Abstract
Background
The protection of canola (Brassica napus) crops against blackleg disease, caused by the fungal pathogen Leptosphaeria maculans, is largely mediated by race-specific resistance genes (R-genes). While many R-genes effective against blackleg disease have been identified in Brassica species, information of the precise genomic locations of the genes is limited.
Results
In this study, the Rlm2 gene for resistance to blackleg, located on chromosome A10 of the B. napus cultivar ‘Glacier’, was targeted for fine mapping. Molecular markers tightly linked to the gene were developed for use in mapping the resistance locus and defining the physical interval in B. napus. Rlm2 was localised to a 5.8 cM interval corresponding to approximately 873 kb of the B. napus chromosome A10.
Conclusion
The recently-cloned B. napus R-gene, LepR3, occupies the same region of A10 as Rlm2 and analysis of the putative B. napus and B. rapa genes in the homologous region identified several additional candidate defense-related genes that may control Rlm2 function.
Keywords: Blackleg, Brassica napus, Leptosphaeria maculans, Marker-assisted breeding, Molecular marker, PGIP-like protein, Receptor-like protein, Resistance
Background
When plants are under attack from fungal pathogens, they can often detect the secretion of fungal effector proteins, either directly or indirectly, by means of plant resistance (R) genes, which initiate a defense response known as effector-triggered immunity (ETI). ETI often causes localized cell death described as a hypersensitive response (HR) and prevents further infection [1,2]. R-genes are of vital importance in providing protection from plant pathogens, and an understanding of the relationship between race-specific R-genes and their corresponding pathogen avirulence (Avr) genes [3] is required for the effective deployment of resistance genetics in crop varieties. The hemibiotrophic fungal pathogen Leptosphaeria maculans (Desmaz.) Ces. & De Not. (anamorph: Phomalingam (Tode ex Fr.) Desmaz.) is the causal agent of blackleg disease; the most economically important disease of Brassica crops worldwide [4]. Eighteen major R-genes for blackleg disease have been identified in Brassica species, though several of these are probably redundant (reviewed in [5]). Most of the R-genes effective against blackleg described to date map to one of two chromosomes in the Brassica A genome; Rlm1, Rlm3, Rlm4, Rlm7 and Rlm9 form a cluster of R-genes on chromosome A07 [6-8], while LepR2 [9], BLMR2/RlmS ([10], Larkan et al. unpublished]) LepR3 [11] and Rlm2 [6] map to chromosome A10. Rlm2 was identified from the European Brassica napus (canola/rapeseed) variety ‘Glacier’ [12], which was later shown to contain two R-genes; Rlm2 and Rlm3 [13]. LepR3 was identified from the B. napus cultivar “Surpass 400” [14] and was reportedly introgressed into B. napus from B. rapa subsp. sylvestris [15,16]. LepR3 is to date the only R-gene for resistance to blackleg to be cloned. It encodes a receptor-like protein (RLP) which conveys resistance via HR during infection by L. maculans isolates expressing the AvrLm1 avirulence gene [11]. The gene ‘BnaA10g20720D’ on chromosome A10 of the newly-sequenced B. napus reference ‘Darmor-bzh’ genome [17], homologous to the B. rapa gene Bra008930 [18], shares 99% identity with the susceptible lepR3 allele of ‘Topas DH16516’ [11].
Rlm2 confers race-specific resistance to L. maculans isolates harbouring the corresponding avirulence gene AvrLm2 [12], which forms part of a genetic cluster of avirulence genes (AvrLm1-2-6) within the L. maculans genome [13]. Previously, the map location of Rlm2 was investigated using two resistant cultivars; ‘Glacier’ and ‘Samouraï’, and shown to be positioned in the same interval of LG16 (corresponding to chromosome A10), confirming the same gene was present in both varieties. The gene was positioned within an interval of 12 cM on the ‘Darmor’ x ‘Samouraï’ LG16 (DS.16) map [6], though the lack of genomic resources available at the time prevented physical definition of the gene interval. Rlm2 has been detected in many other European winter-type canola varieties, including one of the first blackleg-resistant cultivars ‘Ramsès’ [19], which was also used in the development of early Australian blackleg-resistant varieties [20]. Rlm2 has also been detected in B. rapa [21].
The aim of the current study was to define the precise map location of Rlm2, and identify tightly-linked markers for use in marker-assisted breeding programs. By aligning the B. napus map interval containing Rlm2 to the DNA sequence of B. rapa and B. napus for the corresponding region of chromosome A10 [17,18], we were able to physically define the interval containing the Rlm2 locus on chromosome A10, determine the position of Rlm2 relative to LepR3 and identify several candidate defense-related gene homologues.
Results
Phenotypic analysis
The L. maculans isolate ‘165’ was used to inoculate 12 seedlings of each parental line used to construct the BC1F1 population. Infection with the isolate resulted in test scores between 7 and 9 on ‘Topas’ (average score 7.7) and scores of 2 to 5 on ‘Glacier DH24287’ (average score 2.5). Further tests using additional control B. napus lines showed ‘165’ was avirulent on the Rlm2 line ‘Tapidor DH’ (2.1) and virulent on the Rlm3 variety ‘Quantum’ (9.0), the Rlm1/Rlm3 variety ‘Columbus’ (8.9), ‘Topas DH16516’ (9.0) and the Topas DH16516:LepR3 transgenic line ‘NLA8-2’ (9.0). These results confirmed the “avrLm1, AvrLm2, avrLm3” pathotype of the isolate (Figure 1).
Figure 1.

Interaction of L. maculans isolate ‘165’ with differential lines of B. napus . Cotyledons of seven B. napus lines pictured at 14-day post-inoculation. Lines containing Rlm2 (‘Glacier DH24287’ – Rlm2, Rlm3; ‘Tapidor DH’ – Rlm2) showed typical hypersensitive response and restriction of lesions while lines absent Rlm2 (‘Topas’ and ‘Topas DH16516’ – no blackleg resistance; ‘Quantum’- Rlm3; ‘Columbus’ – Rlm1, Rlm3; ‘NLA8-2’ – LepR3) were fully susceptible to infection.
