Abstract
Integrons are natural tools for bacterial evolution and innovation. Their involvement in the capture and dissemination of antibiotic-resistance genes among Gram-negative bacteria is well documented. Recently, massive ancestral versions, the superintegrons (SIs), were discovered in the genomes of diverse proteobacterial species. SI gene cassettes with an identifiable activity encode proteins related to simple adaptive functions, including resistance, virulence, and metabolic activities, and their recruitment was interpreted as providing the host with an adaptive advantage. Here, we present extensive comparative analysis of SIs identified among the Vibrionaceae. Each was at least 100 kb in size, reaffirming the participation of SIs in the genome plasticity and heterogeneity of these species. Phylogenetic and localization data supported the sedentary nature of the functional integron platform and its coevolution with the host genome. Conversely, comparative analysis of the SI cassettes was indicative of both a wide range of origin for the entrapped genes and of an active cassette assembly process in these bacterial species. The signature attC sites of each species displayed conserved structural characteristics indicating that symmetry rather than sequence was important in the recognition of such a varied collection of target recombination sequences by a single site-specific recombinase. Our discovery of various addiction module cassettes within each of the different SIs indicates a possible role for them in the overall stability of large integron cassette arrays.
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. listed in Table 1.]
Natural selection favors the evolution of strategies that increase the rate of adaptation, that is, chance favors the prepared genome (Caporale 1999). Although mutation generally causes only a very small and localized change in a cell, the transfer of genetic material involves much broader changes that may permit the organism to carry out new functions and adapt to environmental changes (Ochman et al. 2000). Integrons are exquisitely suited for this purpose. Integrons are natural cloning and expression systems that incorporate open reading frames (ORFs) and convert them to functional genes (Rowe-Magnus and Mazel 1999, 2001). They have been expansively identified as the constituents of transferable elements responsible for the evolution of multidrug resistance among human, animal, and plant pathogenic isolates during the antibiotic era. More than 70 different antibiotic-resistance genes, covering most antimicrobials used against Gram-negative infections, have been characterized within integrons thus far (Rowe-Magnus et al. 2002a). The substantial impact of integrons on bacterial evolution is underscored by the present dilemma in the treatment of infectious disease, as the development of multiple-antibiotic resistance can often be traced to the stockpiling of resistance loci within integrons to create multiresistance integrons (MRIs). MRIs harboring up to eight resistance cassettes have been isolated from multiresistant clinical isolates (Naas et al. 2001).
The integron platform codes for an integrase (intI) that mediates recombination between a proximal primary recombination site (attI) and a target recombination sequence called an attC site (or 59 base elements; 59 be). The attC site is usually found associated with a single open reading frame in a circularized structure termed a gene cassette (Hall and Stokes 1993; Recchia and Hall 1995; Stokes et al. 1997; Sundstrom 1998). Insertion of the gene cassette at the attI site, which is located downstream of a resident promoter, Pc, internal to the intI gene, drives expression of the encoded proteins (Levesque et al. 1994).
Most of the attC sites of integron gene cassettes identified to date share little homology. Their length and sequence vary considerably (from 57–141 bp) and their sequence similarities are primarily restricted to their boundaries, which correspond to the inverse core site (CS; RYYYAAC) and the core site (CS; G↓TTRRRY, where R is a purine, Y is a pyrimidine, and ↓ is a recombination point; Stokes et al. 1997; Collis et al. 1998). Despite their limited homology, attC sites do share certain structural characteristics (see Fig. 1). Each forms an imperfect inverted repeat, with strong complementarity being particularly apparent over the terminal 25 bp. The CS and ICS are perfectly complementary in the attC site of a circularized cassette. This stretch of complementarity can extend as far as 5 nt downstream of the CS and upstream of the ICS, giving a total of up to 12 consecutive complementary nucleotides in these regions. Two integrase-binding sites, called LH and RH simple sites, are found within each attC site. The LH simple site is composed of the ICS and a second sequence that is related to the CS consensus, called 2L (Stokes et al. 1997; Collis et al. 1998). The ICS and 2L sequences are separated by a 5-bp spacer, and it has been proposed that the integrase binds to the LH simple site either as two monomers or a dimer (Collis et al. 1998). The RH simple site is similarly defined, and it includes the CS and a second sequence that is related to the ICS consensus, called 2R. As seen for the LH simple site, the CS and 2R sequences have a 5–6-bp spacer between them. Because it has been demonstrated that markedly different attC sites are recognized as substrates by the integron-integrase protein (Collis et al. 2001), this indicated that the sequence of the attC site, outside of the ICS and CS, is not as important as its symmetry.
