Abstract
Through phylogeny reconstruction we identified 49 genes with a single copy in man, mouse, and chicken, one or two copies in the tetraploid frog Xenopus laevis, and two copies in zebrafish (Danio rerio). For 22 of these genes, both zebrafish duplicates had orthologs in the pufferfish (Takifugu rubripes). For another 20 of these genes, we found only one pufferfish ortholog but in each case it was more closely related to one of the zebrafish duplicates than to the other. Forty-three pairs of duplicated genes map to 24 of the 25 zebrafish linkage groups but they are not randomly distributed; we identified 10 duplicated regions of the zebrafish genome that each contain between two and five sets of paralogous genes. These phylogeny and synteny data suggest that the common ancestor of zebrafish and pufferfish, a fish that gave rise to ∼22,000 species, experienced a large-scale gene or complete genome duplication event and that the pufferfish has lost many duplicates that the zebrafish has retained.
[Supplemental material is available online at www.genome.org.]
Ohno proposed that without duplicated genes the creation of metazoans, vertebrates and mammals from unicellular organisms would have been impossible (Ohno 1970). Such big leaps in evolution, Ohno argued, required the creation of new gene loci with previously nonexistent functions. Because complete genome duplication increases gene number without upsetting gene dosage, it was advanced as the primary source of redundant genes. Ohno was not the first to suggest that genome-wide redundancy could lead to new evolutionary opportunities. Almost 20 years earlier, Stephens (1951) recognized that mutations were likely to impair original gene function, and he concluded that a mechanism in which a new function could be attained only at the price of discarding an old one would not be an efficient way of effecting evolutionary progress. Stephens proposed that the only way of achieving this evolutionary progress (i.e., the evolution of new species, genera, and “higher categories”) would be by increasing the number of genetic loci, either by the synthesis of new loci from nongenic material or by the duplication and subsequent differentiation of existing loci via genome duplication or unequal recombination.
Genome sequencing projects are now providing evidence that large-scale gene duplication and even complete genome duplication events have contributed significantly to gene family expansion and to genome evolution. For example, in Mycoplasma pneumoniae, >28% of the genome appears to have been produced by lineage-specific duplication events involving about four genes at a time (Jorden et al. 2001). In Mycobacterium tuberculosis >33% of the genome is composed of recently duplicated genes, but in this species some large clusters of between 20 and 90 genes are also involved (Jorden et al. 2001). Intragenome similarity searches have turned up evidence for whole-genome duplication in yeast (Saccharomyces cerevisiae) and in Arabidopsis thalia (Wolfe and Shields 1997; Lynch and Conery 2000; Vision et al. 2000). Goff et al. (2002) estimated the ages of duplicated genes in rice (Oryza sativa japonica) and concluded that the whole rice genome was duplicated between 40 and 50 million years ago. The human genome has also been shaped by a diversity of duplication events including, perhaps, two complete genome duplication events very early during the evolution of vertebrates (Spring 1997; Lynch and Conery 2000; Wang and Gu 2000; Friedman and Hughes 2001; Lynch 2001; Wolfe 2001; Gu and Huang 2002; Valente Samonte and Eichler 2002).
Here, we test the ancient fish-specific genome duplication hypothesis. The discovery that zebrafish possess seven Hox gene clusters, almost twice as many as human and mouse lead to this hypothesis that there was a whole-genome duplication, after the divergence of ray-finned and lobe-finned fishes but before the teleost radiation (Amores et al. 1998). Zebrafish gene-mapping studies (Gates et al. 1999; Barbazuk et al. 2000; Postlethwait et al. 2000; Woods et al. 2000) and phylogenetic analyses of zebrafish genes (Amores et al. 1998; Prince et al. 1998; Meyer and Schartl 1999; Taylor et al. 2001a,b) also support the hypothesis that a genome duplication occurred early during the evolution of ray-finned fishes. Taylor et al. (2001a) estimated that the fish-specific duplication event took place more than 300 million years ago. However, it was impossible to determine precise ages for the zebrafish duplicates because the third codon positions used to estimate their ages were saturated.
In this study, a phylogenetic approach is used to identify zebrafish duplicates and orthologs of these zebrafish duplicates in other fish species including the Japanese pufferfish (Takifugu rubripes). With the release of the pufferfish genome, we anticipated that it would be possible to date the duplication event relative to the speciation event that produced the ancestors of pufferfish and zebrafish.
RESULTS
Identifying Duplicated Fish Genes
In this study, orthologous and paralogous genes were identified using BLAST searches and phylogeny reconstruction. Forty-nine clades of orthologous genes with one copy in human, mouse, and/or chicken, one or two copies in tetraploid Xenopus, and two copies in zebrafish were recovered. For 22 of these genes, both zebrafish sequences also had orthologs in the Japanese pufferfish. For another 20 genes duplicated in zebrafish, we found only one pufferfish ortholog but in each case the pufferfish ortholog was more closely related to one of the zebrafish duplicates than to the other. In one case, HspA1, the two zebrafish sequences, were most closely related to one of two pufferfish genes.
While the aim of this study was to identify duplicated zebrafish genes and orthologs of these genes in other species, some trees that were consistent with the ancient fish-specific genome duplication hypothesis had only one or no zebrafish orthologs. This additional phylogenetic support for genome duplication in fish was uncovered for the following reasons. Our conservative approach to selecting genes for analysis from the results of the BLAST searches meant that many genes used in the first phylogenetic analyses turned out not to be orthologs of the query sequences. Most of these more distantly related sequences were excluded from further analyses, but, for L1Cam, these nonorthologs included human NGCAM and NRCAM (also called CHL1) and pufferfish duplicates of both of these L1Cam-related sequences. Also, human NODAL was included as a BLAST query sequence because Woods et al. (2000) reported that it had been duplicated in zebrafish producing cyclops and squint. The sequences retrieved produced two clades of Nodal genes and showed that cyclops and pufferfish sequence JGI14907 were orthologs of NODAL but that squint was not. However, the monophyletic group that included squint and several “nodal-related” Xenopus sequences also included duplicated pufferfish sequences (JGI12967 and JGI17187). Finally, while only one copy of LdhB was known from zebrafish, human LDHB was included in our list of query sequences because of a report by Stock et al. (1997) showing it occurs twice in killifish (Fundulus heteroclitus). The final phylogeny of LdhB genes included two killifish sequences and two pufferfish sequences, one zebrafish and one eel sequence, and it had a topology consistent with the ancient fish-specific genome duplication hypothesis. All sets of homologous human and fish genes are listed in Table 1. Tree topologies that also include mouse, chicken, and frog sequences are available from www.genome.org as supplementary material. Amino-acid alignments and tree topologies are available at http://www.evolutionsbiologie.uni-konstanz.de/Wanda/.
Table 1.