Phenotypic screening of the 940 BC1F1 (‘Topas’ x ‘Glacier’) individuals showed segregation for the Rlm2 phenotype, with 478 seedlings scored as resistant to ‘165’ (scoring 2–5) and 462 seedlings scored as susceptible (7–9). This conformed to a 1:1 ratio (χ2 = 0.27, P = 0.602) as expected for a single dominant gene.
Rlm2 mapping
Twenty microsatellite markers spanning chromosome A10 were screened and a set of four markers (sR1448, sN8502, sN1982 & sN8474) that closely segregated with the Rlm2 locus were selected for genotyping the Topas x Glacier DH24287 BC1F1 population. These markers spanned the equivalent of 240 genes (Bra008783 - Bra009023) and a physical interval of approximately 1 Mb of the B. rapa genome [18]. The selected microsatellite markers were used in conjunction with the initial 218 BC1F1 individuals to produce a draft map of the Rlm2 interval using MapMaker v3.0b software [22] in order to confirm their linkage to the phenotype (LOD ≥ 4.0) and that they flanked the Rlm2 locus. The remaining BC1F1 individuals were then screened for recombination within the Rlm2 region. A total of 899 BC1F1 individuals where successfully genotyped with the flanking microsatellite markers.
Markers were re-run on the putative recombinant BC1F1 individuals and any non-confirmed recombinants discarded. After rescreening and phenotyping of the BC1F2 generation a total of 61 confirmed recombinants were retained for the informative recombinant mapping subset, each containing a single recombination event within the map interval.
The resulting map of ‘Topas’ x ‘Glacier’ A10 (TG.A10) showed Rlm2 was contained within an interval of 5.8 cM, between sN1982 and sN8474 (Figure 2). This interval corresponded to a collinear span of approximately 926 kb of the B. rapa genome, containing 204 putative genes on chromosome A10 (Bra008819 to Bra009023). The majority of the cross-overs detected in the population (52 of 61) occurred either between sN1982 and the Rlm2 locus (24 cross-overs), or between the Rlm2 locus and sN8474 (28 crossovers). Two additional markers were designed for the Rlm2 interval; one sequence characterised amplified region (SCAR) marker (Ind10-20), targeted to the Bra008930 homologue (previously identified as the LepR3 locus) and one cleaved amplified polymorphic sequence (CAPS) marker (CAPS94), targeted to the Bra008928 homologue. Ind10-20 produced fragments of 142 bp for ‘Topas’ and 138 bp for ‘Glacier’. CAPS94 produces amplicons of approximately 900 bp from both parents. Digestion with BstUI produces fragments of 483 and 410 bp from the ‘Glacier’ A-genome amplicon only. When used to genotype the recombinant subset of the mapping population, both of these markers co-segregated with the Rlm2 phenotype, providing markers tightly-linked to the Rlm2 gene yet failing to reduce the size of the target map interval. An additional nine microsatellite markers positioned within the map interval were tested but also failed to provide additional informative data.
Figure 2.
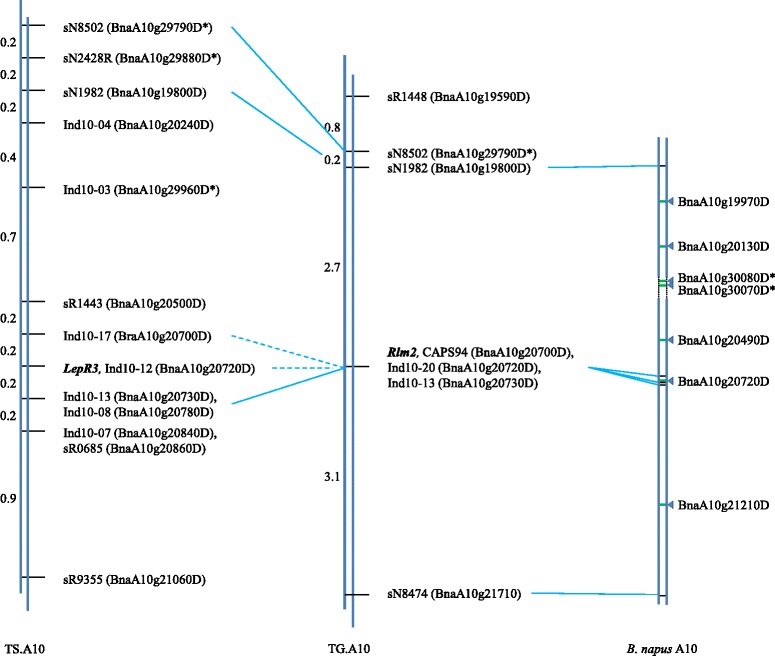
Comparison of genetic maps for Rlm2 and LepR3 to B. napus chromosome A10. Position of Rlm2 relative to microsatellite (prefixed ‘sN’ or ‘sR’), Indel (prefixed ‘Ind’) and CAPS markers on ‘Topas’ x ‘Glacier’ A10 (TG.A10) map. Nearest B. napus gene to each marker given in brackets. Unanchored B. napus genes corresponding to syntenic B. rapa A10 homologues are denoted with “*”. LepR3 ‘Topas’ x ‘Surpass 400’ A10 (TS.A10) map updated from Larkan et al., 2013a. Marker intervals given in centiMorgans. Solid horizontal lines denote same marker, dashed horizontal lines denote markers sharing same nearest B. napus homologue. Approximate physical location of defense-related candidate genes by triangles. Gap in B. napus A10 chromosome build represented by dotted lines.
Comparison of the Rlm2 and LepR3 maps showed that both genes were located within the same genetic interval on chromosome A10. While one additional marker was integrated from the LepR3 map (Ind10-13, corresponding to Bra008931), this also co-segregated with the Rlm2 phenotype. All remaining markers used in the LepR3 map were non-polymorphic in the ‘Topas’ x ‘Glacier’ population. The maps share three common markers and two other markers that have a common closest B. napus gene, and the cluster of markers that co-segregate with Rlm2 span the LepR3 locus (Figure 2).
