Abstract
To identify highly informative markers for a large number of commonly employed murine crosses, we selected a subset of the extant mouse simple sequence length polymorphism (SSLP) marker set for further development. Primer pairs for 314 SSLP markers were designed and typed against 54 inbred mouse strains. We designed new PCR primer sequences for the markers selected for multiplexing using the fluorescent dyes FAM, VIC, NED, and ROX. The number of informative markers for C57BL/6J × DBA/2J is 217, with an average spacing of 6.8 centiMorgans (cM). For all other pairs of strains, the mean number of informative markers per cross is 197.0 (SD 37.8) with a mean distance between markers of 6.8 cM (SD 1.1). To confirm map positions of the 224 markers in our set that are polymorphic between Mus musculus and Mus spretus, we used The Jackson Laboratory (TJL) interspecific backcross mapping panel (TJL BSS); 168 (75%) of these markers had not been previously mapped in this cross by other investigators, adding new information to this community map resource. With this large data set, we sought to reconstruct a phylogenetic history of the laboratory mouse using Wagner parsimony analysis. Our results are largely congruent with the known history of inbred mouse strains.
[The following individuals kindly provided reagents, samples, or unpublished information as indicated in the paper: E. Eicher, T. Golovkina, J. Cheverud, S. Cropp, P. Denny, and A. Southwell.]
Other investigators have developed simple sequence length polymorphism (SSLP) markers for use as genetic markers in many organisms. Short tandem repeat (STR), short tandem repeat polymorphism (STRP), and microsatellite are other names commonly used for this type of marker. A comprehensive genetic linkage map of the mouse has been constructed (Dietrich et al. 1996) using over 6000 SSLP markers with an average spacing of less than 1 cM. However, it is still difficult to identify sufficient markers that are informative for a specific pair of inbred strains, particularly if they are both derived from Mus musculus domesticus. Allele size data are available for only a limited number of inbred mouse strains (Schalkwyk et al. 1999), which makes it difficult to construct a suitable panel of markers that are detectably polymorphic for a selected cross. In addition, many primer pairs have not been extensively characterized or do not yield robust PCR products.
Starting with the CA repeat-containing sequences identified by Dietrich et al. (1996), we assembled a set of marker loci that span the mouse genome and are informative in many common inbred strain combinations, and we designed new optimized primer sequences. Our primers were specifically designed to allow multiplexing by fluorescent dye color and product size. Our current level of multiplexing is 8–9 loci per capillary on the Applied Biosystems 3700 platform, producing ∼750 genotypes per run. The forward primer of each pair was labeled on the 5′ end with the fluorescent dyes FAM, VIC, or NED. We avoided CA sequences on the 3′ end of the forward primers to reduce stuttering artifacts. All reverse primers contain the sequence GTGTCTT (5′–3′) at their 5′ ends to promote nontemplate-directed nucleotide addition (+A; Smith et al. 1995; Brownstein et al. 1996). This simplifies allele calling by reducing stuttering artifacts that arise during amplification of repeat-containing sequences. Theoretically, we could multiplex more loci per lane depending on the sensitivity of the detection platform and the robustness of the PCR reactions. We presently multiplex post-PCR by pooling different ratios of each PCR product.
In order to confirm the map positions identified by our newly designed PCR assays for this set of SSLP markers, we typed 94 animals from TJL BSS interspecific mapping panel [(C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei] to place our markers in this existing comprehensive map that contained over 4500 mapped loci at the time of our analysis (Rowe et al. 1994).
SSLP markers are widely dispersed, selectively neutral, and their alleles typically differ by discrete steps (Weber and Wong 1993). For these reasons, they have proven useful for creating mouse family trees by parsimony analysis. To infer the degree of relatedness between inbred mouse strains, we used the allele size data for all 314 markers that we collected for 54 strains to construct a mouse family tree as described by Acthley and Fitch (1993) and Schalkwyk et al. (1999). We performed Wagner parsimony analysis using the MIX program of PHYLIP v. 3.57 (Felsenstein 1989).
