Abstract
Comparative DNA sequence studies between humans and nonhuman primates will be important for understanding the genetic basis of the phenotypic differences between these species. Here we compare ∼27 Mb of human chromosome 21 with chimpanzee DNA sequences identifying 57 genomic rearrangements (deletions and insertions ranging in size from 0.2 to 8.0 kb) between the two species. These rearrangements are distributed along the entire length of chromosome 21, with ∼35% found in genomic intervals encoding genes (genic intervals), and have occurred in the genomes of both humans and chimpanzees. Comparison of ∼9 Mb of human chromosome 21 with orangutan, rhesus macaque, and woolly monkey DNA sequences identified a combined total of 114 genomic rearrangements between humans and nonhuman primates. Analysis of these rearrangements revealed that they are randomly distributed with respect to genic and nongenic intervals and identified one deletion that has likely resulted in the inactivation of a gene (β1,3-galactosyltransferase) in the woolly monkey. Our data show that genomic rearrangements have occurred frequently during primate genome evolution and significantly contribute to the DNA differences between these species. These DNA rearrangements are commonly found in genic intervals, and thus provide natural starting points for focused investigations of qualitative and quantitative gene expression differences between humans and other primates.
[Supplemental material is available online at www.genome.org.]
Numerous comparative sequence studies have demonstrated that there is more similarity at the nucleotide level between humans and chimpanzees than between humans and any other species (King and Wilson 1975; Hacia 2001). Thus, identifying the types and extent of DNA sequence variation existing between humans and chimpanzees will be important for understanding the genetic basis of recently evolved, human-specific traits (Gagneux and Varki 2001). Previous comparative studies, focused on analyzing the differences between aligned human and chimpanzee sequences (single-nucleotide fixed differences), have indicated that the two species are 98.4%–98.8% identical at the nucleotide level (Koop et al. 1989; Chen and Li 2001; Fujiyama et al. 2002). The ∼1.5% single-nucleotide fixed differences have, to date, been the primary focus of studies aimed at understanding the biological differences, resulting from qualitative and quantitative differential gene expression, between humans and chimpanzees. It was demonstrated previously that large-scale segmental duplications (>400 kb; Bailey et al. 2002), pericentric inversions (Nickerson and Nelson 1998), and chromosomal fusions (Yunis and Prakash 1982) occurring after the separation of humans and chimpanzees from their common evolutionary ancestor resulted in DNA differences between the two species. Several human–chimpanzee comparative sequence analyses have suggested that smaller-sized rearrangements may also exist between human and chimpanzee DNA (Ueda et al. 1990; Fujiyama et al. 2002). However, the extent and significance of DNA sequence differences between humans and chimpanzees due to these smaller-sized genomic rearrangements are poorly characterized. Furthermore, the size, chromosomal distribution across gene-rich and gene-poor intervals, and evolutionary history of these genomic rearrangements have not yet been systematically examined.
We recently demonstrated that human high-density oligonucleotide arrays provide a rapid and effective tool for comparing human sequences with the DNA of other mammalian species (Frazer et al. 2001). We previously performed a cross-species comparative sequence analysis of human chromosome 21 by hybridizing mouse and dog bacterial artificial chromosome (BAC) DNA to human 21q arrays in order to identify evolutionarily conserved elements. In this report we describe the use of human high-density arrays for large-scale comparisons of human sequences with those of nonhuman primates to identify genomic rearrangements that result in DNA sequence differences between the species.
RESULTS AND DISCUSSION
Amplification of Chimpanzee Sequences
Here we compare human chromosome 21 with the syntenic chimpanzee sequences (i.e., chimpanzee chromosome 22) to characterize the genomic rearrangements that contribute to DNA differences between the two species. To obtain chimpanzee sequences, we took advantage of a set of paired polymerase chain reaction (PCR) primers which were designed to amplify minimally overlapping ∼10 kb long-range (LR)-PCR products spanning the entire length (∼32.4 Mb) of human chromosome 21 (Patil et al. 2001). The high level of nucleotide similarity between human and chimpanzee DNA allowed us to use this set of paired PCR primers designed based on human chromosome 21 sequences to efficiently amplify chimpanzee chromosome 22 sequences by LR-PCR (Fig. 1A). Of a total of 3110 paired PCR primers that successfully amplified LR-PCR products from human DNA, 2957 amplified LR-PCR products from chimpanzee DNA, resulting in the comparative analysis of ∼27 Mb of human chromosome 21 and chimpanzee chromosome 22 sequences.