Duplicated Fish Genes and Human Orthologs
| Human Query Sequences | Each Column Lists Members of a Clade That is Orthologous to the Human Query Sequence | NJ | QP | AS | |
| ATP1B1(4502277) | Danio 9789577 | Danio 11096273, Anguilla 1703468, Anguilla 7406523, Takifugu JGI22524 | + | − | + |
| ATP1B2(4502279) | Danio 9789579 | Danio 14150727, Takifugu JGI789 | + | + | + |
| ATP1B3(4502278) | Danio 974774, Anguilla 7406521 | Danio 9837579, Takifugu JGI9802 | + | + | + |
| BMP2 (4557369) | Danio 2149148 | Danio 2804175, Takifugu JGI7838 | + | + | + |
| CYP19 (13904858) | Danio 12655890, Carassius 3913347, Oreochromis 4838530, Oreochromis 4838536, Takifugu JGI6225, Pimephales 14041612 | Danio 12655892, Carassius 2662332, Oreochromis 3913346, Oreochromis 4838538, Takifugu JGI22275, Ictalurus 3913357, Oncoryhnchus 228574, Oryzias 3913355, Dicentrarchus 14589321, Hippoglossus 13620178, Paralicthys4239990 | + | + | + |
| DLL(10518497) | Danio 2809389, Takifugu JGI3940 | Danio 1888392, Takifugu JGI18204 | + | + | − |
| DLX2 (4758168) | Danio 2842748 | Danio 108243, Takifugu JGI119697 | + | + | + |
| DLX4 (4503343) | Danio 2842751 | Danio 2842750 | + | + | + |
| EFNA5(4503487) | Danio 2494365, Takifugu JGI4301 | Danio 2462953, Takifugu JGI34618 | − | − | − |
| EN1 (7710119) | Danio 4322044, Takifugu JGI28510 | Danio 417127, Takifugu JGI32850 | − | − | − |
| EN2(11422302) | Danio 417128 | Danio 417129, Takifugu JGI7515 | − | − | − |
| EPHB4(4758290) | Danio 3163942, Takifugu JGI26074 | Danio 3005901, Takifugu JGI17145 | + | + | + |
| FKH1 (4503735) | Danio 12004940 | Danio 12004938, Takifugu JGI9390 | − | − | − |
| FKH5 (8134472) | Danio 2982347, Takifugu JGI20315 | Danio 2982343, Takifugu JGI3282 | − | − | + |
| FLOT1(5031699) | Danio 12751185, Carassius 2190561, Takifugu JGI8518 | Danio 12751187, Carassius 12751189, Takifugu JGI3374 | − | + | + |
| FZD8 (1033460) | Danio 4164471, Takifugu JGI14550 | Danio 4335927, Takifugu JGI21332 | − | + | + |
| Gdf6(1707885 Bos) | Danio 914116, Takifugu JGI7189 | Danio 1906321, Takifugu JGI32443 | − | + | + |
| HOXB6 (32369) | Danio 62530, Takifugu JGI5208 | Danio 4322075 | − | − | + |
| HOXC6 (4758554) | Danio 4322098, Takifugu 2341089 | Danio 4322100 | − | − | + |
| HSP71 (5729877) | Danio 1865782, Danio 2495341 | Danio 2245606, Oncorhychus 232285, Ictalurus 1346318, Paralichthys 3513540, Oryzias 4589737 | + | − | − |
| HSPA1(5123454) | Takifugu JGI656, Xiphophorus 17061835, Paralichthys 11277120 | Takifugu 1620388, Xiphophorus 17061837, Oncorhychus 17129570, Oncorhychus 2495346, Danio 7061841, Danio 7861932, Oreochromis 3004463, Oryzias 9652348 | + | + | + |
| HUR(4503551) | Danio 6694223, Takifugu JGI20049 | Danio 6694225, Takifugu JGI16540 | − | + | + |
| ISL2 (14755347) | Danio 1708564, Oncorhychus 1708565, Takifugu JGI18627 | Danio 1708561, Oncorhychus 1708557, Oncorhychus 1708558 | − | − | − |
| JAK2(4826776) | Danio 3687398, Takifugu JGI20041 | Danio 3687400, Takifugu JGI19673, Tetraodon 5918520 | − | − | − |
| KAL1(4557683) | Danio 6708056, Takifugu JGI14813 | Danio 6708054, Takifugu JGI16508 | + | + | + |
| L1CAM (4557707) | Danio 1065716, Takifugu 7522081 | Danio 1065714, Takifugu JGI14258, Carassius 11277081 | + | − | − |
| NGCAM(6651380) | Takifugu JGI1459 | Takifugu 2856 | + | + | + |
| NRCAM(5729767) | Takifugu JGI2517 | Takifugu 7031 | + | + | + |
| LDHB(12803117) | Takifugu JGI2932, Fundulus 462491, Anguilla 4321147, Danio 6048361 | Takifugu JGI30368, Fundulus 555487 | + | + | + |
| LHX1 (13652710) | Danio 2155289 | Danio 2497670 | + | − | − |
| MITF(4557755) | Danio 15341251, Takifugu JGI2179 | Danio 5726232, Takifugu JGI24563 | − | − | − |
| Msx3 (6754756 Mus) | Danio 608511 | Danio 62543, Takifugu JGI6688, Tetraodon 8187099 | + | + | + |
| MSX2 (4505269) | Danio 62545, Takifugu JGI20308 | Danio 608509 | − | + | − |
| Nodal2 (897598 Xenopus) | Takifugu JGI17187, Danio 3540235 | Takifugu JGI2967 | + | + | + |
| NOG (4885523) | Danio 4185744 | Danio 5410599, Takifugu 3860047 | + | + | + |
| NOTCH (11275980) | Danio 433867, Takifugu JGI3276 | Danio 2569968, Takifugu JGI22935 | − | + | + |
| NTN1(4758840) | Danio 2327065, Takifugu JGI27841 | Danio 2394302 | − | − | + |
| OPRD(4505509) | Danio 9837282, Takifugu JGI343 | Danio 7441620, Takifugu JGI9982 | + | − | − |
| OTX1 (417425) | Danio 3024327, Takifugu JGI36992 | Danio 3024322 | + | + | + |
| PAX2 (4557820) | Danio 3420030, Oryzias 2344868 | Danio 62550 | + | + | + |
| PAX6 (4505615) | Danio 62549, Astyanax 2369651 | Danio 3779238, Takifugu 3402199, Oryzias 4426551 | + | + | + |
| POU3F3 (5453936) | Danio 1730449, Takifugu JGI15850 | Danio 1730450, Takifugu JGI3511 | − | − | − |
| RARA(4160009) | Danio 704370, Takifugu 4972006, Salmo 9931536 | Danio 215026, Takifugu JGI11888 | + | + | + |
| Rx (6002393 Gallus) | Danio 2240028, Takifugu JGI10186, Oryzias 7635917 | Danio 9903605, Takifugu JGI19484 | − | + | + |
| RXRB (1350911) | Danio 1046297, Takifugu JGI191 | Danio 1046299, Takifugu JGI4030, Scophthalmus 14994052 | + | − | + |
| SHH(4506939) | Danio 6136068 | Danio 6174983, Takifugu JGI13503, Paralichthys 5441265 | + | + | + |
| SPON2(6912682) | Danio 2529223, Takifugu JGI14751 | Danio 2529221, Takifugu JGI7893 | − | − | − |
| SNAIL (5729673) | Danio 545349, Takifugu 5830231 | Danio 841423, Takifugu 5830233 | + | + | + |
| SNAP25(134583) | Danio 3703098, Carassius 548943 | Danio 3703100, Carassius 548945 | − | − | − |
| SOX11 (4507160) | Danio 4099262, Takifugu JGI7177 | Danio 7572946, Oncorhychus 2826913 | + | + | + |
| SPON1(11320819) | Danio 2529224, Takifugu JGI8633 | Danio 2529226 | − | + | + |
| TCF3(11230858) | Danio 5281354 | Danio 3769679 | + | + | + |
| TPI (4507644) | Danio 15149249, Xiphophorus 15149253 | Danio 15149247, Xiphophorus 15149251 | − | − | + |
Paralogous fish genes and their human “pro-ortholog” shown on the same row. Orthologous fish genes listed in the same box. NCIB gi and JGI (for pufferfish) numbers shown. NJ = neighbor-joining. QP = quartet puzzling. AS = ASATURA () analyses. The plus (+) symbol indicates that the topology reflected by the arrangement of genes in the table is the one in the NJ, QP, or ASATURA trees. The minus (−) symbol indicates that this phylogenetic analysis was carried out but that the resulting tree topology was not consistent with the arrangement of the genes in the table.