Candidate gene selection
Inspection of the 204 genes contained within the region of B. rapa A10 collinear to the Rlm2 map interval lead to the identification of seven candidate gene homologues, selected on the basis of their potential roles in resistance to microbial pathogens. Two of the candidates (Bra008836, Bra008851) are homologous to A. thaliana genes involved in disease resistance signaling during infection by the bacterial pathogen Pseudomonas syringae [23]. The interval also contained Bra008930, previously identified as the B. rapa homologue of the B. napus blackleg resistance gene LepR3 [11], and four other genes (Bra008869, Bra008870, Bra008910 & Bra008977) of unknown function in A. thaliana that are members of gene families involved in plant resistance responses (Table 1). Two of these, members of the Leucine Rich-Repeat (LRR) Protein family (Bra008869 & Bra008870) were examined using InterPro 5 and LRRfinder 2.0 and were both predicted to encode small (374 and 373 amino acids, respectively) proteins with predicted primary structures featuring signal peptides, LRR N-terminal domains, seven extracellular LRR domains and potential C-terminal LRR domains, similar to members of the plant defense-related polygalacturonase inhibitor protein (PGIP) family [24,25]. BLAST analysis showed these proteins shared 60% identity with PGIP-like family members from poplar (Populus trichocarpa).
Table 1.
Candidate defense-related B. napus genes identified within A10 region corresponding to the Rlm2 map interval
| B. napus gene | A. thaliana homologue | A. thaliana description | Role |
|---|---|---|---|
| BnaA10g19970D | At5g13320 | PBS3 (avrPphB susceptible 3) | P.syringae defense response |
| BnaA10g20130D | At5g13160 | PBS1 (avrPphB susceptible 1) | P.syringae defense response |
| BnaA10g30070D* | At5g12940 | LRR Family Protein | Suppression of polygalacturonase? |
| BnaA10g30080D* | At5g12940 | LRR Family Protein | Suppression of polygalacturonase? |
| BnaA10g20490D | At5g12180 | CPK17 (Calcium-dependant protein kinase 17) | Unknown |
| BnaA10g20720D (lepR3) | At3g05650 | AtRLP32 (Receptor-like Protein 32) | Homologue of B. napus gene LepR3 |
| BnaA10g21210D | At5g11250 | Disease Resistance Protein (TIR-NBS-LRR) | Unknown |
Genes denoted with “*” are not anchored to B. napus A10 yet correspond to syntenic B. rapa A10 homologues.
Alignment of the Rlm2 map interval to the B. napus after the release of the ‘Darmor-bzh’ reference genome [17] revealed a physical interval of approximately 873 kb spanning 191 genes (BnaA10g19800 – BnaA10g21710) containing syntenic homologues of 5 of the 7 candidate genes previously identified in B. rapa. The two ‘missing’ B. rapa homologues (PGIP-like genes Bra008869 & Bra008870) had been assigned to an unanchored “chrA10_random” pseudo-molecule, with the collinear B. napus A10 region spanning the genes in B. rapa being represented by a gap of approximately 43 kb (Figure 2, Table 1).
Discussion
We were able to define the Rlm2 locus to a map interval of 5.8 cM and a physical interval of 191 predicted B. napus genes, though further dissection of the Rlm2 interval was hampered by a lack of polymorphism, as many additional markers located within the region proved to be monomorphic in our population. We could make efforts to produce additional markers targeted to the interval, however the recent advent of high-density marker systems for B. napus, such as DArT [26], Illumina Infinium 6 K [27,28] or 60 K ([29], Isobel Parkin, AAFC Saskatoon, unpublished data]) SNP arrays, or genotyping-by-sequencing methods [30] makes the pursuit of further specific sequence-characterised markers impractical. While we have provided several markers tightly-linked to the Rlm2 locus which could be of use in blackleg resistance breeding programs, the real value of the work is in defining the physical location of Rlm2 on chromosome A10. With this information any sequence-characterised marker set, including the high-density systems, can be orientated to the map in order to identify markers linked to the physical location of the gene.
The co-localisation of the blackleg resistance genes LepR3 and Rlm2 in the B. napus genome reported here draws an interesting comparison to the clustering of their corresponding avirulence genes, AvrLm1 and AvrLm2, in the L. maculans genome [13]. Molecular characterisation of Rlm2 and AvrLm2 and comparison to the previously-identified LepR3 [11] and AvrLm1 [31] may provide some insight into the evolution of the molecular ‘arms race’ between plant and pathogen. We identified seven potential resistance-related homologues in the regions of B. rapa and B. napus genomes corresponding to the map interval containing Rlm2 (Table 1). One of these genes, BnaA10g20720D, corresponds to the LepR3 blackleg-resistance locus and was targeted in this study by the SCAR marker Ind10-20. This marker co-segregated with the resistance phenotype, as did two other markers corresponding to neighbouring genes (Figure 2). This suggests that Rlm2 is located close to this region of the chromosome however, based on our results, we cannot rule out other candidate genes within the wider map interval (BnaA10g19800 – BnaA10g21710). These include a member of the Calcium-dependant protein kinase (CPK or CDPK) family (BnaA10g20490D). CPKs have been shown to be positive regulators of race-specific pathogen defense in several plant species [32-35]. They act to facilitate HR during ETI-mediated defense responses after translocation from the cytosol to the nucleus, where they activate WRKY transcription factors [36]. Other candidates include a member of the TIR-NBS-LRR class of resistance genes (BnaA10g21210D), which are well established as initiators of plant resistance responses during both direct and indirect interactions with pathogen effectors [37-40], and two genes (BnaA10g20130D & BnaA10g19970D) homologous to A. thaliana genes PBS1 and PBS3, respectively, involved in the P. syringae resistance response [23]. PBS1 encodes a receptor-like cytoplasmic kinase targeted for cleavage by the P. syringae effector AvrPphB, which triggers the CC-NBS-LRR class resistance protein RPS5 [41,42]. The final two candidate genes, BnaA10g30070D and BnaA10g30080D, encode putative PGIP-like proteins. PGIPs are small LRR-containing proteins that are secreted from the host cell into the apoplastic fluid where they inhibit host cell wall degradation by binding to pathogen endopolygalacturonases during infection by fungi [25], nematodes [43] and other plant pathogens. Though BnaA10g30070D and BnaA10g30080D were not incorporated in the main A10 chromosome build of the initial B. napus genome release, their B. rapa (Bra008869 & Bra008870) and A. thaliana (both match At5g12940) homologues are located in regions syntenic to the Rlm2 map interval in their respective genomes [18,44].