Figure 1.
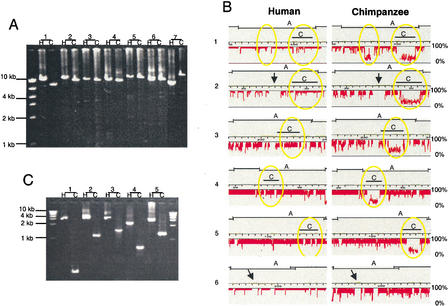
Analysis of syntenic human and chimpanzee LR-PCR products for deletions and insertions. (A) The lengths of syntenic human (H) and chimpanzee (C) LR-PCR products are compared by gel electrophoresis. Syntenic LR-PCR products are the same length (lane 6) indicating that no rearrangement is present, longer in humans than in the chimpanzees (lanes 1–4), indicating that the chimpanzee sequence is deleted with respect to the human sequence, or longer in the chimpanzee than in human (lane 7), indicating that the chimpanzee sequence contains an insertion relative to the human sequence. LR-PCR product #1 corresponds to the rearrangement in Segment 32 at 320 kb as given in Suppl. Table 1, #2 = Segment 36 at 28 kb, #3 = Segment 71 at 234 kb, #4 = Segment 77 at 125 kb, and #7 = Segment 2 at 292 kb. (B) The human and chimpanzee LCR-PCR products shown in (A) were hybridized to the 21q arrays, and their percent conformance (vertical axis), which is a measure of their similarity (Frazer et al. 2001), was plotted relative to their position in the human reference sequence (horizontal axis). Each tick mark in the scale represents a 1-kb interval. The sequence positions of the PCR products in (A) and (C) are indicated by horizontal lines. The overlap of the LR-PCR products in (A) with neighboring chromosome 21 LR-PCR products is shown. The sharp drop in conformance values (yellow circles) indicates the positions of the sequences deleted in the chimpanzee LR-PCR products 1–5 in (A). Sequences with absent conformance information, such as those indicated by black arrows, correspond to interspersed repeats, which were not tiled on the 21q arrays (see Methods). LR-PCR product #1 is the only fragment in which two localized deletions account for a size difference between chimpanzees and humans. For LR-PCR product #5, the variation in the sizes of the human and chimpanzee bands is not detectable on the gel but the deletion is evident in the comparative 21q array data. (C) Paired PCR primers designed to the sequences flanking the deletions in LR-PCR products 1–5 in (A) as shown in (B) were used to amplify human and chimpanzee DNA. The lengths of the human vs. chimpanzee PCR products provide more precise information about the deletion sizes (in bp).
Comparison of Syntenic Human and Chimpanzee LR-PCR Products
Our initial analysis consisted of comparing the lengths of the syntenic human and chimpanzee LR-PCR products by sizing them using agarose gel electrophoresis (Fig. 1A). Although the majority of the syntenic human and chimpanzee LR-PCR products have identical lengths, 33 have a difference in size ranging from ∼1 kb to 8 kb as determined by inspection of the gels (Supplemental Table 1). Of the syntenic LR-PCR products with different lengths, 27 were shorter and six were longer in the chimpanzee, suggesting that the chimpanzee DNA sequences contained deletions and insertions, respectively, relative to the human DNA sequences.
High-Density Arrays Are an Effective Method for Detecting DNA Rearrangements
To determine whether the shorter-length chimpanzee LR-PCR products contain a single localized deletion or numerous small dispersed deletions, we examined the amplified chimpanzee DNA by hybridizing to a series of high-density oligonucleotide arrays containing probes for most of the unique sequences from human chromosome 21 (Hacia et al. 1998, 1999; Frazer et al. 2001). This analysis revealed that the majority of chimpanzee LR-PCR products that are shorter in length than their syntenic human counterparts contain a single localized deletion (Suppl. Table 1). Inspection of the human–chimpanzee sequence deletions, which are detected in the comparative 21q array data by a sharp decrease in the conformance rate within the boundaries of a chimpanzee LR-PCR product (Fig. 1B), determined that they are comprised of varying percentages of unique and repetitive sequences (Fig. 1B), and that most of the deletions result in the loss of some sequences that are unique in the human genome (Suppl. Table 1). These results are the first direct evidence that the human genome contains intervals several kilobases in length that are comprised largely of unique sequences, and which are not present in the syntenic regions of the chimpanzee genome.