RESULTS AND DISCUSSION
Mouse Linkage Mapping Set
We chose a target density of 10 cM for our marker set in order to conduct genome-wide screens for linkage analysis. We first selected ∼160 markers from the Dietrich et al. (1996) set to maximize polymorphism for the cross C57BL/6J × DBA/2J. We then selected an additional ∼160 markers to increase the map density and maximize polymorphism for other pairs of strains. We designed and tested a total of 323 primer pairs for 314 SSLP markers. Twenty-two primer pairs failed or performed poorly. Thirteen of these primer pairs were successfully redesigned, resulting in 305 usable markers. The remaining nine markers were replaced to fill any remaining gaps between markers that were greater than 10 cM. The total number of markers for which primers were successfully designed is 314. Map positions are based on stored values in the Mouse Genome Database (MGD; TJL) which are updated daily.
The result was a set of 314 SSLP markers, which we used to genotype a total of 54 inbred mouse strains. Graphical chromosome maps displaying the cM position of each marker are shown in Figure 1. The number of informative markers for the C57BL/6J × DBA/2J cross is 217, with an average spacing of 6.8 cM. This represents ∼70% of the markers in the mapping set. The largest gap for this cross is 33 cM at the end of chromosome 5. For all other pairs of strains, the mean number of informative markers per cross is 197.0 (SD 37.8). The mean distance between markers is 6.8 cM (SD 1.1), and the mean number of gaps greater than 20 cM is 7.
Figure 1.
Graphical chromosome maps displaying the map positions of 314 newly designed SSLP markers for fluorescent genotyping by PCR analysis. Map positions in centiMorgans (cM) were obtained from the Mouse Genome Database (TJL) with the exception of D3Mit217, D19Mit72, D8Mit46, and D8Mit112, which are displayed relative to their map order on The Jackson Laboratory BSS Mapping Panel.
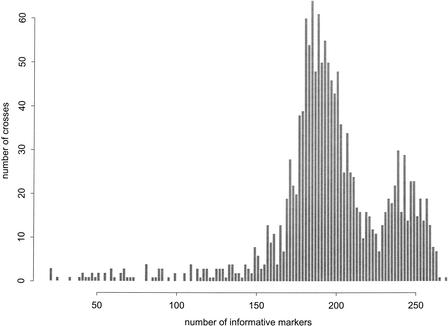
A histogram showing the number of informative markers for each pair of mouse strains we evaluated is shown in Figure 2. The tail on the left side of the histogram displays crosses between closely related substrains. This includes a cluster of six 129 substrains. The genetic variability among 129 substrains was documented by Simpson et al. (1997), who observed differences as high as 22% for alleles at 86 SSLP loci in fifteen 129 substrains. We genotyped ∼3.5-fold more markers and observed differences of 16%–26% among six 129 substrains. We concur with Simpson et al. (1997) that the majority of this variability is most likely the result of unrecognized outcrossing rather than new mutations. The contamination of 129/SvJ has been reported elsewhere (Threadgill et al. 1997). Table 1 indicates the number of informative markers for crosses among 12 inbred mouse strains.
Figure 2.
Number of informative markers per cross. Pairwise combinations of 54 inbred mouse strains were analyzed to determine the number of markers that are polymorphic for each cross. The number of crosses are plotted against the number of informative markers for each pair of strains. The mean number of informative markers per cross for all pairs of strains analyzed is 197.0 (SD 37.8).
Table 1.
