Abstract
Background
X chromosome inactivation is the transcriptional silencing of one X chromosome in the somatic cells of female mammals. In eutherian mammals (e.g. humans) one of the two X chromosomes is randomly chosen for silencing, with about 15% (usually in younger evolutionary strata of the X chromosome) of genes escaping this silencing. In contrast, in the distantly related marsupial mammals the paternally derived X is silenced, although not as completely as the eutherian X. A chromosome wide examination of X inactivation, using RNA-seq, was recently undertaken in grey short-tailed opossum (Monodelphis domestica) brain and extraembryonic tissues. However, no such study has been conduced in Australian marsupials, which diverged from their American cousins ~80 million years ago, leaving a large gap in our understanding of marsupial X inactivation.
Results
We used RNA-seq data from blood or liver of a family (mother, father and daughter) of tammar wallabies (Macropus eugenii), which in conjunction with available genome sequence from the mother and father, permitted genotyping of 42 expressed heterozygous SNPs on the daughter’s X. These 42 SNPs represented 34 X loci, of which 68% (23 of the 34) were confirmed as inactivated on the paternally derived X in the daughter’s liver; the remaining 11 X loci escaped inactivation. Seven of the wallaby loci sampled were part of the old X evolutionary stratum, of which three escaped inactivation. Three loci were classified as part of the newer X stratum, of which two escaped inactivation. A meta-analysis of previously published opossum X inactivation data revealed that 5 of 52 genes in the old X stratum escaped inactivation.
Conclusions
We demonstrate that chromosome wide inactivation of the paternal X is common to an Australian marsupial representative, but that there is more escape from inactivation than reported for opossum (32% v 14%). We also provide evidence that, unlike the human X chromosome, the location of loci within the oldest evolutionary stratum on the marsupial X does not correlate with their probability of escape from inactivation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-014-0267-z) contains supplementary material, which is available to authorized users.
Keywords: X inactivation, Marsupial, Imprinting, X chromosome, Escape, RNA-seq, SNP, Evolution, Strata
Background
Therian (i.e. eutherian and marsupial) mammals have a XX female/XY male sex chromosome system, or some simple variant of it. The X and Y chromosomes evolved from an ordinary pair of autosomes via a process of gene loss on the Y [1], which resulted in an imbalance of X gene dosage with the autosomes (1X: 2A gene dosage) in males. It was proposed that to rebalance transcriptional output between the autosomes and the X chromosome, genes (or a subset of dosage sensitive genes [2]) on the single X in males were upregulated [3]. Carry through of X chromosome upregulation to females would result in functional X tetrasomy (i.e. 4X: 2A gene dosage), which was subsequently balanced by transcriptional silencing of one X in the somatic cells of females – called X chromosome inactivation (XCI) [4].
During early embryogenesis, in eutherian females, the X inactive specific transcript (XIST) is expressed from the X inactivation center (Xic) on the future inactive X chromosome (Xi). XIST, a long non-coding RNA (lncRNA), coats the X chromosome in cis and triggers a cascade of epigenetic events that stably represses transcription of most X loci [5] (reviewed in [6]). In human, genes in older regions of the X chromosome are more likely to be inactivated, whereas genes located in the evolutionarily younger regions of the X chromosome are more likely to be expressed from the Xi (the escaper genes) [7,8]. The choice of which X chromosome (maternal or paternal) becomes inactivate is random [4,9-12]. Although some properties of XCI mechanism vary between species (like timing of XCI initiation [12]), the general epigenetic profile of the Xi appears to be well conserved across eutherian mammals [9,13-15].
Eutherians diverged from the marsupial lineage ~180 million years ago (MYA). The marsupial X is homologous to the long arm and proximal short arm of the human X (denoted as the X conserved region XCR [16]). Like eutherian mammals, one of the two Xs in marsupials is inactivated [17], and although some epigenetic features of the Xi are conserved between clades [18], there are also considerable differences [19,20]. The region homologous to the eutherian Xic has been disrupted on the marsupial X chromosome [21-23] and there is no XIST homologue [24]. Instead, the lncRNA gene RSX (RNA on the silent X) appears to be involved in marsupials XCI [25]. Like XIST, the RSX transcript is exclusively expressed from the Xi and coats it in cis [25].
Early studies of allozyme variants and somatic cell hybrids of four genes in different marsupial species (G6PD, PGK1, GLA and HPRT [26-30]) proposed that marsupial XCI was incomplete and variable between tissues and species (reviewed in [17]). Additionally, the results showed that there was imprinted inactivation of the paternally derived X chromosome (as has been described for extraembryonic tissues of rodents and cow [31-33]). A recent genome-wide study in opossum (Monodelphis domestica) fetal brain and extraembryonic tissues reported complete paternal inactivation for most (86%) genes on the X chromosome, and escape from XCI for 14% of genes [34]. This level of escape is comparable to that reported for the whole human X (15%). However, it is higher than the level of escape on both the mouse X (3%) [35], and the regions of the human X homologous to the marsupial X (6.5%) [7,35].
