36]. Finally, both mass spectrometers, LTQ Orbitrap and QTOF, were used in the integrated platform to overcome the intrinsic weakness of each type of the instruments used alone [20].
As summarized in Table 1, a total of 837 rat platelet proteins were identified in high confidence, including 320 proteins identified by the gel-free 2DLC-LTQ Orbitrap approach and 634 proteins (with redundancy) detected by SDS PAGE-RPLC-LTQ Orbitrap approach. As a widely used pre-separation method, SDS PAGE could effectively fractionate a complex sample, meanwhile, it provides a molecular size window for the proteins of interest [21]. The identified proteins are then readily validated using Western blotting [37]. On the other hand, two dimensional strong cation exchange-reverse phase liquid chromatography (2D SCX-RPLC) approach which is based on the property orthogonality of a variety of peptides is also routinely used by most proteomics labs [38]. Recent data showed that the above two approaches are comparable with respect to the number of identified proteins [39]. Interestingly, our data indicated the SDS PAGE-RPLC-LTQ MS/MS identified more proteins than those identified by SCX-RPLC-LTQ approach. One of the possible reasons is the former approach (14 gel bands) has more fractions than the latter one (7 salt plugs) in the first dimensional separation, which resulted in higher resolution in separation and more protein identifications.
Table 1. The number of identified platelet proteins using each method.
2D LC, 2DLC-LTQ Orbitrap MS/MS; IEFpro, IEF (protein)-LTQ Orbitrap MS/MS; IEFpep, IEF (peptide)-LTQ Orbitrap MS/MS; and SDS-QTOF, SDS PAGE-LC-QTOF MS/MS.
| Species | Total No. (unique) | The identified proteins in each approach | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| 2D LC | SDS-LTQ | IEFpro | IEFpep | SDS-QTOF | Literature a) | ||
| Rat | 837 | 371 | 634 | 220 | 156 | 83 | None |
shows no report result has been found so far for rat platelet proteins.
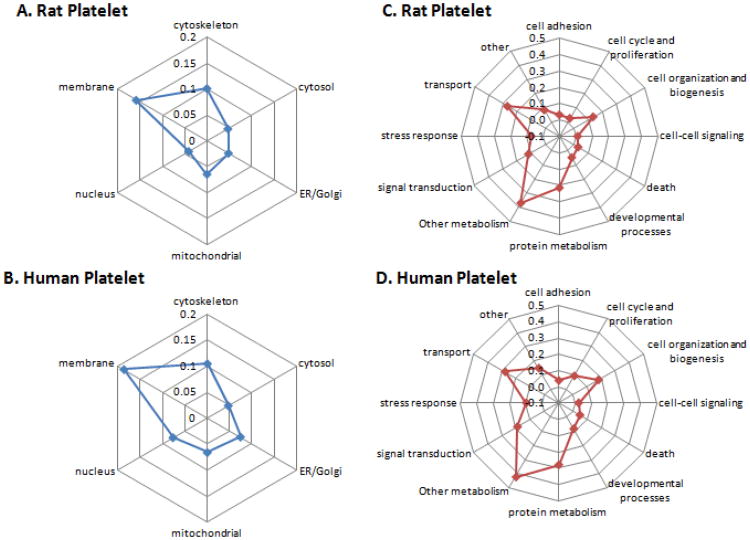
The overall identified proteins were grouped into 6 and12 classes with respect to their subcellular localization and biological function. As shown in Figure 2A & 2C, about 26% of the identified proteins were known as cytoskeleton and membrane proteins, and more than 40% proteins were related to cell organization and transportation, which was consistent with the fundamental roles of platelets in clot formation and wound healing. Meanwhile, up to 23% proteins had the known function directly related to protein metabolism. According to previous findings [21], red blood cells (RBC) have a large number of membrane and cytosolic proteins involving in cellular metabolism, probably due to the relatively short life span of RBCs, which requires fast degradation of organelles or macromolecules [21]. In contrast, platelets have an even shorter life span and may undergo programmed cell death, typically after a week in the blood stream [40]. Therefore, this distribution pattern of functional categories of our identified platelet protein is reasonable. Furthermore, some proteins (4%) were classified as nucleus proteins. Given that platelets are anucleate, these proteins probably derived from megakaryocytes during thrombopoiesis [16, 41].
Figure 2.
(A & B) Radar plots illustrating the distribution of protein subcellular localization from rat and human platelets. Proteins belonging to Other locations were not shown in the radar plots (rat, 20.3%; human, 29.6%). ER, endoplasmic reticulum. (C & D) Radar plots showing the categories of molecular functions and biological processes of the proteins identified in the rat and human platelets.
Interestingly, one of the newly identified human platelet proteins, Calumenin [42], which is responsible for the activation of coagulation protein (such as matrix Gla protein, MGP),[43] was unambiguously identified in the rat platelets. Calumenin was identified in human atherosclerotic lesions but not in normal vasculature, which indicated a potential role in atherosclerosis and thrombosis formation [42]. In our recent study of the rat platelet proteome changes during diethyl nitrosamine (DEN)-induced tumor progression in the mouse liver, calumenin was found to be closely associated with the inflammation or cirrhosis status of the rat liver (Leng TH, et al, unpublished results).
3.2 Minimal contaminations from other blood cells was assayed to ensure a precise profile of platelet proteome
The purity of the isolated platelets was examined by microscopic inspection and only the platelets with 99.9% purity were used for our profiling experiments [26]. Imaging was also performed in a standard scanning electron microscope to confirm the absence of potentially contaminating leukocytes and erythrocytes (data not shown). In general, our platelet sample contained less than 1 red or white blood cell per 10 000 platelets. Furthermore, as the well-known and highly abundant nucleus proteins, either histones or histone fragments were not found, suggesting minimum contamination of nucleated blood cells [44]. The absence of the abundant erythrocyte protein ankyrin in our identification profile also suggested that possible contamination from RBCs was ignorable. However, some representative proteins from RBCs were identified in the rat platelets, such as hemoglobin α/β chain and Spectrin α/β chain,[21] suggesting their potential expressions in rat platelets [44, 45]. In comparison with the published results of human platelets, our protocol of preparing highly purified platelets ensured the data accuracy and consistency of rat or human platelet proteome and made it possible to perform comparison analysis of rat and human platelet proteome based on the same proteomic platform.