Number of Polymorphic Markers Between 12 Pairs of Inbred Mouse Strains
| DBA/2J | 129P3/J | A/J | BALB/cJ | FVB/NJ | NOD/LtJ | CAST/Ei | C3H/HeJ | AKR/J | NZB/B1NJ | SPRET/Ei | |
| DBA/2J | |||||||||||
| 129P3/J | 181 | ||||||||||
| A/J | 177 | 177 | |||||||||
| BALB/cJ | 172 | 186 | 109 | ||||||||
| FVB/NJ | 184 | 196 | 156 | 158 | |||||||
| NOD/LtJ | 185 | 191 | 166 | 178 | 161 | ||||||
| CAST/Ei | 259 | 251 | 263 | 262 | 260 | 255 | |||||
| C3H/HeJ | 163 | 193 | 134 | 158 | 171 | 185 | 259 | ||||
| AKR/J | 188 | 197 | 160 | 167 | 182 | 178 | 258 | 177 | |||
| NZB/B1NJ | 200 | 194 | 210 | 196 | 192 | 193 | 263 | 220 | 200 | ||
| SPRET/Ei | 234 | 231 | 236 | 240 | 238 | 236 | 238 | 234 | 241 | 243 | |
| C57BL/6J | 217 | 197 | 223 | 214 | 217 | 213 | 269 | 246 | 219 | 204 | 244 |
Current, detailed information about this marker set is publicly available on the Web at http://www.cidr.jhmi.edu/mouse/mouse.html. Investigators can submit a query for any pairwise combination of inbred strains to retrieve the informative markers for a cross, or download a spreadsheet with the locus names, map positions, and allele sizes for each marker for all 54 strains. We have incorporated other features into our Web site (http://www.cidr.jhmi.edu/mouse/queries.html), including a panel generator to group primer pairs by dye color and allele sizes for multiplex loading of gels or capillaries, and a query to identify markers for constructing speed congenics by marker-assisted selection (Wakeland et al. 1997).
The panel generator allows the user to (1) select the maximum number of primer pairs in a panel, (2) select the maximum number of primer pairs with the same dye color in a panel, (3) increase the size ranges of each primer pair to avoid overlaps with other primer pairs, and (4) select the minimum number of base pairs between the size ranges of primer pairs with the same dye color in a panel. A list of all of the informative markers is displayed, along with map information, the spacing between markers, PCR performance, and comments about the primer pairs. The user can choose the best markers at the desired spacing to generate the panels. Panels are displayed on a new page with the dye color, size range, allele sizes, and recommended PCR conditions for each primer pair. The user can then click on a link that will display the part numbers for the primer pairs, which can be ordered directly from Applied Biosystems. These marker panels are specific for the amplicon sizes and dye colors in the primer pairs available from Applied Biosystems.
The speed congenics query generates a list of informative markers between the donor and recipient strains such that recipient allele sizes must be different from donor allele sizes. Investigators can select up to three donor strains and up to three recipient strains. We also included a query that only displays markers in which the allele sizes of all of the selected strains are different.
The current marker density is sufficient to conduct 20-cM genome-wide screens in mouse for linkage mapping studies (Silver 1995). For such scans we routinely type approximately 85 markers. We have used this marker set to type 1843 animals for five different studies involving seven different strains. Four studies were genome-wide screens for quantitative trait loci (QTLs), and the fifth study was the first phase of a marker-assisted breeding experiment. The cumulative average missing data rate we have achieved is 5.2%, and the cumulative average error rate as calculated from 5% blind duplicate samples run concurrently with each study is 0.1%.
The primer pairs we designed are different from the primer pairs designed by The Whitehead Institute/MIT Center for Genome Research (WICGR). Few, if any, of the allele sizes observed using these primers will be the same as those observed using the WICGR primers. However, the markers should remain informative for a pair of inbred mouse strains using either set of primers. We developed and characterized all of the markers using Applied Biosystems 377XL sequencers. Running the products on a different instrument platform or with a different size standard will often result in different observed allele sizes, but the size difference between strains should be constant.
Backcross Mapping
We used TJL BSS interspecific backcross mapping panel to confirm the map positions of the markers in our set that were informative between M. musculus and M. spretus. Although 251 markers from our set are polymorphic for this strain combination, 27 of these could not be mapped due to allele dominance (primers designed to C57BL/6J sequence preferentially amplify only from the C57BL/6J allele template in the presence of a highly diverged M. spretus allele sequence). We also observed that a small percentage of marker loci fail to amplify for strains not belonging to the M. m. domesticus subgroup (data not shown).
We mapped the remaining 224 informative markers onto The Jackson Laboratory BSS Mapping Panel; 220 (98%) marker loci mapped to the expected chromosome location. One hundred and sixty-eight of these markers (75%) represent new loci that had not previously been mapped by other investigators onto the BSS panel, contributing new information to this comprehensive genetic map of the mouse. Detailed information for all loci mapped in this cross including the mapping data reported here can be found at http://www.jax.org/resources/documents/cmdata/bkmap/BSS.html.