Comparison of XCI between different tissues and species offers new insights to its mechanism and evolution. In this study we used RNA-Seq data from a captive bred tammar wallaby family (i.e father, mother and daughter), and genome sequencing from the father and the mother [36], to determine the parent of origin of expressed alleles on the daughter’s X chromosome. We confirm paternal inactivation on the tammar wallaby X chromosome, but observed more escape from inactivation than previously reported for human, mouse and opossum.
Results and discussion
Imprinted inactivation of the paternal X chromosome in marsupials was hypothesised more than 20 years ago based on evidence from four genes in six different marsupial species [17]. Consistent with this, a more recent study [34] confirmed preferential paternal inactivation as a chromosome-wide phenomenon in opossum brain and extraembryonic tissues. Here we analysed expression of X-linked genes in liver and blood from an Australian marsupial, the tammar wallaby. We sequenced polyadenylated RNA (RNA-sequencing) from a father and mother blood sample, along with a liver sample from the daughter. To overcome the limitation of some alleles not being present in the mother transcriptome due to silencing via XCI, we also used DNA-sequencing data available from the father and the mother [36].
Mapping results and SNP calling
An average of ~50 million (M) reads were obtained for each of the three samples, of which approximately 26 M, 20 M and 30 M were uniquely mapped for father, mother and daughter respectively (Additional file 1). In total 44,095 SNPs were called between the wallaby reference genome and our three samples. 68% of SNPs were shared between the three individuals (due to being a captive bred colony), so were non-informative when determining parent-of-origin of SNPs in the daughter. A total of 356 SNPs (~1.5% of total) called (in at least one sample) against the reference genome were assigned to the X chromosome (Additional file 2). SNPs that were associated with genes (or scaffolds with genes) that did not have a 1:1 orthologue in human and/or opossum, plus those that we were unable to anchor to the X chromosome, were not included in the analysis (see Methods). Some reported SNPs were located outside wallaby annotated genes and/or exons, likely due to the poor assembly and annotation of the tammar wallaby genome, and were still considered in the analysis.
Assessing allele-specific expression
We calculated relative expression of each allele for heterozygous biallelically expressed SNPs in the daughter’s liver and the mother’s blood (as performed by Wang et al. [34]). In addition, we also included SNPs that were confidently called as heterozygous in daughter (following Mendelian inheritance based on parental genotype), but that were monoallelically expressed (i.e. SNPs that appeared as homozygous in the transcriptome). As such, we could calculate relative expression of each allele for both biallelically and monoallelically expressed heterozygous SNPs.
We observed apparent biallelic expression from the X chromosome in the father, likely because Y-derived reads from the male were reported as best mapped to the X chromosome. This is not surprising since a female individual was sequenced and Y sequence is absent in the tammar wallaby genome assembly [37]. To date, 17 genes have been identified on the tammar wallaby Y chromosome, almost all of which have an X-linked copy [36,38]. We identified 9 X genes with heterozygous SNPs in male including UBA1X and ATRX, both of which have a known copy on the wallaby Y chromosome (UBE1Y and ATRY) [38] (Additional file 3). The remaining genes on the X with apparent biallelic expression in males could represent unidentified homologies between the X and Y in tammar wallaby, or multi copy genes on the X; these SNPs were all removed from the analysis.
SNPs were detected for 200 X-linked loci in daughter (Additional file 2); based on parental transcriptome and genome, as well as the daughter transcriptome, we could infer the daughter’s genotype and determine the parent-of-origin of for 42 SNPs in 34 loci (Additional file 4). Twenty-seven SNPs (representing 23 different loci; 68%) had complete monoallelic expression, all of which were exclusively expressed from the maternal allele (i.e. paternal XCI; Additional file 4).
The remaining 15 SNPs (representing 11 loci; 32%) had biallelic expression and were considered escapers (>10% of reads from the lowly expressed allele); the proportion of escape from XCI that we observed in wallaby was considerably higher than that previously reported for human (15%), mouse (3%) and opossum (14%) [7,34,35]. Wang et al [34] focused on heterozygous biallelically expressed SNPs, which could potentially underestimate the proportion of inactive loci because heterozygous SNPs expressed from a single allele would be overlooked. When analysing only heterozygous biallelically expressed SNPs on the X, we observed that all loci (29 out of 29) escaped X inactivation in liver (Additional file 2). In addition, we examined heterozygous biallelically expressed X-borne SNPs in the mother’s blood and observed that 2 out of 19 loci escaped inactivation (Additional file 2). This frequency is much higher than the reported for opossum brain and extraembryonic tissues using equivalent methods (14% and 18% respectively).