3.3 Proteomes with high dynamic range were detected by using the integrated platform
We utilized a semi-quantitation analysis tool, the exponentially modified protein abundance index (emPAI) which proved to be effective in human cancer cell proteome [46, 47], to estimate the relative abundance of our identified platelet proteins. All emPAI indexes were calculated through the exponentially modified number of observed peptides divided by the number of observable peptides for each individual protein [48]. As a result, the relative abundance of 837 rat platelet proteins was quantified by emPAI. According to the order of emPAI, these proteins were classified as low (emPAI<0.1 and without emPAI annotated), medium (0.1≤emPAI<1) and high (emPAI≥1) abundance proteins, respectively. The number of proteins in each category was 86 for low-abundance, 624 for medium-abundance, and 127 for high-abundance, respectively. All the quantitative information of the overall proteins was presented in Supplementary Table 1.
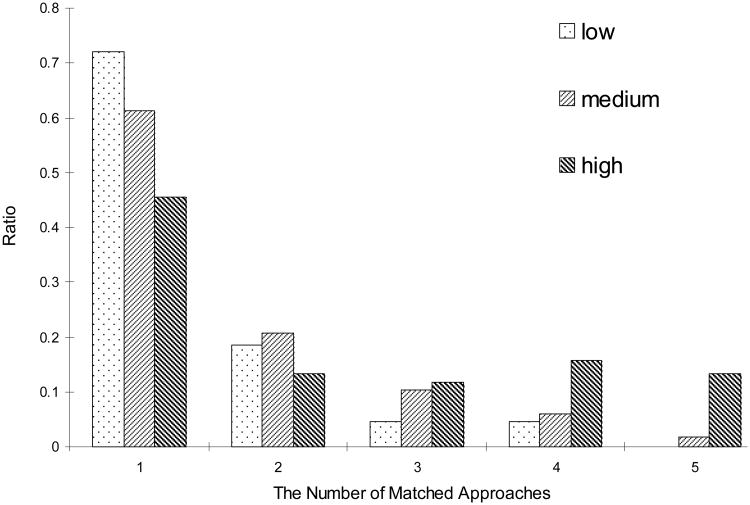
According to the semi-quantitative analysis, more than 72% of low-abundance proteins and 61% of medium-abundance proteins were detected by a single approach (Figure 3), which implicated the necessity of combined approaches. But for high-abundance proteins, more than 54% were detected by at least two approaches; moreover, 13% were detected by all five approaches. For instance, thrombospondin 1 (TSP1), defined as a highly abundant protein by emPAI index (emPAI=1.183), was identified by all five approaches mentioned above. TSP1 is secreted from platelet α-granules and interacts with collagens and fibrinogen in the extracellular matrix [49]. So, modulation of activity of various thrombospondins might be therapeutically effective [50]. In our study, the correlation between protein abundance and the number of approaches used in combination coincided well. The combination of multiple strategies was effective to identify maximum numbers of the proteins expressed in a wide dynamic range.
Figure 3. The distribution of quantified proteins versus the number of matched approaches used.
Low, emPAI<0.1 and without emPAI annotated; medium, 0.1≤emPAI<1; high, emPAI≥1.
By using this integrated platform, we also identified six isoforms of 14-3-3 proteins in rat platelets, ε, τ, γ, β/α, ζ/δ and η. Even through there are seven known mammalian isoforms (14-3-3σ is expressed in epithelial cells only) [51], only five of them have been previously found in human platelets, which contain high levels of the ζ, β, and γ isoforms and lower levels of the ε and η [52]. By using the integrated approach, we detected almost all the 14-3-3 isomers with varied abundance in rat platelets.
3.4 Combination of multiple search engines provided more protein identifications
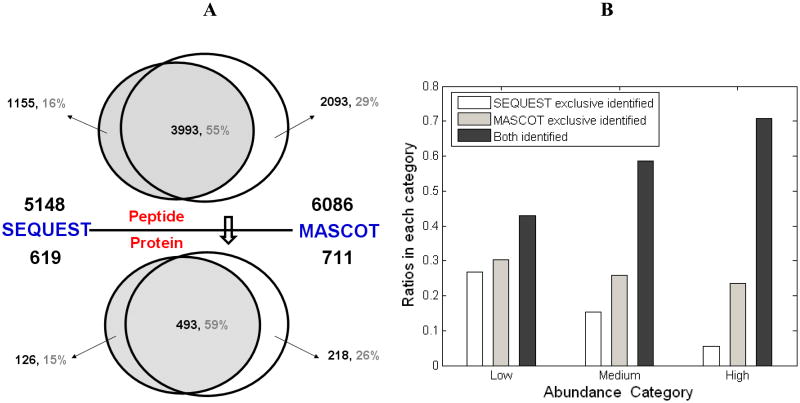
We also demonstrated that the coverage of the MS-detectable platelet proteome could be dramatically improved when MS/MS spectra were analyzed with multiple search algorithms in a complementary way, which was in agreement with previous reports [20]. For the spectra acquired by using LTQ Orbitrap, a total of 7241 peptides were assigned by either Mascot or Sequest (Figure 4A); 55% of them were confirmed by both search engines, 16% were exclusively interpreted by Sequest and 29% by Mascot, respectively. The overlap in our studies was slightly different from the previous study [20], in which 68% of the assigned spectra were matched by dual search engines, 22% and 9% were exclusively matched by Mascot and Sequest, respectively. Considering the complexity of large-scale protein composition in platelets, such diversity suggested that the combination of multiple search engines can be exploited to increase the coverage of protein identification for a proteome.
Figure 4. Results of the database search using dual engines including Mascot and Sequest.