Four (2%) of our mapped marker loci did, however, map to unexpected positions. D3Mit217 and D19Mit72 mapped to chromosome 1, whereas D8Mit46 and D8Mit112 both mapped to chromosome 9. All of these new chromosomal assignments are corroborated by independent data sets from other laboratories submitted to The Jackson Laboratory Mapping Panels, using the interspecific backcross panels or the mouse T31 Radiation Hybrid mapping panel (http://www.jax.org/resources/documents/cmdata/rhmap/RHIntro.html) or both. Based on this evidence, these four markers have been renamed by the Mouse Locus Nomenclature Committee as of May 2002 as D1Mit1000 for the old D3Mit217, D1Mit1001 for the former D19Mit72, D9Mit1000 for the locus previously named D8Mit46, and D9Mit1001 for the old D8Mit112.
We compared our BSS mapping results to the composite maps available from the MGD (http://www.informatics.jax.org). We used the MGD values to generate the chromosome maps in Figure 1. Composite chromosome maps are longer than the BSS maps, because composite maps are based on many types of crosses and other mapping methods. Recombination frequencies vary from one cross to another, and the number of commonly mapped anchor markers in some combinations of crosses can be scarce, and because creating composite maps is an inherently inaccurate process, particularly for closely linked loci mapped in independent crosses or systems. In addition, any data errors expand the apparent distances between markers. Although these problems can and do lead to order inaccuracies in composite maps, we observe no differences between the order of our markers in the TJL BSS map and MGD composite maps.
We were unable to confirm the map positions of 90 markers using the backcross panel. For 79 of these markers, the physical location was determined using the Mouse Genome Sequencing Consortium Assembly 3b (http://www.genome.wi.mit.edu/cgi-bin/mouse/index#genetic). For seven of the remaining 11 markers, we used published polymorphism data (WICGR) to verify that the same locus was being amplified with our primer pairs. The map positions of the remaining markers (D3Mit51, D13Mit119, D14Mit74, D19Mit90) could not be substantiated with either resource at this time.
Mouse Family Tree
Information about the relatedness of inbred mouse strains is useful in determining the extent of polymorphism between strains when designing crosses and evaluating the genetic background of mice when accessing phenotypes. Atchley and Fitch (1993) performed a phylogenetic analysis employing classical genotypes from 144 gene loci in 24 inbred mouse strains, using parsimony analysis. Schalkwyk et al. (1999) performed a similar analysis using data from 128 SSLP loci in 11 inbred strains. We sought to reconstruct a phylogenetic history of the laboratory mouse using a larger data set. To infer the degree of relatedness between inbred mouse strains, we used the allele size data for all 314 markers that we collected for 54 strains to construct a mouse family tree using Wagner parsimony analysis (Atchley and Fitch 1993; Schalkwyk et al. 1999). We did not consider Y chromosome (Tucker et al. 1992) or mtDNA data (Ferris et al. 1983) in our analysis. Most new mutations in SSLP markers are thought to result from gains or losses of a single repeat unit (Weber and Wong 1993). However, multiple repeat units can be added or deleted between alleles, and sometimes the entire repeat is duplicated. Without a defined model for the process of addition or deletion of multiple repeat units, it is not clear how this process might affect the analysis. Larger data sets should minimize any distortion of results due to this type of variation. We coded allele sizes as n-1 integers, where n is the number of alleles, and allele sizes differ by one step (i.e., two base pairs). This gives order to the mutational changes between alleles but does not assume that a particular allele is ancestral. We performed a Wagner parsimony analysis using the MIX program of PHYLIP v. 3.57 (Felsenstein 1989). User options were selected to generate an unrooted tree after the input order of allele sizes was randomized three times. No ancestral state for allele sizes was assumed for analysis. A consensus family tree is shown in Figure 3.
