Abstract
Background
Pathogenic chlamydiae are obligate intracellular pathogens and have adapted successfully to human cells, causing sexually transmitted diseases or pneumonia. Chlamydial outer protein N (CopN) is likely a critical effector protein secreted by the type III secretion system in chlamydiae, which manipulates host cells. However, the mechanisms of its action remain to be clarified. In this work, we aimed to identify previously unidentified CopN effector target in host cells.
Results
We first performed a pull-down assay with recombinant glutathione S-transferase (GST) fusion CopN proteins (GST–CpCopN: Chlamydia pneumoniae TW183, GST–CtCopN: Chlamydia trachomatis D/UW-3/CX) as “bait” and soluble lysates obtained from human immortal epithelial HEp-2 cells as “prey”, followed by SDS-PAGE with mass spectroscopy (MS). We found that a host cell protein specifically bound to GST–CpCopN, but not GST–CtCopN. MS revealed the host protein to be fructose bisphosphate aldolase A (aldolase A), which plays a key role in glycolytic metabolism. We also confirmed the role of aldolase A in chlamydia-infected HEp-2 cells by using two distinct experiments for gene knockdown with an siRNA specific to aldolase A transcripts, and for assessment of glycolytic enzyme gene expression levels. As a result, both the numbers of chlamydial inclusion-forming units and RpoD transcripts were increased in the chlamydia-infected aldolase A knockdown cells, as compared with the wild-type HEp-2 cells. Meanwhile, chlamydial infection tended to enhance expression of aldolase A.
Conclusions
We discovered that one of the C. pneumoniae CopN targets is the glycolytic enzyme aldolase A. Sequestering aldolase A may be beneficial to bacterial growth in infected host cells.
Keywords: Chlamydia pneumoniae, Type III secretion, Effectors, Chlamydial outer protein N
Background
Obligate intracellular pathogens Chlamydia pneumoniae and Chlamydia trachomatis are major clinical and public health concerns, and cause pneumonia [1,2] and the most prevalent sexually transmitted diseases worldwide [3,4], respectively. Although C. pneumoniae is also associated with chronic inflammatory diseases such as asthma [5] and atherosclerosis [2], its etiological role remains to be clarified.
Chlamydiae undergo a unique biphasic developmental cycle, with a morphological change between the elemental body (EB), which is an infectious form, and reticulate body (RB), which is a replicative form, into the surrounding plasma membrane, the so-called inclusion membrane [6-8]. To complete their developmental cycle, chlamydiae utilize the type III secretion system (T3SS) to translocate several effector proteins across the inclusion membrane [9]. Accumulating evidence from experiments with Shigella or Salmonella have shown that T3SS effectors play a critical role in bacterial invasion and maturation, and maintenance of host cell viability, which is directly related to bacterial pathogenesis [10,11]. Although several chlamydial T3SS effectors such as translocated actin recruiting phosphoprotein (TARP) [12], inclusion membrane protein A (IncA) [13], and chlamydial outer protein N (CopN) [14] have been documented, mechanisms of host cell manipulation evoked by these effectors remain to be clarified.
Gram-negative bacterial T3SS is a complex of multiple proteins including tip, needle, rod and plug proteins, and ATPase [15], and the effector proteins translocating across the plasma membrane are strictly regulated by the plug proteins. For instance, MxiC, a plug protein of Shigella flexneri, regulates effector secretion [16,17]. The activation of Yersinia T3SS effector secretion is controlled by YopN/TyeA as a plug complex of Yersinia T3SS in response to extracellular calcium and/or contact with eukaryotic cells [18,19]. Also, translocation of some plug proteins such as YopN or MxiC into eukaryotic host cells has been documented [16,20]. Thus, these bacterial plug molecules, possibly with effector function, have a central role into controlling T3SS effector secretion.
Recent work has shown that chlamydial CopN, which is homologous to YopN and MxiC but with low similarity, is a T3SS plug protein, and is secreted by a Yersinia type III apparatus [14,21,22]. Also, CopN domain structure analysis using gel filtration assay with His-tagged recombinant proteins has revealed that CopN binds to tubulin, which prevents microtubule assembly [23]. Furthermore, yeast two-hybrid analysis has shown the essential role of the CopN effector protein in C. pneumoniae intracellular growth [24]. Thus, these findings support that chlamydial CopN has a critical role as a T3SS plug with effector function in the chlamydial host manipulation system. Whether the CopN targets tubulin alone remains to be determined.
Here, we showed that C. pneumoniae CopN interacted with a glycolytic enzyme, fructose bisphosphate aldolase A (aldolase A). Our data also suggested that sequestering aldolase A was beneficial to bacterial growth in infected cells.
Methods
Human cell lines
The immortal human epithelial cell lines HEp-2 and HeLa were purchased from American Type Culture Collection (Manassas, VA, USA) and Riken Cell Bank (Tsukuba, Japan), respectively. Both the cell lines were cultured at 37°C in 5% CO2 in Dulbecco’s Modified Eagle’s Medium (DMEM; Sigma, St. Louis, MO, USA) containing 10% heat-inactivated fetal calf serum (FCS), and antibiotics [10-μg/ml gentamicin, 10-μg/ml vancomycin and 0.5-μg/ml amphotericin B (Sigma)] (GVA antibiotics) [25,26].