Mapping Duplication Events onto Phylogenies
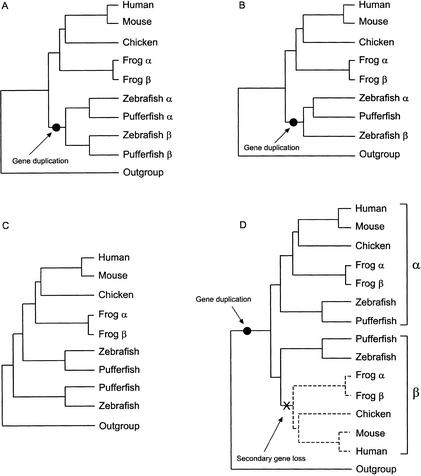
For 24 of the 53 genes with duplicates in fish (49 with duplicates in zebrafish and the four additional genes described above), neighbor-joining (NJ) and quartet puzzling (QP) phylogenetic methods produced trees consistent with the ancient fish-specific genome duplication event (Table 1), that is, trees in which all fish sequences formed a monophyletic group and, when sequences from more than one species were available, a branching order showing that the duplication event was not specific to one fish lineage (Fig. 1A,B). For five of these genes (Epbh4, HspA1, Kal1, LdhB, and RarA) the “duplication” topology was significantly better (had a significantly higher likelihood) than alternative topologies according to the Kishino-Hasagawa test (Kishino and Hasegawa 1989). Fourteen of the remaining 29 trees had NJ or QP-based topologies that were consistent with an ancient fish-specific genome duplication event.
Figure 1.
Phylogenetic representations of data shown in Table 1. A. Topology predicted for genes duplicated in ray-finned fish before the divergence of the zebrafish and pufferfish lineages with both copies retained and sequenced in both species. B. Topology predicted for genes duplicated in fish before the divergence of zebrafish and pufferfish but with one copy lost or not sequenced in pufferfish. C. “Out-group topology”: Topology often recovered when a gene/phylogenetic method combination did not produce the topology predicted by the fish-specific genome duplication hypothesis (e.g., “−“ in Table 1). For many genes, we suspect that the recovery of an out-group topology is an artefact produced by Long Branch Attraction (see Discussion). D. Biologically plausible explanation for the out-group topology: Loss of tetrapod orthologs of “basal” fish lineage.
Using ASATURA (Van de Peer et al. 2002) we removed amino-acid positions that appeared to be saturated from each pairwise sequence comparison prior to genetic distance estimation and phylogeny reconstruction. In most cases, saturation, which was qualitatively identified using a substitution frequency versus genetic distance plot in ASATURA, occurred for amino-acid positions with substitution frequencies of 696, 722, or 831 in the PAM1 matrix (Dayhoff 1978). Overall, ASATURA produced a tree consistent with the ancient fish-specific genome duplication hypothesis for 37 genes (more genes than any other method) including five of the 15 genes that did not have the duplication topology using either NJ or QP. Interestingly, Hox genes were among those that had the duplication topology only when ASATURA was used suggesting that neither Poisson-correction nor the default amino-acid substitution model used by TREEPUZZLE for QP adequately reflected the evolution of these sequences.
This left 10 genes with two copies in fish but without the topology predicted by the ancient fish-specific genome duplication hypothesis. Among these 10 genes were four (Efna5, En1, Fkh1, and Isl2) for which user-defined trees with the “duplication topology” had the highest likelihood. These are cases where the duplication topology must not have been considered during the QP search (a search that is heuristic and not exhaustive). The remaining six genes (En2, Jak2, Spon2, MitF, Pou3F3, and Snap25) are duplicated in zebrafish (there are also two Jak2, two Spon2, and two Pou3F3 genes in pufferfish) but the trees suggested that the mutations producing these duplicates were independent of the proposed whole-genome duplication event.
When more than one or all phylogenetic methods failed to produce a topology consistent with the ancient fish-specific genome duplication hypothesis, the usual pattern was the reconstruction of a tree showing that one of the duplicated fish sequences (or one clade of duplicated fish sequences) was sister to the remaining actinopterygian and sarcopterygian sequences (Fig. 1C). Although this “out-group topology” has several biologically plausible explanations, we suspected that failure to find fish monophyly was usually because of tree reconstruction artifacts such as long-branch attraction (see Discussion).
For 18 genes duplicated in zebrafish, pufferfish, or both species, orthologs from other fish species were also uncovered. In addition to providing support for an ancient duplication event, these trees showed evidence for lineage-specific gene duplication of Atp1B1 in European eels (Anguilla anguilla), Isl2 in Chinook salmon (Onchorynchus tshawytscha) and HspA1 in zebrafish (Table 1).
In summary, at least one of the three phylogenetic methods used produced a tree with the topology predicted by the ancient fish-specific genome duplication hypothesis for 38 of 49 genes with duplicates in zebrafish. For four additional genes the topology consistent with the ancient fish-specific genome duplication hypothesis had the highest likelihood despite the failure of NJ, QP, or ASATURA to recover it. In 22 cases, both zebrafish duplicates had a pufferfish ortholog (e.g., Fig. 1A) and in 20 cases one of the two zebrafish duplicates had a pufferfish ortholog (e.g., Fig. 1B). In addition to the zebrafish duplicates, we found four genes (LdhB, NgCam, Nodal, and NrCam) that also appear to have been duplicated early during the evolution of ray-finned fish. The HspA1 duplicates in zebrafish are the products of a lineage-specific duplication event, however, data from pufferfish and swordtails (Xiphophorus) show that HspA1 was also duplicated in fish before the zebrafish and pufferfish lineages diverged.
Synteny
Next we asked how the duplicated pairs of zebrafish genes, those with and without the predicted topology, are distributed among the 25 zebrafish linkage groups. Forty-four pairs of zebrafish duplicates have been mapped (Table 2); 10 chromosome pairs have two or more (up to five) sets of gene duplicates. This number is significantly higher than expected (P < .01) assuming duplicates were distributed among the zebrafish chromosomes according to a Poisson distribution.
Table 2.