The genomic interval containing Rlm2 in the B. napus variety ‘Samouraï’ [6] was also shown to harbour a QTL for L. maculans resistance during field trials in France [6,45] despite the absence of the corresponding avirulence gene AvrLm2 in French isolates [19]. It was suggested that either Rlm2 has a residual effect on avrLm2 L. maculans isolates, or that another genetic factor limiting growth of the pathogen was linked to the Rlm2 locus [6]. A number of the candidate defense-related genes identified here as being linked to the Rlm2 locus could also be of interest as candidates in non-race specific adult-plant resistance. In particular, the interaction of plant PGIPs with pathogen polygalacturonases inhibits the degradation of the host cell wall and can also lead to accumulation of non-specific defense responses such as lignification and the production of reactive oxygen species [46]. Fungal polygalacturonases have been shown to accumulate in B. napus stems during infection by L. maculans and may play an important role in the development of canker lesions [47] though expression of the L. maculans polygalacturonase-encoding pg1 gene was not detected during cotyledon and leave infection [48]. If the candidate genes we identified do indeed encode functional PGIP proteins then they could potentially play a role in the suppression of blackleg disease in the adult plant.
The detailed genetic map and physical location of the Rlm2 gene presented here should aid in the marker-assisted breeding of the gene into modern B. napus varieties. While Rlm2 is of little use in Europe, due to the absence of the matching AvrLm2 avirulence gene in most European L. maculans populations [49,50], it is a valuable source of resistance to blackleg disease in Canada. A survey of western Canadian isolates showed that 100% of isolates collected between 1997 and 2000, and 93.9% of isolates collected between 2003 and 2005, harboured AvrLm2. This high frequency was unexpected, as Rlm2 has been available to Canadian plant breeders for some time and a greater adaptation by the pathogen was expected [51] suggesting Rlm2 has not yet been widely deployed in Canadian B. napus varieties.
While Rlm2 is potentially valuable in developing blackleg resistance canola varieties in North America, breeders must be cautious in its deployment. The lack of AvrLm2 in current European L. maculans populations stands in stark contrast to the effectiveness of Rlm2 in controlling blackleg in Europe several decades ago. High selection pressure has resulted in a dynamic evolution of race structure within the pathogen populations of Europe, leading to the sequential loss of Rlm2, Rlm4 and Rlm1-mediated resistance [19,51,52]. A similar situation could develop in North America; an earlier survey of L. maculans isolates in southern Ontario, a winter-type canola growing region geographically isolated from the prairies of western Canada, found no AvrLm2 isolates [53]. Kutcher et al. (2010) noted that the only avrLm2 isolates detected in their survey were collected in southern Manitoba in 2003–2005. Another survey also found avrLm2 isolates both in Alberta, Manitoba and directly south across the Canada/US border in North Dakota [54] and a recent report suggests nearly all L. maculans in North Dakota would be virulent on Rlm2 varieties [55]. Clearly relying on Rlm2 as the single resistance source for a canola variety would be foolhardy. Proper stewardship of the gene would entail pyramiding the resistance by combining Rlm2 with other effective R-genes and/or quantitative resistance genetics [56].
Conclusions
We are presented with three scenarios as to the identity of Rlm2; 1) the co-localisation of Rlm2 and LepR3 is due to the genes being allelic variants of the same gene locus (BnaA10g20720D), 2) Rlm2 corresponds to one of the other B. napus candidate defense-related homologues identified within the syntenic map interval, or 3) Rlm2 is a gene specific only to certain varieties of B. napus and not represented in the B. napus ‘Darmor-bzh’ or B. rapa var. ‘Chiifu’ genome sequences. Investigation of the candidate genes identified in this study is currently underway. Regardless of the molecular identity of Rlm2, delimiting the physical region of the B. napus genome that harbours the gene provides the information required for the efficient marker-assisted selection of Rlm2 in modern canola breeding programs.
Methods
Mapping population
For the mapping of Rlm2, a BC1F1 population segregating for the Rlm2 phenotype was produced by first creating F1 plants via a cross between the susceptible B. napus variety ‘Topas’ and the resistant B. napus doubled-haploid line ‘Glacier DH24287’ (Rlm2, Rlm3). A single F1 seedling was vernalised (4°C) for 8 weeks to ensure flowering, then backcrossed to ‘Topas’ to produce BC1F1 seeds.
Phenotypic analysis
Seedlings were germinated in 96-cell trays containing an artificial soil mix [57] in a controlled growth chamber (20°C, 16 h days, light intensity c. 450 μmol m−2 s−1 at bench level, and 18°C, 8 h nights). The cotyledons of 7 day-old seedlings were inoculated with a pycnidiospore suspension of L. maculans isolate ‘165’ (avrLm1, AvrLm2, avrLm3) from the Rimmer Collection, AAFC Saskatoon, which is virulent on ‘Topas’ (no effective blackleg resistance) and avirulent on ‘Glacier DH24287’ (Rlm2, Rlm3). A small wound was made in the centre of each cotyledon lobe and 10 μL of 2×107 spores/mL suspension was applied to each wound (4 infection sites per seedling). The resistance phenotype of the seedlings was rated at 14 days post-infection using a 0–9 scale [58]; where ratings of 0–4 (induced HR) are classified as ‘resistant’, 5 as ‘intermediate’ and 6–9 (no HR) as ‘susceptible’. To confirm that the resistance screening of the BC1F1 population was detecting the Rlm2 gene and not Rlm3 or LepR3, the “avrLm1, Avrlm2, avrLm3” pathotype of ‘165’ was tested using 4–8 seedlings each (8–16 cotyledons per test) of the additional B. napus control lines ‘Topas DH16516’ (a doubled-haploid line of ‘Topas’), ‘Tapidor DH’ (Rlm2), ‘Columbus’ (Rlm1, Rlm3), ‘Quantum’ (Rlm3) and the transgenic line ‘NLA8-2’ (LepR3).