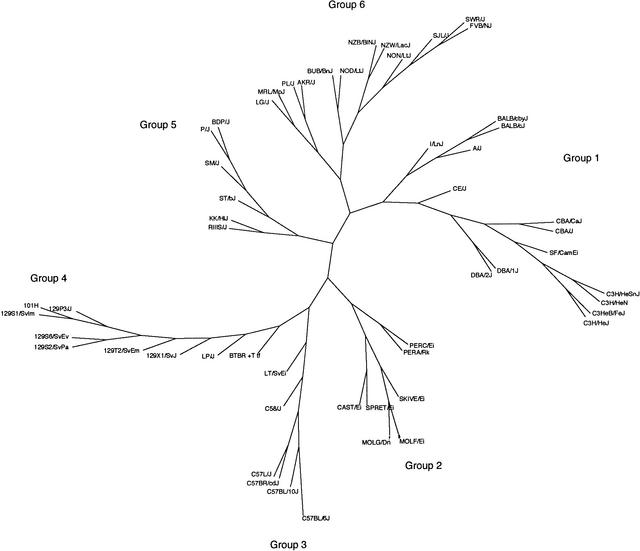
Figure 3.
Mouse family tree. Mouse strains are organized into six major groups of branches. Group 1: C3H/HeJ, C3H/HeN, C3H/HeSnJ, C3HeB/FeJ, SF/CamEi, CBA/CaJ, CBA/J, DBA/2J, DBA/lJ, CE/J, A/J, BALB/cByJ, BALB/cJ, and I/LnJ. Group 2: MOLF/Ei, MOLG/Dn, SKIVE/Ei, SPRET/Ei, CAST/Ei, PERA/Rk, and PERC/Ei. Group 3: C57BL/6J, C57BL/10J,C57BR/cdJ, C57L/J, C58/J, and LT/SvEi. Group 4: 101/H, 129P3/J, 129S1/SvlmJ, 129T2/SvEmsJ, 129X1/SvJ, 129S2/SvPas, 129S6/SvEv, LP/J, and BTBR +T tf/tf. Group 5: BDP/J, PL/J, SM/J, ST/J, KK/H1J, and RIIIS/J. Group 6: LG/J, MRL/MpJ, P/J, AKR/J, BUB/BnJ, NOD/LtJ, NON/LtJ, NZB/BlNJ, NZW/LacJ, SJL/J, SWR/J, and FVB/NJ. The length and angles of the branches are optimized for printing and are not quantitative measures of evolutionary distance between strains.
This tree has six major groups of branches, labeled 1 to 6. If recent inbreeding occurred between the strains, we would observe a net rather than a tree structure. Group 1 is comprised of Bagg albino derivatives and includes I/LnJ mice. Strong originally derived these mice from an unpedigreed stock. Group 2 is made up of strains derived from species of wild mice not belonging to M. m. domesticus (Bonhomme and Guenet 1996). Group 3 contains C57 and C58 animals from Miss Abbie Lathrop's stock, which unlike most “old” inbred strains carry a Y chromosome of Asian M. m. musculus origin (Tucker et al. 1992). Group 4 contains Castle's 129 and LP mice, which are known to be similar. 101H has a common origin with 129. Groups 3 and 4 appear to be parts of a large limb, and their members may be related. In light of the fact that both sets of animals were generated from coat color stocks provided by English fanciers, this seems reasonable (Potter 1978). Groups 5 and 6 hold animals that appear to derive from European (“Swiss”) M. m. domesticus. BDP, P, and SM mice (Group 5) are grouped separately from DBA (Group 1). BDP was produced by crossing Strong's pink-eyed and Little's DBA. P was derived by Snell from a cross involving BDP. SM was produced by crossing seven strains, including DBA, and selecting for small body size. These strains appear to have diverged significantly from the Bagg albino group. The BUB/BnJ inbred strain (Group 6) originated from randomly bred albino mice of unknown ancestry.
We considered the possibility that our results would be biased by the selection of markers informative for the C57BL/6J × DBA/2J strain combination. However, parsimony analysis considers specific mutation steps rather than the simple proportion of alleles that are the same size or different for a given marker. Further, it assumes that species and characters evolve independently. It does not assume that the ancestral state is known or that mutation in one direction is more probable than the other. Nonetheless, our results are largely congruent with the known history of inbred mouse strains (Festing 1996; Beck et. al 2000). Discretion should be used, however, when considering this information to ascertain the effects of genetic background when assessing phenotypes.