Average expression from the paternal allele of SNPs that escape XCI was 46% (Additional file 4), whereas in opossum expression from the paternal allele (Xi) is significantly lower than the maternal allele (average of 28%) for most escaper genes [34]. Although different tissues were studied (liver and blood versus fibroblast, brain and extraembryonic tissues), this pronounced escape is likely a reflection of the relative “leakiness” of the tammar wallaby XCI system, which was also observed in tammar wallaby cultured fibroblasts [39].
Generally, SNPs within the same gene or in nearby regions exhibited very similar allelic bias, with the exception of three SNPs mapped to Scaffold28735: two SNPs (X-SNP-I and X-SNP-E) were biallelically expressed, and the nearby SNP (X-SNP-V) ~1.3 kb down stream had paternal inactivation (Additional file 4).
Inactive status of annotated genes
To determine inactivation status for tammar wallaby X genes we examined only the SNPs within annotated genes. Complete paternal inactivation was observed for ten genes (Table 1) and unbiased biallelic expression for seven genes (Table 1). SNPs in EBP, RPS4X and VAMP7 were validated as heterozygous in daughter by conventional sequencing (Additional file 5).
Table 1.
Expression status for the 17 wallaby annotated X-linked genes for which parent-of-origin could be determined
| Ensembl ID | Gene name | Coverage | Father | Mother | Inactive status | Escapee | Inactive |
|---|---|---|---|---|---|---|---|
| ENSMEUG00000006054 | NAA10 | 8 | 62% | 38% | Biallelic | - | - |
| ENSMEUG00000012279 | SMC6 | 39 | 54% | 46% | Biallelic | - | - |
| ENSMEUG00000005589 | CHM | 6 | 33% | 67% | Biallelic | - | - |
| ENSMEUG00000008176 | HCFC1 | 10 | 30% | 70% | Biallelic | Mdo/Hsa | Mmu |
| ENSMEUG00000004133 | HTATSF1 | 10 | 40% | 60% | Biallelic | - | Hsa/Mmu |
| ENSMEUG00000002761 | MECP2 | 7 | 57% | 43% | Biallelic | Mdo | Hsa/Mmu |
| ENSMEUG00000015172 | VAMP7 | 30 | 40% | 60% | Biallelic | - | - |
| ENSMEUG00000015485 | RAS11LC | 30 | 0% | 100% | PI | - | - |
| ENSMEUG00000015892 | RBMX | 6 | 0% | 100% | PI | - | - |
| ENSMEUG00000002871 | EBP | 50 | 0% | 100% | PI | - | - |
| ENSMEUG00000012762 | LAS1L | 5 | 0% | 100% | PI | Mdo/Hsa | Mmu |
| ENSMEUG00000007878 | OPHN1 | 7 | 0% | 100% | PI | - | Mdo/Hsa/Mmu |
| ENSMEUG00000011740 | PFKFB1 | 26 | 0% | 100% | PI | - | Hsa |
| ENSMEUG00000007969 | RPS4X | 23 | 0% | 100% | PI | Mdo/Hsa | Mmu |
| ENSMEUG00000003387 | HAUS7 | 20 | 0% | 100% | PI | - | Mmu |
| ENSMEUG00000003603 | - | 6 | 0% | 100% | PI | - | - |
| ENSMEUG00000000692 | TSPAN6 | 5 | 0% | 100% | PI | Hsa | Mmu |
PI denotes paternal inactivation. Escapee column denotes genes that are confirmed as escaping X inactivation in other mammalian species; inactive column denotes genes that are confirmed as inactive in other mammalian species (Mdo: Monodelphis domestica [34]; Hsa: Homo sapiens [7]; Mmu: Mus musculus [35]; -: not information available).
Finally, we attempted to determine parent of origin for expression of RSX (which is expressed exclusively from the inactive X chromosome [25]) to better elucidate which X chromosome was inactivated. In daughter no biallelic expression (heterozygous SNPs in the transcriptome data) of RSX was observed, and because there was no coverage of RSX in either of the parental genomes the daughter’s genotype could not be inferred. Genotyping (via Sanger sequencing; data not shown) 4.4 kb of the non-repetitive regions of RSX in each individual revealed no heterozygous SNPs. It was, therefore, impossible to determine whether RSX had monoallelic expression from the paternal allele in the daughter.