The Venn plot demonstrates the overlap and exclusive matched proteins or peptides of MASCOT and SEQUEST. The number of peptides or proteins was marked in the corresponding areas. (B) The histogram shows the matched ratios of search engines in each abundance category. All of the peptides and proteins were confirmed through bioinformatics analysis.
Notably, one more interesting finding can also be illustrated in terms of the relationship between the search engines and protein-abundance distributions (Figure 4B). For highly abundant proteins, more than 70% were assigned by both search engines with extremely high confidence. This percentage dramatically decreased for low-abundance proteins as only 43% could be confirmed by both engines, indicating that single algorithm is not sufficient enough to assign each spectrum with proper protein entries, especially those low abundance proteins. Furthermore, we also found that Sequest matched more low-abundance proteins than Mascot. On the contrary, the exclusive identifications from Mascot were significantly higher than those from Sequest for high-abundance proteins. Due to the inherent nature of these two different algorithms, the probability-based algorithm, Mascot, may be more sensitive than the empirical and correlative measurements provided by Sequest. Thereby, the complementary use of dual search engines and algorithms was helpful in obtaining a higher coverage of platelet proteome.
3.5 The quantitative analysis of the thrombin-inducible proteins in the MS-identified platelet proteome further validates the accuracy of our platelet proteome dataset
Given the fact that the response to thrombin stimulation is a characteristic function of platelets, we reason the expression of those platelet-characteristic proteins should be thrombin-inducible. Therefore, by using thrombin to stimulate the rat platelets we examined the functional relevance of our platelet proteome dataset and further analyzed the accuracy of our identification of platelet proteins. We then applied iTRAQ-based quantitative proteomic approach to identify those proteins showing the thrombin-inducible changes in their expressions. Through iTRAQ-labeling the paired stimulated versus non-stimulated platelets were analyzed through a 2D-LC-MS/MS approach. In total, 415 non-redundant proteins were confidently identified in more than 3 duplicated runs. Using the thresholds previously set [53], i.e. 20% abundance changes or the isotope ratio larger than 1.2 or smaller than 0.8, measured by iTRAQ signal peaks, the expression level of 16 proteins in the profile of identified platelet proteins were found elevated while 17 proteins in the profile showed decreased expression level (Supplementary Table 6).All of these differentially expressed proteins were classified according to the biological processes and pathways using PANTHER classification system (http://www.pantherdb.org/).
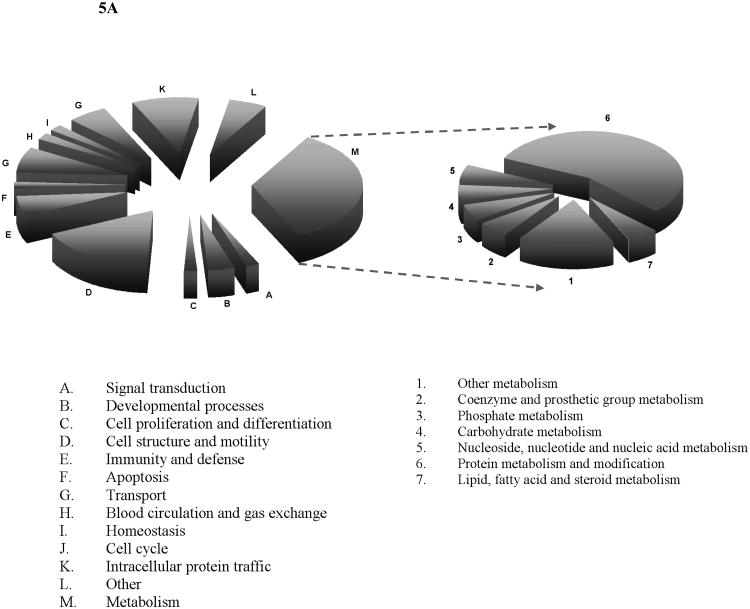
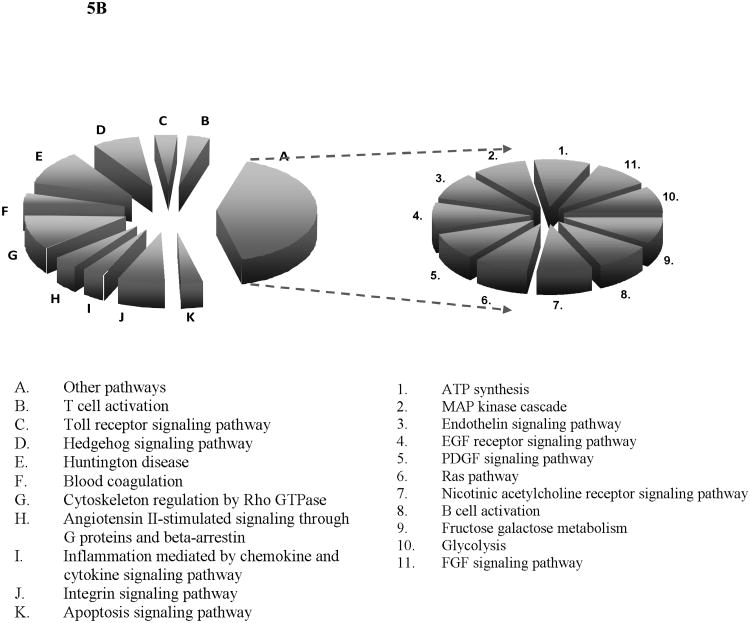
As shown in Figure 5, most of the thrombin-inducible proteins were related to metabolic processes wherein over 50% of them were directly involved in protein metabolism and modifications, also many of them were involved in the household metabolisms for nucleotide or carbohydrate, or fatty acid, etc. This observation suggested the metabolic activities in platelets were enhanced following thrombin stimulation. Another prominent group of the thrombin-inducible proteins were related to cell structure and motility and intracellular protein trafficking, which was unsurprising in accordance with the known physiological role of activated platelets. For instance, through a study using a knockout mouse model calpain-1 was found as a positive regulator in platelet aggregation and clot retraction [54]. Our quantitative proteomics data indicated that upon thrombin stimulation calpain-1 was found up-regulated by 1.4 fold. Meanwhile, the thrombin-inducible proteins are linked to a variety of signaling pathways including blood coagulation, apoptosis, integrin signaling pathway and toll receptor signaling pathway. Furthermore, a EF-hand domain containing 2 (EFHD2) protein was recently found involving in protein synthesis in the encephalomyelitis (EAE) rats with autoimmune diseases[55]. Although the role of EFHD2 in platelets and its contribution to thrombin stimulation remain undefined, we found the expression level of EFHD2 was up-regulated by 1.5 fold upon stimulation, a sign indicating that protein synthesis may occur in platelets after thrombin treatment. Calpain 1 is one of the ubiquitously expressed isoforms of calpain family, which is a highly conserved group of intracellular calcium-dependent cysteine proteases[56, 57]. Calpain 1 was shown to be part of the integrin signal-transduction apparatus. It associates with focal adhesion proteins in platelets and regulates the attachment of αIIbβ3 to the cytoskeleton[57]. Recent evidences suggested that inhibition of calpain using overexpressed calpastatin (its highly specific endogenous inhibitor) prevents thrombin-stimulated α-granule secretion and platelet aggregation[58]. While we have identified this protein in the resting platelets, the expression of calpain 1 was also found up-regulated by 1.4, which is in line with the previous finding of its function