Chlamydiae and growth conditions
C. pneumoniae TW183 strain was kindly provided by G. Byrne, University of Tennessee (Memphis). C. trachomatis D/UW3 CX strain (VR-885) was purchased from ATCC. Chlamydiae were propagated in HEp-2 and HeLa cells, as described previously [25,26]. DMEM supplemented with 20% FCS and GVA antibiotics was used for cell culture. The infected HeLa and HEp-2 cells were harvested at 48 and 72 h after infection, respectively, and stored at −80°C. The cell suspensions were disrupted by freezing–thawing and centrifugation at 400 × g to remove cell debris. The bacteria were collected by centrifugation at 1000 × g and resuspended in sucrose–phosphate–glutamic acid buffer (0.2-M sucrose, 3.8-mM KH2PO4, 6.7-mM Na2HPO4 and 5-mM l-glutamic acid). The bacteria were stored at −80°C until use. The number of bacterial infectious particles equal to EBs was determined by inclusion-forming unit (IFU) assay [27] as described below. Both bacterial genomic DNAs were extracted with High Pure PCR Template Preparation Kit (Roche, Indianapolis, IN, USA) and used as a template DNA for copN genes cloning.
Construction of recombinant CopNs
Full-length copN genes [CpcopN: C. pneumoniae TW183, CpB0334 (NC_005043.1); CtcopN: C. trachomatis UW-3/CX serovar D, CT_089 (NC_000117.1)] were amplified from each of the genomic DNA templates. The amplified DNA products were cloned to the downstream of glutathione S-transferase (GST) gene region into pGEX-6P-1 plasmid (GE Healthcare Bio-Sciences AB, Uppsala, Sweden). pGEX-6P-1 plasmid was kindly provided by I. Hirai, University of the Ryukyus (Okinawa, Japan). GST-fused recombinant proteins were finally expressed in Escherichia coli BL21 (DE3) in Luria–Bertani medium with 0.1-mM isopropyl-1-thio-β-d-galactopyranoside for 4 h at 37°C. The bacteria were collected by centrifugation and then lysed chemically using bacterial protein extraction reagent (Thermo Fisher Scientific, Waltham, MA, USA). After centrifugation to remove bacterial debris, GST-fusion proteins were prepared from the supernatant by using glutathione–agarose beads (Thermo Fisher Scientific) and the protein concentrations were determined using the Bradford assay. The solutions were adequately dispensed and stored at −80°C until use.
Pull-down assay with mass spectrometry (MS)
GST-fusion proteins were mixed with the soluble fraction obtained from HEp-2 cells, and then incubated for 4–12 h at 4°C in cell lysis buffer containing 40-mM Tris–HCl (pH 7.5), 150-mM sodium chloride, 0.5% Triton-X-100, and 1% protease inhibitor cocktail (Nacalai Tesque, Kyoto, Japan) in the presence of glutathione–agarose beads (Thermo Fisher Scientific). After washing with 50-mM Tris–HCl (pH 6.8), the captured proteins on beads were eluted with sample buffer for SDS-PAGE. Protein profiles into the eluted solutions were visualized by SDS-PAGE with Coomassie brilliant blue staining, and the specific band presumably captured by CopN was cut out from the gel, and then used for nano-liquid chromatography tandem MS (nano-LC-MS/MS) analysis (Japan Bio Services, Saitama, Japan). The MS analysis was carried out by the Japan Bio Services.
RNA extraction and conventional or quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted from chlamydia-infected or uninfected HEp-2 cells using High Pure RNA Isolation Kit (Roche) according to the manufacturer’s protocol. Reverse transcription of 500-ng total RNA was performed with ReverTraAce qPCR RT Master Mix (Toyobo, Osaka, Japan). Resultant cDNAs were used for PCR amplification with the following primers: (gapdh) (forward 5′-AAC GGG AAG CTC ACT GGC ATG-3′; reverse 5′-TCC ACC ACC CTG TTG CTG TAG-3′) [28], aldolase A (forward 5′-CGC AGA AGG GGT CCT GGT GA-3′; reverse 5′-CAG CTC CTT CTT CTG CTC CGG GGT-3′) [29], enolase (forward 5′-GAG CTC CGG GAC AAT GAT AA-3′; reverse 5′-CTG TTC CA TCC ATC TCG ATC-3′) [30], β-actin (forward 5′-GAC CAC ACC TTC TAC AAT GAG-3′; reverse 5′-GCA TAC CCC TCG TAG ATG GG-3′) [31] and chlamydial 16S rRNA (forward 5′-CGG CGT GGA TGA GGC AT-3′; reverse 5′-TCA GTC CCA GTG TTG GC-3′) [32]. PCR conditions were 95°C for 10 min, followed by 25–40 cycles of 95°C for 30 s, 55 or 60°C for 30 s and 72°C for 45 s, followed by 72°C for 10 min. PCR products were visualized on 2% agarose gels with ethidium bromide. For qRT-PCR, resultant cDNAs were also amplified by SYBR Green Realtime PCR Master Mix (Toyobo) using the above primer sets. The transcript amount of target gene products was expressed as a ratio to that of β-actin.
Establishment of aldolase A knockdown cells and chlamydial infection
Transient aldolase A knockdown HEp-2 cells were established by 24-h transfection of cells with siRNA as follows: passenger strand, 5′-CCG AGA ACA CCG AGG AGA A dTdT-3′; guide strand, 5′-UUC UCC UCG GUG UUC UCG G dTdT-3′. Non-targeting scramble RNA was also constructed by Hokkaido System Science (Sapporo, Japan) as follows: passenger strand, 5′-UUC UCC GAA CGU GUC ACG UdTdT-3′; guide strand, 5′-ACG UGA CAC GUU CGG AGG AGA AdTdT-3′. Transfection of siRNA (or scramble RNA) to cells was performed with a transfection reagent, Multifectam (Promega, Madison, WI, USA), according to the manufacturer’s protocol as below. Transfection complexes were prepared by incubating 20 μL of the prepared siRNA [or scramble RNA (1.0 pmol/μl)] with 10-μl Multifectam for 30 min at room temperature. After incubation, total transfection complexes were added to HEp-2 cells adjusted to 1 × 105 cells, and then incubated for 24 h at 37°C in 5% CO2. Expression of aldolase A was confirmed by Western blotting. At 24 h after transfection, HEp-2 cells were infected with chlamydiae at MOI 5 by centrifugation at 400 × g for 1 h. After washing uninfected bacteria with DMEM, chlamydia-infected cells were cultured for up to 72 h, and bacterial growth was monitored by IFU assay and Western blotting.