We next examined the comparative human–chimpanzee 21q array data to determine whether we could identify the presence of additional deletions in the amplified chimpanzee sequences. We searched for the deletion signature in the array data—a sharp decrease in the conformance rate—and found 24 such intervals (∼0.2–3.0 kb in length) that had not been detected by LR-PCR product size variations on gels (Suppl. Table 1). To demonstrate that these intervals of low conformance on the human chromosome 21 arrays correspond to sequence deletions on chimpanzee chromosome 22, we designed paired PCR primers to sequences bordering five of the intervals and compared the lengths of PCR products amplified from human and chimpanzee genomic DNA (Fig. 1C). In all cases, the syntenic human PCR product was longer than the chimpanzee PCR product by the approximate basepair amount predicted by the comparative 21q array data. These data indicate that comparative analysis of human and chimpanzee DNA using high-density arrays is an effective method for identifying intervals in the human genome containing unique sequences that are missing in the syntenic regions of the chimpanzee genome.
Genomic Rearrangements Account for a Significant Fraction of the DNA Sequence Differences Between Humans and Chimpanzees
In the ∼27 Mb segment of chromosome 21 analyzed, genomic DNA rearrangements account for ∼0.6% (161 kb) basepair differences between the human and chimpanzee syntenic sequences (Suppl. Fig. 1), of which ∼82% correspond to 51 sequence deletions and ∼18% correspond to six sequence insertions in the chimpanzee DNA (Suppl. Table 1). Our observation that deletions are more prevalent than insertions is due at least in part to an ascertainment bias, based on the fact that insertions are only detectable by variation in the size of the LR-PCR products on gels, whereas deletions are identified by both size variations on gels and the comparative 21q array data. Rearrangements smaller and larger in size than the detectable range in this study (0.1–10.0 kb, see Methods) are likely to also be present, and thus our data represent the minimal amount of basepair differences between the syntenic human chromosome 21 and chimpanzee chromosome 22 sequences due to insertions and deletions. These results suggest that genomic rearrangements are responsible for a significant fraction of the DNA sequence differences between humans and chimpanzees, accounting for ∼50% as much DNA variation as single-nucleotide fixed differences.
Characterization of Human Sequences at the Boundaries of DNA Rearrangements
We inspected the nucleotide sequences at the boundaries of the human–chimpanzee deletions to ascertain whether a molecular mechanism could be proposed to explain the frequency and distribution of these rearrangements. This analysis revealed that both unique as well as a variety of repetitive sequences are present at the deletion boundaries in the human genome (Suppl. Table 2). These data neither implicate a particular class of sequences nor indicate an obvious mechanism that gives rise to these rearrangements, but suggest that they may be the result of a stochastic process.
DNA Rearrangements Have Occurred in Both Humans and Chimpanzees
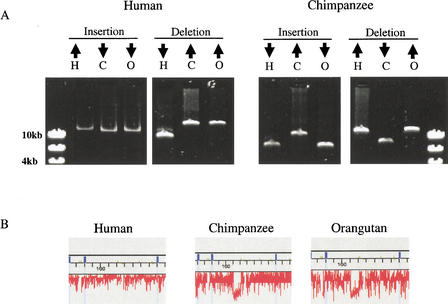
To elucidate whether the 57 genomic rearrangements observed in the human–chimpanzee comparative analysis are the result of deletions and insertions occurring predominately in one or the other of these primates, we used the orangutan as an outgroup. For 16 of the human–chimpanzee rearrangements, we were able to ascertain whether or not the orangutan also had the rearrangement by examining both relative sizes of the corresponding syntenic human, chimpanzee, and orangutan LR-PCR products and comparative human–orangutan 21q array data (Suppl. Table 3). Based on these data we determined for each of these rearrangements whether it occurred in the human genome (the chimpanzee and orangutan LR-PCR products are the same) or the chimpanzee genome (the human and orangutan LR-PCR products are the same; Fig. 2). Of the 16 rearrangements that we examined, five occurred in the human genome (three insertions and two deletions) and 11 occurred in the chimpanzee genome (10 deletions and one insertion; Suppl. Table 3). The fact that a greater number of deletions than insertions are identified in the chimpanzee lineage is likely to be at least in part due to our ascertain bias for finding deletions with respect to human chromosome 21 sequences. These data indicate that genomic DNA deletions and insertions have occurred in both the human and chimpanzee genomes.