in platelets. Apolipoprotein E (ApoE) was previously known to inhibit platelet aggregation through the L-Arginine:Nitric Oxide Pathway[59]. In our quantitative analysis, ApoE indeed showed a trend of down-regulation following thrombin activation, i.e., with an iTRAQ ratio < 0.7. Tubulin is the major component of microtubules which are involved in numerous processes such as cell division and migration[60]. In an effort to characterize its function, the mice lacking β1-tubulin was found to produce approximately 60% less platelets than their wild-type littermates, and these platelets show an attenuated response to thrombin[61]. While the responses of β2- and β5-tubulin to thrombin stimulation are still unknown, our results indicated that these two tubulin isoforms were down-regulated after thrombin stimulation.
Figure 5. Biological process (A) and pathway (B) analyses of the differentially expressed proteins after thrombin stimulation.
Metabolism group (in A) and other pathways group (in B) were further classified into 7 and 11 groups, respectively, according to the bioinformatics analysis results.
In general, because thrombin is a characteristic stimulus which activates platelets, all thrombin-induced differentially expressed proteins were found in our dataset of platelet proteome, suggesting the accurate content of functionally related platelet proteome identified by our proteomic platform.
3.6 Generation of a human platelet proteome database and comparison with the rat platelet proteome
We have combined some of the platelet datasets available so far with our human proteomic dataset to construct a comprehensive human platelet protein database. Those datasets derived from pathological, toxicological or pharmacological platelet samples or individual protein analysis was not included in the final list. As a result, 1053 human platelet proteins were summarized from several reported results (Supplementary Table 2) [44, 62-65]. After comparison with our database, 114 novel proteins candidates were confidently identified using our integrated strategies (Supplementary Table 3 and 4).
A global analysis on the proteome level of the rat and human platelets was then performed, which mainly focused on physicochemical characteristics including hydrophobicity (HP) and pI distribution[66] and Gene Ontology (GO) annotations [67, 68]. Firstly, HP and pI distributions showed almost the same patterns (Supplementary Figure 3). According to previous findings[69], the multi-modality of pI distribution is a common feather in several known proteomes (e.g. E.coli, C. elegans, H. sapiens and M. musculus)[70]. Our data confirmed the modality, furthermore, revealed that the protein pIs in a sub-proteome or a specific organelle also follows the similar tend. The subcellular localization and functional distribution patterns (Figure 2) also showed high similarities between rat and human platelet proteins. To highlight the comparative profile, two more tissues, human plasma and heart (Supplementary S1, Supplementary Figure 4 and 5), were introduced into the analysis. Biologically, it was not surprising that the protein localization and functions showed clear tissue-specific distributions. No significant species-specific differences were observed between the human and rat platelet proteins.
Furthermore, we performed the pathway mapping analysis in comparison with human and rat platelet proteins as all identified platelet proteins were mapped to the pathway database by using a bioinformatics platform of KEGG (Kyoto Encyclopedia of Genes and Genomes) [71], which allows user input genes on static pathway maps generated by BioCarta and KEGG with correspondingly calculated p-value [72]. The pathways were ranked in the order of the number of mapped platelet proteins. The top 10 mapped pathways in human platelet were then compared with those mapped in rat platelets (Table 2). Eight of the top 10 significantly mapped pathways in human platelets were also found in the top 10 pathways in rat, except for the proteasome and apoptosis pathway. The high similarity between human and rat with minimal differences was a reflection of platelet evolution: ‘the stable core, and the variable shell’ [73].
Table 2. The top ten pathways mapped in the human platelet versus the corresponding ranking in the rat platelet proteome.
Rank denoted the mapped rank of the pathway in human or rat platelet; Count denoted the number of proteins matched to the particular pathway;
| Pathways | human | rat | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Rank | Count | p value a) | Rank | Count | p value | |
| Focal adhesion | 1 | 53 | 2.3E-08 | 2 | 36 | 7.8E-05 |
| Regulation of actin cytoskeleton | 2 | 50 | 2.0E-07 | 1 | 42 | 1.6E-07 |
| Leukocyte transendothelial migration | 3 | 28 | 2.3E-04 | 3 | 26 | 2.8E-05 |
| Insulin signaling pathway | 4 | 25 | 2.0E-02 | 4 | 21 | 8.4E-03 |
| Tight junction | 5 | 23 | 1.5E-02 | 6 | 19 | 2.8E-02 |
| GAP junction | 6 | 22 | 1.7E-03 | 9 | 14 | 5.8E-02 |
| Glycolysis/Gluconeogenesis | 7 | 22 | 1.8E-06 | 5 | 19 | 1.4E-07 |
| Proteasome | 8 | 20 | 3.7E-10 | 17 | 10 | 1.4E-04 |
| Apoptosis | 9 | 17 | 4.7E-02 | N/Ab) | N/A | N/A |
| Adherens junction | 10 | 17 | 1.3E-02 | 8 | 15 | 4.9E-03 |
value reflects a modified Fisher exact P value, the smaller means the more enriched.