Chromosomal Locations of Duplicated Zebrafish Genes and Human Orthologs
| Human Ortholog | Chr. | Zebrafish Duplicates | Map Position |
| DLX2 | 2 | dlx5 | LG1 |
| dlx2 | LG9 | ||
| EN1 | 2 | en1b | LG1 |
| en1a | LG9 | ||
| DLL | 6 | dla | LG1 |
| dld | LG13 | ||
| Msx3 | Mus | msxb | LG1 |
| msxc | LG13 | ||
| EN2 | 7 | en2b | LG2 |
| en2a | LG7 | ||
| SHH | 7 | twhh | LG2 |
| shh | LG7 | ||
| HOXB5 | 16 | hoxB5a | LG3 |
| hoxB5b | LG12 | ||
| HOXB6 | 16 | hoxB6a | LG3 |
| hoxB6b | LG12 | ||
| NOG | 17 | nog1 | LG3 |
| noggin | LG12 | ||
| RARA | 17 | rara2b | LG3 |
| rara2a | LG12 | ||
| DLX4 | 17 | dlx8 | LG3 |
| dlx7 | LG12 | ||
| NOTCH | 9 | notch1a | LG5 |
| notch1b | LG21 | ||
| JAK2 | 9 | jak2b | LG5 |
| jak2a | LG21 | ||
| ISL2 | 15 | isl3 | LG7 |
| isl2 | LG25 | ||
| PAX6 | 11 | pax6.2 | LG7 |
| pax6.1 | LG25 | ||
| FKH5 | 15 | fkd5 (foxb1.1) | LG7 |
| fkd3 (foxb1.2) | LG25 | ||
| HOXC6 | 12 | hoxC6b | LG11 |
| hoxC6a | LG23 | ||
| SNAIL | 20 | snail1 | LG11 |
| snail2 | LG23 | ||
| Gdf6 | Bos | radar | LG16 |
| dynamo | LG19 | ||
| TPI | 1 | tpiB | LG16 |
| tpiA | LG19 | ||
| OPRD | 1 | or4 | LG16 |
| oprd1 | LG19 | ||
| RXRB | 6 | rxrD | LG16 |
| rxrE | LG19 | ||
| BMP2 | 20 | bmp2a | LG17 |
| bmp2b | LG20 | ||
| SNAP25 | 20 | snap25.2 | LG17 |
| snap25.1 | LG20 | ||
| SOX11 | 2 | sox11a | LG17 |
| sox11b | LG20 | ||
| CYP19 | 15 | cyp19a | LG18 |
| cyp19b | LG25 | ||
| SPON1 | 11 | spon1a | LG18 |
| spon1b | LG25 | ||
| ATP1B1 | 1 | atp1b1b | LG1 |
| atp1b1a | LG6 | ||
| ATP1B2 | 17 | atp1b2b | LG5 |
| atp1b2a | LG23 | ||
| ATP1B3 | 3 | atp1b3a | LG2 |
| atp1b3b | LG15 | ||
| EFNA5 | 5 | efna5a | LG8 |
| efna5b | LG21 | ||
| EPHB4 | 7 | rtk5 | — |
| rtk8 | LG23 | ||
| FKH1 | 13 | foxc1.1 | LG2 |
| foxc1.2 | LG20 | ||
| FLOT1 | 6 | reggie2a | — |
| reggies2b | — | ||
| FZD8 | 10 | fz8b | LG2 |
| fz8a | LG24 | ||
| HSP71 (HSPA8) | 11 | hsc70 (1865782) | LG10 |
| hsc70(2495341) | — | ||
| hsp70 (2245606) | — | ||
| HSPA1 | 6 | hsp70(7861932) | — |
| hsp70(17061841) | — | ||
| HUR | 19 | hua | LG2 |
| hug | LG11 | ||
| KAL1 | X | kal | — |
| kal1b | LG22 | ||
| L1CAM | X | nadl1.1 | LG23 |
| nadl1.2 | LG23 | ||
| LHX1 | 11 | lim6 | LG5 |
| lim1 | LG15 | ||
| SPON2 | 4 | spon2a | LG14 |
| spon2b | LG14 | ||
| MITF | 3 | mitfa | LG6 |
| mitfb | LG13 | ||
| MSX2 | 5 | msxa | LG14 |
| msxd | LG21 | ||
| NTN1 | 17 | ntn1 | LG3 |
| ntn1a | LG6 | ||
| OTX1 | 2 | otx3 | LG1 |
| otx1 | LG17 | ||
| PAX2 | 10 | pax2.2 | — |
| pax2a | LG13 | ||
| POU3F3 | 2 | pou23 (zp23, brn1.2) | LG6 |
| pou12 (zp12, brn1.1) | LG9 | ||
| RX | 18 | rx2 | LG2 |
| rx1 | LG22 | ||
| TCF3 | 2 | tcf3 | LG10 |
| tcf3b | LG15 |
LG = linkage group. “—” = unmapped. Gene location data obtained from ZFIN (http://zfin.org/ZFIN/), LocusLink and Map Viewer (http://www.ncbi.nlm.nih.gov/Tools/index.html).
The L1Cam duplicates both occur on LG23 and Spon2 duplicates occur on LG14. These are two of the genes that did not have the duplication topology suggesting that lineage-specific tandem duplication events produced them. Interestingly, three other genes that did not have the predicted topology regardless of the phylogenetic method used (En2, Jak2, and Snap25) occur on what appeared to be paralogous chromosome segments (Table 2).
DISCUSSION
Large-Scale Gene Duplication
We identified 49 genes that occur once in human, mouse, chicken, once or twice in tetraploid X. laevis, and twice in zebrafish. Orthologs of 42 of these 49 genes were also uncovered in pufferfish and the phylogenies of these 42 genes show that in all but one case (HspA1), the gene duplication event occurred before the ancestors of zebrafish and pufferfish lineages diverged from one another. Even in HspA1, where the zebrafish duplicates are likely to be the products of a more recent lineage-specific duplication event, we found ancient duplicates in pufferfish and swordtails and reconstructed a topology consistent with an ancient fish-specific duplication event.
For many genes, not all phylogenetic methods produced the duplication topology. Where the topology predicted by the ancient genome duplication hypothesis was not recovered, our analyses frequently produced a tree with one of the zebrafish duplicates (or one of the clades of duplicated fish genes) as the sister group to the remaining set of fish and tetrapod genes. This “out-group topology” (Fig. 1C) has several plausible explanations. It is possible that the tetrapod orthologs of this basal sequence or set of fish sequences have been lost (Fig. 1D). Alternatively, the basal fish genes might be orthologs of the human sequence used to root the tree. Finally, the basal fish sequences might be duplicates that were erroneously “pushed” to the base of the tree by long-branch attraction (LBA). LBA can occur when the same traits (e.g., amino acid residues) evolve independently in the out-group and in a member (or members) of the in-group and it is most likely when one or more of the in-group sequences have an accelerated evolutionary rate (Felsenstein 1978).
