Abstract
Background
Long intervening non-coding RNAs (lincRNAs) are a recently discovered subclass of non-coding RNAs. LincRNAs are expressed across the mammalian genome and contribute to the pervasive transcription phenomenon. They display a tissue-specific and species-specific mode of expression and are present abundantly in the brain.
Results
Here, we report the expression patterns of oligodendrocyte maturation-associated long intervening non-coding RNA (OLMALINC), which is highly expressed in the white matter (WM) of the human frontal cortex compared to the grey matter (GM) and peripheral tissues. Moreover, we identified a novel isoform of OLMALINC that was also up-regulated in the WM. RNA-interference (RNAi) knockdown of OLMALINC in oligodendrocytes, which are the major cell type in the WM, caused significant changes in the expression of genes regulating cytostructure, cell activation and membrane signaling. Gene ontology enrichment analysis revealed that over 10% of the top 25 up- and down-regulated genes were involved in oligodendrocyte maturation. RNAi experiments in neuronal cells resulted in the perturbation of genes controlling cell proliferation. Furthermore, we identified a novel cis-natural antisense non-coding RNA, which we named OLMALINC-AS, which maps to the first exon of the dominant isoform of OLMALINC.
Conclusions
Our study has demonstrated for the first time that a primate-specific lincRNA regulates the expression of genes critical to human oligodendrocyte maturation, which in turn might be regulated by an antisense counterpart.
Electronic supplementary material
The online version of this article (doi:10.1186/s13041-014-0091-9) contains supplementary material, which is available to authorized users.
Keywords: Long intervening non-coding RNA, OLMALINC, Human brain, Frontal cortex, White and grey matter, Antisense RNA
Background
There is a growing comprehension that the majority of the transcribed genome does not code for proteins but comprises a variety of non-coding RNAs of different properties, such as length, as well as functionality in transcriptional and epigenetic control [1]. In particular, long non-coding RNAs (lncRNAs) seem to be a very recent evolutionary development, and this is supported by the observation that primates, and humans in particular, express most of the currently characterized lncRNAs. A subcategory of lncRNAs, termed long intervening non-coding RNAs (lincRNAs), show a tissue-specific mode of expression and substantially contribute to the pervasive transcription that is observed in the mammalian genome. LincRNAs are defined as intervening (relative to the current gene annotations) transcripts that are longer than 200 nucleotides in length and lack protein-coding capacity [2,3].
The role of lincRNAs is thought to be in gene regulation, and in the brain it has been proposed that non-coding RNAs may play a role in regulating axon myelination in WM and glial cell differentiation [4]. It is thought that lincRNAs act as scaffolds to allow for epigenetic changes to occur within the genome, such as increasing or decreasing mRNA levels through sequestration or stabilization, thus regulating the transcription of target genes. Such a process is of the utmost importance in the development of the complicated array of cells that is observed within the brain.
There is a growing body of evidence that the human brain, as the most complex organ in the body, is the richest source of lncRNAs, reflecting the demand for highly complex control mechanisms for the differentiation and development of numerous cell types, including neurons and oligodendroglia [4]. Indeed, our recent RNA sequencing (RNA-Seq) study has revealed vast differences between the transcriptomes of the GM and WM in the human superior frontal cortex [5]. While not unexpected, given the differences in cell type populations within these two structures, this study has demonstrated that a significant number of annotated and unidentified lincRNAs are expressed at high levels and disparate quantities between GM and WM.
One of the most noticeable examples of differentially expressed lincRNAs between WM and GM was linc00263 [5]. Compared to most lincRNAs, which remain at low expression levels, i.e. < 2 fragments per kilobase of exon per million fragments mapped (fpkm), linc00263 was expressed at 16.2 and 71.5 fpkm in GM and WM, respectively [5]. The high levels of expression in WM suggest that linc00263 may have a functional importance in oligodendrocytes, which constitute the majority of cells in the WM. Here, we validated the differential expression of linc00263 in GM and WM using RT-qPCR. Furthermore, we performed a comparative analysis of linc00263 using vertebrate genomes and found that this gene is specific to primates. Finally, RNAi knockdown of linc00263 in human neuronal and oligodendrocyte cell lines followed by transcriptome sequencing demonstrated that silencing of linc00263, which we renamed to OLMALINC (oligodendrocyte maturation-associated long intervening non-coding RNA), results in coordinated changes in the expression of genes controlling the cytoskeleton and maturation of glial cells.
Results
OLMALINC is highly expressed in the white matter of the human frontal cortex
In our recently published global analysis of the transcriptome of the WM and GM of the human frontal cortex, we detected 4.4-fold overexpression of OLMALINC in WM versus GM [5]. The overall expression of OLMALINC was 16.2 fpkm in GM tissue and 71.5 fpkm in WM (Additional file 1: Figure S1A). RNA-Seq analysis revealed that OLMALINC is expressed as two isoforms (Figure 1). The alignment pattern of RNA-Seq reads in the WM samples shows equal amounts of reads aligning to each exon of OLMALINC-002 (Figure 1). This read distribution excludes the possibility of the long terminal repeat (LTR) element, that covers exon 3, might artificially inflate the calculated expression level of OLMALINC. Neither of the isoforms represented OLMALINC-001, which is annotated in Ensembl [Ensembl ID: ENSG00000235823]. The most highly expressed transcript in our comparative analysis represented the NCBI annotated isoform [NCBI ID: NR_026762.1], which for clarity, we called OLMALINC-002. The isoform is 988-nucleotides (nts) long, consists of three exons (Figure 1), and is overexpressed 4.5-fold in WM (62.5 fpkm) compared to GM (14 fpkm) (Additional file 1: Figure S1B). Thus, OLMALINC-002 is a major contributor to the observed overexpression of the OLMALINC gene in WM. The second alternatively spliced isoform was not annotated in any of the reference databases. We named this novel isoform OLMALINC-003. It is composed of two exons that form a 1517-nts-long transcript (Figure 1). This novel transcript does not encode any protein, as determined by an open reading frame (ORF) finder. OLMALINC-003 is expressed at residual levels in GM (1.6 fpkm) and 10 fpkm in WM (Additional file 1: Figure S1B).
Figure 1.