N/A indicates the corresponding pathway was not significantly mapped.
3.7 Comparison of orthologous proteins
Orthologs are sequences of genes that evolved from a common ancestor and can be traced evolutionarily across different species [73]. Highly conserved sequences may suggest similar functions that are regulated by similar biochemical pathways and playing similar roles in different species [73, 74]. Therefore, with the availability of the rat and human platelet proteome, it is now possible to use orthologous proteins to inspect proteome evolution between species and to annotate newly discovered proteins.
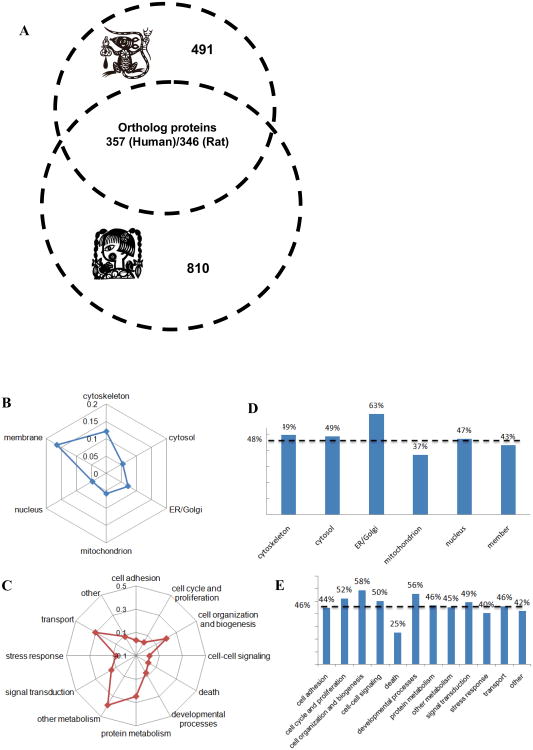
In the present study, based on the OrthoMCL database which contains the grouped ortholog protein sequences in a genome-scale across different species [75, 76], we extracted all orthologous pairs from human and rat platelet proteome. Such highly conserved protein sets represented the essential function and structural similarities of platelets between both species. As shown in Figure 6A, 386 orthologous pairs were found in human and rat platelet datasets, including 327 and 346 unique orthologous proteins for human and rat, respectively. According to the semi-quantitative results, more than 58% of orthologous pairs were high abundant, which indicated that the abundant proteins also play some fundamental roles even though they're almost always discarded by proteomics researchers. Furthermore, all orthologous proteins were categorized according to their subcellular localizations, molecular functions and cellular processes as mentioned above (Figure 6B and C). At least two interesting conclusions could be made according to the comparison of the orthologous with overall proteins. First, the distribution patterns based on localization and function of orthologous proteins were coincident with the distributions of the global profiles of platelet proteins (Figure 2). This raised the possibility that orthologous proteins represent the ‘core proteome’ within different species, which could conduct almost all the fundamental functions in platelets; second, the orthologous proteins in each category have similar percentages (versus the overall proteins in the same group, Figure 6D and E). These relatively even distributions (percentage of subcellular localizations, 48.0%±8.5%; percentage of molecular functions, 46.2%±8.6%) of orthologs in each locational and functional group provide us with new knowledge of natural selection, if any, that the ‘evolutionary proteome’ is a type of static proteome.
Figure 6. Bioinformatics analysis of orthologous proteins.
(A) The Venn plot (left) demonstrates the orthologous proteins between human and rat platelets. (B, C) All orthologous proteins in rat platelets are classified according to their subcellular localization and molecular functions. (D, E) The histogram demonstrates the ratios of the orthologous proteins in each category versus the overall proteins in the same category. The dotted lines show the average value of the distributions.
4 Concluding remarks
Due to the abundance-based nature of MS-based proteomics approaches, we selected rat as the model species for obtaining much more materials of platelets. Actually, compared with mouse, rat is a pathologically relevant model for investigating human cardiovascular disease [77, 78]. Meanwhile, rat platelet proteome hasn't been fully investigated yet since the availability of rat genome sequence from 2004 [79]. Here, we integrated a variety of approaches including multi-step protein extraction, various combinations of multidimensional LC-MS/MS schemes, and two MS/MS spectra interpretation algorithms to maximum the coverage of the rat platelet proteome. This integrated platform, validated by its successful application to the human platelets study, provided the scientific community an experimental and informational resource, especially a high quality rat platelet protein database and a combined human platelet protein database. Impressively, the newly constructed human platelet protein database demonstrated multidimensional information and new insights not visible within a single data set [80]. The first global comparison of human and rat platelet proteomes presented interesting knowledge of evolutionary proteomics, revealed some important features of basic cellular processes, and thus improve our understanding of the anucleated platelet proteins conserved among mammals. The data also show significant similarities between the rat and human platelet proteins, which highlights the use of the rat model for studying platelet-related thrombosis and cardiovascular diseases. Although our analysis of the platelet proteome was conducted for the resting state of platelets, our profiling datasets served as a starting point for molecular understanding the complex functions of platelets in mammals [80].
Supplementary Material
Acknowledgments
This work was supported by National Natural Science Foundation of China (20328508 and 90408023), National HighTec Research Developing Programme (02BAC11A11), Shanghai Sci.&Tech. Research Program (03DZ14024 and 04DZ14005). This research was also supported by US NIH 1R01AI064806-01A2, and the Office of Science (BER), U.S. Department of Energy, Grant No. DE-FG02-07ER64422. The authors thank Ye Lu and Chen Zhang for the discussion of animal experiments and platelet separation. Special thanks to Dr. Leslie Parise for her critical comments. Also the authors thank Andy Maglione and David Robinette for their helpful discussions.