Marker selection
An initial group of 218 BC1F1 individuals were assessed for their reaction to the L. maculans isolate ‘165’. DNA was extracted using DNeasy 96 Plant Kits (QIAGEN Inc., USA) or as described by Larkan et al. (2013). Thirty BC1F1 individuals (15 resistant, 15 susceptible) were selected in order to screen microsatellite markers (http://aafc-aac.usask.ca/BrassicaMAST/) spanning chromosome A10, based on the previously described map location of Rlm2 [6]. This identified markers that were both polymorphic in the population and linked to the Rlm2 locus. Microsatellite marker reactions were performed as described by [59] and genotyping was performed using a MegaBACE capillary sequencer (GE Health, Canada).
Fine mapping
Further screening was performed to expand the mapping population; in total of 940 BC1F1 seedlings were phenotyped for their reaction to ‘165’and screened for recombination between the Rlm2-flanking markers. Putative recombinant individuals were selected, vernalised and allowed to set selfed seed (BC1F2).
To confirm the Rlm2 phenotype and marker genotypes for the recombinant BC1F1 selections, between 8 and 12 BC1F2 seedlings per line were infected with ‘165’, and DNA produced from bulked BC1F2 tissue was used to confirm the genotypes for each marker. To enrich the map, an additional SCAR marker (“Ind10-20”), designed and run as previously described [11], was targeted to the LepR3 locus (Bra008890) using the following primers; F - TTCGTGATGAGTTTGCGGTTC, R -CAGTCGCTGTTATTCACCCATGA. Additionally, a single CAPS marker, “CAPS94”, was designed and added to the map, taking advantage of a differential restriction site for the enzyme BstUI in the Glacier DH24287 homologue of Bra008928. CAPS alleles were PCR amplified (95°C, 5:00; (95°C, 0:30; 55°C, 0:30; 72°C, 1:00) x 35 cycles; 72°C, 10:00; F primer – CCTTCCTGGGGGAAAACTAA, R primer - TCGTAGCGTTTCCTCCAAAC) using AmpliTaq Gold Master Mix (Life Technologies, USA) before digestion with BstUI restriction enzyme (New England Biolabs, USA). Digested CAPS products were resolved on 1.5% agarose gels.
A final linkage map of the Rlm2 interval was constructed after placing markers in order based on their homologous B. rapa position and integrating the Rlm2 phenotypic data. Map distances were calculated manually (cMBC = (x/n) *100, where x = recombination events and n = total population size) and parsimonious marker order was confirmed using the MAP function of QTL IciMapping v3.2 software [60].
Comparison to LepR3 map and identification of Rlm2 candidate homologues in B. rapa and B. napus
During the mapping process, performed concurrently with the mapping of LepR3 [11], it became apparent that both genes mapped to a similar region of chromosome A10. Additional markers from the LepR3 map were tested for polymorphism in the ‘Topas’ x ‘Glacier’ population and integrated into the Rlm2 map. Markers common to both studies were used to align the maps and assess the relative positions of the genes.
For each marker placed on the map, the sequence used to create the marker was initially matched to its homologous region of the B. rapa genome [18] using BLASTN [61] in the BRAD Brassica Database portal [62] and the matching or closest B. rapa gene recorded. Homologous genes occurring within the Rlm2 map interval were examined and pathogen defense-related genes or gene family members were considered as candidates. Marker positions and candidate genes were later reassessed in relation to the recently-released B. napus genome [17]. Additional annotation of candidate proteins was performed using the online tools InterProScan 5 [63] and LRRfinder 2.0 [64].
Acknowledgements
The authors would like to thank S. Kuzmicz, C. Hammond, C. Guenther, S. McMillan, K. Tomporowski and J. Albert for technical assistance. This work was funded by the AAFC-Industry Blackleg Consortium II (partners include Bayer CropScience, Crop Production Services, Department of Environment and Primary Industries Victoria, Lantmännen Lantbruk, Pacific Seeds and Rapool-Ring GmbH).
Abbreviations
- CAPS
Cleaved amplified polymorphic sequence
- LRR
Leucine-rich repeat
- NBS
Nucleotide binding site
- PCR
Polymerase chain reaction
- PGIP
Polygalacturonase inhibitor protein
- SCAR
Sequence characterised amplified region
Footnotes
S Roger Rimmer deceased.
Competing interests
The authors declare no competing interests in regards to the work presented here.
Authors’ contributions
DL, FY and SR conceived of the study. NL and FY carried out the molecular genetic studies. NL performed the genomic analysis and drafted the manuscript. DL, SR and MB participated in the project coordination. DL and MB helped to draft the manuscript. All authors except SR (deceased) read and approved the final manuscript.
Contributor Information
Nicholas J Larkan, Email: Nicholas.Larkan@agr.gc.ca.
Derek J Lydiate, Email: Derek.Lydiate@agr.gc.ca.
Fengqun Yu, Email: Fengqun.Yu@agr.gc.ca.
S Roger Rimmer, Email: Hossein.Borhan@agr.gc.ca.
M Hossein Borhan, Email: Hossein.Borhan@agr.gc.ca.
References
- 1.Ali S, Bakkeren G. Fungal and oomycete effectors - strategies to subdue a host. Can J Plant Pathol. 2011;33(4):425–446. doi: 10.1080/07060661.2011.625448. [DOI] [Google Scholar]
- 2.Catanzariti AM, Jones DA. Effector proteins of extracellular fungal plant pathogens that trigger host resistance. Funct Plant Biol. 2010;37(10):901–906. doi: 10.1071/FP10077. [DOI] [Google Scholar]
- 3.Flor HH. Current status of the gene-for-gene concept. Annu Rev Phytopathol. 1971;9:275–296. doi: 10.1146/annurev.py.09.090171.001423. [DOI] [Google Scholar]
- 4.Fitt BDL, Brun H, Barbetti MJ, Rimmer SR. World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus) Eur J Plant Pathol. 2006;114(1):3–15. doi: 10.1007/s10658-005-2233-5. [DOI] [Google Scholar]
- 5.Raman H, Raman R, Larkan N: Genetic dissection of blackleg resistance loci in rapeseed (Brassica napusL.). In: Plant Breeding from Laboratories to Fields. Edited by Andersen SB. Rijeka, Croatia: InTech; 2013: 85–120.