IFU assay and inclusion images
The number of chlamydial infectious particles (EBs) was determined by IFU assay by counting chlamydial inclusions formed in HEp-2 cells using fluorescein-isothiocyanate-conjugated monoclonal anti-Chlamydia antibody specific for Chlamydia lipopolysaccharide (with Evans Blue) (Denka Seiken, Tokyo, Japan), as described previously [27]. In some experiments, the cells were grown on cover glass, and chlamydial inclusion images were captured using a digital camera with a fluorescence microscope.
Western blotting
Infected and uninfected cells collected from each culture were boiled for 5 min in SDS sample buffer containing 2-mercaptoethanol. After boiling, each sample was sonicated to remove viscosity. Samples were loaded and separated by 10% (w/v) SDS-PAGE. Separated proteins were transferred to polyvinylidene difluoride membranes by semi-dry electroblotting. The membrane was blocked with 5% skimmed milk in Tris-buffered saline including Tween-20 (TBST). After washing the blocked membrane with TBST, the membrane was incubated with a primary antibody [mouse anti-GST monoclonal antibody (Nacalai Tesque), rabbit anti-human aldolase A polyclonal antibody (Cell Signaling Technology, Danvers, MA, USA), mouse anti-α-tubulin monoclonal antibody (Sigma–Aldrich) and rabbit anti-RpoD polyclonal antibody (prepared by Sigma–Aldrich)] for 1 h at room temperature. This was followed by a secondary antibody [horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG and IgM antibody (Jackson ImmunoResearch, West Grove, PA, USA) and HRP-conjugated goat anti-rabbit IgG antibody (KPL, Gaithersburg, MD, USA)] for 1 h at room temperature. Labeled proteins were visualized with Pierce Western Blotting Substrate (Thermo Fisher Scientific).
Statistical analysis
Data were expressed as mean ± standard deviation (SD). Comparison of data was performed using an unpaired Student t test. A value of p < 0.05 was considered significant. All experiments were repeated at least three times.
Results and discussion
Chlamydiae are obligate intracellular pathogens infecting various vertebrate cells, including human. To optimize the intracellular niche for their propagation, chlamydiae utilize several T3SS effector proteins. Several chlamydial T3SS effectors including TARP [12], IncA [13] and CopN [14] have been reported with detailed protein structures, and these provide hints for elucidating how chlamydiae manipulate host cells, resulting in successful adaptation. CopN effector is well conserved among chlamydiae, functioning both as the T3SS plug and a secreted effector protein [23]. The effector protein is responsible for a central role in chlamydial adaptation to host cells. Recent studies have elegantly demonstrated that CopN manipulates host cells, causing mitotic arrest [33], and that CopN interacts with tubulin, preventing microtubule assembly [23,24]. However, whether the CopN targets tubulin alone remains undetermined. In this work, we therefore attempted to find previously unidentified chlamydial CopN effector targets in host cells.
C. pneumoniae CopN interacts with human aldolase A
The CopN protein family, which functions as T3SS plug proteins with effector functions, is strictly conserved with weak sequence homology to chlamydiae and other Gram-negative bacteria [9], and the sequence similarity of CopNs used for this study was 48% (Figure 1A). To identify the host proteins that interact with chlamydial CopN, we performed a pull-down assay with GST-fused recombinant CopN proteins (C. pneumoniae: GST–CpCopN, C. trachomatis: GST–CtCopN) and HEp-2 cell lysates. Coomassie brilliant blue staining revealed that unrelated contaminants in the recombinant fractions were minimal in purified GST-fused protein fractions (Figure 1B). As a result, a specific band with ~40-kDa molecular size was detected in the eluted pull-down fraction with GST–CpCopN, but not in that with GST–Ctr CopN or GST alone (Figure 1C, arrowhead). It was a possible candidate for CopN target molecule, thus, the band cut out from the gel was analyzed by nano-LC/MS/MS. As a result, MS revealed two distinct eukaryotic proteins to be aldolase A, which is involved in glycolytic metabolism, and lancC-like protein, which is a lanthionine synthetase component (Figure 2A). The peptide hit numbers and coverage against aldolase A were significantly high compared with those for lancC-like protein (Figure 2A); therefore, we concluded that GST–CpCopN target protein might be a glycolytic enzyme, aldolase A. To confirm this, we performed Western blotting with a specific antibody against human aldolase A in pull-down eluted fractions. As expected, Western blotting clearly indicated a specific band reacting with the specific antibody, demonstrating that the GST-fused CpCopN target molecule was aldolase A, which is an important enzyme in glycolysis and glyconeogenesis, and affects actin polymerization via the Wiskott–Aldrich syndrome protein (WASP)/Arp2/3 pathway [34] (Figure 2B).
Figure 1.