Footnotes
Conflict of interest statement: The authors have declared no competing financial interests.
References
- 1.Ruggeri ZM. Platelets in atherothrombosis. Nat Med. 2002;8:1227–1234. doi: 10.1038/nm1102-1227. [DOI] [PubMed] [Google Scholar]
- 2.Mason KD, Carpinelli MR, Fletcher JI, Collinge JE, et al. Programmed Anuclear Cell Death Delimits Platelet Life Span. Cell. 2007;128:1173–1186. doi: 10.1016/j.cell.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 3.von Hundelshausen P, Weber C. Platelets as Immune Cells: Bridging Inflammation and Cardiovascular Disease. Circ Res. 2007;100:27–40. doi: 10.1161/01.RES.0000252802.25497.b7. [DOI] [PubMed] [Google Scholar]
- 4.Sambrano GR, Weiss EJ, Zheng Y, Huang W, Coughlin SR. Role of thrombin signalling in platelets in haemostasis and thrombosis. Nature. 2001;413:74–78. doi: 10.1038/35092573. [DOI] [PubMed] [Google Scholar]
- 5.Jackson SP. The growing complexity of platelet aggregation. Blood. 2007;109:5087–5095. doi: 10.1182/blood-2006-12-027698. [DOI] [PubMed] [Google Scholar]
- 6.Nash GF, Turner LF, Scully MF, Kakkar AK. Platelets and cancer. Lancet Oncol. 2002;3:425–430. doi: 10.1016/s1470-2045(02)00789-1. [DOI] [PubMed] [Google Scholar]
- 7.Andonegui G, Kerfoot SM, McNagny K, Ebbert KVJ, et al. Platelets express functional Toll-like receptor-4. Blood. 2005;106:2417–2423. doi: 10.1182/blood-2005-03-0916. [DOI] [PubMed] [Google Scholar]
- 8.Clark SR, Ma AC, Tavener SA, McDonald B, et al. Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat Med. 2007;13:463–469. doi: 10.1038/nm1565. [DOI] [PubMed] [Google Scholar]
- 9.Denis MM, Tolley ND, Bunting M, Schwertz H, et al. Escaping the nuclear confines: Signal-dependent Pre-mRNA splicing in anucleate platelets. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lesurtel M, Graf R, Aleil B, Walther DJ, et al. Platelet-Derived Serotonin Mediates Liver Regeneration. Science. 2006;312:104–107. doi: 10.1126/science.1123842. [DOI] [PubMed] [Google Scholar]
- 11.Kuebler WM. Selectins revisited: the emerging role of platelets in inflammatory lung disease. J Clin Invest. 2006;116:3106–3108. doi: 10.1172/JCI30664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shiraki R, Inoue N, Kawasaki S, Takei A, et al. Expression of Toll-like receptors on human platelets. Thromb Res. 2004;113:379–385. doi: 10.1016/j.thromres.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 13.Gupta GP, Massagué J. Cancer Metastasis: Building a Framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Boucharaba A, Serre CM, Gres S, Saulnier-Blache JS, et al. Platelet-derived lysophosphatidic acid supports the progression of osteolytic bone metastases in breast cancer. J Clin Invest. 2004;114:1714–1725. doi: 10.1172/JCI22123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boucharaba A, Serre CM, Guglielmi J, Bordet JC, et al. The type 1 lysophosphatidic acid receptor is a target for therapy in bone metastases. Proc Natl Acad Sci U S A. 2006;103:9643–9648. doi: 10.1073/pnas.0600979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gnatenko DV, Perrotta PL, Bahou WF. Proteomic approaches to dissect platelet function: half the story. Blood. 2006;108:3983–3991. doi: 10.1182/blood-2006-06-026518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macaulay IC, Carr P, Gusnanto A, Ouwehand WH, et al. Platelet genomics and proteomics in human health and disease. J Clin Invest. 2005;115:3370–3377. doi: 10.1172/JCI26885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia A, Watson SP, Dwek RA, Zitzmann N. Applying proteomics technology to platelet research. Mass Spectrom Rev. 2005;24:918–930. doi: 10.1002/mas.20047. [DOI] [PubMed] [Google Scholar]
- 19.Zahedi RP, Lewandrowski U, Wiesner J, Wortelkamp S, et al. Phosphoproteome of Resting Human Platelets. J Proteome Res. 2008;7:526–534. doi: 10.1021/pr0704130. [DOI] [PubMed] [Google Scholar]
- 20.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 21.Pasini EM, Kirkegaard M, Mortensen P, Lutz HU, et al. In-depth analysis of the membrane and cytosolic proteome of red blood cells. Blood. 2006;108:791–801. doi: 10.1182/blood-2005-11-007799. [DOI] [PubMed] [Google Scholar]
- 22.Pasini EM, Kirkegaard M, Salerno D, Mortensen P, et al. Deep Coverage Mouse Red Blood Cell Proteome: A First Comparison with the Human Red Blood Cell. Mol Cell Proteomics. 2008;7:1317–1330. doi: 10.1074/mcp.M700458-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Schrimpf SP, Weiss M, Reiter L, Ahrens CH, et al. Comparative Functional Analysis of the Caenorhabditis elegans and Drosophila melanogaster Proteomes. PLoS Biology. 2009;7:e48. doi: 10.1371/journal.pbio.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marcus K, Moebius J, Meyer HE. Differential analysis of phosphorylated proteins in resting and thrombin-stimulated human platelets. Anal Bioanal Chem. 2003;376:973–993. doi: 10.1007/s00216-003-2021-z. [DOI] [PubMed] [Google Scholar]
- 25.Garcia A, Prabhakar S, Hughan S, Anderson TW, et al. Differential proteome analysis of TRAP-activated platelets: involvement of DOK-2 and phosphorylation of RGS proteins. Blood. 2004;103:2088–2095. doi: 10.1182/blood-2003-07-2392. [DOI] [PubMed] [Google Scholar]
- 26.Claeys D, Geering K, Meyer BJ. Two-dimensional Blue Native sodium dodecyl sulfate gel electrophoresis for analysis of multimeric proteins in platelets. Electrophoresis. 2005;26:1189–1199. doi: 10.1002/elps.200406196. [DOI] [PubMed] [Google Scholar]