- 6.Delourme R, Pilet-Nayel ML, Archipiano M, Horvais R, Tanguy X, Rouxel T, Brun H, Renard M, Balesdent MH. A cluster of major specific resistance genes to Leptosphaeria maculans in Brassica napus. Phytopathology. 2004;94(6):578–583. doi: 10.1094/PHYTO.2004.94.6.578. [DOI] [PubMed] [Google Scholar]
- 7.Raman R, Taylor B, Lindbeck K, Coombes N, Barbulescu D, Salisbury P, Raman H. Molecular mapping and validation of Rlm1 gene for resistance to Leptosphaeria maculans in canola (Brassica napus L.) Crop Pasture Sci. 2012;63(10):1007–1017. doi: 10.1071/CP12255. [DOI] [Google Scholar]
- 8.Raman R, Taylor B, Marcroft S, Stiller J, Eckermann P, Coombes N, Rehman A, Lindbeck K, Luckett D, Wratten N, Batley J, Edwards D, Wang X, Raman H. Molecular mapping of qualitative and quantitative loci for resistance to Leptosphaeria maculans causing blackleg disease in canola (Brassica napus L.) Theor Appl Genet. 2012;125(2):405–418. doi: 10.1007/s00122-012-1842-6. [DOI] [PubMed] [Google Scholar]
- 9.Yu F, Lydiate DJ, Gugel RK, Sharpe AG, Rimmer SR. Introgression of Brassica rapa subsp. sylvestris blackleg resistance into B. napus. Mol Breed. 2012;30(3):1495–1506. doi: 10.1007/s11032-012-9735-6. [DOI] [Google Scholar]
- 10.Long Y, Wang Z, Sun Z, Fernando DWG, McVetty PBE, Li G. Identification of two blackleg resistance genes and fine mapping of one of these two genes in a Brassica napus canola cultivar 'Surpass 400'. Theor Appl Genetics. 2011;122(6):1223–1231. doi: 10.1007/s00122-010-1526-z. [DOI] [PubMed] [Google Scholar]
- 11.Larkan NJ, Lydiate DJ, Parkin IAP, Nelson MN, Epp DJ, Cowling WA, Rimmer SR, Borhan MH. The Brassica napus blackleg resistance gene LepR3 encodes a receptor-like protein triggered by the Leptosphaeria maculans effector AVRLM1. New Phytol. 2013;197(2):595–605. doi: 10.1111/nph.12043. [DOI] [PubMed] [Google Scholar]
- 12.Ansan-Melayah D, Balesdent MH, Delourme R, Pilet ML, Tanguy X, Renard M, Rouxel T. Genes for race-specific resistance against blackleg disease in Brassica napus L. Plant Breed. 1998;117(4):373–378. doi: 10.1111/j.1439-0523.1998.tb01956.x. [DOI] [Google Scholar]
- 13.Balesdent MH, Attard A, Kuhn ML, Rouxel T. New avirulence genes in the phytopathogenic fungus Leptosphaeria maculans. Phytopathology. 2002;92(10):1122–1133. doi: 10.1094/PHYTO.2002.92.10.1122. [DOI] [PubMed] [Google Scholar]
- 14.Li CX, Cowling WA. Identification of a single dominant allele for resistance to blackleg in Brassica napus 'Surpass 400'. Plant Breed. 2003;122(6):485–488. doi: 10.1111/j.1439-0523.2003.00863.x. [DOI] [Google Scholar]
- 15.Buzza G, Easton A: A new source of blackleg resistance fromBrassica sylvestris. In GCIRC Technical Meeting: June 2002. Poznan, Poland: GCIRC Bulletin; 2002:B18.
- 16.Crouch JH, Lewis BG, Mithen RF. The effect of A genome substitution on the resistance of Brassica napus to infection by Leptosphaeria maculans. Plant Breed. 1994;112:265–278. doi: 10.1111/j.1439-0523.1994.tb00684.x. [DOI] [Google Scholar]
- 17.Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science. 2014;345(6199):950–953. doi: 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- 18.Wang X, Wang H, Wang J, Sun R, Wu J, Liu S, Bai Y, Mun J, Bancroft I, Cheng F, Huang S, Li X, Hua W, Wang J, Wang X, Freeling M, Pires JC, Paterson AH, Chalhoub B, Wang B, Hayward A, Sharpe AG, Park B, Weisshaar B, Liu B, Li B. The genome of the mesopolyploid crop species Brassica rapa. Nat Genet. 2011;43(10):1035–1039. doi: 10.1038/ng.919. [DOI] [PubMed] [Google Scholar]
- 19.Rouxel T, Willner E, Coudard L, Balesdent MH. Screening and identification of resistance to Leptosphaeria maculans (stem canker) in Brassica napus accessions. Euphytica. 2003;133(2):219–231. doi: 10.1023/A:1025597622490. [DOI] [Google Scholar]
- 20.Salisbury P, Wratten N: Brassica napusBreeding. In: Canola in Australia - The First Thirty Years. Edited by Salisbury PA, Potter TD, McDonald G, Green AG. Gosford, Australia: The Regional Institute Ltd.; 1999: 29–36.