Several observations suggest that LBA was to blame for gene/method combinations that produced topologies with one set of duplicates as sister to all remaining orthologs. If the basal fish genes were members of a clade that currently has no orthologs in tetrapods (Fig. 1D), then these sequences should be equally related to all members of the in-group and we would not expect any of the phylogenetic methods employed to reconstruct a tree with fish sequence monophyly. Yet, for most genes, at least one of the methods recovered a tree with good support for a monophyletic group that included all of the fish sequences. The hypothesis that the fish genes that form a sister group to the remaining fish plus tetrapod sequences are orthologs of the human out-group sequence can be tested by reconstructing trees with more distantly related sequences. Our final analyses were restricted to sets of orthologous genes and a single, usually human, out-group sequence because the inclusion of distantly related sequences almost always meant that unambiguous alignments were shorter. However, the preliminary trees did include many more distantly related genes and these analyses provided no support for the hypothesis that any fish genes in Table 1 were in fact orthologs of the human out-group sequence; in most cases a different zebrafish or pufferfish ortholog of the out-group sequence was identified in the preliminary tree reconstruction step. Finally, the observation that ASATURA produced the duplication topology where other methods did not suggests that the fast-evolving, amino-acid positions (i.e., those most likely to lead to LBA) were often responsible for the “basal” position of one set of duplicates.
Whole-Genome Duplication?
Our results indicate that a large number of fish genes were duplicated before the divergence of the ancestors of the zebrafish and the pufferfish. Although it is possible that these duplicates were formed by multiple independent gene duplication events after the divergence of Sarcopterygii (lobed-finned fish and tetrapods) and Actinopterygii but before the divergence of the zebrafish and pufferfish lineages, independent gene duplication events would not be expected to produce multiple, multigene blocks of paralogy. Most of the zebrafish duplicates we identified using a phylogenetic approach have been mapped. We used a radiation hybrid panel to map some genes (hua, hug, oprd1, or1, tpiA, tpiB, foxc1.1, foxc1.2) that had not been mapped and we looked to see if the duplicates were members of paralogous genome regions. Fifty-four genes (27 paralogous pairs) map to 10 paralogous synteny groups that each contain between two and five sets of duplicates (Table 2). For these 27 pairs of genes, we used a test described by Gates et al. (1999) to show that there are significantly fewer chromosome pairs with a single set of duplicates and significantly more chromosome pairs with two or more sets of duplicates than expected by chance. Thirteen different duplicated (paralogous) chromosomal regions have been previously identified in zebrafish (Amores et al. 1998; Gates et al. 1999; Barbazuk et al. 2000; Postlethwait et al. 2000) including all but one of the chromosome pairs identified in our study; our conclusion that LG18 and LG25 might be paralogous based upon the co-occurrence of Cyp19 and Spon1 duplicates is an addition to the list. Also, our conclusion that previously unmapped duplicates of Tpi and Oprd map to LG16 and LG19 (HoxAa and HoxAb cluster bearing chromosomes) increases the number of duplicates that co-occur on this pair of zebrafish chromosomes. Thus, the hypothesis that the paralogous regions of the zebrafish genome identified in the studies cited above were produced by a fish-specific duplication event was supported by these phylogenetic analyses.
Synteny was not limited to zebrafish chromosomes but also occurred among the zebrafish paralogs and human chromosomes. For example, duplicated Distal-less 2 and Engrailed 1 genes occur on zebrafish LG1 and LG9, and in human, DLX2 and EN1 both occur on chromosome 2. Duplicates of Engrailed 2 and Sonic hedgehog occur on linkage groups 2 and 7 in zebrafish and EN2 and SHH occur on human chromosome 7. Such a pattern would not be expected if these zebrafish duplicates were products of independent duplication events.
The ages and locations of the zebrafish duplicates can also be used to generate hypotheses about the genomic structure of ancestral vertebrates. For example, synteny between LG3 and LG12 within the zebrafish genome (i.e., the co-occurrence of HoxB, Nog, Rara and DLX4 duplicates on these two chromosomes) suggests that human chromosome 16, which contains HoxB genes, and human chromosome 17, which contains NOG, RARA and DLX4 were once linked. Synteny between LG16 and LG19, which contain duplicates of Gdf6, Tpi, Oprd and RxrB, suggests that chromosome 1 (with TPI and OPRD) and human chromosome 6 (with RXRB) might also have been linked.
These intra- and interspecific synteny data support the ancient fish-specific genome duplication hypothesis and provide insight into the origin of duplicates that have ambiguous phylogenies (e.g., the three genes that occur on duplicated chromosomes but do not have the predicted topology). However, it is possible for genes that have experienced independent duplication events to have their paralogs end up on the same two chromosomes. For example, MSX2, CNOT8, and TAF2 occur on the long arm of human chromosome five. There are two copies of each of these genes in the zebrafish genome and the duplicates all map to LG14 and LG21. From these observations, Liu et al. (2002) proposed portions of LG14 and LG21 were orthologous to the one region of human chromosome five. Our phylogenetic analyses support the hypothesis that a fish-specific duplication event produced msxd and msxa, zebrafish “co-orthologs” of MSX2 (a hypothesis first proposed by Barbazuk et al. 2000); however, we also found that genes that Liu et al. (2002) considered to be CNOT8 duplicates (fd59c07 and fd17b08) each have a different human ortholog. Zebrafish sequence fd17b08 is orthologous to CNOT8 (gi 15213949), mouse Ccr4NOT (gi 13386186) and pufferfish sequence JGI13618 whereas, fd59c07 is orthologous to CCR4NOT (gi 18595912), mouse Ccr4 (gi 6755126), and pufferfish sequence JGI4519. Thus, fd59c07 and fd17b08 (on LG14 and LG21) are not CNOT8 duplicates after all. Also, Woods et al. (2000) concluded that zebrafish genes bmpr1ab and bmpr1a were duplicates of BMPR1A because they were found on LG12 and LG13 along with duplicates of PAX2 and ADK. However, bmpr1ab is an ortholog of human BMPR1B (gi 4502431) and bmpr1a is orthologous to BMPR1A (gi 4757854). Thus, while synteny data can help with the interpretation of phylogenetic data and should aid in the search for duplicates that have diverged to the extent that they are difficult to identify using similarity-based approaches, a combination of phylogenetic analyses and gene mapping appear to be the best approach to reconstructing genome evolution.
A phylogenetic approach has been used to find evidence for genome duplication and to date duplication events relative to speciation events in several taxonomic groups (Wolfe and Shields 1997; Friedman and Hughes 2001; Robinson-Rechavi et al. 2001). Robinson-Rechavi et al. (2001) argued that an ancestral, whole-genome duplication may not have been responsible for the abundance of duplicated fish genes. Their phylogenetic analyses show that fish have more copies of many genes than humans and mice, but the duplicated fish genes Robinson-Rechavi et al. (2001) studied were frequently the products of lineage-specific gene duplication events. These results are consistent with the observation that gene duplication occurs at a very high frequency for a diversity of species (Lynch and Conery 2000). These results are also consistent with the observation that lineage-specific, whole-genome duplication is common among ray-finned fishes (e.g., Uyeno and Smith 1972; Ferris and Whitt 1977; Allendorf 1978; Schmidtke et al. 1979; Ludwig et al. 2001) and that some actinopterygian groups (e.g., families Salmonidae and Catastomidae) appear to retain more genes produced during lineage-specific duplication events than theory predicts they should (Bailey et al. 1978; Li 1980). However, the discovery that genes have been duplicated in one taxon (e.g., Salmonidae) but not in another (e.g., the family Cyprinidae) reveals little about the events that shaped the genome of the ancestor of these two lineages. Thus, the data discussed by Robinson-Rechavi et al. (2001) are not evidence against the ancient fish-specific genome duplication hypothesis (Taylor et al. 2001b).