- 27.Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Mater. 2007;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 28.Cargile BJ, Talley DL, Stephenson JL. Immobilized pH gradients as a first dimension in shotgun proteomics and analysis of the accuracy of pI predictability of peptides. Electrophoresis. 2004;25:936–945. doi: 10.1002/elps.200305722. [DOI] [PubMed] [Google Scholar]
- 29.Krijgsveld J, Gauci S, Dormeyer W, Heck AJR. In-Gel Isoelectric Focusing of Peptides as a Tool for Improved Protein Identification. J Proteome Res. 2006;5:1721–1730. doi: 10.1021/pr0601180. [DOI] [PubMed] [Google Scholar]
- 30.Keller A, Eng J, Zhang N, Li X, Aebersold R. A uniform proteomics MS/MS analysis platform utilizing open XML file formats. Mol Syst Biol. 2005;1:E1–E8. doi: 10.1038/msb4100024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu CC, MacCoss MJ, Howell KE, Yates JR. A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003;21:532–538. doi: 10.1038/nbt819. [DOI] [PubMed] [Google Scholar]
- 32.He P, He HZ, Dai J, Wang Y, et al. The human plasma proteome: Analysis of Chinese serum using shotgun strategy. Proteomics. 2005;5:3442–3453. doi: 10.1002/pmic.200401301. [DOI] [PubMed] [Google Scholar]
- 33.Ross PL, Huang YLN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Garcia A, Senis YA, Antrobus R, Hughes CE, et al. A global proteomics approach identifies novel phosphorylated signaling proteins in GPVI-activated platelets: Involvement of G6f, a novel platelet Grb2-binding membrane adapter. Proteomics. 2006;6:5332–5343. doi: 10.1002/pmic.200600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Winkler W, Zellner M, Diestinger M, Babeluk R, et al. Biological Variation of the Platelet Proteome in the Elderly Population and Its Implication for Biomarker Research. Mol Cell Proteomics. 2008;7:193–203. doi: 10.1074/mcp.M700137-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Chen EI, Cociorva D, Norris JL, Yates JR. Optimization of Mass Spectrometry-Compatible Surfactants for Shotgun Proteomics. J Proteome Res. 2007;6:2529–2538. doi: 10.1021/pr060682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gao BB, Stuart L, Feener EP. Label-free Quantitative Analysis of One-dimensional PAGE LC/MS/MS Proteome: Application on Angiotensin II-Stimulated Smooth Muscle Cells Secretome. Mol Cell Proteomics. 2008;7:2399–2409. doi: 10.1074/mcp.M800104-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gilar M, Olivova P, Daly AE, Gebler JC. Orthogonality of Separation in Two-Dimensional Liquid Chromatography. Anal Chem. 2005;77:6426–6434. doi: 10.1021/ac050923i. [DOI] [PubMed] [Google Scholar]
- 39.Hojlund K, Yi Z, Hwang H, Bowen B, et al. Characterization of the Human Skeletal Muscle Proteome by One-dimensional Gel Electrophoresis and HPLC-ESI-MS/MS. Mol Cell Proteomics. 2008;7:257–267. doi: 10.1074/mcp.M700304-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qi B, Hardwick JM. A Bcl-xL Timer Sets Platelet Life Span. Cell. 2007;128:1035–1036. doi: 10.1016/j.cell.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 41.Patel SR, Hartwig JH, Italiano JE., Jr The biogenesis of platelets from megakaryocyte proplatelets. J Clin Invest. 2005;115:3348–3354. doi: 10.1172/JCI26891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coppinger JA, Cagney G, Toomey S, Kislinger T, et al. Characterization of the proteins released from activated platelets leads to localization of novel platelet proteins in human atherosclerotic lesions. Blood. 2004;103:2096–2104. doi: 10.1182/blood-2003-08-2804. [DOI] [PubMed] [Google Scholar]
- 43.Luo G, Ducy P, McKee MD, Pinero GJ, et al. Spontaneous calcification of arteries and cartilage in mice lacking matrix GLA protein. Nature. 1997;386:78–81. doi: 10.1038/386078a0. [DOI] [PubMed] [Google Scholar]
- 44.Martens L, Van Damme P, Van Damme J, Staes A, et al. The human platelet proteome mapped by peptide-centric proteomics: A functional protein profile. Proteomics. 2005;5:3193–3204. doi: 10.1002/pmic.200401142. [DOI] [PubMed] [Google Scholar]
- 45.Gevaert K, Goethals M, Martens L, Van Damme J, et al. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- 46.Ishihama Y, Oda Y, Tabata T, Sato T, et al. Exponentially Modified Protein Abundance Index (emPAI) for Estimation of Absolute Protein Amount in Proteomics by the Number of Sequenced Peptides per Protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-Scale Proteomic Analysis of the Human Spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 49.Schreiweis MA, Butler JP, Kulkarni NH, Knierman MD, et al. A proteomic analysis of adult rat bone reveals the presence of cartilage/chondrocyte markers. J Cell Biochem. 2007 doi: 10.1002/jcb.21196. [DOI] [PubMed] [Google Scholar]
- 50.Bhatt DL, Topol EJ. Scientific and therapeutic advances in antiplatelet therapy. Nat Rev Drug Discov. 2003;2:15–28. doi: 10.1038/nrd985. [DOI] [PubMed] [Google Scholar]
- 51.Hermeking H. The 14-3-3 cancer connection. Nat Rev Cancer. 2003;3:931–943. doi: 10.1038/nrc1230. [DOI] [PubMed] [Google Scholar]
- 52.Wheeler Jones C, Learmonth M, Martin H, Aitken A. Identification of 14-3-3 proteins in human platelets: effects of synthetic peptides on protein kinase C activation. Biochem J. 1996;315:41–47. doi: 10.1042/bj3150041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unwin RD, Smith DL, Blinco D, Wilson CL, et al. Quantitative proteomics reveals posttranslational control as a regulatory factor in primary hematopoietic stem cells. Blood. 2006;107:4687–4694. doi: 10.1182/blood-2005-12-4995. [DOI] [PubMed] [Google Scholar]