Figure 1.
The sequence of the aadB attC site (Stokes et al. 1997). Inverted repeats are indicted with black arrowheads (◂ and ▸), and the asterisk (*) indicates the position of the protruding C present in 2L relative to 2R. The extents of the LH and RH sites as defined by Stokes et al. (1997) are bracketed. White arrows denote the extended regions of complementarity, beyond the ICS and CS, over the terminal 25 bp of the attC site. (ICS) inverse core site; (CS) core site; ( ↓ ) recombination point within the CS.
Five classes of MRIs have been reported based on the divergence of their integrase genes, which share between 39% and 58% identity. The evolutionary divergence among the integrase genes indicated that the activity of this system has extended far beyond the 60 yr of the antibiotic era. This postulate was confirmed by the recent discovery of massive ancestral versions, the superintegrons (SIs), in the genomes of diverse proteobacterial species. The first was discovered in the Vibrio cholerae genome (Mazel et al. 1998). Located on the smaller of the two circular V. cholerae chromosomes (Heidelberg et al. 2000), this SI spanned 126 kb and harbored 179 cassettes of mainly unassigned function (Rowe-Magnus et al. 1999), dwarfing any previously described MRI. Strikingly, the attCsites associated with each captured cassette, termed VCRs (Vibrio cholerae repeated sequences; Barker et al. 1994), were highly homologous in length and sequence, unlike their counterparts of MRIs. In addition, the cassettes of the V. cholerae SI were demonstrated to be substrates for the class 1 integrase of MRIs (Mazel et al. 1998; Rowe-Magnus et al. 2002b). These observations led to the proposal that each distinct attC site of MRI gene cassettes was representative of a distinct SI. Using a systematic search, similar SIs were identified in the genomes of diverse proteobacteria (Rowe-Magnus et al. 2001), and they all shared the same general characteristics as the V. cholerae SI. Other integron-integrase genes and hundreds of gene cassettes have also been directly isolated from a variety of soil samples (Nield et al. 2001). Although it is not known if these soil-derived integrases and cassettes are of SI or MRI origin, it is now evident that SIs, like MRIs, are widespread among bacterial populations.
Phylogenetic analysis based on the comparison of the 16S RNA and SI-integrase genes revealed that the evolutionary history of the integron systems paralleled that of the radiation, indicating that integrons are ancient structures (Rowe-Magnus et al. 2001). Furthermore, we observed from the limited sample of available cassettes that their attC sites (1) appeared to be species-specific and (2) highly conserved within each SI. We have now performed an in-depth analysis of the SIs identified within divergent members of the Vibrio lineage. These analyses confirmed the sedentarity of the integron platform (intI gene and attI site). A comparison of subsets of cassettes from the SIs of Vibrio metschnikovii, Vibrio fischeri, and V. cholerae indicated that the majority of the cassettes found in the SI of one species were not found in that of another. Such cassettes may have important roles in the exploitation of a particular niche by a particular species; they lend credence to the hypothesis of an in vivo cassette assembly process that is independently active within each species. Although the signature attC sites of each species showed limited sequence homology, they all displayed conserved structural characteristics that support the idea that symmetry, rather than sequence, is a key feature in the recognition of such a varied collection of target recombination sequences by a single site-specific recombinase. Finally, our demonstration of an active toxin/antidote system encoded in a V. fischeri SI gene cassette that is homologous to the control of cell death (ccdAB) system of the F plasmid and the presence of similar systems as gene cassettes in the V. choleraeN16961 and V. metschnikovii SIs might provide an explanation for the apparent stability of the massive cassette arrays in these species.