- 21.Leflon M, Brun H, Eber F, Delourme R, Lucas MO, Vallée P, Ermel M, Balesdent MH, Chèvre AM. Detection, introgression and localization of genes conferring specific resistance to Leptosphaeria maculans from Brassica rapa into B. napus. Theor Appl Genet. 2007;115(7):897–906. doi: 10.1007/s00122-007-0616-z. [DOI] [PubMed] [Google Scholar]
- 22.Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L. MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics. 1987;1(2):174–181. doi: 10.1016/0888-7543(87)90010-3. [DOI] [PubMed] [Google Scholar]
- 23.Warren RF, Holub E, Innes RW. Identification of three putative signal transduction genes involved in R gene-specified disease resistance in Arabidopsis. Genetics. 1999;152(1):401–412. doi: 10.1093/genetics/152.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Matteo A, Bonivento D, Tsernoglou D, Federici L, Cervone F. Polygalacturonase-inhibiting protein (PGIP) in plant defence: a structural view. Phytochemistry. 2006;67(6):528–533. doi: 10.1016/j.phytochem.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 25.Toubart P, Desiderio A, Salvi G, Cervone F, Daroda L, De Lorenzo G, Bergmann C, Darvill AG, Albersheim P. Cloning and characterization of the gene encoding the endo polygalacturonase-inhibiting protein (PGIP) of Phaseolus vulgaris L. Plant J. 1992;2(3):367–373. doi: 10.1046/j.1365-313x.1992.t01-35-00999.x. [DOI] [PubMed] [Google Scholar]
- 26.Raman H, Raman R, Kilian A, Detering F, Long Y, Edwards D, Parkin IAP, Sharpe AG, Nelson MN, Larkan N, Zou J, Meng J, Aslam MN, Batley J, Cowling WA, Lydiate D. A consensus map of rapeseed (Brassica napus L.) based on diversity array technology markers: Applications in genetic dissection of qualitative and quantitative traits. BMC Genomics. 2013;14(1):277. doi: 10.1186/1471-2164-14-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delourme R, Falentin C, Fomeju BF, Boillot M, Lassalle G, André I, Duarte J, Gauthier V, Lucante N, Marty A, Pauchon M, Pichon JP, Ribière N, Trotoux G, Blanchard P, Rivière N, Martinant JP, Pauquet J: High-density SNP-based genetic map development and linkage disequilibrium assessment inBrassica napusL.BMC Genomics 2013, 14(1). [DOI] [PMC free article] [PubMed]
- 28.Raman H, Dalton-Morgan J, Diffey S, Raman R, Alamery S, Edwards D, Batley J. SNP markers-based map construction and genome-wide linkage analysis in Brassica napus. Plant Biotechnol J. 2014;12(7):851–860. doi: 10.1111/pbi.12186. [DOI] [PubMed] [Google Scholar]
- 29.Clarke WE, Parkin IA, Gajardo HA, Gerhardt DJ, Higgins E, Sidebottom C, Sharpe AG, Snowdon RJ, Federico ML, Iniguez-Luy FL: Genomic DNA enrichment using sequence capture microarrays: a novel approach to discover sequence nucleotide polymorphisms (SNP) inBrassica napusL.PLoS ONE 2013, 8(12):e81992. [DOI] [PMC free article] [PubMed]
- 30.Raman H, Raman R, Kilian A, Detering F, Carling J, Coombes N, Diffey S, Kadkol G, Edwards D, McCully M, Ruperao P, Parkin IAP, Batley J, Luckett DJ, Wratten N: Genome-wide delineation of natural variation for pod shatter resistance inBrassica napus. PLoS ONE 2014, 9(7):e101673. [DOI] [PMC free article] [PubMed]
- 31.Gout L, Fudal I, Kuhn M-L, Blaise F, Eckert M, Cattolico L, Balesdent M-H, Rouxel T. Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol. 2006;60(1):67–80. doi: 10.1111/j.1365-2958.2006.05076.x. [DOI] [PubMed] [Google Scholar]
- 32.Fu L, Yu X, An C. OsCPK20 positively regulates Arabidopsis resistance against Pseudomonas syringae pv. tomato and rice resistance against Magnaporthe grisea. Acta Physiol Plant. 2014;36(2):273–282. doi: 10.1007/s11738-013-1408-5. [DOI] [PubMed] [Google Scholar]
- 33.Gao X, Chen X, Lin W, Chen S, Lu D, Niu Y, Li L, Cheng C, McCormack M, Sheen J, Shan L, He P: Bifurcation ofArabidopsisNLR Immune Signaling via Ca2 + −Dependent Protein Kinases. PLoS Pathogens 2013, 9(1):e1003127. [DOI] [PMC free article] [PubMed]
- 34.Kobayashi M, Yoshioka M, Asai S, Nomura H, Kuchimura K, Mori H, Doke N, Yoshioka H. StCDPK5 confers resistance to late blight pathogen but increases susceptibility to early blight pathogen in potato via reactive oxygen species burst. New Phytol. 2012;196(1):223–237. doi: 10.1111/j.1469-8137.2012.04226.x. [DOI] [PubMed] [Google Scholar]
- 35.Romeis T, Ludwig AA, Martin R, Jones JDG. Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 2001;20(20):5556–5567. doi: 10.1093/emboj/20.20.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gao X, He P. Nuclear dynamics of Arabidopsis calcium-dependent protein kinases in effector-triggered immunity. Plant Signal Behav. 2013;8(4):e238681–e238685. doi: 10.4161/psb.23868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belkhadir Y, Subramaniam R, Dangl JL. Plant disease resistance protein signaling: NBS-LRR proteins and their partners. Curr Opin Plant Biol. 2004;7(4):391–399. doi: 10.1016/j.pbi.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Borhan MH, Holub EB, Kindrachuk C, Omidi M, Bozorgmanesh-Frad G, Rimmer SR. WRR4, a broad-spectrum TIR-NB-LRR gene from Arabidopsis thaliana that confers white rust resistance in transgenic oilseed brassica crops: short communication. Mol Plant Pathol. 2010;11(2):283–291. doi: 10.1111/j.1364-3703.2009.00599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deslandes L, Olivier J, Peeters N, Feng DX, Khounlotham M, Boucher C, Somssich I, Genin S, Marco Y. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A. 2003;100(13):8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dodds PN, Lawrence GJ, Catanzariti A-M, Teh T, Wang C-IA, Ayliffe MA, Kobe B, Ellis JG. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci U S A. 2006;103(23):8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A. 2007;104(7):2531–2536. doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi D, Dubiella U, Kim SH, Isaiah Sloss D, Dowen RH, Dixon JE, Innes RW. Recognition of the protein kinase AVRPPHB SUSCEPTIBLE1 by the disease resistance protein RESISTANCE TO PSEUDOMONAS SYRINGAE5 Is dependent on S-acylation and an exposed loop in AVRPPHB SUSCEPTIBLE. Plant Physiol. 2014;164(1):340–351. doi: 10.1104/pp.113.227686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahalingam R, Wang G, Knap HT. Polygalacturonase and polygalacturonase inhibitor protein: gene isolation and transcription in Glycine max-Heterodera glycines interactions. Mol Plant Microbe Interact. 1999;12(6):490–498. doi: 10.1094/MPMI.1999.12.6.490. [DOI] [PubMed] [Google Scholar]