Our previous work, using RNA-FISH to conduct single-cell expression assays in tammar wallaby fibroblast nuclei, provided a similar result to this study where all loci tested escaped XCI to some degree [39]. Only two genes sampled in the present study were assayed previously: 1) MECP2 was expressed from both X chromosomes in 41% of female nuclei, and in the present study we observed nearly equal expression from the paternal and maternal alleles (Table 1). And 2) HCFC1 was expressed from both X chromosomes in <10% nuclei (considered inactive), whereas in the present study it was expressed at 30% from paternal allele and 70% from the maternal allele (Table 1). Contrary to our findings, none of the genes studied in [39] completely escaped XCI (i.e. expressed from both X chromosomes in all nuclei), a discrepancy likely due to the different cell types studied.
Inactivation does not correlate with location on the X in marsupials
Biallelic expression ratios of close to 1:1 were observed along the length of the X chromosome with no apparent correlation between inactivation and locus location, or correlation with proximity to RSX (Figure 1). This is in stark contrast to human, where X-linked genes located in newer strata are more likely to escape inactivation than genes in the older strata. In the human XCR (homologous to the marsupial X) 30 of 462 (6.5%) sampled genes escape inactivation [7]. This is significantly fewer (Fisher’s exact; p = 6.2e-06) than the expected number of escapee genes (69 of 462) should the XCR have the same frequency of escape from inactivation as the whole X.
Figure 1.
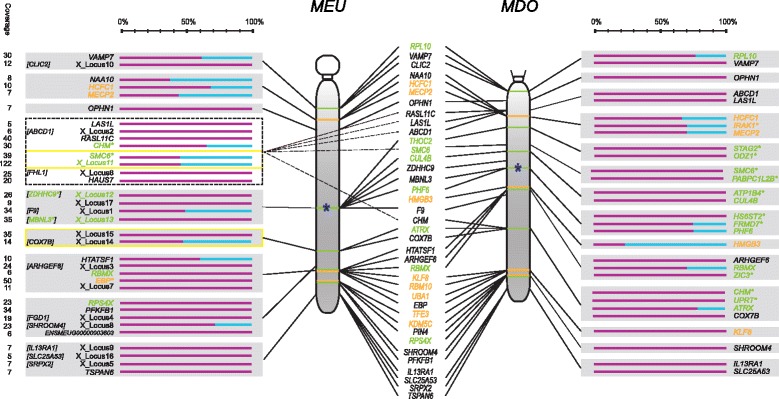
Parent of origin expression status of X-linked loci on the wallaby and opossum X chromosome. The position of SNPs on the wallaby X chromosome was predicted using the physical map [40] and opossum sytenic blocks with 1:1 orthologous genes. SNPs in wallaby intergenic regions are labelled as X_Locus 1-17, with the name of the neighbouring gene in brackets. SNPs for which position could not be resolved in wallaby are shown in dotted lines. Read depth at each position is given in the ‘Coverage’ column. Yellow boxes identify SNPs within the same scaffold. Orange represents genes in the newest X stratum in marsupials, genes highlighted in green are in the oldest stratum, genes highlighted in black could not be assigned a stratum, and the stars (*) beside gene names denote predicted new or old genes (see Methods). Parent of origin expression of the opossum genes were obtained from [34]. Bars represent the percentage of reads derived from the maternal (pink) or paternal (blue) allele. The dark blue stars are the predicted locations of RSX in each species.
Two evolutionary strata were recently identified on the marsupial X (here called ‘old’ and ‘new’ [36]), into which X-linked loci were binned when possible (see Methods). We did not observe a correlation with escape from inactivation and location in either of the two marsupial X evolutionary strata [36]. Seven of the 34 wallaby X loci sampled could be classified as members of the oldest X stratum, with three escaping XCI. Although the number of genes sampled in the old stratum was low, the frequency of escape remained similar to the frequency of escape on the whole X (32%). Likewise, of the three loci assigned to the newer tammar wallaby X stratum, two escaped inactivation.
After examination of previously published opossum X inactivation data ([34]; see Methods) just five genes could be assigned to the newer stratum, of which four escaped inactivation. However, 52 genes with known XCI status were placed in the oldest marsupial X stratum, of which five escaped XCI. In contrast to the different inactivation levels between human X strata, the number of escapee genes in the opossum old X stratum was not statistically different (Fisher’s exact; p = 0.7602) from the expected number of escapee genes (7 of 52; calculated from the whole X frequency of escape: 24 of 176).
Conclusions
Our results demonstrate that chromosome wide inactivation of the paternally inherited X is common to both Australian and American marsupials. Also, in stark contrast to human where just 6.5% of genes located in the two oldest X chromosome evolutionary strata escape inactivation and 40.7% escape inactivation in the three youngest strata, we show that a higher probability of inactivation does not correlate with location in the oldest marsupial X stratum.