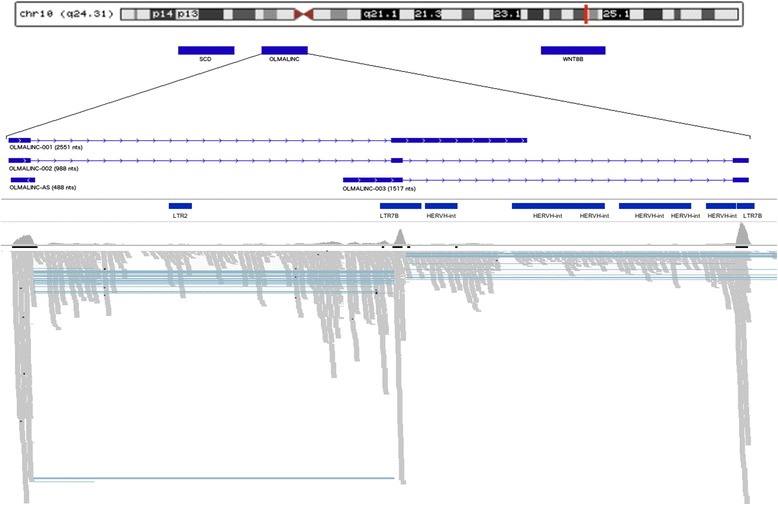
Genomic context splice of variants and genomic features of OLMALINC . Track 1 represents the chromosomal positioning of OLMALINC. Track 2 shows OLMALINC in genomic context. OLMALINC is located downstream of the gene stearoyl-CoA desaturase (delta-9-desaturase) (SCD) and up-stream of wingless-type MMTV integration site family, member 8B (WNT8B). Track 3 is a schematic representation of the exon/intron structure of the OLMALINC-001, −002 and −003 isoforms as well as OLMALINC-AS. Track 4 is a schematic of the repeat elements that appears in this region of the genome including long terminal repeats LTR2, LTR7B and HERVH-int. A LTR 7B over laps with a large portion of the final exon of OLMALINC-002 and OLMALINC-003. Track 5 show the read alignment in the WM samples carried out by TopHat, demonstrating that the repeat elements have not artificially inflated the RNA-Seq fpkm values.
To confirm the specificity of the short sequence read alignment and assembly of the data, both OLMALINC isoforms were reverse transcribed and amplified using isoform specific primers, and the PCR products were Sanger sequenced. Sequence alignment of the annotated isoform and the sequence extracted from the RNA-Seq data for the novel isoform to the reference genome confirmed the sequence specificity of the RT-PCR results (Additional file 2: Figure S2A and B).
Next, we validated the differential expression patterns of the OLMALINC using RT-qPCR of independent sets of samples representing the GM and WM of the frontal cortex. The high expression of OLMALINC, including both isoforms, was confirmed (Figure 2), with a 12.05-fold higher expression level in WM (p-value< 0.05). The OLMALINC primers, used for RT-qPCR, spanned exons 2 and 3 of OLMALINC-002 and exons 1 and 2 of OLMALINC-003. This primer design ruled out the possibility of the high FPKM value attributed to OLMALINC by RNA-Seq as a result of additional alignment of reads to the LTR element that covers a large portion of the final exon in both OLMALINC isoforms (Figure 1).
Figure 2.
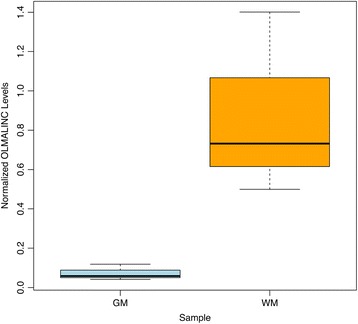
RT-qPCR validation of the OLMALINC gene expression patterns in GM and WM samples. This boxplot shows OLMALINC was up-regulated 12.05-fold in WM when compared to GM (p-value < 0.05).
Comparative sequence analysis and expression of OLMALINC in mammals
Sequence conservation level of OLMALINC locus was analyzed using blastn searches, DNA dot plots (https://www.sanger.ac.uk/resources/software/seqtools/) and chain blastz alignments on the UCSC genome browser (https://genome.ucsc.edu/). In primates, homology was detected to all human exons in chimpanzee, gorilla, orangutan and rhesus monkey. In bushbaby, marmoset, mouse, elephant, opossum and platypus homology was only observed with human exon 1 (Figure 3), and in each case in the same genomic context (downstream of the SCD gene). The UCSC phyloP base-wise conservation across 100 vetebrates shows a peak in conservation 5′ of OLMALINC-001 exon 1, suggesting a possible conserved promoter region (Additional file 3: Figure S3).
Figure 3.
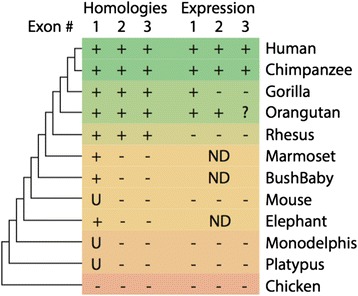
Summary of homology and expression of OLMALINC (UCSC uc001kqz.4) exons in representative vertebrate genomes. Homology and expression of all three exons is only detected in human and chimpanzee. Exons 2 and 3 are not detected outside of old world monkeys. There is low expression of exon 3 in orangutan. Outside of great apes there is no expression of any exon. ND = no data. U = homology was only detected in the UCSC genome browser and not by any other method (blastn/blat/dotplots). - = no expression detected. + = expression detected. ? = low levels of expression detected.
Analysis of published RNA-Seq data [6,7] revealed expression of all exons in chimpanzee and orangutan brain, although relative expression of exon 3 was lower than in human (Additional file 4: Figure S4). In gorilla brain, only expression of exon 1, and neighboring regions (perhaps the antisense transcript), was detected. RNA-Seq reads did not map to expected exons in any other species examined (Additional file 4: Figure S4), suggesting that OLMALINC-001 is not expressed outside of great apes. This information is summarized in Figure 3.
OLMALINC expression profiles in non-brain tissues
To determine whether OLMALINC was likely to be brain-specific or commonly expressed in all tissues, a meta-analysis was performed. This analysis utilized publicly available RNA-Seq datasets from the Illumina Transcriptome BodyMap 2 project, which included samples from 16 human tissues, including brain, using humans of varying ages and sex. The 15 tissues in the Illumina data sets were compared with the brain transcriptome data. The non-brain tissues comprised liver, kidney, heart, lung, skeletal muscle, breast, adrenal, thyroid, prostate, ovary, testes, adipose, colon, lymph node, and white blood cells. The analysis was carried out using the Tuxedo protocol [8] and showed that brain expression level of OLMALINC was higher than the expression levels in any other tissue type (Figure 4 and Additional file 5: Figure S5). Liver, ovary and breast tissue had the next highest levels of OLMALINC expression; these levels were approximately half of the OLMALINC expression seen in the brain. On average OLMALINC was expressed 7.5-fold higher in the brain when compared to other sources of tissue. To validate the expression levels from the Illumina Transcriptome BodyMap 2 project, the expression level of OLMALINC was analysed in another publicly available RNA-Seq dataset produced by The Human Protein Atlas project. The tissue types assessed were brain, liver, kidney, skin and bone marrow (Additional file 5: Figure S5). OLMALINC was expressed at 3-fold higher level in the brain when compared to liver and on average 20-fold higher than the other tissue types. Usage of this independent set of tissue-specific transcriptome sequence data thus corroborates the pattern seen in the Illumina dataset.
Figure 4.
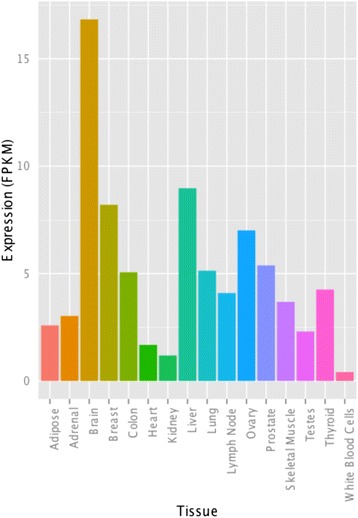
Comparative analysis of the OLMALINC expression levels in brain and 15 other human tissues. The bar graph shows the difference in expression levels of OLMALINC in varying tissues compared to the brain. The RNA-Seq datasets used for this analysis were taken from Illumina’s BodyMap2 project. The brain has at least a greater than 1.9x expression level of OLMALINC than any other tissue. The y-axis is expression in fpkm.