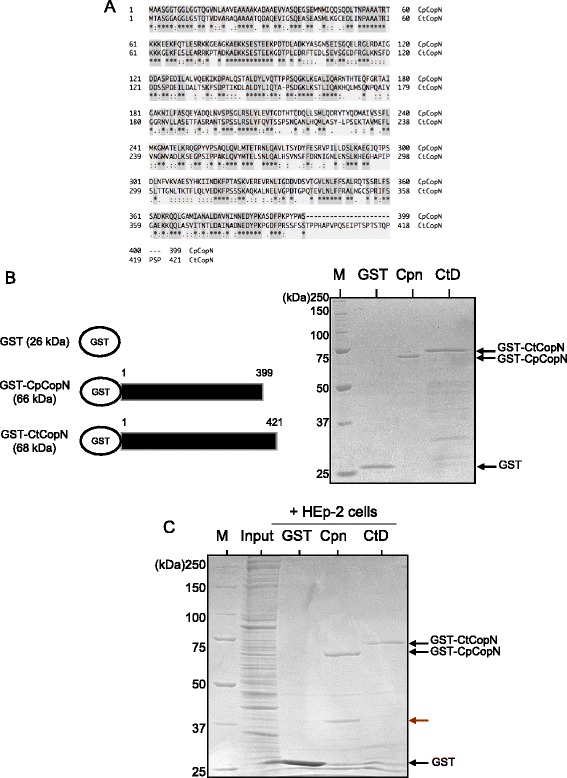
Sequence similarity between C. pneumoniae and C. trachomatis CopNs and representative pull-down images. (A) Sequence similarity between C. pneumoniae and C. trachomatis CopNs. Amino acid sequence data were obtained from NCBI genome site (http://www.ncbi.nlm.nih.gov/genome/). C. pneumoniae TW183 CopN, CpB0334 (accession number: NC_005043.1). C. trachomatis UW-3/CX serovar D CopN, CT_089 (accession number: NC_000117.1). Alignment was constructed by web software, UniProt (http://www.uniprot.org/). C. pneumoniae CopN had 48% homology as compared with C. trachomatis CopN. Stars highlighted gray indicate identical amino acids. (B) Protein expression profiles of constructed recombinant GST-fused CopNs. Cpn, C. pneumoniae CopN full length (399 aa) (GST-CpCopN). CtD, C. trachomatis CopN full length (421 aa) (GST-CtCopN). GST, GST alone. (C) Representative pull-down images with GST-fused chlamydial CopNs in HEp-2 cell lysates. Red arrow shows a specific band captured by GST–CpCopN, but not GST–CtCopN, in the HEp-2 cell lysates.
Figure 2.
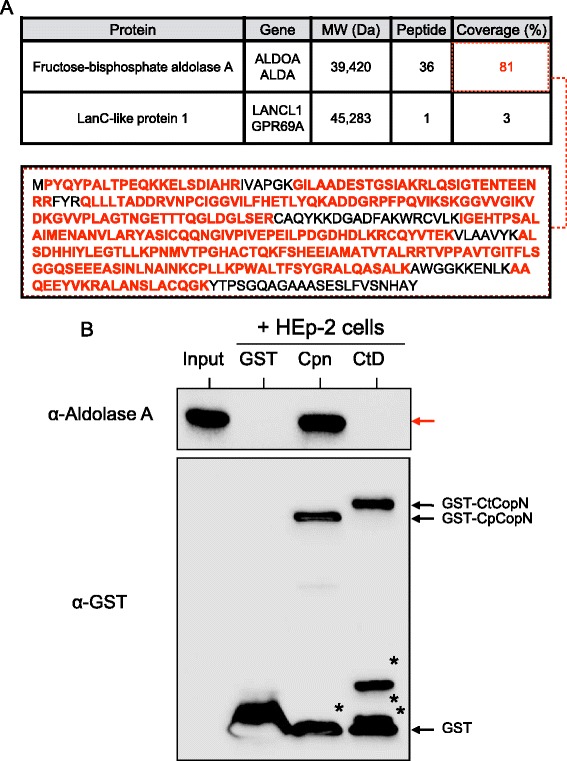
Pull-down assay with MS analysis and Western blotting showing that C. pneumoniae CopN interacts with human aldolase A. (A) MS data with nano-LC-MS/MS. Bold red amino acid sequences, peptide matched sequences. (B) Representative Western blotting pattern with a specific aldolase A antibody for elution obtained from pull-down assay with GST-fused chlamydial CopNs and HEp-2 cell lysates. Input, HEp- 2 cell lysates. GST, GST alone. Cpn, GST-fused C. pneumoniae CopN (GST-CpCopN). CtD, GST-fused C. trachomatis CopN (GST-CtCopN). Red arrow, aldolase A. Asterisk, fragmented proteins.
To the best of our knowledge, we are the first to show that chlamydial CopN could capture human aldolase A. Intriguingly, because no specific protein was seen in the case of C. trachomatis CopN, this capturing was specific to C. pneumoniae CopN effector. The exact reason for specific binding remains unclear, but it is likely associated with the low amino acid sequence similarity (48%) between the CopNs. Furthermore, previous work has indicated that while C. pneumoniae directly secretes CopN into the host cytosol across the inclusion membrane, C. trachomatis CopN is embedded in the inclusion membrane, and is never translocated to the host cell cytosol, suggesting that the target molecule of C. trachomatis CopN may be buried in the membrane fraction, without high solubility [14]. Taken together, the findings imply a possible different effector target and function between C. pneumoniae and C. trachomatis CopNs.
Our pull-down system with GST-fused CopN proteins and cell lysates unfortunately could not capture microtubulin, which is a cytoskeletal fiber, as described previously [23]. That research group used a gel filtration assay with microtubulin pelleting required for large amounts of recombinant proteins [23]; thus, there might have been a critical difference in detection sensitivity. Alternatively, the previous study also indicated that, in contrast to tubulin, C. pneumoniae CopN could not bind directly to microtubulin [23]; therefore, a possible explanation may be in our pull-down system with whole cell lysates, which naturally conserves cellular molecular structures.