Elgar et al. (1999) analyzed 25 Mb (>6%) of random sequence from the T. rubripes genome and did not find large numbers of duplicated genes where there is only one copy in mammals. This observation was recently reinforced by comparisons between the most recent release of the pufferfish genome and the human genome (Aparicio et al. 2002). These observations are not consistent with our phylogeny and synteny-based conclusion that an ancient fish-specific genome duplication event preceded the divergence of the ancestors of zebrafish and pufferfish. However, gene loss in pufferfish can reconcile our observations with those of the Elgar et al. (1999) and Aparicio et al. (2002). If the pufferfish has not retained as many duplicates as zebrafish, as is suggested by the large number of trees with topologies consistent with ancient tetraploidy but with only one pufferfish sequence, then the discovery of duplicates in random fragments of the pufferfish genome or in comparisons with the human genome will be less likely.
Evolutionary Consequences of Genome Duplication
Zebrafish and pufferfish are distant relatives within Euteleostei (Nelson 1994; Arratia 1997), a subdivision that includes ∼22,000 species. Our conclusion that the ancestor of these two species experienced a genome duplication event lends support to the idea that genome duplication and speciation might be causally linked (Amores et al. 1998; Wittbrodt et al. 1998; Meyer and Schartl 1999; Taylor et al. 2001a,c). An intuitive link between extra genes and speciation is the one proposed by Stephens, Ohno, and many others, that is, the evolution of beneficial new gene functions in redundant genes. The number of examples of the evolution of new, potentially adaptive functions in duplicated genes is growing but still quite small, e.g., antifreeze proteins in Antarctic fishes (Cheng and Chen 1999), color vision in new-world monkeys (Dulai et al. 1999), thermal adaptation in Escherichia coli (Riehle et al. 2001) and RNA digestion in colobine monkeys (Zhang et al. 2002) and complete genome duplication would provide an enormous number of genes with the potential to evolve new functions.
Divergent resolution or reciprocal silencing is another possible link between genome duplication and speciation in Actinopterygii. Divergent resolution occurs when different allopatric populations lose different copies of duplicated genes. Hybridization between such populations produces an F1 generation with one functional allele and one pseudogene at each of the duplicated loci and crosses between F1 individuals produce individuals with between zero and four alleles at the duplicated loci (Werth and Windham 1991; Lynch and Force 2000; Taylor et al. 2001c). Genome duplication produces an enormous number of gene duplicates that could be divergently resolved. Selection against F2 individuals with more or less than two alleles per locus might provide a genetic environment in which speciation alleles (i.e., alleles for assortative mating) would be favored.
Summary
The zebrafish is a model organism, used largely for the study of gene expression during development (Westerfield 1993) and the pufferfish genome sequence is facilitating the identification of regulatory elements that influence gene expression (Yu et al. 2001). Other fishes such as the Japanese medaka (Oryzias latipes) and the platyfish (Xiphophorus maculatus) are also being developed as “complementary” model organisms to the zebrafish and pufferfish (Wittbrodt et al. 2002). Medaka and platyfish are more closely related to the pufferfish than to the zebrafish and, therefore, our phylogenies indicate that all four model species differ from human with respect to ancestral ploidy. This means that comparative studies will have to be designed that, as a starting point, do not assume a 1:1 ratio of “orthologous” genes between human and model fish species.
For a diversity of studies, polyploidy in model fish species might be advantageous. For example, it should be possible to identify regulatory elements in each of the zebrafish duplicates by comparing orthologous sequences in zebrafish and pufferfish. A given human gene often has many expression domains, and if these expression domains have been subdivided between the fish duplicates (Force et al. 1999), then by comparing the zebrafish and pufferfish sequences it might be possible to identify the regulatory elements associated with expression domains in zebrafish. These data might then be used to associate regulatory elements with expression domains in humans.
Furthermore, sequence-level studies on species that experienced genome duplication may help us to determine whether our own genome is the product of an ancient genome duplication event because they indicate what the evolutionary products of genome duplication look like (Wolfe 2001).
METHODS
Identifying Duplicated Zebrafish Genes
Sets of orthologous genes were obtained by reconstructing phylogenetic trees from sequences obtained through BLASTp searches (Altschul et al. 1990). Query sequences for BLAST searches included 174 human genes identified as duplicates of Drosophila genes (Spring 1997) and human orthologs of genes that occur on what appear to be duplicated zebrafish chromosomes (Gates et al. 1999; Barbazuk et al. 2000; Woods et al. 2000).
BLASTp searches of the NCBI database were carried out one species at a time for Homo sapiens, Mus musculus, Gallus gallus, Xenopus laevis, and Danio rerio. The BLAST e-values, which estimate the likelihood of alignment scores occurring by chance, were used to determine which genes to include in the phylogenetic analyses. Several different potential e-value cut-off points were often noticeable in the list of genes retrieved by BLAST, especially for members of large gene families such as homeobox-containing genes. When this occurred, a cut-off was selected that included genes that we suspected might not be orthologs of the query sequences in order to avoid excluding genes that might be orthologous.
Protein sequences identified by this method were aligned using CLUSTAL (Thompson et al. 1997) in BIOEDIT (Hall 1999). Manual editing of the alignments (e.g., removal of large gaps and removal of long stretches of sequence without counterparts in other species) was carried out also using BIOEDIT. TREECON (Van de Peer and De Wachter 1994) was used to calculate Poisson-corrected genetic distances and to reconstruct neighbor-joining (NJ) trees (Saitou and Nei 1987). These preliminary phylogenetic analyses identified sequences that differed only in length or by few amino-acid substitutions (e.g., allelic variation or very recent duplications) and highly divergent genes (e.g., genes that were retrieved in BLAST searches because they shared a conserved domain with the query sequence but differed to a large extent elsewhere), which were usually excluded from further analyses. Especially important for this study, these preliminary trees identified genes that appeared to be duplicated in zebrafish, orthologs of these duplicates in human, mouse, chicken, and frog, and the most closely related nonortholog in human, which was used to root subsequent phylogenetic analyses. The zebrafish duplicates were then used as query sequences for BLAST searches of the October, 2001, release of the Japanese pufferfish (T. rubripes) genome (http://www.jgi.doe.gov/fugu/index.html) and of all actinopterygian protein sequences in the NCBI nonredundant database (http://www.ncbi.nlm.nih.gov). The final set of orthologous genes from zebrafish, other fish species, human, mouse, chicken, and Xenopus, and the most closely related human out-group sequence were realigned. Nucleotide sequences for these final sets of genes were also obtained from the NCBI database. Nucleotide sequences were translated and aligned in BIOEDIT. By toggling between nucleotide and amino-acid format, it was sometimes possible to improve the amino-acid alignments (i.e., using information from third codon positions).