The OLMALINC locus co-expresses cis-natural antisense RNA
Recently, we performed strand-specific RNA-Seq analysis of combined RNA samples derived from the GM and WM tissue from the frontal cortex (Mills et al., unpublished data). Bioinformatics analysis of this dataset revealed the presence of an antisense RNA that mapped to the first exon of the OLMALINC-002 isoform (Figure 1). This antisense transcript is 488-nts long, consists of one exon and is expressed at 18 fpkm in the whole brain tissue. We named this RNA OLMALINC-AS to emphasize its cis-natural feature in regard to its overlap with the OLMALINC locus.
To validate the sensitivity of the strand-specific RNA-Seq analysis, we amplified and Sanger sequenced a fragment that was unique to the OLMALINC-AS transcript. Alignment to the reference genome confirmed expression of the OLMALINC-AS in the human frontal cortex (Additional file 2: Figure S2C).
To further explore the pattern of OLMALINC-AS expression in specific histological structures of the cortex, we quantified the levels of this transcript in GM and WM samples using RT-qPCR. Figure 5 shows 11.7-fold up-regulation (p-value = 0.07) of OLMALINC-AS in WM. This fold-change did not reach the p-value cut-off level for statistical significances.
Figure 5.
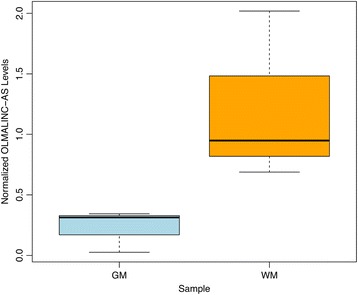
Quantification of OLMALINC-AS in GM and WM of the frontal cortex. This boxplot shows OLMALINC-AS was up-regulated 11.7-fold in WM (p-value = 0.07).
OLMALINC knockdown in oligodendrocytes and neurons
To provide insight into the influence of OLMALINC on gene expression and regulation of transcription in cortical tissue, we performed RNAi-driven silencing of OLMALINC in the human MO3.13 oligodendrocytes and SK-N-SH neurons. The efficiency of the down-regulation of OLMALINC expression in the two cell lines was estimated by RT-qPCR. We were able to specifically reduce OLMALINC levels in oligodendrocytes and neurons by 4.5- and 3.5-fold, respectively (p-values < 0.05) (Additional file 6: Figure S6).
Next, to assess the impact of OLMALINC depletion at the level of the whole transcriptome, we performed RNA-Seq analysis on the OLMALINC silenced cells. In MO3.13 oligodendrocytes, 81 genes were up-regulated and 41 were down-regulated (Additional file 7: Table S1). Notably, RNAi-driven reduction of OLMALINC expression led to twice as many genes being up-regulated as down-regulated in these cells.
In SK-N-SH neurons, 29 genes showed a significant increase in expression after knockdown of both OLMALINC isoforms, whereas 17 genes were down-regulated (Additional file 8: Table S2). Again, amongst the genes affected by OLMALINC knockdown, nearly twice as many genes showed increased rather than decreased expression in neurons.
OLMALINC knockdown affects genes regulating cell activation and membrane signaling in oligodendrocytes
Gene ontology analysis of differentially expressed genes (DEGs) and differentially expressed isoforms (DEIs) (Additional file 9: Table S3) in oligodendrocytes revealed two superclusters of genes and isoforms that were involved in the regulation and maintenance of cytostructure and cellular adhesion. Moreover, there were four individual clusters related to heart development, cell activation, cell surface receptor-linked signal transduction and positive regulation of cell adhesion (Figure 6). Overall 56 DEGs and DEIs were clustered in the pathway analysis, comprising 34% of the 162 unique DEGs and DEIs that underwent analysis.
Figure 6.
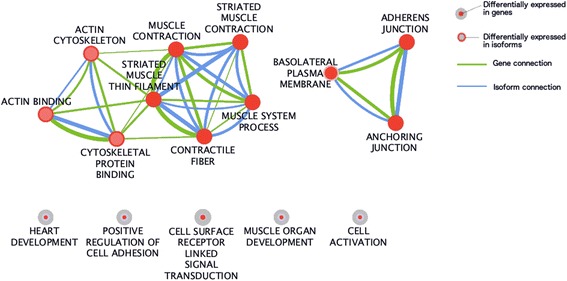
Enrichment map of the Gene Ontology clusters derived from the DEGs and DEIs in oligodendrocytes following silencing of OLMALINC . The size of the node relates to the number of genes in each term. Lack of branching in the bottom clusters indicates that genes contributing to these clusters are not present in other clusters and thus are unique for particular pathway.
Interestingly, amongst the top 25 genes that were up- and down-regulated as a result of OLMALINC silencing in oligodendrocytes (Additional file 7: Table S1), 12 genes contributed to the pathway clusters that were linked to the regulation of cell activation and cell surface receptor-linked signal transduction. In contrast, only 2 of the top 25 DEGs contributed to the cytostructure supercluster, and none contributed to the cell adhesion supercluster (Table 1). Several genes of the subset, that are shown in Table 1, caught our attention due to their involvement in the physiology of oligodendrocytes. The expression profiles for these selected genes are shown in Figure 7A.
Table 1.
Expression levels for selected DEGs enriched in gene ontology analysis following OLMALINC silencing in oligodendrocytes
| Gene | Chr. | Description | GO Clusters | FPKM Control | FPKM RNAi | Fold Change | q-value |
|---|---|---|---|---|---|---|---|
| SAA1 | chr11 | Serum amyloid A1 | Cell activation | 12.64 | 2.20 | −5.73 | 0.0014 |
| IL7R | chr5 | Interleukin 7 receptor | Cell activation, cell surface receptor linked signal transduction | 1.60 | 0.42 | −3.83 | 0.0194 |
| APLN | chrX | Apelin | Cell surface receptor linked signal transduction | 17.51 | 5.38 | −3.25 | 2.99E-07 |
| GPR126 | chr6 | G protein-coupled receptor 126 | Cell surface receptor linked signal transduction | 2.67 | 0.88 | −3.04 | 0.0116 |
| CXCL5 | chr4 | Chemokine (C-X-C motif) ligand 5 | Cell surface receptor linked signal transduction | 8.84 | 2.97 | −2.98 | 4.01E-04 |
| AXL | chr19 | AXL receptor tyrosine kinase | Cell surface receptor linked signal transduction | 4.23 | 1.75 | −2.42 | 0.0114 |
| ANXA1 | chr5 | Annexin A1 | Cell surface receptor linked signal transduction | 4.80 | 2.06 | −2.33 | 4.41E-04 |
| COL3A1 | chr2 | Collagen, type III, alpha 1 | Cell activation, cell surface receptor linked signal transduction | 380.21 | 191.13 | −1.99 | 0.0396 |
| SOX4 | chr6 | SRY (sex determining region Y)-box 4 | Cell activation, cell surface receptor linked signal transduction | 11.85 | 33.56 | 2.83 | 6.13E-11 |
| EGR1 | chr5 | Early growth response 1 | Cell activation | 1.81 | 5.41 | 2.99 | 3.92E-06 |
| HDAC9 | chr7 | Histone deacetylase 9 | Cell activation | 0.83 | 3.33 | 4.02 | 0.0486 |
| WIPF3 | chr7 | WAS/WASL interacting protein family, member 3 | Cytostructure supercluster | 0.79 | 3.44 | 4.37 | 4.09E-08 |
| TRIM63 | chr1 | Tripartite motif containing 63, E3 ubiquitin protein ligase | Cytostructure supercluster | 0.35 | 1.59 | 4.62 | 0.0128 |
| WNT11 | chr11 | Wingless-type MMTV integration site family, member 11 | Cell surface receptor linked signal transduction | 0.13 | 0.93 | 7.38 | 0.0439 |
Chr. - chromosome.