Chlamydial growth is stimulated in aldolase A knockdown cells
Although it is not clear why chlamydiae should sequester host glycolytic enzymes such as aldolase A into infected cells, it is possible that glycolytic enzymes may be signaling molecules that sense viral or bacterial infection in the host cells, and that diminishing the signals may benefit chlamydial survival in the host cells. To confirm this hypothesis, we constructed transient aldolase A knockdown cells using siRNA, and assessed whether chlamydial growth could change in these cells. First, successful silencing of aldolase A was confirmed by western blotting with a specific antibody against aldolase A (Figure 3A, upper panel right). Chlamydial growth was carefully monitored in the knockdown cells by observation of inclusion morphology, bacterial count (EB numbers) with IFU assay, and western blotting for bacterial RpoD expression. In contrast to the immunofluorescence staining patterns of chlamydial inclusions (Figure 3B), the number of C. pneumoniae IFUs significantly increased in the knockdown cells (Figure 3C), and such a tendency to increase bacterial numbers was also seen in the C. trachomatis-infected knockdown cells, but with no significance (Figure 3D). Furthermore, changes in expression of RpoD, which is a core component of RNA polymerase and constitutively expressed in chlamydiae [34], were monitored using western blotting with a specific antibody against RpoD. Similar to IFU assessment data, the expression of RpoD tended to increase in the knockdown cells with either C. pneumoniae or C. trachomatis infection (Figure 4). We concluded that aldolase A silencing enhanced chlamydial growth. In addition, we confirmed by trypan blue staining that viability of aldolase A knockdown cells was maintained, indicating that the influence of dead cells was minimal in chlamydial growth (data not shown).
Figure 3.
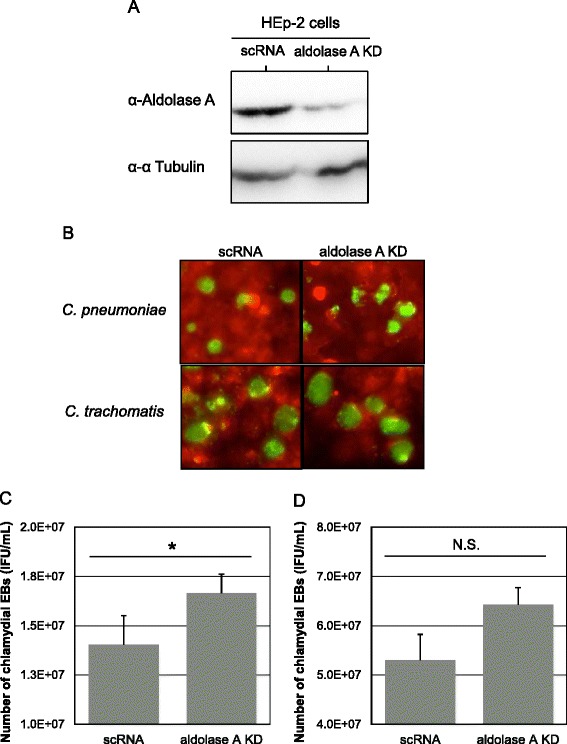
Changes in chlamydial infectious particle numbers in aldolase A knockdown cells. (A) Representative aldolase A protein expression images in knockdown HEp-2 cells. scRNA, scramble RNA transfected cells. ALDOA, aldolase A targeting siRNA transfected cells. (B) Representative images of inclusions formed in the knockdown HEp-2 cells. Chlamydiae infected knockdown cells at MOI 5, and were incubated for up to 3 days. Morphological changes in the inclusions were assessed under fluorescence microscopy at 3 (for C. pneumoniae) or 2 (for C. trachomatis) days after infection. Green, inclusions. (C, D) Changes in chlamydial infectious particle (IFU) numbers in the knockdown cells. Bacterial IFUs were evaluated by IFU assay. The IFU numbers were also assessed at the same time as above. Data show mean ± SD obtained from at least three independent experiments. *p < 0.05 versus value into scRNA-transfected cells. N.S., not significant. C. pneumoniae (C). C. trachomatis (D).
Figure 4.
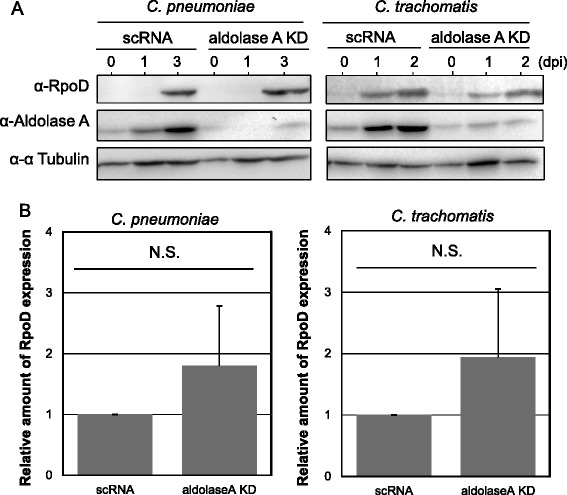
Representative images showing chlamydial RpoD protein expression and its kinetic change into aldolase A knockdown cells. (A) Representative images showing chlamydial RpoD protein expression in knockdown cells. Aldolase A knockdown cells infected with chlamydiae at MOI 5 were incubated for up to 3 days. RpoD protein expression was evaluated using Western blotting with a specific antibody against chlamydial RpoD. (B) Kinetic change of chlamydial RpoD protein expression in aldolase A knockdown cells at 3 (for C. pneumoniae) or 2 (for C. trachomatis) days after infection. Band-density values were evaluated by NIH Image J software. Data show mean ± SD obtained from at least three independent experiments. N.S., not significant.