RESULTS
The Chromosomal Location of the Vibrio SI Loci
The sequence of the loci carrying the previously characterized integrases from V. metschnikovii A267, Vibrio parahaemolyticus 75.2, V. fischeri 103203, and Listonella pelagia 10276.2 (Rowe-Magnus et al. 2001) were extended downstream of the intIA genes. Novel SIs were also identified in Vibrio natriegens 103193T, Listonella anguillarum 63.36, and Vibrio vulnificus 75.4. Each of the SIs identified among all the Vibrionaceae in this work were estimated to be at least 100 kb by Southern analysis using the attC site sequences as probes (see below). Sequence analysis showed that, as in V. cholerae and Vibrio mimicus, the V. metschnikovii, V. vulnificus, and L. anguillarum intIA genes were located adjacent to the rpmL–rplT ribosomal protein operon (Fig. 2). The L. pelagia, V. natriegens, and V. parahaemolyticus intIA genes were located downstream of a cluster of conserved ORFs different from those in the V. cholerae clade. The closest homologs of two of these, a rimJ ortholog and ORF474, were on Chromosome 1 of V. cholerae (VC1309 and VC1310, respectively; Fig. 2). However, a homolog of ORF588 could not be identified on either of the two chromosomes in V. cholerae. ORF588 did show significant homology (40% identity) to an ORF of unknown function in Deinococcus radiodurans (accession no. E75485). The V. fischeri integrase gene, VfiintIA, was located next to an ORF that was most closely related (at 39% identity) to VCA0034 of V. cholerae N16961. VCA0034 resides on chromosome 2 of V. cholerae, 250 kb away from the V. cholerae SI-integrase gene, VchintIA. Comparison of the intergenic sequence between these intIAgenes and the neighboring genes or ORFs did not reveal any conserved motifs.
Figure 2.
Chromosomal location of the Vibrio superintegrons. Representation of the genetic context in which each of the SIs was found relative to their phylogenetic distribution according to their IntIA genes is shown. SIs in identical locations are grouped within the same phylogenetic clade. The corresponding orthologs in V. cholerae are marked with VC(A) followed by the number designation for the ORF. The attI site and VXRs are also indicated. For clarity, only the first cassettes within each SI are shown. The integrases intI1 (Liebert et al. 1999), intI2(Sundstrom et al. 1991), intI3 (Hall et al. 1999), intI9 SXT (Hochhut et al. 2001), and intIHS (H. Sorum, K. Dommarsnes, K. Sandersen, L. Sundstrom, M. Gullberg, and A. Solberg, 2001, GenBank accession no. AJ277063) are found associated with mobile DNA elements. The integron-interases I8-2, I7-2, and I6-2 were amplified from DNA soil samples (Nield et al. 2001). The integrases (IntIA) of V. cholerae, V. mimicus, V. metschnikovii, V. parahaemolyticus, V. fischeri (Vfi), L. pelagia (Lpe), Shewanella oneidensis (Son), Shewanella putrefaciens (Spu), Xanthomonas campestris (Xca), Xanthomonas species(Xsp), Nitrosomonas europaea (Neu), P. alcaligenes (Pal), and Pseudomonas mendocina (Pm) have been previously described (Rowe-Magnus et al. 2001; Vaisvila et al. 1999, 2001). The recombinases XerC and XerD are from E. coli (Eco); (rplT and rpmL) ribosomal genes; (dark gray, striped, or black boxes) adjacent gene(s) that are not part of the SI structure.