Figure 7.
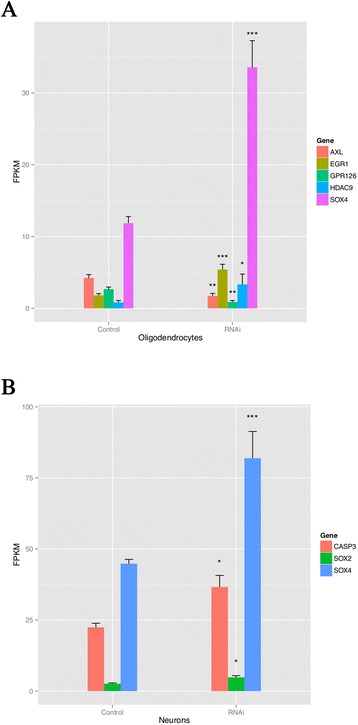
Expression levels of selected DEGs in OLMALINC -depleted oligodendrocytes (A) and neurons (B). In oligodendrocytes the EGR1, HDAC9 and SOX4 were all up-regulated after RNAi of OLMALINC. AXL and GPR126 was down-regulated in oligodendrocytes after RNAi of OLMALINC. The expression levels are in fpkm as calculated by RNA-Seq. In neurons CASP3, SOX2 and SOX4 were up-regulated after RNAi of OLMALINC. Level of significance: *q-value < 0.05, **q-value < 0.02, ***q-value < 0.01.
The histone deacetylase 9 (HDAC9) was up-regulated 4.1-fold, after knockdown of OLMALINC. The protein product of this gene belongs to class II Hdacs and is expressed mainly in post-mitotic, mature neurons in the murine cerebral cortex [9,10]. Studies of HDAC9 have revealed its association with medulloblastoma [11] and schizophrenia [9].
SRY (Sex-Determining Region Y)-Box 4 (SOX4) was up-regulated 2.8-fold in OLMALINC-depleted oligodendrocytes. SRY (Sex-Determining Region Y) (Sox) proteins of group C are strongly expressed in the developing nervous system and have been associated with the maturation of neurons and glia. Prolonged SOX4 expression in cells of the oligodendrocyte lineage is incompatible with the acquisition of a fully mature phenotype, which indicates that the presence of SOX4, and possibly SRY (Sex-Determining Region Y)-Box 11 (SOX11), in oligodendrocyte precursors may normally prevent premature differentiation. SOX4 transgenic mice develop the full spectrum of phenotypic traits that are associated with severe hypomyelination during the first postnatal weeks. In these mice, myelin gene expression is severely reduced, and myelin dramatically thinned in several central nervous system (CNS) regions [12]. SOX4 expression counteracts the differentiation of radial glia and must be down-regulated before full maturation can occur [13].
G protein-coupled receptor 126 gene (GPR126) was down-regulated 3.1-fold in RNAi-treated MO3.13 oligodendrocytes. In Schwann cells, GPR126 controls proper development and myelination [14]. A mutation in GPR126 causes severe congenital hypomyelinating peripheral neuropathy in mice and the expression of differentiated Schwann cell markers [15].
AXL receptor tyrosine kinase (AXL) is a member of the Tyro3-Axl-Mer (TAM) receptor tyrosine kinase subfamily and was down-regulated 2.5-fold in our RNAi experiments. The TAM receptors are widely expressed in the nervous system, including oligodendrocytes [16]. It has been shown that growth arrest-specific 6 (Gas6), a ligand for the tyro3/Axl/Mer (TAM) receptors, can affect the severity of demyelination in mice, and a loss of signaling via Gas6 leads to decreased oligodendrocyte survival and increased microglial activation during cuprizone-induced demyelination [17]. Gas6 significantly increased myelination in a dose-dependent manner, suggesting that TAM receptor signaling could be directly involved in myelination by oligodendrocytes [18].
Early growth response gene (EGR1) is a zinc-finger transcription factor that was 3-fold up-regulated in OLMALINC-silenced oligodendrocytes. EGR1 down-regulation is critical for oligodendrocyte progenitor cell differentiation, and the gene was recently included in a de-repression model of oligodendrocyte lineage progression that relied on the concurrent down-regulation of several inhibitors of differentiation [19]. The expression pattern of selected genes, described above, was confirmed in an independent OLMALINC silencing experiment using individual siRNAs followed by RT-qPCR analysis (Additional file 10: Figure S7).
The cell proliferation pathway is affected by knockdown of OLMALINC in neurons
GO enrichment analysis of genes whose expression had been affected by OLMALINC knockdown in neurons grouped GO terms for cell proliferation into a single cluster. This cluster was composed of 11 genes that comprised 24% of the all DEGs resulting from OLMALINC knockdown in neurons (Additional file 11: Table S4). Several genes within this subset have been previously described to be involved in neuronal biology.
SRY (Sex-Determining region Y)-box 2 gene (SOX2) was 1.8-fold up-regulated following OLMALINC knockdown. Sox2 plays a role in the maturation and survival of embryonic and adult neurons, and Sox2 expression is high in undifferentiated neurons, but declines upon differentiation [20]. Sox2 deficiency results in decreased precursor cell proliferation and the generation of new neurons in adult mouse neurogenic regions [21].
Caspase-3 (CASP3) was up-regulated 1.6-fold following knockdown of OLMALINC. CASP3 is a key mediator of apoptosis in neuronal cells. The functions of non-apoptotic caspase-3 in neuronal cells include synaptic plasticity, dendrite pruning, as well as learning and memory processes [22].
Finally, SOX4, which was up-regulated 1.8-fold in neurons, promotes neuronal differentiation and has a central regulatory role during neuronal maturation. It mechanistically separates cell cycle withdrawal from the establishment of neuronal properties [23]. Figure 7B presents differences in the expression levels of the SOX2, CASP3 and SOX4 genes as a result of OLMALINC silencing.