METHODS
Primers
Oligonucleotides were synthesized by Applied Biosystems. Primers were designed using DNA sequences from The Whitehead Institute/MIT Center for Genome Research (WICGR) Genetic Map of the Mouse Database Release 10 (http://www.ensembl.org/Mus_musculus). The forward primer of each pair was labeled on the 5′ end with the fluorescent dye FAM, VIC, or NED. To reduce stuttering artifacts, CA dinucleotide sequences were avoided on the 3′ end of the forward primers. All reverse primers contain the sequence GTGTCTT (5′–3′) at their 5′ ends to promote nontemplate-directed nucleotide addition (+A) to the PCR products. Primer sequences are proprietary. Primer pairs can be purchased individually or as a complete set directly from Applied Biosystems.
DNA
Genomic DNA for all inbred mouse strains was obtained from The Jackson Laboratory with the exception of 129S2/SvPas (Charles River Laboratories) and 129S6/SvEv (Taconic). PL/J, LT/SvEi, C3H/HeN, C3H/HeSnJ, BALB/cByJ, and 129/Sv (agouti) DNA were a gift from E. Eicher and T. Golovkina (TJL). LG/J DNA was a gift from J. Cheverud and S. Cropp (Washington University). 101H DNA was a gift from P. Denny and A. Southwell (Harwell, UK). DNA from the BSS interspecific backcross panel [(C57BL/6JEi × SPRET/Ei)F1 × SPRET/Ei] was obtained from L. Rowe (TJL).
PCR
PCR reactions were performed using PE 9700 thermal cyclers in a volume of 10 μL. Final reaction conditions were 50 ng DNA, 250 μM each dNTP, 2.5 mM MgCl2, 0.4 μM each primer, and 0.25 U Taq Gold DNA Polymerase (Applied Biosystems). PCR was initiated with an automatic hot start for 15 min at 95°C to activate the Taq Gold polymerase. Standard cycling conditions were carried out at 94°C for 45 sec, 55°C for 60 sec, and 72°C for 60 sec (30 cycles) with a final extension at 72°C for 10 min. Modified PCR programs, including touchdown PCR, were used to optimize some primer pairs when necessary. Products were pooled post-PCR and resolved on an ABI 377XL sequencer using GS-500 ROX (Applied Biosystems) internal size standard in each lane. Gel image data were processed and alleles scored using GeneScan version 3.1 and Genotyper version 2.1 software (Applied Biosystems).
Chromosome Maps
SSLP markers were selected from the Dietrich et al. (1996) set. Map positions were obtained from the Mouse Genome Database (TJL). Graphical linkage maps for each chromosome was generated using Encyclopedia of the Mouse Genome 3.0, Version 1.0a17 for the Macintosh (TJL).
WEB SITE REFERENCES
http://www.jax.org/resources/documents/cmdata/bkmap/BSS.html; TJL BSS Panel: Index.
http://www.jax.org/resources/documents/cmdata/rhmap/RHIntro.html; TJL Mouse RH Panel Introduction.
http://www.cidr.jhmi.edu/mouse/mouse.html; CIDR-Mouse Genotyping Home Page.
http://www.informatics.jax.org; Mouse Genome Database Home Page.
http://www.genome.wi.mit.edu/cgi-bin/mouse/index#genetic; WICGR Resources, Genetic and Physical Maps of the Mouse Genome.
http://www.ensembl.org/Mus_musculus/; Ensembl Genome Browser, Mouse Genome Server.
Acknowledgments
The Center for Inherited Disease Research (CIDR) is fully funded through a federal contract from the NIH to The Johns Hopkins University, contract number N01-HG-65403. The Jackson Laboratory Mapping Panels are supported by NIH grant HG00941 to Janan T. Eppig. We also acknowledge the expert technical assistance of Mary Barter.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL dwitmer@cidr.jhmi.edu; FAX (410) 550-3559.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.717903.
REFERENCES
- 1.Atchley W.R. and Fitch, W.M. 1993. Genetic affinities of inbred mouse strains of uncertain origin. Mol. Biol. Evol. 10: 1150-1169. [DOI] [PubMed] [Google Scholar]
- 2.Beck J.A., Lloyd, S., Hafezparast, M., Lennon-Pierce, M., Eppig, J.T., Festing, M.F., and Fisher, E.M. 2000. Genealogies of mouse inbred strains. Nature Genet. 24: 23-25. [DOI] [PubMed] [Google Scholar]
- 3.Bonhomme F., Guenet, J.-L., et al. 1996. The laboratory mouse and its wild relatives. In Genetic variants and strains of the laboratory mouse (ed. M.F. Lyon), pp. 1577–1595. Oxford University Press, Oxford, UK.