Characterization of the V. metschnikovii Cassette Array
Southern hybridization of BglII- or EcoRI-digested V. metschnikovii genomic DNA with a VMeR probe (Vibrio metschnikovii repeats, the signature attC site of the V. metschnikovii cassettes) revealed a series of fragments totaling >100 kb for the VMeR cassette array (data not shown). Four of these nonoverlapping fragments (Table 1) were cloned and sequenced, providing a total of 26 cassettes (Table 2). An alternative PCR strategy, using VMeR1 and VMeR2 as primers, allowed the retrieval of 6 additional cassettes and recloning of the c253-5 cassette (Table 2). Among the total of 32 cassettes recovered, two were found to be repeated twice and six times, respectively, reducing the pool to a total of 26 unique cassettes. Interestingly, six of these unique cassettes were found to have counterparts in the V. cholerae SI (see section below). Twenty-three were found to carry a plausible ORF that was preceded by a potential ribosome-binding site (RBS). The putative products of seven cassettes did not show any significant similarity to any characterized or hypothetical proteins found in the databases (Table 2). The potential ORFs encoded in all but two cassettes were found in the classical (positive) orientation for integron cassettes, that is, the 3′ end of the ORF was located just upstream of or within the ICS of the downstream VMeR. The likely start codons of most of these classically orientated ORFs were located within the first 40 nt of the cassettes, leaving little space for a potential promoter. The ORFs carried by c374-2 and c374-4 were identical, and both copies were in the inverse (negative) orientation with respect to the VMeR. Interestingly, the start codons for the ORFs inside these negatively oriented cassettes were located 89 and 109 bp upstream of the VMeR, which provides sufficient space to encoded potential promoter sequences. Four cassettes, of sizes varying from 464–690 nt, including the cassette present in six copies, did not contain a recognizable ORF.
Table 1.
Constructions and Plasmids
| Plasmids | Description | Reference or source (GenBank accession number) |
| pNOT218 | pTZ18R containing two in-frame NotI sites boarding the MCS polylinker, ApR | (AF480833) |
| pTZ19R | ColE1 replicon ApR | Pharmacia (Y14835) |
| pSU19 | p15A replicon CmR | Bartolome et al. 1991 (X53939) |
| pUC18 | ColE1 replicon ApR | Norrander et al. 1983 (L08752) |
| pCR2.1 | ColE1 replicon ApR KmR | Invitrogen |
| Vibrio metschnikovii inserts | ||
| p253 | pUC18 BamHI (genomic BglII 4.0-kb fragment) | This work (AF525313) |
| p273 | pUC18 EcoRI (genomic EcoRI 4.6-kb fragment) | This work (AF530480) |
| P372 | pUC18 BamHI (genomic BglII 4.3-kb fragment) | This work (AY141193) |
| P374 | pNOT218 (genomic EcoRV–BspEI 6.4-kb fragment) | This work (AY014398) |
| V. fischeri inserts | ||
| p641a | pUC18 BamHI (genomic BglII 2.1-kb fragment) | This work (AF177199) |
| p667a | pUC18 HindIII (genomic HindIII 3.3-kb fragment) | This work (AF177199) |
| p668 | pUC18 HindIII (genomic HindIII 3.9-kb fragment) | This work (AY178758) |
| p669 | pUC18 XbaI (genomic XbaI 4.5-kb fragment) | This work (AY181031) |
| p672 | pUC18 XbaI (genomic XbaI 3.5-kb fragment) | This work (AY181032) |
| p789 | pMTL22 NsiI (genomic NsiI 6.0-kb fragment) | (AY014400) |
| p1357 | pNOT218 PstI (p669 NsiI 739-bp fragment carrying ccdABVfi in anti-lacZ orientation) | This work |
| p1400 | pTZ19R EcoRI-HindIII (p1357 EcoRI-HindIII insert; ccdABVfi in lacZ orientation) | This work |
| p1446 | pSU19 AccI–EcoRI (p1400 AclI–EcoRI 590-bp fragment, carrying ccdBVfi* R35opal) | This work (AY181033) |
| V. natriegens insert | ||
| p1216b | pCR2-1 ([LPR1-2 + INT8-2] PCR product 1.8 kb) | This work (AY181034) |
| p2018b | pCR2-1 ([Vna1 + ORF474] PCR product 1.25 kb) | This work (AY181034) |
| V. vulnificus insert | ||
| p1184c | pCR2-1 ([LPR1 + I456/5] PCR product 2.7 kb) | This work (AF539751) |
| p2029c | pCR2-1 ([RPLT + Vvu1] PCR product 1.4 kb) | This work (AF539751) |
| Listonella anguillarum insert | ||
| p1456d | pUC18 HindIII (genomic HindIII 2.7-kb fragment) | This work (AY126447) |
| p2010d | pCR2-1 ([RPLT + Lang1] PCR product 0.63 kb) | This work (AY126447) |
p641 and p667 inserts have a 1354-nt overlap and have been deposited as a single file in GenBank.