Discussion
This study shows that OLMALINC is overexpressed in the human brain compared to peripheral tissue and that WM is a major source of this overexpression. Moreover, silencing of OLMALINC in oligodendrocytes and neurons affects the expression of functionally related gene sets. Furthermore, this lincRNA is co-expressed with an antisense counterpart, OLMALINC-AS, which shows a similar expression pattern, i.e., up-regulation in WM compared to GM. Such co-expression of sense and antisense RNAs with overlapping loci has been previously observed in the case of the BACE1 mRNA and its natural antisense non-coding partner BACE1-AS [24]. Comparative analysis of the OLMALINC sequence demonstrates its unique expression in primates.
Recent genome-wide surveys on lincRNAs allow us to draw some general observations about this particular RNA species [25]. First, it has been suggested that most lincRNAs are expressed at low levels, usually below 2 fpkm. In contrast to this observation, OLMALINC and its antisense counterpart, OLMALINC-AS, remain at levels of at least 16.2 fpkm and reach 71.5 fpkm in WM. Moreover, our meta-analysis of OLMALINC expression in peripheral tissues revealed at least 1.9-fold higher expression of this transcript in brain when compared to 15 other cell types of the body. Indeed, lincRNAs have also been shown to exhibit tissue-specific patterns in model organisms such as zebrafish [26]. In particular, the overexpression of OLMALINC in the WM corroborated the outcome of OLMALINC silencing, which affected a number of genes involved in oligodendrocyte maturation (see below).
Our comparative analysis of the OLMALINC sequence with vertebrate genomes revealed a remarkably high degree of conservation in great ape and old world monkey genomes (Figure 3). In all other mammal species examined only homology with exon 1 was observed. There was also evidence for conservation of the region 5′ of exon 1, which was detected as far back as opossum, and even platypus (Figure 3). Although all exons are present in old world monkeys, detectable expression of OLMALINC (and its antisense) was only observed in great apes, with exon 3 under expressed outside of humans (Additional file 4: Figure S4). Loss of expression of exons 2 and 3 in gorilla must have occurred after divergence from the human/chimp lineage.
These findings suggests that the building blocks for OLMALINC exon 1 are ancient (>200 mya, being common to all mammals), whereas exons 2 and 3 only arose in the ancestor of old world moneys (~30-45 mya) [27]. The evolution of exons 2 and 3 was followed by the expression of OLMALINC in at least the Hominidae ancestor (>17 mya), with further specialization (high expression of exon 3) only humans (<6 mya). Although the original building blocks of OLMALINC appear ancient, its recently evolved expression suggests that it is a young gene. Indeed, less than 6% of zebrafish lincRNAs have detectable sequence conservation with mouse or human lincRNAs [28]. Moreover, merely 12% of the human and mouse lincRNAs are conserved in other species [29,30].
It has been previously suggested that lincRNAs tend to act in cis, thus affecting genes located in a 10-300-kb vicinity of the particular lincRNA locus [31,32]. Interestingly, we have not observed elevated expression of the protein-coding genes located in the 300-kb vicinity of the OLMALINC locus (data not shown), which is in contrast one study [33], but corroborates observations of others showing that only 3% of the human lincRNAs have expression profiles correlated with their neighboring genes [34].
Comparative analysis of the OLMALINC sequence corroborated earlier observations on its fast evolution [29,30]. Indeed, OLMALINC is only conserved within primate genomes, and its homology abruptly drops for rodents and other non-primate vertebrates. The presence of the human-specific exon 3 in the OLMALINC-002 isoform further supports the thesis that lincRNAs belong to rapidly evolving segments of the primate genome [35].
Along with metastasis associated lung adenocarcinoma transcript 1 (MALAT-1) [36] and polyadenylated nuclear RNA (PAN) [37], OLMALINC belongs to the exceptionally abundantly expressed lincRNAs. High expression levels of some lincRNAs may facilitate their trans function as a decoy to titrate proteins from their potential targets, as reported for the growth arrest-specific 5 (Gas5) and P21 associated ncRNA DNA damage activated (PANDA) lincRNAs [38,39]. Hence, preservation of a titration mechanism will require high numbers of lincRNA molecules interacting with numerous proteins [28].
The RNAi silencing efficiency of OLMALINC was less effective in neurons than in an oligodendrocytic cell line, and this could be a result of cell type-specific differences in the uptake capacity of the siRNA between neurons and oligodendrocytes [40]. Second, the initial levels of OLMALINC were much below its expression level in oligodendrocytes, which further led to diminution of the silencing effect in neurons.
Silencing of OLMALINC expression in oligodendrocytes affected the expression of genes, such as SOX4, GPR126 and EGR1, which are involved in the differentiation of these cells from the neuronal stem lineage and in the control of myelination processes. Several non-coding RNAs (ncRNA) have been shown to exhibit dynamic expression patterns during neuronal and oligodendrocyte lineage specification, neuronal-glial fate transitions, and myelination, and they include ncRNAs associated with differentiation-specific nuclear subdomains, such as Gomafu and Neat1, ncRNAs associated with developmental enhancers, and genes encoding important transcription factors and homeotic proteins [41,42]. Up-regulation of the deacetylase 5 and 9 genes (Additional file 7: Table S1) by depletion of OLMALINC further underpins the previous observations of the involvement of lincRNAs in chromatin remodeling-controlling cellular differentiation, as has been shown in lineage-specific gene expression programs in mouse embryonic stem cells [43].
The number of genes whose expression was affected by OLMALINC silencing remains similar to the average of 175 protein-coding genes that changed their expression pattern in 137 individual lincRNA knockdowns in Guttman’s study (2011). Moreover, changes in expression were characterized by up- and down-regulation, suggesting a versatile impact of OLMALINC on transcription that leads to the activation or repression of transcription. This remains in contrast to previous suggestions that the action of lincRNAs mainly leads to repression of gene expression [44].
Conclusions
Our evidence indicates that the recently evolved OLMALINC is a primate specific transcript that is a major contributor to the maintenance of oligodendrocyte maturation. This conclusion remains in line with previous observations that lincRNAs are involved in development of the nervous system [45]. However, it should be emphasized that further characterization of OLMALINC function will require systematic studies, including defining all protein complexes with which the lincRNA possibly interacts, determining where these protein interactions assemble on the RNA, and ascertaining whether they bind simultaneously or alternatively. Moreover, understanding how OLMALINC–protein or –DNA interactions give rise to specific patterns of gene expression will require determination of the functional contribution of each interaction and possible localization of the complex to its genomic targets.
Methods
Brain tissue and RNA extraction
Samples representing the GM and WM of the human superior frontal gyrus (SFG) were obtained from the Sydney Brain Bank following ethical approval from the Human Research Ethics Committee of the University of New South Wales. The GM and WM tissues were obtained from three individuals (aged 79, 94 and 98) without significant neuropathology. The post mortem interval (PMI) of the samples ranged from 8–24 h, and the pH ranged from 5.77-6.65. For RT-qPCR experiments, another two SFG samples were used that were matched to the previous samples regarding age, PMI and pH.
Total RNA was extracted from approximately 100 mg for each case using Qiagen’s RNeasy Lipid Tissue RNA Extraction Kit. The RNA integrity numbers (RINs) were determined using an Agilent 2100 BioAnalyzer RNA Nano Chip. The RIN values ranged from 4.9-7.2. This RIN range was previously shown to have little effect on relative gene expression ratios [46].