- 4.Brownstein M.J., Carpten, J.D., and Smith, J.R. 1996. Modulation of non-templated nucleotide addition by Taq DNA polymerase: Primer modifications that facilitate genotyping. Biotechniques 20: 1004-1006. [DOI] [PubMed] [Google Scholar]
- 5.Dietrich W.F., Miller, J., Steen, R., Merchant, M.A., Damron-Boles, D., Husain, Z., Dredge, R., Daly, M.J., Ingalls, K.A., O'Conner, T.J., et al. 1996. A comprehensive genetic map of the mouse genome. Nature 380: 149-152. [DOI] [PubMed] [Google Scholar]
- 6.Felsenstein J. 1989. PHYLIP—Phylogeny inference package (v. 3.2). Cladistics 5: 164-166. [Google Scholar]
- 7.Ferris S.D., Sage, R.D., Prager, E.M., Ritte, U., and Wilson, A.C. 1983. Mitochondrial DNA evolution in mice. Genetics 105: 681-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Festing M.F.W., et al. 1996. Origins and characteristics of inbred strains of mice. In Genetic variants and strains of the laboratory mouse (ed. M.F. Lyon), pp. 1537–1576. Oxford University Press, Oxford, UK.
- 9.Potter M. 1978. Comments on the relationship of inbred strains to the genus Mus. In Origins of inbred mice. (ed. H.C. Morse III), pp. 497–509. Academic Press, New York, NY.
- 10.Rowe L.B., Nadeau, J.H., Turner, R., Frankel, W.N., Letts, V., Eppig, J.T., Ko, M.S.H., Thurston, S.J., and Birkenmeier, E.H. 1994. Maps from two interspecific backcross DNA panels available as a community genetic mapping resource. Mamm. Genome 5: 253-274. [DOI] [PubMed] [Google Scholar]
- 11.Schalkwyk L.C., Jung, M., Daser, A., Weiher, M., Walter, J., Himmelbauer, H., and Lehrach, H. 1999. Panel of microsatellite markers for whole-genome scans and radiation hybrid mapping and a mouse family tree. Genome Res. 9: 878-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Silver L.M., 1995. Mouse genetics: Concepts and applications, pp. 240–244. Oxford University Press, Oxford, UK.
- 13.Simpson E.M., Linder, C.C., Sargent, E.E., Davisson, M.T., Mobraaten, L.E., and Sharp, J.J. 1997. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat. Genet. 16: 19-27. [DOI] [PubMed] [Google Scholar]
- 14.Smith J.R., Carpten, J.D., Brownstein, M.J., Ghosh, S., Magnuson, V.L., Gilbert, D.A., Trent, J.M., and Collins, F.S. 1995. Approach to genotyping errors caused by nontemplated nucleotide addition by Taq DNA polymerase. Genome Res. 5: 312-317. [DOI] [PubMed] [Google Scholar]
- 15.Threadgill D.W., Yee, D., Matin, A., Nadeau, J.H., and Magnuson, R. 1997. Genealogy of the 129 inbred strains: 129/SvJ is a contaminated inbred strain. Mamm. Genome 8: 390-393. [DOI] [PubMed] [Google Scholar]
- 16.Tucker P.K., Lee, B.K., Lundrigen, B.L., and Eicher, E.M. 1992. Geographic origin of the Y chromosome in “old” inbred strains of mice. Mamm. Genome 3: 254-261. [DOI] [PubMed] [Google Scholar]
- 17.Wakeland E., Morel, L., Achey, K., Yui, M., and Longmate, J. 1997. Speed congenics: A classic technique in the fast lane (relatively speaking). Immunol. Today 18: 472-477. [DOI] [PubMed] [Google Scholar]
- 18.Weber J. and Wong, C. 1993. Mutation of human short tandem repeats. Hum. Mol. Genet. 2: 1123-1128. [DOI] [PubMed] [Google Scholar]