Figure 2.
The relative sizes of the syntenic human (H), chimpanzee (C), and orangutan (O) LR-PCR products were used to determine whether the rearrangement occurred in the human or chimpanzee genome and whether it was an insertion or deletion event. (A) The relative sizes of syntenic human, chimpanzee, and orangutan LR-PCR products compared using gels. (B) The size variation of the human insertion in (A) is barely detectable on gels but is evident in the comparative 21q array data. These LR-PCR products correspond to the rearrangements in Suppl. Table 2 as follows: human insertion = Segment 10 at 161 kb, human deletion = Segment 2 at 301 kb, chimpanzee insertion = Segment 12 at 278 kb, chimpanzee deletion = Segment 76 at 51 kb.
Distribution of Human–Chimpanzee DNA Rearrangements on Chromosome 21
To determine the spatial distribution of the human–chimpanzee rearrangements, we divided chromosome 21 into 132 adjacent 250-kb intervals and determined the number of genomic rearrangements mapping within each interval (Fig. 3). A statistical analysis revealed that the rearrangements are randomly distributed along the entire length of chromosome 21, except for one 250-kb interval which contains an increased number of rearrangements (P < 0.01). To further investigate the spatial distribution of diverged human–chimpanzee sequences, we looked at the distribution of 76 paired PCR primers that amplified LR-PCR products from human but not chimpanzee DNA. The fact that these paired PCR primers (designed based on human sequence) specifically fail to amplify LR-PCR products from chimpanzee DNA suggests that their corresponding sequences have either been rearranged or have significantly diverged in the chimpanzee genome. Thus, looking at the distribution of these chimpanzee-specific LR-PCR failures is an indirect way of identifying regions containing either a genomic rearrangement or high sequence divergence. In agreement with the distribution analysis of the genomic rearrangements, the paired PCR primers corresponding to chimpanzee-specific LR-PCR failures are randomly distributed in the 250-kb intervals on chromosome 21, except for two intervals that contain increased LR-PCR failures (P < 0.0005; Fig. 3). Interestingly, the three 250-kb intervals that we identified which contain an increased number of rearrangements and/or an increased amount of sequence divergence are clustered within an ∼1-Mb gene-poor region on chromosome 21 (Hattori et al. 2000). These findings are consistent with a previous comparative analysis of human chromosome 21 with chimpanzee sequences (based on analyzing paired PCR primers that amplify human but not chimpanzee DNA; Fujiyama et al. 2002). Our data indicate that the majority of genomic DNA rearrangements are randomly distributed; however, a gene-poor region on chromosome 21 contains an increased number of rearrangements and/or a greater amount of sequence divergence than is expected by chance.
Figure 3.
Distribution of 57 human–chimpanzee rearrangements (black) and 76 human-specific LR-PCR products (red) in 250-kb adjacent intervals along the length of human chromosome 21. The position on chromosome 21 (about 10.0 to 11.4 Mb from the centromeric end; Hattori et al. 2000) containing an increased number of rearrangements and/or an increased amount of sequence divergence is indicated (blue bar). The positions of intervals A (purple bar) and B (green bar), each of which are ∼4.5 Mb in length and contain human chromosome 21 sequences compared with orangutan, rhesus macaque, and woolly monkey DNA (see Methods), are shown.
DNA Rearrangements Are Commonly Located in Genic Intervals
To identity human–chimpanzee rearrangements with possible functional consequences, we examined their distribution with respect to chromosome 21 genic and nongenic intervals. Genic intervals (∼13.0 Mb) were defined as all sequences contained within 10 kb upstream to 10 kb downstream of the 216 annotated genes (Frazer et al. 2001), and nongenic sequences (∼20.9 Mb) were defined as all other sequences on human chromosome 21. Twenty of the rearrangements mapped into genic intervals, of which 13 deleted intronic sequences of known genes, and 37 mapped into nongenic intervals. These data indicate that deletions and insertions occur at relatively equal frequencies in genic and nongenic intervals. The fact that the observed genomic rearrangements are commonly located within and near the vicinity of genes implicates that they may play a larger role than previously recognized in the differential expression of certain genes between humans and chimpanzees.