Cell lines
MO3.13 oligodendrocytes and SK-N-SH neurons were obtained from the ATCC (Manassas, VA) and were cultured in 12-well plates in Dulbecco’s modified Eagle’s medium containing 10% fetal calf serum, 2 mM glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin at 37°C in humidified air containing 5% CO2. Cell culture media and additives were obtained from Invitrogen (Melbourne, Australia) unless stated otherwise.
Reverse transcription and PCR
Using total RNA samples previously used for the RNA-Seq analysis [5], the OLMALINC-002, −003 and -AS transcripts were reverse transcribed and amplified using the Qiagen One-Step RT-PCR kit. The forward and reverse primers used to amplify OLMALINC-002 were TGTGGTACTAAGCTTGACAGC and TCATAGGTGGATCTCCTCACG; for OLMALINC-003, TAGACCTTGCTAACCAGGACG and TGGTATCAGTTAGCGTGGGGC; and for OLMALINC-AS, CCCGAGATTCTTTGTGGGCT and CTCTCCCACCACACACCAC. Standard RT-PCR conditions, as recommended by Qiagen, were used. PCR products of 560, 1100 and 280 bp were purified and Sanger sequenced.
Quantitative PCR
Primers were designed for the OLMALINC, OLMALINC-AS and proteasome (prosome, macropain) subunit beta type 4 (PSMB4) genes. For quantification of EGR1, HDAC9, SOX4, GPR126 and AXL expression in MO3.13 oligodendrocytes primers were purchased from Qiagen. We used PSMB4 as a housekeeping gene, as described previously [47]. The PSMB4 forward and reverse primers were ordered from Qiagen (HS_PSMB4_1_SG QuantiTect primer assay). The sequence for the OLMALINC forward primer was GACTCCTTTGGGAGACCAGTG, and that of the reverse primer was AGGTCACAGGGGATTTGATGG. The OLMALINC primers spanned the fragment of the transcript that was common for the OLMALINC-002 and −003 isoforms. The sequence for the OLMALINC-AS forward primer was GTCACTGGGGAGAACGTGAC, and that for the reverse primer was CTCTCCCACCACACACCAC. The OLMALINC-AS primers were unique for the –AS transcript. All of the primers used for RT-qPCR had an efficiency of between 90%-110%. The RNA samples were reverse-transcribed using the Qiagen QuantiTect Reverse Transcription (RT) Kit, and the gene expression was quantified using the QuantiTect SYBR green PCR master mix (Qiagen). The PCR reaction was performed on a Rotor-Gene 6000 (Qiagen) using three independent GM and WM samples and MO3.13 oligodendrocytes, and each reaction was performed in triplicate. PSMB4 was used to normalize the results from each RT-qPCR run to reduce batch effects and correct any variation in template input [48]. Expression levels were calculated using 2-ΔCt and fold-change was calculated using the 2-ΔΔCt method [49]. Statistically significant changes in gene expression were assessed using the R project for statistical computing (www.r-project.org).
RNA interference
RNAi knockdown of OLMALINC was carried out using four commercially prepared siRNA oligonucleotides (Qiagen) specific to different fragments of the OLMALINC transcript. Briefly, MO3.13 and SK-N-SH cells were seeded at 40% confluence in 6-well plates in antibiotic-free media and equimolar mix of four oligonucleotides was transfected using Lipofectamine 2000 and Opti-MEM I (Invitrogen) following the manufacturer’s protocol. For validation experiments MO3.13 oligodendrocytes were transfected with two individual siRNAs used previously in the four siRNA mix. Transfection efficiency was confirmed by substituting the siRNA with fluorescently labelled BLOCK-iT reagent (Invitrogen), using the same transfection procedure and this confirmed >95% transfection efficiency was achieved as assessed by fluorescence microscopy.
RNA isolation, library preparation and sequencing
After 48 hrs of culture total RNA was isolated using RNeasy Mini Kit (Qiagen) followed by RNase-free DNase treatment to remove traces of genomic DNA. The RNA quality of the total RNA was assessed using the Agilent 2100 BioAnalyser RNA Nano Chip and the RIN values ranged between 8.0 and 9.0. Six RNA samples (three MO3.13 and three SK-N-SH replicates) were prepared for sequencing according to the Illumina TruSeq RNA sample preparation guide and subjected to 100 bp paired-end sequencing using Illumina HiSeq1000.
Meta-analysis of Illumina expression data of non-brain tissues
Meta-analysis of Illumina BodyMap2 transcriptome files was carried out to determine the expression distribution of OLMALINC across tissues at the gene level. The BodyMap2 project was carried out by Illumina to provide a sample RNA-Seq dataset of 16 individual and mixed tissues. A second analysis was performed on five independent RNA-Seq datasets produced by The Human Protein Atlas (http://www.proteinatlas.org/tissue). The meta-analysis was carried out on Galaxy using the Tuxedo protocol [13], and files from Illumina’s BodyMap2 project and the Human protein Atlas project were imported into Galaxy from the NCBI sequence read archive (SRA) (project accession number: ERP000546, http://www.ncbi.nlm.nih.gov/sra/?term=ERP000546, project accession number: ERP003613, http://www.ncbi.nlm.nih.gov/sra/?term=ERP003613). The selected SRA file reads were assembled with TopHat, which utilizes Bowtie to align short sequence reads to the H. sapiens reference genome (build hg19). The default setting for TopHat was used. Cufflinks was then used to assemble the aligned reads into individual transcripts by inferring splicing structure and provided a minimal number of predicted transcripts through parsimonious assembly. Cufflinks also normalized the read count of each input file to allow for calculation of the relative abundance of each transcript in fpkm. To guide the assembly process, the iGenomes UCSC hg19 full annotation GTF file was used. These steps were performed for all 16 tissue types that were assessed. Subsequently, the results for each tissue type were merged with the files for the brain transcriptome and the iGenomes reference annotation via Cuffmerge and then passed through Cuffdiff.
Comparative analysis with vertebrate genomes
The UCSC Genome Browser website (http://genome.ucsc.edu/index.html) contains the reference sequences and working draft assemblies of a large collection of genomes from a variety of organisms. Using the UCSC Genome Browser, the level of sequence conservation of OLMALINC across numerous species was analyzed.
Gene ontology enrichment analysis
The lists of DEGs and DEIs were entered into the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) [50]. DAVID can only utilize annotated genes/isoforms; thus, all un-annotated genes/isoforms and indecisively annotated genes/isoforms were removed from the lists. DAVID tests GO terms for over representation in each of the DEG and DEI lists. The GO terms lost produced by DAVID, were processed using the ‘Enrichment Map’ plug in for ‘Cytoscape’ (http://www.cytoscape.org/) [51]. This produces a visual putput of the text based GO term lists.
Data access
The sequence data have been submitted to the NCBI Short Read Archive with accession number SRA602249.