Methods
Sample collection
Samples were collected and held under the Australian National University’s Animal Experimentation Ethics Committee proposal number A2011/57, and the CSIRO Sustainable Ecosystems Animal Ethics Committee application number 10-02. Blood samples were obtained from the mother and the father, and liver tissue sample was obtained from a 12 week old female pouch young and snap frozen in liquid nitrogen.
RNA extraction and sequencing
Total RNA from parental blood was extracted from 400 uL starting material using the RiboPure Blood™ Kit (Ambion) following manufacturer’s instructions. RNA from liver was extracted using the GenElute® Total Mammalian RNA Extraction Kit (Sigma-Aldrich) following the manufacturer’s guidelines. RNA integrity (RIN) and quantity was checked on an Agilent 2100 Bioanalyzer (Agilent Technologies). RNA (RIN >7) was shipped in RNAstable® storage tubes (Biometrica). A single run for each sample was performed at the Beijing Genomics Institute (BGI), Tai Po, Hong Kong. Poly(A) mRNA was isolated utilizing oligo(dT) beads, fragmented and random hexamer-primers were used to synthesize single-strand cDNA, followed by a second round of cDNA synthesis. Fragments were purified and concentrated with the QiaQuick® PCR Extraction Kit, eluted with EB buffer for end repair followed by addition of poly(A). Resulting fragments were ligated with sequencing adaptors, size-selected and single end 90 bp reads were generated using the Illumina HiSeq™ 2000 platform. These sequences are available at the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra; accession number: PRJNA258238).
Read mapping
Raw reads were processed using the web-based Galaxy server [41] (https://usegalaxy.org). Read quality was assessed using the FastQC software available at the Galaxy server (v0.52) and processed with the FASTX-Toolkit (v.0.0.13). Reads were mapped using TopHat v1.1.2 [42] to the wallaby genome release 74 (MacEug1.0.74) (ftp://ftp.ensembl.org/pub/release-74/fasta/macropus_eugenii/dna/), allowing a maximum of one mismatch and reporting only uniquely aligned reads. Post-mapping processing and duplicate removal was done with SAMTools (v.0.1.18) [43]. Reads were visualized using Integrative Genome Viewer (IGV) [44].
Single nucleotide polymorphism (SNP) calling and filtering
For SNP calling, SAMTools [43] and VarScan (v2.3.5) [45] was utilised, high coverage bases (≥5 at each SNP position) were reported, and were manually curated. Heterozygous SNPs with a single read supporting either allele were compared to SNPs in the vicinity (i.e. SNPs in the same gene; or in a ~20 kb window for SNPs in intergenic regions) and were ignored if at least three neighbour SNPs were homozygous and no heterozygous SNPs were present in the vicinity.
Parent of origin
Parent of origin for each allele in the daughter was determined by comparing each SNP to the same position in each parent where information was available from the transcriptome (from this study) and/or genome [36], with three possible outcomes: complete inactivation, ambiguous or biallelic expression (Additional file 6). For SNPs with biallelic expression, we quantified allele-specific expression as the ratio of the number of reads supporting a particular allele divided by the total coverage at that position; SNPs were considered escapers if more than 10% of reads support the lowly expressed allele [7].
Assigning SNPs to chromosomes and annotated gene exons
Wallaby annotated protein-coding genes with 1:1 human orthologues were obtained from Ensembl74. Gene scaffolds in the tammar wallaby were assigned to the X chromosome when at least 1:1 orthologue on the human XCR and/or the opossum X was observed. Genes were assigned as autosomal if there was 1:1 orthologue on the remainder of human X chromosome, or on human autosomes. SNPs in exonic regions were annotated based on exon genomic coordinates available from Ensembl74. For X-assigned SNPs mapped to intergenic regions in X scaffolds, a 1000 bp window (500 bp upstream and 500 bp downstream) was search with BLAST (discontiguous megablast) against the NCBI nucleotide collection, to identify potential unannotated tammar wallaby genes that were orthologues to known human or opossum X-linked genes.
Assigning loci to X chromosome strata
Opossum X-linked loci were classified as ‘old’ or ‘new’ genes according to the strata information in Cortez et al. [36]. Loci that mapped between two X genes in the old stratum were considered old loci. Similarly, X-linked loci located between two new genes were considered to be in the new stratum. All loci located between an old and a new gene were excluded from the strata analysis. Conserved syntenic blocks between opossum and wallaby [40] were used to assign genes to the new or old strata in tammar wallaby.