Comparison of Human Chromosome 21 With Other Nonhuman Primate Sequences
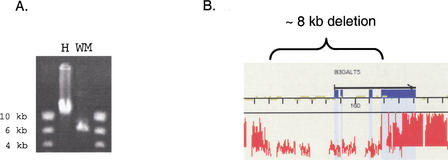
To further examine the frequency of deletions and insertions between humans and nonhuman primates and the percentage of these rearrangements located within genes, we targeted two intervals, each ∼4.5 Mb in length and combined representing ∼27% of chromosome 21 sequences, for comparative analysis with three additional primates (Fig. 3). Our efforts resulted in the comparison of ∼5.7 Mb (63%), ∼4.2 Mb (46%), and ∼3.7 Mb (41%) of the targeted human chromosome 21 sequences with orangutan (a great ape), rhesus macaque (an old world monkey), and woolly monkey (a new world monkey) syntenic sequences, respectively (Suppl. Fig. 1A; Suppl. Table 4). Interestingly, analysis of the 114 identified genomic DNA rearrangements (the combined total for the chimpanzee, orangutan, rhesus macaque, and woolly monkey comparative analyses in the two targeted regions) revealed that ∼9% of chromosome 21 DNA is deleted in at least one nonhuman primate (Suppl. Fig. 1B). Of these genomic rearrangements, ∼35% are located in genic regions, and one deletion observed in the human–woolly monkey comparative analysis results in the loss of three alternatively spliced exons of a gene encoding an enzyme, β1,3-galactosyltransferase-T5 (β3Gal-T5; Fig. 4). β3Gal-T5 is involved in the synthesis of a cell-surface epitope which is frequently used for the clinical diagnosis of cancer in humans (Isshiki et al. 1999). These results indicate that genomic DNA rearrangements occur frequently between humans and nonhuman primates, and further support the supposition that they may play a larger role than previously recognized in gene expression differences between the species.
Figure 4.
Deletion in woolly monkey genomic DNA affecting the β3GalT5 gene. (A) The length of the LR-PCR product containing the β3GalT5 gene is 14 kb in humans and 6 kb in woolly monkeys. (B) Human–woolly monkey comparative 21q array data reveal that the ∼8-kb rearrangement in the LR-PCR product in (A) results in the deletion of three alternatively spliced noncoding exons of the β3GalT5 gene. The effect of deleting these three alternatively spliced noncoding exons on the expression of the single coding exon of β3Gal-T5 is not known. Analysis of the woolly monkey β3Gal-T5 coding exon (by examining the 21q array data and dideoxysequencing of ∼50% of the 933 translated bp) did not reveal the presence of stop codons.
Conclusions
It has long been postulated that qualitative and quantitative expression differences of genes will be found responsible for the major biological differences between humans and chimpanzees (King and Wilson 1975). To date, it has commonly been thought that single-basepair changes between the human and chimpanzee genomes would underlie the majority of these postulated regulatory differences. However, the data we present in this study demonstrate that genomic rearrangements are a significant source of DNA variation between humans and chimpanzees, as well as other nonhuman primates. These rearrangements provide excellent starting points for focused studies of gene expression differences in humans and chimpanzees as part of an effort to identify the genetic differences responsible for the biological, physiological, and behavior differences between these species.
METHODS
Amplification of Nonhuman Primate Sequences by LR-PCR
LR-PCR reactions were performed using genomic chimpanzee DNA (Coriell Repository No. NG06939), orangutan DNA (Coriell Repository No. NG12256), rhesus macaque DNA (Coriell Repository No. NG07109), and woolly monkey DNA (Coriell Repository No. NG05356) as described (Patil et al. 2001) using PCR primer pairs designed based on human chromosome 21 sequences. Initially, 153 of the 3110 paired chromosome 21 PCR primers amplified LR-PCR products from human but not chimpanzee DNA. After retesting, 76 of these primer pairs were again only successful for human DNA, indicating that they specifically fail to amplify LR-PCR products from chimpanzee DNA. For the orangutan, rhesus macaque, and woolly monkey comparative analysis, 429 paired PCR primers were used to amplify Region A (located on chromosome 21 from ∼0 to 4.6 Mb from the centromeric end), and 432 paired PCR primers were used to amplify Region B (located ∼22.6 to 27.1 Mb from the centromeric end).