p1216 and p2018 inserts overlap and have been deposited as a single file in GenBank.
p1184 and p2029 inserts overlap and have been deposited as a single file in GenBank.
p1456 and p2010 inserts overlap and have been deposited as a single file in GenBank.
Table 2.
Properties of Vibrio metschnikovii Superintegron Cassettes
| Gene cassettea | Cassette coordinatesb (bp) | Length of attC sitec (bp) | Named/length of ORF (bp) | G + C content of ORF (%) | Sequence similarity,eE-value,f and motifsg |
| p253 insert | |||||
| c253-1* | <1–987h | 117 | ND | — | (ISVme1 insertion) |
| c253-2 | 988–1531 | 116 | Orfc253-2/387 | 37.2 | 70% identity to the V. cholerae VCA0890 Glyoxylase I family protein |
| c253-3 | 1532–2036 | 118 | Orfc253-3/360 | 40.1 | 94% identity to the V. choleraeVCA0338 and VC0415 |
| c253-4 | 2037–2825 | 117 | Orfc253-4/642 | 36.6 | NH—1 transmembrane helix |
| c253-5 | 2826–3373 | 118 | Orfc253-5/396 | 39.4 | 66% identity to the V. choleraeVCA0414 and VC0425 signal peptide sequence |
| c253-6 | 3374–3926> | — | Orfc253-6a/171 | 39.3 | 100% identity to the V. choleraeVCA0474 C-terminal 56 aa, see text. |
| Orfc253-6b/320> | 42.5 | 99% identity to the V. choleraeVCA0475 (30% identity to Phage P1Doc) | |||
| p273 insert | |||||
| c273-1 | <1–490 | 118 | Orfc273-1/<375 | 38.7 | 48% identity to the C-terminal part of Bacillus halodurans hypothetical protein BH3804 (E = 7e − 6) |
| c273-2* | 491–992 | 117 | ND | — | — |
| c273-3 | 993–1697 | 118 | Orfc273-3/558 | 38.0 | NH |
| c273-4 | 1698–2385 | 118 | Orfc273-4/336 | 49.4 | NH |
| c273-5 | 2386–3367 | 117 | Orfc273-5/837 | 36.9 | 22% identity to HphI restriction endonuclease (E = 4e − 8) |
| c273-6* | 3368–4055 | 117 | ND | — | — |
| c273-7 | 4056–4557> | — | Orfc273-7/361> | 31.7 | NH—signal peptide sequence |
| p372 insert | |||||
| c372-1 | <1–821 | 118 | Orfc372-1/<689 | 31.7 | NH |
| c372-2 | 822–1316 | 117 | Orfc372-2/291 | 34.7 | 34% identity to Salmonella enterica hypothetical protein CAD05348 (E = 0.01)—signal peptide sequence |
| c372-3 | 1317–2077 | 117 | Orfc372-3/609 | 35.3 | NH—signal peptide sequence—6 transmembrane helices |
| c372-4 | 2078–2931 | 118 | Orfc372-4/708 | 36.6 | 40% identity to the Salmonella typhimurium LT2 putative aspartate racemase AAL21891 (E = 7e − 41) |
| c372-5* | 2932–3621 | 117 | ND | — | — |
| c372-6 | 3622–4242>i | — | ND | — | (ISVme1 insertion) |
| p374 insert | |||||
| c374-1 | 658–1711 | 117 | Orfc374-1/879 | 34.0 | NH |
| c374-2** | 1712–2607 | 118 | Orfc374-2/669 | 38.9 | 20.5% identity to Lactococcus lactis methyltransferase CAA68045 (E = 0.001) |