As mentioned above, the exact reason why sequestering aldolase A is required for stimulating chlamydial growth in infected HEp-2 cells remains undetermined. Recent work has shown that multifunctional glycolytic enzymes including aldolase A have so-called “moonlight functions” [35,36]. For example, it is well known that aldolase A binds to F-actin [37], and this binding step is important for secondary moonlight functions, such as vesicle trafficking [37], or cell motility [38]. Furthermore, aldolase A sequesters WASP and affects WASP/Arp2/3 complex associating with actin remodeling [39,40]. WASP/Scar proteins stimulated by Rho family small GTPases in turn activate the Arp2/3 complex to create new actin filaments [41]. More importantly, a recent study has indicated that caspase-1 digestome senses fragmented glycolytic enzymes, including aldolase A, digested in infected cells, resulting in cell death; a novel host defense system [42]. Based on these findings, we now speculate that sequestering aldolase A by CopN may be a survival strategy of chlamydiae in infected host cells, via preventing host cell death. Similar to C. pneumoniae, C. trachomatis growth was stimulated in aldolase A knockdown cells.
Aldolase A gene expression tends to increase in C. pneumoniae-infected cells
To understand the possible fate of aldolase A sequestered by CopN effector in C. pneumoniae-infected cells, the changes in expression of glycolytic enzyme genes including aldolase A, gapdh and enolase were monitored in parallel using qRT-PCR in infected and uninfected HEp-2 cells. Chlamydial infection to HEp-2 cells tended to enhance aldolase A gene expression as compared with the uninfected cells (Figure 5A). The slight increases in gapdh and enolase expression were also seen in the infected cells (Figure 5B, middle and right panels). Although further study is needed to clarify this, there may be a backup system to recover unexpected damage with abnormal metabolic pathways, occurring by sequestering glycolytic enzymes into host cells. It has been reported that changes in expression of glycolytic enzymes by bacterial infections have been shown in Drosophila melanogaster [43]. This finding presumably supports our hypothesis.
Figure 5.
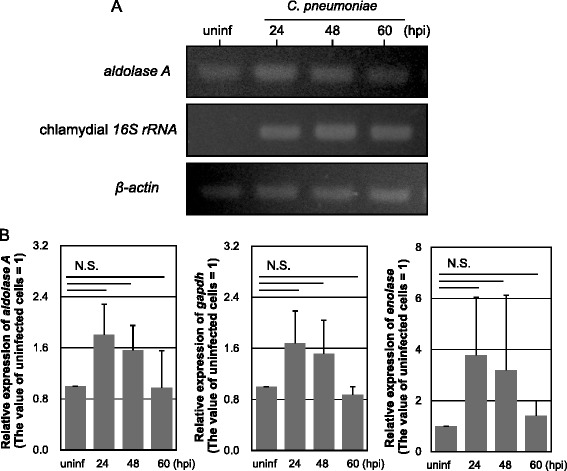
Expression of genes encoding glycolytic enzymes including aldolase A, GAPDH and enolase, and the kinetic changes in HEp-2 cells with chlamydial infection. (A) Representative RT-PCR images showing expression of genes encoding aldolase A, chlamydial 16S rRNA and β-actin in infected HEp-2 cells. C. pneumoniae infected cells at MOI 5 were collected at 24, 48 and 60 h. uninf, uninfected HEp-2 cells. (B) Kinetic changes in glycolytic enzyme gene expression in HEp-2 cells with chlamydial infection. The gene expression changes were evaluated by qRT-PCR. The values were normalized to that of β-actin. Data show mean ± SD from at least three independent experiments. N.S., not significant.
Conclusions
This is believed to be the first study to show the interaction between C. pneumoniae T3SS effector CopN and human aldolase A. Although more experiments are needed to reveal why CopN should bind to aldolase A, sequestering aldolase A may be beneficial to bacterial growth in infected host cells, contributing to our understand of chlamydial pathogenesis and complicated host–parasite relationships.
Acknowledgments
We thank Dr. Itaru Hirai who provided us with a plasmid vector, pGEX-6P-1 plasmid. This study was supported by Grants-in-Aid for Japan Society for the Promotion Science (JSPS) fellows (12 J03692) and scientific research from KAKENHI (21590474, 24659194, and 24117501 “Innovation Areas: Matryoshka-type evolution”).
Footnotes
Competing interests
The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript. The authors declare that no competing interests exist.
Authors’ contributions
KI performed the research and data analysis. KI, JM, YY and HY designed the experiments. KI and HY wrote the paper. All authors read and approved the final manuscript.
Contributor Information
Kasumi Ishida, Email: kasumi_ishida@ec.hokudai.ac.jp.
Junji Matsuo, Email: matsuo@hs.hokudai.ac.jp.
Yoshimasa Yamamoto, Email: yyamamoto@iph.pref.osaka.jp.
Hiroyuki Yamaguchi, Email: hiroyuki@med.hokudai.ac.jp.