- 54.Azam M, Andrabi SS, Sahr KE, Kamath L, et al. Disruption of the Mouse m-Calpain Gene Reveals an Essential Role in Platelet Function. Mol Cell Biol. 2001;21:2213–2220. doi: 10.1128/MCB.21.6.2213-2220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu T, Donahue KC, Hu J, Kurnellas MP, et al. Identification of Differentially Expressed Proteins in Experimental Autoimmune Encephalomyelitis (EAE) by Proteomic Analysis of the Spinal Cord. J Proteome Res. 2007;6:2565–2575. doi: 10.1021/pr070012k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Franco SJ, Huttenlocher A. Regulating cell migration: calpains make the cut. J Cell Sci. 2005;118:3829–3838. doi: 10.1242/jcs.02562. [DOI] [PubMed] [Google Scholar]
- 57.Glading A, Lauffenburger DA, Wells A. Cutting to the chase: calpain proteases in cell motility. Trends Cell Biol. 2002;12:46–54. doi: 10.1016/s0962-8924(01)02179-1. [DOI] [PubMed] [Google Scholar]
- 58.Croce K, Flaumenhaft R, Rivers M, Furie B, et al. Inhibition of Calpain Blocks Platelet Secretion, Aggregation, and Spreading. J Biol Chem. 1999;274:36321–36327. doi: 10.1074/jbc.274.51.36321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Riddell DR, Graham A, Owen JS. Apolipoprotein E Inhibits Platelet Aggregation through the L-Arginine:Nitric Oxide Pathway. J Biol Chem. 1997;272:89–95. doi: 10.1074/jbc.272.1.89. [DOI] [PubMed] [Google Scholar]
- 60.Verdier-Pinard P, Pasquier E, Xiao H, Burd B, et al. Tubulin proteomics: Towards breaking the code. Anal Biochem. 2009;384:197–206. doi: 10.1016/j.ab.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwer HD, Lecine P, Tiwari S, Italiano JE, Jr, et al. A lineage-restricted and divergent beta.-tubulin isoform is essential for the biogenesis, structure and function of blood platelets. Curr Biol. 2001;11:579–586. doi: 10.1016/s0960-9822(01)00153-1. [DOI] [PubMed] [Google Scholar]
- 62.Senis YA, Tomlinson MG, Garcia A, Dumon S, et al. A comprehensive proteomics and genomics analysis reveals novel transmembrane proteins in human platelets and mouse megakaryocytes including G6b-B, a novel ITIM protein. Mol Cell Proteomics. 2007;6:548–564. doi: 10.1074/mcp.D600007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.García A, Prabhakar S, Brock CJ, Pearce AC, et al. Extensive analysis of the human platelet proteome by two-dimensional gel electrophoresis and mass spectrometry. Proteomics. 2004;4:656–668. doi: 10.1002/pmic.200300665. [DOI] [PubMed] [Google Scholar]
- 64.O'Neill EE, Brock CJ, von Kriegsheim AF, Pearce AC, et al. Towards complete analysis of the platelet proteome. Proteomics. 2002;2:288–305. doi: 10.1002/1615-9861(200203)2:3<288::aid-prot288>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 65.Garcia BA, Smalley DM, Cho HJ, Shabanowitz J, et al. The platelet microparticle proteome. J Proteome Res. 2005;4:1516–1521. doi: 10.1021/pr0500760. [DOI] [PubMed] [Google Scholar]
- 66.Taylor SW, Fahy E, Zhang B, Glenn GM, et al. Characterization of the human heart mitochondrial proteome. Nat Biotechnol. 2003;21:281–286. doi: 10.1038/nbt793. [DOI] [PubMed] [Google Scholar]
- 67.Ashburner M, Ball CA, Blake JA, Botstein D, et al. Gene Ontology: tool for the unification of biology. Nature Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Blake JA, Richardson JE, Bult CJ, Kadin JA, et al. The Mouse Genome Database (MGD): the model organism database for the laboratory mouse. Nucl Acids Res. 2002;30:113–115. doi: 10.1093/nar/30.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schwartz R, Ting CS, King J. Whole Proteome pI Values Correlate with Subcellular Localizations of Proteins for Organisms within the Three Domains of Life. Genome Res. 2001;11:703–709. doi: 10.1101/gr.gr-1587r. [DOI] [PubMed] [Google Scholar]
- 70.Wu S, Wan P, Li J, Li D, et al. Multi-modality of pI distribution in whole proteome. Proteomics. 2006;6:449–455. doi: 10.1002/pmic.200500221. [DOI] [PubMed] [Google Scholar]
- 71.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, et al. From genomics to chemical genomics: new developments in KEGG. Nucl Acids Res. 2006;34:D354–357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 73.Makarova KS, Aravind L, Galperin MY, Grishin NV, et al. Comparative Genomics of the Archaea (Euryarchaeota): Evolution of Conserved Protein Families, the Stable Core, and the Variable Shell. Genome Res. 1999;9:608–628. [PubMed] [Google Scholar]
- 74.Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- 75.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of Ortholog Groups for Eukaryotic Genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucl Acids Res. 2006;34:D363–368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abbott A. Laboratory animals: The Renaissance rat. Nature. 2004;428:464–466. doi: 10.1038/428464a. [DOI] [PubMed] [Google Scholar]
- 78.Russell JC. Of mice and men, rats and atherosclerosis. Cardiovasc Res. 2003;59:810–811. doi: 10.1016/s0008-6363(03)00530-3. [DOI] [PubMed] [Google Scholar]
- 79.Consortium RGSP. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- 80.Huh WK, Falvo JV, Gerke LC, Carroll AS, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.