RSX
Homology to the opossum RSX was previously reported for three wallaby ESTs: EX198340.1, FY484131.3 and FY484132.2 [25], all of which map to a ~2 kb region at the end of wallaby Scaffold61551. To detect further regions of homology, the RSX complete sequence (X:35,605,415-35,651,609; coordinates from [25]) and individual exons [25] were searched for (using BLAST) in the wallaby reference genome (MacEug1.0.70).
PCR and DNA sequencing
The DNeasy® Blood and Tissue Kit (Qiagen) were used to extract genomic DNA from each individual, following the manufacturer’s instructions. Primers were designed using Primer3 [46] (Additional file 7). PCR reactions were performed using 100 ng/uL template DNA, MyTaq™ Red Mix (Bioline) and 1 pmol of primers, with initial denaturation at 94°C for 2 minutes, followed by 36 cycles of: 30 seconds at 94°C, 30 seconds at 50°C-65°C, 60 seconds at 72°C; then 72°C for 10 minutes. Resulting products were separated and purified using the PureLink® Kit (Invitrogen). DNA sequencing was performed by the Biomolecular Resource Facilty (The Australian National University), or the Ramaciotti Centre (University of New South Wales). Sequence trace files were visualised using 4 Peaks software from Nucleobytes (www.nucleobytes.com) and sequence alignments were performed in ClustalX 2.1 with default parameters.
Availability of supporting data
Transcriptome sequences generated in this analysis are available in the NCBI Sequence Read Archive (SRA) database, (http://www.ncbi.nlm.nih.gov/sra), with accession number: PRJNA258238 (http://www.ncbi.nlm.nih.gov/bioproject/258238). Genome sequences are available from [36].
Acknowledgements
We thank Prof Henrik Kaessmann and Dr Diego Cortez for providing mapped parental genomic sequence data. CLRD was funded by the Mexican government branch Consejo Nacional de Ciencia y Tecnología (CONACyT). PDW was supported by an ARC Australian Research Fellowship.
Abbreviations
- XCI
X chromosome inactivation
- XIST
X inactive specific transcript
- Xic
X inactive centre
- Xi
inactive X chromosome
- MYA
Million years ago
- XCR
X conserved region
- RSX
RNA-on the silent X
- SNP
Single nucleotide polymorphism
- M
Million
Additional files
Mapping statistics.
The 23,613 SNPs that were assigned to an autosome or X chromosome based on 1:1 orthology between wallaby and human autosomes, and wallaby and human/opossum X chromosomes.
X-linked SNPs in father with biallelic expression (concluded from transcriptome or genome) that were removed from the analysis.
SNPs on the X chromosome for which parent of origin could be determined in the daughter.
Genotype validation of three X-linked SNPs via conventional Sanger sequencing.
Criteria (and examples) followed to determine parent-of-origin for X-linked SNPs in the daughter. Pink represents maternal allele; blue represents paternal allele; red represents unknown origin. The transcriptome is representative of genotype for X-linked SNPs in the father.
Primers used to amplify regions spanning SNPs selected for genotype validation, and amplify random regions in the RSX transcript.
Footnotes
Competing interests
The authors have declared that they have no competing interests.
Author’s contributions
CLRD performed extractions and expression analysis. CLRD and SAW performed PCR analysis. CLRD drafted the manuscript. PDW conceived, designed and directed the study. PDW and SAW contributed to revision of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Claudia L Rodríguez-Delgado, Email: claudia.rodriguez@anu.edu.au.
Shafagh A Waters, Email: shafagh.waters@unsw.edu.au.
Paul D Waters, Email: p.waters@unsw.edu.au.