References
- 1.Blasi F, Tarsia P, Aliberti S. Chlamydophila pneumoniae. Clin Microbiol Infect. 2009;15(1):29–35. doi: 10.1111/j.1469-0691.2008.02130.x. [DOI] [PubMed] [Google Scholar]
- 2.Belland RJ, Ouellette SP, Gieffers J, Byrne GI. Chlamydia pneumoniae and atherosclerosis. Cell Microbiol. 2001;6(2):117–127. doi: 10.1046/j.1462-5822.2003.00352.x. [DOI] [PubMed] [Google Scholar]
- 3.Gottlieb SL, Berman SM, Low N. Screening and treatment to prevent sequelae in women with Chlamydia trachomatis genital infection: how much do we know? J Infect Dis. 2010;201(Suppl 2):S156–S167. doi: 10.1086/652396. [DOI] [PubMed] [Google Scholar]
- 4.Malik A, Jain S, Rizvi M, Shukla I, Hakim S. Chlamydia trachomatis infection in women with secondary infertility. Fertil Steril. 2009;91(1):91–95. doi: 10.1016/j.fertnstert.2007.05.070. [DOI] [PubMed] [Google Scholar]
- 5.Hahn DL, Schure A, Patel K, Childs T, Drizik E, Webley W. Chlamydia pneumoniae-specific IgE is prevalent in asthma and is associated with disease severity. PLoS One. 2012;7(4):e35945. doi: 10.1371/journal.pone.0035945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abdelrahman YM, Belland RJ. The chlamydial developmental cycle. FEMS Microbiol Rev. 2005;29(5):949–959. doi: 10.1016/j.femsre.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 7.Abromaitis S, Stephens RS. Attachment and entry of Chlamydia have distinct requirements for host protein disulfide isomerase. PLoS Pathog. 2009;5(4):e1000357. doi: 10.1371/journal.ppat.1000357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hybiske K, Stephens RS. Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc Natl Acad Sci U S A. 2007;104(27):11430–11435. doi: 10.1073/pnas.0703218104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betts-Hampikian HJ, Fields KA. The chlamydial type III secretion mechanism: revealing cracks in a tough Nut. Front Microbiol. 2010;1:114. doi: 10.3389/fmicb.2010.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carayol N, Tran Van Nhieu G. Tips and tricks about Shigella invasion of epithelial cells. Curr Opin Microbiol. 2013;16(1):32–37. doi: 10.1016/j.mib.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 11.van der Heijden J, Finlay BB. Type III effector-mediated processes in Salmonella infection. Future Microbiol. 2012;7(6):685–703. doi: 10.2217/fmb.12.49. [DOI] [PubMed] [Google Scholar]
- 12.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci U S A. 2004;101(27):10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Subtil A, Parsot C, Dautry-Varsat A. Secretion of predicted Inc proteins of Chlamydia pneumoniae by a heterologous type III machinery. Mol Microbiol. 2001;39(3):792–800. doi: 10.1046/j.1365-2958.2001.02272.x. [DOI] [PubMed] [Google Scholar]
- 14.Fields KA, Hackstadt T. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol. 2000;38(5):1048–1060. doi: 10.1046/j.1365-2958.2000.02212.x. [DOI] [PubMed] [Google Scholar]
- 15.Enninga J, Rosenshine I. Imaging the assembly, structure and activity of type III secretion systems. Cell Microbiol. 2009;11(10):1462–1470. doi: 10.1111/j.1462-5822.2009.01360.x. [DOI] [PubMed] [Google Scholar]
- 16.Botteaux A, Sory MP, Biskri L, Parsot C, Allaoui A. MxiC is secreted by and controls the substrate specificity of the Shigella flexneri type III secretion apparatus. Mol Microbiol. 2009;71(2):449–460. doi: 10.1111/j.1365-2958.2008.06537.x. [DOI] [PubMed] [Google Scholar]
- 17.Martinez-Argudo I, Blocker AJ. The Shigella T3SS needle transmits a signal for MxiC release, which controls secretion of effectors. Mol Microbiol. 2010;78(6):1365–1378. doi: 10.1111/j.1365-2958.2010.07413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plano GV, Schesser K. The Yersinia pestis type III secretion system: expression, assembly and role in the evasion of host defenses. Immunol Res. 2013;57(1–3):237–245. doi: 10.1007/s12026-013-8454-3. [DOI] [PubMed] [Google Scholar]
- 19.Cornelis GR, Boland A, Boyd AP, Geuijen C, Iriarte M, Neyt C, Sory MP, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62(4):1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferracci F, Schubot FD, Waugh DS, Plano GV. Selection and characterization of Yersinia pestis YopN mutants that constitutively block Yop secretion. Mol Microbiol. 2005;57(4):970–987. doi: 10.1111/j.1365-2958.2005.04738.x. [DOI] [PubMed] [Google Scholar]
- 21.Herrmann M, Schuhmacher A, Mühldorfer I, Melchers K, Prothmann C, Dammeier S. Identification and characterization of secreted effector proteins of Chlamydophila pneumoniae TW183. Res Microbiol. 2006;157(6):513–524. doi: 10.1016/j.resmic.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 22.Ho TD, Starnbach MN. The Salmonella enterica serovar typhimurium-encoded type III secretion systems can translocate Chlamydia trachomatis proteins into the cytosol of host cells. Infect Immun. 2005;73(2):905–911. doi: 10.1128/IAI.73.2.905-911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Archuleta TL, Du Y, English CA, Lory S, Lesser C, Ohi MD, Ohi R, Spiller BW. The Chlamydia effector chlamydial outer protein N (CopN) sequesters tubulin and prevents microtubule assembly. J Biol Chem. 2011;286(39):33992–33998. doi: 10.1074/jbc.M111.258426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang J, Lesser CF, Lory S. The essential role of the CopN protein in Chlamydia pneumoniae intracellular growth. Nature. 