Acknowledgements
Tissues were received from the Sydney Brain Bank at Neuroscience Research Australia and the New South Wales Tissue Resource Centre at the University of Sydney which are supported by the National Health and Medical Research Council of Australia (NHMRC), University of New South Wales, Neuroscience Research Australia, Schizophrenia Research Institute and National Institute of Alcohol Abuse and Alcoholism (NIH (NIAAA) R24AA012725). This research was supported by the National Health & Medical Research Council of Australia (Project grant #1022325 to WSK and Fellowship #630434 to GMH) and Brain Foundation Australia (to MJ). Authors would like to thank Caroline Janitz for her expert advice regarding Illumina sequencing.
Abbreviations
- AXL
AXL receptor tyrosine kinase
- BACE1
Beta-site APP-cleaving enzyme 1
- BACE1-AS
Beta-site APP-cleaving enzyme 1 antisense RNA
- CASP3
Caspase-3
- CNS
Central nervous system
- EGR1
early growth response 1
- Fpkm
Fragments per kilobase of exon per million fragments mapped
- Gas5
Growth arrest-specific 5 (non-protein coding)
- Gas6
Growth arrest-specific 6
- GM
Grey matter
- Gomafu
myocardial infarction associated transcript (non-protein coding)
- GPR126
G protein-coupled receptor 126
- Hdac
Histone deacetylase
- HDAC9
Histone deacetylase 9
- HIF-1alpha
Hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
- ING4
Inhibitor of growth family, member 4
- OLMALINC
Oligodendrocyte maturation-associated long intervening non-coding RNA
- OLMALINC-AS
Oligodendrocyte maturation-associated long intervening non-coding RNA
- lincRNAs
long non-coding intervening RNAs
- lncRNAs
Long non-coding RNAs
- LTR
long terminal repeat
- Malat1
Metastasis associated lung adenocarcinoma transcript 1
- Neat1
Nuclear paraspeckle assembly transcript 1 (non-protein coding)
- ncRNA
Non-coding RNA
- nts
Nucleotides
- ORF
Open reading frame
- PAN
Polyadenylated nuclear RNA
- PANDA
P21 associated ncRNA DNA damage activated
- PMI
Post mortem interval
- PSMB4
Proteasome (prosome, macropain) subunit, beta type, 4
- RIN
RNA integrity number
- RNAi
RNA-interference
- RNA-Seq
RNA-Sequencing
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- SCD
Stearoyl-CoA desaturase (delta-9-desaturase)
- SFG
Superior frontal gyrus
- Sox
SRY (sex-determining region Y)
- SOX2
SRY (sex-determining region Y)-box 2
- SOX4
SRY (Sex-determining region Y)-box 4
- SOX11
SRY (sex-determining region Y)-box 11
- TAM
tyro3/Axl/Mer
- TFs
Transcription factors
- WM
White matter
Additional files
Differential expression of the OLMALINC gene and its isoforms as revealed by RNA-Seq analysis of GM and WM samples from the human frontal cortex. (A) Expression levels of the OLMALINC gene in GM and WM. (B) Expression levels of the OLMALINC-002 and −003 isoforms in GM and WM; bars represent SD. Level of significance: **q-value< 0.02, ***q-value<0.01.
Sequence alignment of Sanger sequenced RT-PCR OLMALINC products with sequences derived from RNA-Seq data. The query sequence is the sequence derived from Sanger sequencing and the subject sequence is derived from RNA-Seq data. A. Sequence alignment of OLMALINC-002. B. Sequence alignment of OLMALINC-003 C. Sequence alignment of OLMALINC-AS. The letter N marks nucleotides where sequencing failed to develop a consensus sequence.
Homology detected to OLMALINC in the chimpanzee, gorilla, orangutan, rhesus monkey, marmoset and opossum genomes with blat searches on the UCSC genome browser. All homologies are displayed relative to the SCD gene. In the opossum genome (reverse orientation) homology to exon 1 was not detected with blat searches, but was detected in the multiZ alignments track. In rhesus monkey, exon 3 is not detected in the correct position with balt searches, but is detected with blastz. Exon 1 homologies are boxed in red, exon 2 in green, and exon 3 in blue. Basewise conservation calculated by PhyloP across 100 vertebrates shows a peak in conservation upstream of OLMALINC exon 1 indicated a possible promoter region.
Read coverage from male brain RNA-seq data across the OLMALINC homologous regions in human, chimpanzee, gorilla, orangutan, rhesus monkey and opossum. Outside of great apes there is no evidence for expression of any exon. Exon 1 homologies are boxed in red, exon 2 in green, and exon 3 in blue. Read depth bedGraph files were available for human, gorilla, rhesus monkey and opossum [11]. For chimpanzee and orangutan, RNA-seq reads [12] were mapped with TopHat v2.0.9 with the command line options: tophat -p 4 -a 8 -i 40 -m 1 -I 1000000 --coverage-search --microexon-search. Read depth coverage bedGraph files were generated with the samtools depth utility.
Comparative analysis of OLMALINC expression across five tissue samples. The RNA-Seq data sets were taken from the Human Protein Atlas project (http://www.proteinatlas.org/). Again, OLMALINC is expressed at its highest level in brain tissue and it is expressed 3-fold higher than liver, the tissue source with the next highest level of expression. The independent dataset confirms the results from Figure 4. The y-axis is expression in fpkm.
Quantification of the OLMALINC transcript following its knockdown in oligodendrocytes and neurons using RT-qPCR. OLMALINC levels in oligodendrocytes and neurons were reduced by 4.5- and 3.5-fold, respectively (p-values<0.05). N – neurons; O – oligodendrocytes.
Full list of differentially expressed genes and isoforms between the oligodendrocyte cell controls and the oligodendrocyte cells that underwent RNAi.
Full list of differentially expressed genes and isoforms between the neuronal cell controls and the neuronal cells that underwent RNAi.
Enriched gene ontology terms for genes and isoforms differentially expressed in the oligodendrocyte cell line.
RT-qPCR validation of the EGR1, HDAC9, SOX4, AXL and GPR126 genes expression pattern in MO3.13 oligodendrocytes silenced with individual OLMALINC siRNAs. EGR1, HDAC9 and SOX4 genes were up-regulated 4-, 4.4-, 3.6-fold in RNAi-treated oligodendrocytes when compared to control (p-value<0.05), respectively. The GPR126 and AXL genes were down-regulated 3.5- and 7.5-, respectively. Con – control.
Enriched gene ontology terms for genes and isoforms differentially expressed in the neuronal cell line.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JDM, WSK, PDW, GMH and MJ conceived and designed the experiments. JDM, TK and WSK performed the experiments. JDM, TK, BJC and PDW analyzed the data. JDM, WSK, PDW, GMH and MJ wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
James D Mills, Email: j.mills@unsw.edu.au.
Tomas Kavanagh, Email: t.kavanagh@student.unsw.edu.au.
Woojin S Kim, Email: w.kim@neura.edu.au.
Bei Jun Chen, Email: b.j.chen@student.unsw.edu.au.
Paul D Waters, Email: p.waters@unsw.edu.au.
Glenda M Halliday, Email: g.halliday@neura.edu.au.
Michael Janitz, Email: m.janitz@unsw.edu.au.