References
- 1.Muller HJ. A gene for the fourth chromosome of drosophila. J Exp Zool. 1914;17(3):325–336. doi: 10.1002/jez.1400170303. [DOI] [Google Scholar]
- 2.Pessia E, Makino T, Bailly-Bechet M, McLysagh A, Marias ABG. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012;109(14):5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohno S. Sex Chromosomes and Sex Linked Genes. Berlin: Springer; 1967. [Google Scholar]
- 4.Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- 5.Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991;349(6304):38–44. doi: 10.1038/349038a0. [DOI] [PubMed] [Google Scholar]
- 6.Heard E, Clerc P, Avner P. X-chromosome inactivation in mammals. Annu Rev Genet. 1997;31:571–610. doi: 10.1146/annurev.genet.31.1.571. [DOI] [PubMed] [Google Scholar]
- 7.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 8.Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008;4(1):e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X, Miller DC, Clark AG, Antczak DF. Random X inactivation in the mule and horse placenta. Genome Res. 2012;22(10):1855–1863. doi: 10.1101/gr.138487.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- 11.de Mello JC M, de Araujo ES, Stabellini R, Fraga AM, de Souza JE, Sumita DR, Camargo AA, Pereira LV. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS One. 2010;5(6):e10947. doi: 10.1371/journal.pone.0010947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, Heard E. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature. 2011;472(7343):370–374. doi: 10.1038/nature09872. [DOI] [PubMed] [Google Scholar]
- 13.Waters PD, Ruiz-Herrera A, Dobigny G, Garcia Caldes M, Robinson TJ. Sex chromosomes of basal placental mammals. Chromosoma. 2007;116(6):511–518. doi: 10.1007/s00412-007-0116-6. [DOI] [PubMed] [Google Scholar]
- 14.Waters PD, Dobigny G, Pardini AT, Robinson TJ. LINE-1 distribution in Afrotheria and Xenarthra: implications for understanding the evolution of LINE-1 in eutherian genomes. Chromosoma. 2004;113(3):137–144. doi: 10.1007/s00412-004-0301-9. [DOI] [PubMed] [Google Scholar]
- 15.Bell RJ, Lees GE, Murphy KE. X chromosome inactivation patterns in normal and X-linked hereditary nephropathy carrier dogs. Cytogenet Genome Res. 2008;122(1):37–40. doi: 10.1159/000151314. [DOI] [PubMed] [Google Scholar]
- 16.Graves JA, Watson JM. Mammalian sex chromosomes: evolution of organization and function. Chromosoma. 1991;101(2):63–68. doi: 10.1007/BF00357055. [DOI] [PubMed] [Google Scholar]
- 17.Cooper DW, Johnston PG, Vandeberg JL, Robinson ES. X-Chromosome Inactivation in Marsupials. Aust J Zool. 1990;37(2–4):411–417. [Google Scholar]
- 18.Mahadevaiah SK, Royo H, VandeBerg JL, McCarrey JR, Mackay S, Turner JM. Key features of the X inactivation process are conserved between marsupials and eutherians. Curr Biol: CB. 2009;19(17):1478–1484. doi: 10.1016/j.cub.2009.07.041. [DOI] [PubMed] [Google Scholar]
- 19.Chaumeil J, Waters PD, Koina E, Gilbert C, Robinson TJ, Graves JA. Evolution from XIST-independent to XIST-controlled X-chromosome inactivation: epigenetic modifications in distantly related mammals. PLoS One. 2011;6(4):e19040. doi: 10.1371/journal.pone.0019040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rens W, Wallduck MS, Lovell FL, Ferguson-Smith MA, Ferguson-Smith AC. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc Natl Acad Sci U S A. 2010;107(41):17657–17662. doi: 10.1073/pnas.0910322107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hore TA, Koina E, Wakefield MJ, Marshall Graves JA. The region homologous to the X-chromosome inactivation centre has been disrupted in marsupial and monotreme mammals. Chromosome Res. 2007;15(2):147–161. doi: 10.1007/s10577-007-1119-0. [DOI] [PubMed] [Google Scholar]
- 22.Shevchenko AI, Zakharova IS, Elisaphenko EA, Kolesnikov NN, Whitehead S, Bird C, Ross M, Weidman JR, Jirtle RL, Karamysheva TV, Rubtsov NB, VandeBerg JL, Mazurok NA, Nesterova TB, Brockdorff N, Zakian SM. Genes flanking Xist in mouse and human are separated on the X chromosome in American marsupials. Chromosome Res. 2007;15(2):127–136. doi: 10.1007/s10577-006-1115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidow LS, Breen M, Duke SE, Samollow PB, McCarrey JR, Lee JT. The search for a marsupial XIC reveals a break with vertebrate synteny. Chromosome Res. 2007;15(2):137–146. doi: 10.1007/s10577-007-1121-6. [DOI] [PubMed] [Google Scholar]
- 24.Duret L, Chureau C, Samain S, Weissenbach J, Avner P. The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science. 2006;312(5780):1653–1655. doi: 10.1126/science.1126316. [DOI] [PubMed] [Google Scholar]