Identification of DNA Rearrangements by Agarose Gel Electrophoresis
Visual inspection of the gels allowed us to detect deletions and insertions ranging from ∼1 kb to 10 kb in size. Due to the large size of the LR-PCR products (average length ∼10 kb), genomic rearrangements smaller than ∼1 kb in length would result in size variations between the syntenic human and nonhuman primate sequences too small to detect by gels. In contrast, deletions in the nonhuman primate genomes greater than ∼10 kb in length are not detected because the LR-PCR products are not amplified (the paired PCR primers are designed based on human sequence).
Identification of DNA Rearrangements Using Human 21q High-Density Arrays
The 21q high-density arrays consist of a series of eight wafer designs, on which each of the unique chromosome 21 bases is interrogated by eight unique oligonucleotides (25-mers) as described (Frazer et al. 2001; Patil et al. 2001). The nonhuman primate LR-PCR products were pooled based on the syntenic human chromosome 21 sequences represented on each of the 21q high-density arrays, and hybridized as a single reaction as described (Patil et al. 2001). When the probe complementary to the human reference sequence had greater fluorescent intensity than the corresponding noncomplementary probes (these probes differ from the complementary probe only at the 13th position basepair), the nucleotide under interrogation was referred to as conforming to the human reference sequence. To calculate the “conformance rate”, we looked at 30-nucleotide (nt)-length windows and averaged the conformance of the individual nucleotides. For example, if 27 of the 30 nucleotides conformed to the human reference sequence, the window would have a 90% conformance rate (Frazer et al. 2001). The ability to confidently correlate a decreased “conformance rate” within a LR-PCR product with a sequence deletion results in the inability to detect deletions less than ∼100 bp in length. Because only unique human sequences are tiled on the 21q arrays, sequence deletions solely encompassing interspersed repeats are not detected in the comparative 21q array data. DNA rearrangements identified by LR-PCR products that are longer in nonhuman primates than in humans are also not detected by analysis of the comparative 21q array data.
Chromosomal Distribution Analysis of DNA Rearrangements
To determine the expected distribution of the 57 human–chimpanzee rearrangements, we performed 10,000 simulations in which we randomly distributed 57 rearrangements among our 250-kb intervals, and counted the frequency of multiple rearrangements in the same interval. In our data, there is one 250-kb interval containing five rearrangements. In the simulations, the probability of seeing five or more rearrangements in one interval was about 0.01. A similar analysis for the distribution of the 76 human-specific LR-PCR products was performed. We observed two 250-kb intervals containing seven human-specific LR-PCR products; the probability of seeing this in the simulations was 0.0005.
Characterization of Human–Chimpanzee DNA Rearrangements in Genic Intervals
Of the 20 rearrangements mapping within genic intervals, 13 were located in the introns of 12 genes (USP25, NCAM2, PRED16, APP, PRED29, PRED33, IL10RB, KCNJ15, DSCAM, ERG, C21orf1, and ADARB1) and deleted exonic sequences of two predicted genes (PRED16 and PRED58), whereas seven were located within 10 kb upstream or downstream of seven genes (CLDN17, KCNE2, C21orf5, PRED47, CSTB, COL6A1, and COL6A2).
Analysis of DNA Rearrangements in Targeted Chromosome 21 Intervals A and B
For two chromosome 21 intervals, each ∼4.5 Mb in length, human sequences were compared with chimpanzee, orangutan, rhesus macaque, and woolly monkey DNA. The total number of chromosome 21 basepairs in these two intervals that are present in humans and absent in one of the nonhuman primates or vice versa was calculated as follows: the lengths of the deletions and insertions (in bp) observed in the nonhuman primate divided by the total number of bp analyzed in the nonhuman primate (Suppl. Figure 1). To calculate the percent of chromosome 21 bp that are present in human but absent in at least one nonhuman primate or vice versa (the combined total for the chimpanzee, orangutan, rhesus macaque, and woolly monkey comparative analyses), we adjusted for deletions and insertions that are shared.
Acknowledgments
We thank Curtis Kautzer for expert technical assistance and Dennis Ballinger for critical reading of the manuscript. This work was supported in part by the following grant: NIH GM-5748202 (K.A.F.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL kelly_frazer@perlegen.com; FAX (650) 625-4510.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.554603. Article published online before print in February 2003.