| c374-3 | 2608–3383 | 117 | Orfc374-3/633 | 36.8 | 26.3% identity to Agrobacterium tumefaciens hypothetical methyltransferase AAK87648 (E = 3e − 13) |
| 374-4** | 3384–4279 | 118 | Orfc374-4/669 | 39.3 | 98% identity to c374-2 |
| c374-5 | 4280–5137 | 118 | Orfc374-5/705 | 36.3 | NH |
| c374-6 | 5138–5601 | 116 | ND | — | — |
| c374-7 | 5602–6437> | — | Orfc374-7/771 | 40.6 | 29% identity to Sinorhizobium meliloti hypothetical oxydoreductase CAC46573 (E = 7e − 22) |
| PCR (VMR1 + VMR2) | |||||
| Vme1 | 1–437> | / | OrfVme1/396 | 39.4 | 100% identity to c253-5 |
| Vme2 | 1–578> | / | OrfVme2a/267 OrfVme2b/243 | 39.1 40.0 | 92% identity to the V. choleraeVCA0332 82% identity to V. choleraeVCA0333 |
| Vme4* | 1–393> | / | ND | — | — |
| Vme9 | 1–502> | / | OrfVme9/375 | 33.6 | 46% identity between the last 40 C-terminal aa and a central segment of chicken paxillin B55933—signal peptide sequence—2 transmembrane helices |
| Vme11 | 1–496> | / | OrfVme11/414 | 41.3 | 91% identity to the V. choleraeVCA0476 |
| Vme12 | 1–335> | / | OrfVme12/198 | 34.9 | 36% identity to the V. choleraeVCA0426 (E = 4e − 6) |
| Vme23* | 1–394> | / | ND | — | — |
Cassettes (c) have been named according to their plasmid number (see Table 1) and their position in the insert, or for the cassette obtained from (VMR1 + VMR2) PCR by the prefix Vme followed of a number; the two families of repeated cassettes are indicated by * and **, respectively.
Sequences missing 5′ or 3′ in incomplete cassettes are indicated by < and >, respectively.
The given attC site length is from the last Y of the inverse core site (RYYYAAC) to the G located upstream of the recombination point in the core site of the integrated cassette (GTTRRRY).
ORFs are in classical positive orientation that is in the same direction as their associated attC site. ORFs in the opposite orientations are underlined; ND, no ORF > 150-bp detected.
NH, no homologous protein detected by BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/). When related to a V. cholerae SI cassette, the corresponding VCAxxx name is underlined.
Number of equal scoring matches expected by chance, results with value ≤10−2 have been considered.
Motifs have been evidenced through the CDD search option in BLAST analysis and by using the signal peptide and transmembrane segment prediction programs SignalP and TMHMM (Center for Biological Sequence Analysis; http://www.cbs.dtu.dk/services/).
The sequence from 1 to 540 corresponds to a never described IS, ISVme1.
The sequence from 3703 to the end corresponds to an ISVme1, identical to the one found in cassette c253-1.