- 25.Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, Royo H, Duckworth J, McCarrey JR, VandeBerg JL, Renfree MB, Taylor W, Elgar G, Camerini-Otero RD, Gilchrist MJ, Turner JM. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature. 2012;487(7406):254–258. doi: 10.1038/nature11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston PG, Sharman GB. Studies on metatherian sex chromosomes. I. Inheritance and inactivation of sex-linked allelic genes determining glucose-6-phosphate dehydrogenase variation in kangaroos. Aust J Biol Sci. 1975;28(5-6):567–574. doi: 10.1071/bi9750567. [DOI] [PubMed] [Google Scholar]
- 27.Samollow PB, Ford AL, VandeBerg JL. X-linked gene expression in the Virginia opossum: differences between the paternally derived Gpd and Pgk-A loci. Genetics. 1987;115(1):185–195. doi: 10.1093/genetics/115.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper DW, Edwards C, James E, Sharman GB, Vandeberg JL, Marshall Graves JA. Studies on metatherian sex chromosomes VI. A third state of an X-linked gene: partial activity for the paternally derived Pgk-A allele in cultured fibroblasts of Macropus giganteus and M. parryi. Aust J Biol Sci. 1977;30:431–443. [Google Scholar]
- 29.Cooper DW, Woolley PA, Maynes GM, Sherman FS, Poole WE. Studies on metatherian sex chromosomes. XII. Sex-linked inheritance and probable paternal X-inactivation of alpha-galactosidase A in Australian marsupials. Aust J Biol Sci. 1983;36(5–6):511–517. doi: 10.1071/bi9830511. [DOI] [PubMed] [Google Scholar]
- 30.Graves JA, Dawson GW. The relationship between position and expression of genes on the kangaroo X chromosome suggests a tissue-specific spread of inactivation from a single control site. Genet Res. 1988;51(2):103–109. doi: 10.1017/S0016672300024113. [DOI] [PubMed] [Google Scholar]
- 31.Wake N, Takagi N, Sasaki M. Non-random inactivation of X chromosome in the rat yolk sac. Nature. 1976;262(5569):580–581. doi: 10.1038/262580a0. [DOI] [PubMed] [Google Scholar]
- 32.West JD, Papaioannou VE, Frels WI, Chapman VM. Preferential expression of the maternally derived X chromosome in extraembryonic tissues of the mouse. Basic Life Sci. 1978;12:361–377. doi: 10.1007/978-1-4684-3390-6_25. [DOI] [PubMed] [Google Scholar]
- 33.Dindot SV, Kent KC, Evers B, Loskutoff N, Womack J, Piedrahita JA. Conservation of genomic imprinting at the XIST, IGF2, and GTL2 loci in the bovine. Mamm Genome. 2004;15(12):966–974. doi: 10.1007/s00335-004-2407-z. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Douglas KC, Vandeberg JL, Clark AG, Samollow PB. Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res. 2014;24(1):70–83. doi: 10.1101/gr.161919.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20(5):614–622. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grutzner F, Kaessmann H. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508(7497):488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- 37.Renfree MB, Papenfuss AT, Deakin JE, Lindsay J, Heider T, Belov K, Rens W, Waters PD, Pharo EA, Shaw G, Wong ES, Lefèvre CM, Nicholas KR, Kuroki Y, Wakefield MJ, Zenger KR, Wang C, Ferguson-Smith M, Nicholas FW, Hickford D, Yu H, Short KR, Siddle HV, Frankenberg SR, Chew KY, Menzies BR, Stringer JM, Suzuki S, Hore TA, Delbridge ML, et al. Genome sequence of an Australian kangaroo, Macropus eugenii, provides insight into the evolution of mammalian reproduction and development. Genome Biol. 2011;12(8):R81. doi: 10.1186/gb-2011-12-8-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murtagh VJ, O’Meally D, Sankovic N, Delbridge ML, Kuroki Y, Boore JL, Toyoda A, Jordan KS, Pask AJ, Renfree MB, Fujiyama A, Graves JA, Waters PD. Evolutionary history of novel genes on the tammar wallaby Y chromosome: Implications for sex chromosome evolution. Genome Res. 2012;22(3):498–507. doi: 10.1101/gr.120790.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al Nadaf S, Waters PD, Koina E, Deakin JE, Jordan KS, Graves JA. Activity map of the tammar X chromosome shows that marsupial X inactivation is incomplete and escape is stochastic. Genome Biol. 2010;11(12):R122. doi: 10.1186/gb-2010-11-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deakin JE, Koina E, Waters PD, Doherty R, Patel VS, Delbridge ML, Dobson B, Fong J, Hu Y, van den Hurk C, Pask AJ, Shaw G, Smith C, Thompson K, Wakefield MJ, Yu H, Renfree MB, Graves JA. Physical map of two tammar wallaby chromosomes: a strategy for mapping in non-model mammals. Chromosome Res. 2008;16(8):1159–1175. doi: 10.1007/s10577-008-1266-y. [DOI] [PubMed] [Google Scholar]
- 41.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J: Galaxy: a web-based genome analysis tool for experimentalists. Curr Protoc Mol Biol 2010, 0 19:Unit-19.1021. [DOI] [PMC free article] [PubMed]
- 42.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thorvaldsdottir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14(2):178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25(17):2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3–new capabilities and interfaces. Nucleic Acids Res. 2012;40(15):e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]