References
- 1.Mattick JS. The central role of RNA in human development and cognition. FEBS Lett. 2011;585:1600–16. doi: 10.1016/j.febslet.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338:1435–9. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 4.Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, et al. Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation. BMC Neurosci. 2010;11:14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mills JD, Kavanagh T, Kim WS, Chen BJ, Kawahara Y, Halliday GM, et al. Unique transcriptome patterns of the white and grey matter corroborate structural and functional heterogeneity in the human frontal lobe. PLoS One. 2013;8:e78480. doi: 10.1371/journal.pone.0078480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–40. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 7.Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–8. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 8.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang B, Alrahbeni TM, Clair DS, Blackwood DH, International Schizophrenia C, McCaig CD, et al. HDAC9 is implicated in schizophrenia and expressed specifically in post-mitotic neurons but not in adult neural stem cells. Am J Stem Cells. 2012;1:31–41. [PMC free article] [PubMed] [Google Scholar]
- 10.Broide RS, Redwine JM, Aftahi N, Young W, Bloom FE, Winrow CJ. Distribution of histone deacetylases 1–11 in the rat brain. J Mol Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 11.Milde T, Oehme I, Korshunov A, Kopp-Schneider A, Remke M, Northcott P, et al. HDAC5 and HDAC9 in medulloblastoma: novel markers for risk stratification and role in tumor cell growth. Clin Cancer Res. 2010;16:3240–52. doi: 10.1158/1078-0432.CCR-10-0395. [DOI] [PubMed] [Google Scholar]
- 12.Potzner MR, Griffel C, Lutjen-Drecoll E, Bosl MR, Wegner M, Sock E. Prolonged Sox4 expression in oligodendrocytes interferes with normal myelination in the central nervous system. Mol Cell Biol. 2007;27:5316–26. doi: 10.1128/MCB.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoser M, Baader SL, Bosl MR, Ihmer A, Wegner M, Sock E. Prolonged glial expression of Sox4 in the CNS leads to architectural cerebellar defects and ataxia. J Neurosci. 2007;27:5495–505. doi: 10.1523/JNEUROSCI.1384-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mogha A, Benesh AE, Patra C, Engel FB, Schoneberg T, Liebscher I, et al. Gpr126 functions in Schwann cells to control differentiation and myelination via G-protein activation. J Neurosci. 2013;33:17976–85. doi: 10.1523/JNEUROSCI.1809-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monk KR, Oshima K, Jors S, Heller S, Talbot WS. Gpr126 is essential for peripheral nerve development and myelination in mammals. Development. 2011;138:2673–80. doi: 10.1242/dev.062224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Binder MD, Cate HS, Prieto AL, Kemper D, Butzkueven H, Gresle MM, et al. Gas6 deficiency increases oligodendrocyte loss and microglial activation in response to cuprizone-induced demyelination. J Neurosci. 2008;28:5195–206. doi: 10.1523/JNEUROSCI.1180-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsiperson V, Li X, Schwartz GJ, Raine CS, Shafit-Zagardo B. GAS6 enhances repair following cuprizone-induced demyelination. PLoS One. 2010;5:e15748. doi: 10.1371/journal.pone.0015748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binder MD, Xiao J, Kemper D, Ma GZ, Murray SS, Kilpatrick TJ. Gas6 increases myelination by oligodendrocytes and its deficiency delays recovery following cuprizone-induced demyelination. PLoS One. 2011;6:e17727. doi: 10.1371/journal.pone.0017727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swiss VA, Nguyen T, Dugas J, Ibrahim A, Barres B, Androulakis IP, et al. Identification of a gene regulatory network necessary for the initiation of oligodendrocyte differentiation. PLoS One. 2011;6:e18088. doi: 10.1371/journal.pone.0018088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavallaro M, Mariani J, Lancini C, Latorre E, Caccia R, Gullo F, et al. Impaired generation of mature neurons by neural stem cells from hypomorphic Sox2 mutants. Development. 2008;135:541–57. doi: 10.1242/dev.010801. [DOI] [PubMed] [Google Scholar]
- 21.Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–19. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 22.D’Amelio M, Cavallucci V, Cecconi F. Neuronal caspase-3 signaling: not only cell death. Cell Death Differ. 2010;17:1104–14. doi: 10.1038/cdd.2009.180. [DOI] [PubMed] [Google Scholar]
- 23.Bergsland M, Werme M, Malewicz M, Perlmann T, Muhr J. The establishment of neuronal properties is controlled by Sox4 and Sox11. Genes Dev. 2006;20:3475–86. doi: 10.1101/gad.403406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hangauer MJ, Vaughn IW, McManus MT. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013;9:e1003569. doi: 10.1371/journal.pgen.1003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaushik K, Leonard VE, Kv S, Lalwani MK, Jalali S, Patowary A, et al. Dynamic expression of long non-coding RNAs (lncRNAs) in adult zebrafish. PLoS One. 2013;8:e83616. doi: 10.1371/journal.pone.0083616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perelman P, Johnson WE, Roos C, Seuanez HN, Horvath JE, Moreira MA, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7:e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 2011;147:1537–50. doi: 10.1016/j.cell.2011.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–27. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Church DM, Goodstadt L, Hillier LW, Zody MC, Goldstein S, She X, et al. Lineage-specific biology revealed by a finished genome assembly of the mouse. PLoS Biol. 2009;7:e1000112. doi: 10.1371/journal.pbio.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ponjavic J, Oliver PL, Lunter G, Ponting CP. Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet. 2009;5:e1000617. doi: 10.1371/journal.pgen.1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kutter C, Watt S, Stefflova K, Wilson MD, Goncalves A, Ponting CP, et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;8:e1002841. doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22:1775–89. doi: 10.1101/gr.132159.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 37.Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc Natl Acad Sci U S A. 1996;93:11883–8. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3:ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–9. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim JB, Choi JS, Nam K, Lee M, Park JS, Lee JK. Enhanced transfection of primary cortical cultures using arginine-grafted PAMAM dendrimer, PAMAM-Arg. J Control Release. 2006;114:110–7. doi: 10.1016/j.jconrel.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 41.Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- 42.Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–26. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Young RS, Marques AC, Tibbit C, Haerty W, Bassett AR, Liu JL, et al. Identification and properties of 1,119 candidate lincRNA loci in the Drosophila melanogaster genome. Genome Biol Evol. 2012;4:427–42. doi: 10.1093/gbe/evs020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ho-Pun-Cheung A, Bascoul-Mollevi C, Assenat E, Boissiere-Michot F, Bibeau F, Cellier D, et al. Reverse transcription-quantitative polymerase chain reaction: description of a RIN-based algorithm for accurate data normalization. BMC Mol Biol. 2009;10:31. doi: 10.1186/1471-2199-10-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eisenberg E, Levanon EY. Human housekeeping genes are compact. Trends Genet. 2003;19:362–5. doi: 10.1016/S0168-9525(03)00140-9. [DOI] [PubMed] [Google Scholar]
- 48.Forlenza M, Kaiser T, Savelkoul HF, Wiegertjes GF. The use of real-time quantitative PCR for the analysis of cytokine mRNA levels. Methods Mol Biol. 2012;820:7–23. doi: 10.1007/978-1-61779-439-1_2. [DOI] [PubMed] [Google Scholar]
- 49.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 50.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 51.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]