- 44.Kaul S, Koo HL, Jenkins J, Rizzo M, Rooney T, Tallon LJ, Feldblyum T, Nierman W, Benito MI, Lin X, Town CD, Venter JC, Fraser CM, Tabata S, Nakamura Y, Kaneko T, Sato S, Asamizu E, Kato T, Kotani H, Sasamoto S, Ecker JR, Theologis A, Federspiel NA, Palm CJ, Osborne BI, Shinn P, Dewar K, Kim CJ, Buehler E, et al. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 45.Pilet ML, Duplan G, Archipiano M, Barret P, Baron C, Horvais R, Tanguy X, Lucas MO, Renard M, Delourme R. Stability of QTL for field resistance to blackleg across two genetic backgrounds in oilseed rape. Crop Sci. 2001;41(1):197–205. doi: 10.2135/cropsci2001.411197x. [DOI] [Google Scholar]
- 46.Federici L, Di Matteo A, Fernandez-Recio J, Tsernoglou D, Cervone F. Polygalacturonase inhibiting proteins: players in plant innate immunity? Trends Plant Sci. 2006;11(2):65–70. doi: 10.1016/j.tplants.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 47.Easton CJ, Rossall S. The production of certain cell wall-degrading enzymes by Leptosphaeria maculans in culture and in stem canker lesions of oilseed rape. Physiol Plant Pathol. 1985;26(2):185–197. doi: 10.1016/0048-4059(85)90019-0. [DOI] [Google Scholar]
- 48.Sexton AC, Paulsen M, Woestemeyer J, Howlett BJ. Cloning, characterization and chromosomal location of three genes encoding host-cell-wall-degrading enzymes in Leptosphaeria maculans, a fungal pathogen of Brassica spp. Gene. 2000;248(1–2):89–97. doi: 10.1016/S0378-1119(00)00142-6. [DOI] [PubMed] [Google Scholar]
- 49.Balesdent MH, Louvard K, Pinochet X, Rouxel T. A large-scale survey of races of Leptosphaeria maculans occurring on oilseed rape in France. Eur J Plant Pathol. 2006;114(1):53–65. doi: 10.1007/s10658-005-2104-0. [DOI] [Google Scholar]
- 50.Stachowiak A, Olechnowicz J, Jedryczka M, Rouxel T, Balesdent MH, Happstadius I, Gladders P, Latunde-Dada A, Evans N. Frequency of avirulence alleles in field populations of Leptosphaeria maculans in Europe. Eur J Plant Pathol. 2006;114(1):67–75. doi: 10.1007/s10658-005-2931-z. [DOI] [Google Scholar]
- 51.Kutcher HR, Balesdent MH, Rimmer SR, Rouxel T, Chevre AM, Delourme R, Brun H. Frequency of avirulence genes in Leptosphaeria maculans in western Canada. Can J Plant Pathol. 2010;32(1):77–85. doi: 10.1080/07060661003594109. [DOI] [Google Scholar]
- 52.Rouxel T, Penaud A, Pinochet X, Brun H, Gout L, Delourme R, Schmit J, Balesdent MH. A 10-year survey of populations of Leptosphaeria maculans in France indicates a rapid adaptation towards the Rlm1 resistance gene of oilseed rape. Eur J Plant Pathol. 2003;109(8):871–881. doi: 10.1023/A:1026189225466. [DOI] [Google Scholar]
- 53.Mahuku GS, Goodwin PH, Hall R, Hsiang T. Variability in the highly virulent type of Leptosphaeria maculans within and between oilseed rape fields. Can J Bot. 1997;75(9):1485–1492. doi: 10.1139/b97-863. [DOI] [Google Scholar]
- 54.Chen Y, Fernando WGD. Prevalence of pathogenicity groups of Leptosphaeria maculans in western Canada and North Dakota. USA Canadian J Plant Pathol. 2006;28(4):533–539. doi: 10.1080/07060660609507331. [DOI] [Google Scholar]
- 55.Nepal A, Markell S, Knodel J, Bradley CA, del Río Mendoza LE. Prevalence of blackleg and pathogenicity groups of Leptosphaeria maculans in North Dakota. Plant Dis. 2014;98(3):328–335. doi: 10.1094/PDIS-04-13-0458-RE. [DOI] [PubMed] [Google Scholar]
- 56.Brun H, Chèvre AM, Fitt BD, Powers S, Besnard AL, Ermel M, Huteau V, Marquer B, Eber F, Renard M, Andrivon D. Quantitative resistance increases the durability of qualitative resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 2010;185(1):285–299. doi: 10.1111/j.1469-8137.2009.03049.x. [DOI] [PubMed] [Google Scholar]
- 57.Larkan NJ, Ruzicka DR, Edmonds-Tibbett T, Durkin JMH, Jackson LE, Smith FA, Schachtman DP, Smith SE, Barker SJ. The reduced mycorrhizal colonisation (rmc) mutation of tomato disrupts five gene sequences including the CYCLOPS/IPD3 homologue. Mycorrhiza. 2013;23(7):573–584. doi: 10.1007/s00572-013-0498-7. [DOI] [PubMed] [Google Scholar]
- 58.Koch E, Song K, Osborn TC, Williams PH. Relationship between pathogenicity and phylogeny based on restriction fragment length polymorphism in Leptosphaeria. Mol Plant-Microbe Interact. 1991;4(4):341–349. doi: 10.1094/MPMI-4-341. [DOI] [Google Scholar]
- 59.Hughes SL, Hunter PJ, Sharpe AG, Kearsey MJ, Lydiate DJ, Walsh JA. Genetic mapping of the novel Turnip mosaic virus resistance gene TuRB03 in Brassica napus. Theor Appl Genetics. 2003;107(7):1169–1173. doi: 10.1007/s00122-003-1363-4. [DOI] [PubMed] [Google Scholar]
- 60.Li H, Ribaut JM, Li Z, Wang J. Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet. 2008;116(2):243–260. doi: 10.1007/s00122-007-0663-5. [DOI] [PubMed] [Google Scholar]
- 61.Altschul SF, Madden TL, Schaeffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheng F, Liu S, Wu J, Fang L, Sun S, Liu B, Li P, Hua W, Wang X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011;11:136. doi: 10.1186/1471-2229-11-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Offord V, Werling D. LRRfinder2.0: a webserver for the prediction of leucine-rich repeats. Innate Immunity. 2013;19(4):398–402. doi: 10.1177/1753425912465661. [DOI] [PubMed] [Google Scholar]