2008;456(7218):112–115. doi: 10.1038/nature07355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishida K, Kubo T, Saeki A, Yamane C, Matsuo J, Yimin, Nakamura S, Hayashi Y, Kunichika M, Yoshida M, Takahashi K, Hirai I, Yamamoto Y, Shibata K, Yamaguchi H. Chlamydophila pneumoniae in human immortal Jurkat cells and primary lymphocytes uncontrolled by interferon-γ. Microbes Infect. 2013;15(3):192–200. doi: 10.1016/j.micinf.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 26.Yamazaki T, Matsuo J, Nakamura S, Oguri S, Yamaguchi H. Effect of Ureaplasma parvum co-incubation on Chlamydia trachomatis maturation in human epithelial HeLa cells treated with interferon-γ. J Infect Chemother. 2014;20(8):460–464. doi: 10.1016/j.jiac.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi M, Ishida K, Matsuo J, Nakamura S, Nagasawa A, Motohashi K, Yao T, Hirai I, Yamamoto Y, Suzuki H, Shimizu C, Matsuno K, Yamaguchi H. Chlamydophila pneumoniae attachment and infection in low proteoglycan expressing human lymphoid Jurkat cells. Microb Pathog. 2011;51(3):209–216. doi: 10.1016/j.micpath.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 28.Gnoatto N, Lotufo RF, Matsuda M, Penna V, Marquezini MV. Expression of cell-surface heparan sulfate proteoglycans in human cyclosporin-induced gingival overgrowth. J Periodontal Res. 2007;42(6):553–558. doi: 10.1111/j.1600-0765.2007.00981.x. [DOI] [PubMed] [Google Scholar]
- 29.Marone M, Mozzetti S, De Ritis D, Pierelli L, Scambia G. Semiquantitative RT-PCR analysis to assess the expression levels of multiple transcripts from the same sample. Biol Proced Online. 2001;3:19–25. doi: 10.1251/bpo20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamaguchi T, Iizuka N, Tsunedomi R, Hamamoto Y, Miyamoto T, Iida M, Tokuhisa Y, Sakamoto K, Takashima M, Tamesa T, Oka M. Glycolysis module activated by hypoxia-inducible factor 1alpha is related to the aggressive phenotype of hepatocellular carcinoma. Int J Oncol. 2008;33(4):725–731. [PubMed] [Google Scholar]
- 31.Kim SH, Kim KH, Yoo BC, Ku JL. Induction of LGR5 by H2O2 treatment is associated with cell proliferation via the JNK signaling pathway in colon cancer cells. Int J Oncol. 2012;41(5):1744–1750. doi: 10.3892/ijo.2012.1596. [DOI] [PubMed] [Google Scholar]
- 32.Everett KD, Bush RM, Andersen AA. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;Pt 2:415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 33.Nawrotek A, Guimarães BG, Velours C, Subtil A, Knossow M, Gigant B. Biochemical and Structural Insights into Microtubule Perturbation by CopN from Chlamydia pneumoniae. J Biol Chem. 2014;pii:jbc.M114.568436. doi: 10.1074/jbc.M114.568436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saka HA, Thompson JW, Chen YS, Kumar Y, Dubois LG, Moseley MA, Valdivia RH. Quantitative proteomics reveals metabolic and pathogenic properties of Chlamydia trachomatis developmental forms. Mol Microbiol. 2011;82(5):1185–1203. doi: 10.1111/j.1365-2958.2011.07877.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Morris AJ, Tolan DR, Pagliaro L. The molecular nature of the F-actin binding activity of aldolase revealed with site-directed mutants. J Biol Chem. 1996;271(12):6861–6865. doi: 10.1074/jbc.271.12.6708. [DOI] [PubMed] [Google Scholar]
- 36.O'Reilly G, Clarke F. Identification of an actin binding region in aldolase. FEBS Lett. 1993;321(1):69–72. doi: 10.1016/0014-5793(93)80623-3. [DOI] [PubMed] [Google Scholar]
- 37.Merkulova M, Hurtado-Lorenzo A, Hosokawa H, Zhuang Z, Brown D, Ausiello DA, Marshansky V. Aldolase directly interacts with ARNO and modulates cell morphology and acidic vesicle distribution. Am J Physiol Cell Physiol. 2011;300(6):C1442–C1455. doi: 10.1152/ajpcell.00076.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Du S, Guan Z, Hao L, Song Y, Wang L, Gong L, Liu L, Qi X, Hou Z, Shao S. Fructose-bisphosphate aldolase a is a potential metastasis-associated marker of lung squamous cell carcinoma and promotes lung cell tumorigenesis and migration. PLoS One. 2014;9(1):e85804. doi: 10.1371/journal.pone.0085804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ritterson Lew C, Tolan DR. Aldolase sequesters WASP and affects WASP/Arp2/3-stimulated actin dynamics. J Cell Biochem. 2013;114(8):1928–1939. doi: 10.1002/jcb.24538. [DOI] [PubMed] [Google Scholar]
- 40.Buscaglia CA, Penesetti D, Tao M, Nussenzweig V. Characterization of an aldolase-binding site in the Wiskott-Aldrich syndrome protein. J Biol Chem. 2006;281(3):1324–1331. doi: 10.1074/jbc.M506346200. [DOI] [PubMed] [Google Scholar]
- 41.Pollard TD. The cytoskeleton, cellular motility and the reductionist agenda. Nature. 2003;422(6933):741–745. doi: 10.1038/nature01598. [DOI] [PubMed] [Google Scholar]
- 42.Shao W, Yeretssian G, Doiron K, Hussain SN, Saleh M. The caspase-1 digestome identifies the glycolysis pathway as a target during infection and septic shock. J Biol Chem. 2007;282(50):36321–36329. doi: 10.1074/jbc.M708182200. [DOI] [PubMed] [Google Scholar]
- 43.Chambers MC, Song KH, Schneider DS. Listeria monocytogenes infection causes metabolic shifts in Drosophila melanogaster. PLoS One. 2012;7(12):e50679. doi: 10.1371/journal.pone.0050679. [DOI] [PMC free article] [PubMed] [Google Scholar]