TREECON and TREEPUZZLE (Strimmer and Von Haeseler 1996) were used to reconstruct genetic distance-based trees and maximum likelihood trees from these final alignments, respectively. TREEPUZZLE was also used to calculate the likelihoods of user-defined topologies. For example, for genes with two copies in zebrafish and one in pufferfish, we compared the likelihood of the topology showing a sister sequence relationship between one zebrafish and the pufferfish sequence (i.e., the topology expected if the gene duplication event preceded the speciation event) to the likelihood of the topology showing a sister sequence relationship for the zebrafish duplicates (i.e., the topology expected if the duplication event was specific to the zebrafish lineage). We also used ASATURA (Van de Peer et al. 2002) to remove frequently substituted amino-acid positions from each pairwise comparison prior to genetic distance estimation and phylogeny reconstruction.
Locating Duplicates on Chromosomes
Map data for duplicated zebrafish genes were obtained from Woods et al. (2000) and from ZFIN (http://zfin.org/ZFIN/). Also, a zebrafish radiation hybrid panel (Kwok et al. 1998) was used to experimentally map genes. We then compared the number of chromosome pairs with more than one set of duplicates to the number of chromosomes pairs expected to have more than one set of duplicates assuming a Poison distribution of duplicates (see Gates et al. 1999). For this calculation, the HoxB5 and HoxB6 duplicates were treated as a single locus. The chromosomal locations of human orthologs were obtained from LocusLink and Map Viewer (http://www.ncbi.nlm.nih.gov/Tools/index.html).
WEB SITE REFERENCES
http://www.evolutionsbiologie.uni-konstanz.de/Wanda/; WANDA—database of genes duplicated in fish.
http://www.jgi.doe.gov/fugu/index.html; The Fugu rubripes genome project Web site at the Joint Genome Institute.
http://www.ncbi.nlm.nih.gov; National Center for Biotechnology Information (NCBI).
http://zfin.org/ZFIN/; The Zebrafish Information Network.
http://www.ncbi.nlm.nih.gov/Tools/index.html: NCBI Tools for Data Mining Web site.
Acknowledgments
We thank Henner Brinkmann for comments on the manuscript. JST is supported by a Postdoctoral Fellowship from the Natural Sciences and Engineering Research Council of Canada. We thank the Deutsche Forschungsgemeinschaft for the Schwerpunkt program: Informatikmethoden zur Analyse und Interpretation großer Datenmengen, grant number SPP 1063.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL: axel.meyer@uni-konstanz.de; FAX: 0049 7531 883018.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.640303.
REFERENCES
- 1.Allendorf F.W. 1978. Protein polymorphism and the rate of loss of duplicate gene expression. Nature 272: 76-78. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amores A., Force, A., Yan, Y.-L., Joly, L., Amemiya, C., Frity, A., Ho, R.K., Langeland, J., Prince, V., Wang, Y.-L., et al. 1998. Zebrafish hox clusters and vertebrate genome evolution. Science 282: 1711-1714. [DOI] [PubMed] [Google Scholar]
- 4.Aparicio S., Chapman, J., Stupka, E., Putnam, N., Chia, J.M., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- 5.Arratia G. 1997. The monophyly of Teleostei and stem-group teleosts. Consensus and disagreements. In Mesozoic fishes 2—systematics and fossil record. (eds. G. Arratia & H.-P. Schultze), pp. 265–334. Verlag Dr. Friedrich Pfeil, München, Germany.
- 6.Bailey G.S., Poulter, R.T., and Stockwell, P.A., 1978. Gene duplication in tetraploid fish: Model for gene silencing at unlinked duplicated loci. In Proc. Natl. Acad. Sci. 5575–5579 11:. [DOI] [PMC free article] [PubMed]
- 7.Barbazuk W.B., Korf, I., Kadavi, C., Heyen, J., Tate, S., Wun, E., Bedell, J.A., McPherson, J.D., and Johnson, S.L. 2000. The syntenic relationship of the zebrafish and human genomes. Genome Res. 10: 1351-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng C.-H. C. and Chen, L. 1999. Evolution of an antifreeze glycoprotein. Nature 401: 443-444. [DOI] [PubMed] [Google Scholar]
- 9.Dayhoff M., 1978. Atlas of Protein Sequence and Structure, pp. 345–358. National Biomedical Research Foundation, Washington D.C.
- 10.Dulai K.S., von Dornum, M., Mollon, J.D., and Hunt, D.M. 1999. The evolution of trichromatic colour vision by opsin gene duplication in New World and Old World primates. Genome Res. 9: 629-638. [PubMed] [Google Scholar]
- 11.Elgar G., Clark, M.S., Meek, S., Smith, S., Warner, S., Edwards, Y.J.K., Bouchireb, N., Cottage, A., Yeo, G.S.H., Umrania, Y., et al. 1999. Generation and analysis of 25 Mb of genomics DNA from the pufferfish Fugu rubripes by sequence scanning. Genome Res. 9: 960-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein J. 1978. Cases in which parsimony or compatibility methods will be positively misleading. Systematic Zoology 27: 401-410. [Google Scholar]
- 13.Ferris S.D. and Whitt, G.S. 1977. Loss of duplicate gene expression after polyploidization. Nature 265: 258-260. [DOI] [PubMed] [Google Scholar]
- 14.Force A., Lynch, M., Pickett, F.B., Amores, A., Yan, Y.-I., and Postlethwait, J. 1999. The preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman R. and Hughes, A.L. 2001. Pattern and timing of gene duplication in animal genomes. Genome Res. 11: 1842-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gates M.A., Kim, L., Egan, E.S., Cardozo, T., Sirotkin, H.I., Dougan, S.T., Lashkari, D., Abagyan, R., Schier, A., and Talbot, W.S. 1999. A genetic linkage map for zebrafish: Comparative analysis and localization of genes and expressed sequences. Genome Res. 9: 334-347. [PubMed] [Google Scholar]
- 17.Goff S.A., Ricke, D., Lan, T.-H., Presting, G., Wang, R., Dunn, M., Glazebrook, J., Sessions, A., Oeller, P., Varma, H., et al. 2002. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92-100. [DOI] [PubMed] [Google Scholar]
- 18.Gu X. and Huang, W. 2002. Testing the parsimony test of genome duplications: A counter example. Genome Res. 12: 1-2. [DOI] [PubMed] [Google Scholar]
- 19.Hall T.A. 1999. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 41: 95-98. [Google Scholar]
- 20.Jorden I.K., Makarova, K.S., Spouge, J.L., Wolf, Y.I., and Koonin, E.V. 2001. Lineage-specific gene expansions in bacterial and archaeal genomes. Genome Res. 11: 555-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kishino S. and Hasegawa, M. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29: 170-179. [DOI] [PubMed] [Google Scholar]
- 22.Kwok C., Korn, R.M., Davis, M.E., Burt, D.W., Critcher, R., McCarthy, L., Paw, B.H., Zon, L.I., Goodfellow, P.N., and Schmitt, K. 1998. Characterization of whole genome radiation hybrid mapping resources for non-mammalian vertebrates. Nucleic Acids Res. 26: 3562-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li W-H. 1980. Rate of gene silencing at duplicated loci: A theoretical study and interpretation of data from tetraploid fishes. Genetics 95: 237-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu T. X., Kanki, J. P., Deng, M., Rhodes, J., Yang, H.W., Sheng, X.M., Zon, L.I., and Look, A.T. 2002. Evolutionary conservation of zebrafish linkage group 14 with frequently deleted regions of human chromosome 5 in myeloid malignancies. Proc. Natl. Acad. Sci. 99: 6136-6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ludwig A., Belifiore, N.M., Pitra, C., Svirsky, V., and Jenneckens, I. 2001. Genome duplication events and functional reduction of ploidy levels in sturgeon (Acipenser, Huso and Scaphirhynchus). Genetics 158: 1203-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lynch M. 2001. The molecular natural history of the human genome. Trends Ecol. Evol. 16: 420-422. [Google Scholar]
- 27.Lynch M. and Conery, J.S. 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151-1155. [DOI] [PubMed] [Google Scholar]
- 28.Lynch M. and Force, A.G. 2000. The origin of interspecific genomic incompatibility via gene duplication. Am. Nat. 156: 590-605. [DOI] [PubMed] [Google Scholar]
- 29.Meyer A. and Schartl, M. 1999. Gene and genome duplications in vertebrates: The one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 11: 699-704. [DOI] [PubMed] [Google Scholar]
- 30.Nelson J.S., 1994. Fishes of the world Wiley & Sons, New York, NY.