REFERENCES
- 1.Bailey J.A., Yavor, A.M., Viggiano, L., Misceo, D., Horvath, J.E., Archidiacono, N., Schwartz, S., Rocchi, M., and Eichler, E.E. 2002. Human-specific duplication and mosaic transcripts: The recent paralogous structure of chromosome 22. Am. J. Hum. Genet. 70: 83-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen F.C. and Li, W.H. 2001. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am. J. Hum. Genet. 68: 444-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frazer K.A., Sheehan, J.B., Stokowski, R.P., Chen, X., Hosseini, R., Cheng, J.F., Fodor, S.P., Cox, D.R., and Patil, N. 2001. Evolutionarily conserved sequences on human chromosome 21. Genome Res. 11: 1651-1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujiyama A., Watanabe, H., Toyoda, A., Taylor, T.D., Itoh, T., Tsai, S.F., Park, H.S., Yaspo, M.L., Lehrach, H., Chen, Z., et al. 2002. Construction and analysis of a human-chimpanzee comparative clone map. Science 295: 131-134. [DOI] [PubMed] [Google Scholar]
- 5.Gagneux P. and Varki, A. 2001. Genetic differences between humans and great apes. Mol. Phylogenet. Evol. 18: 2-13. [DOI] [PubMed] [Google Scholar]
- 6.Hacia J.G. 2001. Genome of the apes. Trends Genet. 17: 637-645. [DOI] [PubMed] [Google Scholar]
- 7.Hacia J.G., Makalowski, W., Edgemon, K., Erdos, M.R., Robbins, C.M., Fodor, S.P., Brody, L.C., and Collins, F.S. 1998. Evolutionary sequence comparisons using high-density oligonucleotide arrays. Nat. Genet. 18: 155-158. [DOI] [PubMed] [Google Scholar]
- 8.Hacia J.G., Fan, J.B., Ryder, O., Jin, L., Edgemon, K., Ghandour, G., Mayer, R.A., Sun, B., Hsie, L., Robbins, C.M., et al. 1999. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 22: 119-120. [DOI] [PubMed] [Google Scholar]
- 9.Hattori M., Fujiyama, A., Taylor, T.D., Watanabe, H., Yada, T., Park, H.S., Toyoda, A., Ishii, K., Totoki, Y., and Choi, D.K. 2000. The DNA sequence of human chromosome 21. Nature 405: 311-319. [DOI] [PubMed] [Google Scholar]
- 10.Isshiki S., Togayachi, A., Kudo, T., Nishihara, S., Watanabe, M., Kubota, T., Kitajima, M., Shiraishi, N., Sasaki, K., Andoh, T., et al. 1999. Cloning, expression, and characterization of a novel UDP-galactose:β-N-acetylglucosamine β1,3-galactosyltransferase (β3Gal-T5) responsible for synthesis of type 1 chain in colorectal and pancreatic epithelia and tumor cells derived therefrom. J. Biol. Chem. 274: 12499-12507. [DOI] [PubMed] [Google Scholar]
- 11.King M.C. and Wilson, A.C. 1975. Evolution at two levels in humans and chimpanzees. Science 188: 107-116. [DOI] [PubMed] [Google Scholar]
- 12.Koop B.F., Tagle, D.A., Goodman, M., and Slightom, J.L. 1989. A molecular view of primate phylogeny and important systematic and evolutionary questions. Mol. Biol. Evol. 6: 580-612. [DOI] [PubMed] [Google Scholar]
- 13.Nickerson E. and Nelson, D.L. 1998. Molecular definition of pericentric inversion breakpoints occurring during the evolution of humans and chimpanzees. Genomics 50: 368-372. [DOI] [PubMed] [Google Scholar]
- 14.Patil N., Berno, A.J., Hinds, D.A., Barrett, W.A., Doshi, J.M., Hacker, C.R., Kautzer, C.R., Lee, D.H., Marjoribanks, C., McDonough, D.P., et al. 2001. Blocks of limited haplotype diversity revealed by high-resolution scanning of human chromosome 21. Science 294: 1719-1723. [DOI] [PubMed] [Google Scholar]
- 15.Ueda S., Washio, K., and Kurosaki, K. 1990. Human-specific sequences: Isolation of species-specific DNA regions by genome subtraction. Genomics 8: 7-12. [DOI] [PubMed] [Google Scholar]
- 16.Yunis J.J. and Prakash, O. 1982. The origin of man: A chromosomal pictorial legacy. Science 215: 1525-1530. [DOI] [PubMed] [Google Scholar]