- 31.Ohno S., 1970. Evolution by gene duplication. Spinger-Verlag, New York, NY.
- 32.Postlethwait J.H., Woods, I.G., Ngo-Hazelett, P., Yan, Y.-L., Kelly, P.D., Chu, F., Huang, H., Hill-Force, A., and Talbot, W.S. 2000. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10: 1890-1902. [DOI] [PubMed] [Google Scholar]
- 33.Prince V.E., Joly, L., Ekker, M., and Ho, R.K. 1998. Zebrafish hox genes: Genomics organization and modified colinear expression patterns in the trunk. Development 125: 407-420. [DOI] [PubMed] [Google Scholar]
- 34.Riehle M.M., Bennette, A.F., and Long, A.D. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc. Natl. Acad. Sci. 98: 525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson-Rechavi M., Marchand, O., Escriva, H., and Laudet, V. 2001. An ancestral whole-genome duplication may not have been responsible for the abundance of duplicated fish genes. Curr. Biol. 11: R458-R459. [DOI] [PubMed] [Google Scholar]
- 36.Saitou N. and Nei, M. 1987. The neighbor-joining methods: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406-425. [DOI] [PubMed] [Google Scholar]
- 37.Schmidtke J., Schmitt, E., Matzke, E., and Engel, W. 1979. Non-repetitive DNA sequence divergence in phylogenetically diploid and tetraploid teleostean species of the family Cyprinidae and the order Isopondyli. Chromosoma 75: 185-198. [DOI] [PubMed] [Google Scholar]
- 38.Spring J. 1997. Vertebrate evolution by interspecific hybridization—are we polyploid? FEBS Lett. 400: 2-8. [DOI] [PubMed] [Google Scholar]
- 39.Stephens S.G. 1951. Possible significance of duplications in evolution. Adv. Genet. 4: 247-265. [DOI] [PubMed] [Google Scholar]
- 40.Stock D.W., Quattro, J.M., Whitt, G.S., and Powers, D.A. 1997. Lactate dehydrogenase (LDH) gene duplication during chordate evolution: The cDNA sequence of LDH of the tunicate Styela plicata. Mol. Biol. Evol. 14: 1273-1284. [DOI] [PubMed] [Google Scholar]
- 41.Strimmer K. and Von Haeseler, A. 1996. Quartet puzzling: A quartet maximum likelihood methods for reconstructing tree topologies. Mol. Biol. Evol. 13: 964-969. [Google Scholar]
- 42.Taylor J.S., Van de Peer, Y., Braasch, I., and Meyer, A. 2001a. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R. Soc. 356: 1661-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Taylor J.S., Van de Peer, Y., and Meyer, A. 2001b. Revisiting recent challenges to the ancient fish-specific genome duplication hypothesis. Curr. Biol. 11: R1005-1007. [DOI] [PubMed] [Google Scholar]
- 44.Taylor J.S., Van de Peer, Y., and Meyer, A. 2001c. Genome duplication, divergent resolution and speciation. Trends Genet. 17: 299-301. [DOI] [PubMed] [Google Scholar]
- 45.Thompson J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The Clustal_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uyeno T. and Smith, G.R. 1972. Tetraploid origin of the karyotype of catastomid fishes. Science 175: 644-646. [DOI] [PubMed] [Google Scholar]
- 47.Valente Samonte R. and Eichler, E. 2002. Segmental duplications and the evolution of the primate genome. Nat. Genet. Rev. 3: 65-72. [DOI] [PubMed] [Google Scholar]
- 48.Van de Peer Y. and De Wachter, Y. 1994. TREECON for Windows: A software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10: 569-570. [DOI] [PubMed] [Google Scholar]
- 49.Van de Peer Y., Frickey, T., Taylor, J.S., and Meyer, A. 2002. Dealing with saturation at the amino acid level: A case study based on anciently duplicated zebrafish genes. Gene 295: 205-211. [DOI] [PubMed] [Google Scholar]
- 50.Vision T.J., Brown, D.G., and Tanksley, S.D. 2000. The origins of genomic duplications in Arabidopsis. Science 290: 2114-2117. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y. and Gu, X. 2000. Evolution patterns of gene families generated in the early stages of vertebrates. J. Mol Evol. 51: 88-96. [DOI] [PubMed] [Google Scholar]
- 52.Werth C.R. and Windham, M.D. 1991. A model for divergent, allopatric speciation of polyploidy pteridophytes resulting from silencing of duplicate-gene expression. Am. Nat. 137: 515-526. [Google Scholar]
- 53.Westerfield M., 1993. The zebrafish book. University of Oregon Press, Eugene, OR.
- 54.Wittbrodt J., Meyer, A., and Schartl, M. 1998. More genes in fish? Bioessays 20: 511-512. [Google Scholar]
- 55.Wittbrodt J., Shima, A., and Schartl, M. 2002. Medaka—a model organism from the far east. Nat. Genet. Rev. 3: 53-64. [DOI] [PubMed] [Google Scholar]
- 56.Wolfe K.H. 2001. Yesterday's polyploids and the mystery of diploidization. Nat. Genet. Rev. 2: 333-341. [DOI] [PubMed] [Google Scholar]
- 57.Wolfe K.H. and Shields, D.C. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387: 708-713. [DOI] [PubMed] [Google Scholar]
- 58.Woods I.G., Kelly, P.D., Chu, F., Ngo-Hazelett, P., Yan, Y.-L., Huang, H., Postlethwait, J.H., and Talbot, W.S. 2000. A comparative map of the zebrafish genome. Genome Res. 10: 1903-1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu W.P., Pallen, C.J., Tay, A., Jirik, F.R., Brenner, S., Tan, Y.H., and Venkatesh, B. 2001. Conserved synteny between the Fugu and human PTEN locus and the evolutionary conservation of vertebrate PTEN function. Oncogene 20: 5554-5561. [DOI] [PubMed] [Google Scholar]
- 60.Zhang J., Zhang, Y.-p., and Rosenberg, H.F. 2002. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 30: 411-415. [DOI] [PubMed] [Google Scholar]