Characterization of the V. fischeri Cassette Array
Southern hybridization of BamHI- or HindIII-digested V. fischeri genomic DNA with a VFR probe (Vibrio fischeri repeats, the V. fischeri cassette attC sites) revealed a series of fragments equaling >100 kb for the VFR SI cassette array (data not shown). Six fragments were cloned and sequenced (Table 1), and two were found to overlap. These fragments provided a total of 27 cassettes. Two cassettes were found to be repeated twice and eight times, respectively, giving a total of 19 unique cassettes. Sixteen of these were found to carry an ORF larger than 250 bp that was preceded by a potential RBS. As seen in V. cholerae and V. metschnikovii, the majority of the ORFs were found in the classical orientation, and a single cassette, c669-2, was found to carry two ORFs in inverse orientation. This cassette encoded a functional cytotoxic protein and its antidote (see below). The putative products encoded in seven cassettes, totaling eight ORFs, were found to have similarity to previously characterized proteins (Table 3) that are related to simple functions or previously described ORFs. Analysis of the coding potential of the cassette repeated eight times, c667-2, revealed no significant ORF larger than 150 bp; however, BLASTX analysis revealed a region with significant similarity to a 40-amino-acid domain of intron maturases. None of the V. fischeri cassettes showed any similarity to any of the SI cassettes found in the SIs of the other Vibrio species examined.
Table 3.
Properties of Vibrio fischeri Superintegron Cassettes
| Gene cassettea | Cassette coordinatesb (bp) | Length of attC sitec (bp) | Named/length of ORF (bp) | G + C content of ORF (%) | Sequence similarity,e E-value,f and motifs/activityg |
| p667 + p641 insertsh | |||||
| c667-1 | <1–39 | — | —i | — | — |
| c667-2* | 40–601 | 98 | ND | — | BLASTX detects a 56-codon seqment that shows homology to intron maturase |
| c667-3* | 602–1161 | 99 | ND | — | — |
| c667-4* | 1162–1724 | 98 | ND | — | — |
| c667-5/c641-1 | 1725–3415 | 116 | Orfc667-5/1539 | 29.4 | NH—3 transmembrane helices |
| c641-2 | 3416–4070 | 116 | Orfc641-2/426 | 35.7 | 40% identity to the Yersinia pestis putative acetyltransferase CAC93527 (E = 4e − 24) |
| c641-3 | 4071–4123> | — | ND | — | NH |
| p668 insert | |||||
| c668-1 | 1–686 | 118 | Orfc668-1/513 | 39.0 | 39% identity to the V. cholerae VCA1017methylated- DNA–protein-cysteine S-methyltransferase hypothetical OGT (E = 3e − 22) |
| c668-2* | 687–1264 | 117 | ND | — | |
| c668-3 | 1265–1780 | 116 | Orfc668-3j/147 | 34.0 | NH |
| c668-4* | 1781–2339 | 97 | ND | — | — |
| c668-5 | 2340–3251 | 116 | Orfc668-5/756 | 31.7 | NH—signal peptide—2 transmembrane helices |
| c668-6 | 3552–4003> | — | Orfc668-6/711> | 30.2 | NH |
| p669 insert | |||||
| c669-1 | <1–404 | 116 | Orfc669-1/<280 | 33.3 | NH—1 transmembrane helix |
| c669-2 | 405–1145 | 118 | CcdB/315 CcdA/246 | 33.3 34.2 | Gyrase inhibiting protein, 42% identity to the F plasmid CcdB CcdB antidote, 42% identity to Escherichia coli O157:H7 CcdA (AE005182.1) |
| c669-3 | 1146–1910 | 118 | Orfc669-3/615 | 33.3 | 40% identity to Bacillus halodurans transcriptional repressor of sporulation and degradative enzymes production NP_241278 (E = 2e − 35) |
| c669-4 | 1911–3268 | 116 | Orfc669-4/1200 | 34.9 | 37% identity to the N-terminal 240 aa of Streptomyces coelicolor Serine/threonine protein kinase T36502 (E = 1e − 30) |
| c669-5* | 3269–3829 | 98 | — | — | — |
| c669-6** | 3830–4483> | — | Orfc669-6/632> | 34.3 | NH |
| p672 insert | |||||
| c672-1 | <1–79 | — | —i | — | — |
| c672-2 | 80–682 | 116 | Orfc672-2/444 | 35.6 | 28% identity to the S. coelicolor putative acetyl transferase (E = 1e − 07) |
| c672-3* | 683–1260 | 117 | ND | — | — |
| c672-4 | 1261–2254 | 118 | Orfc672-4/843> | 39.3 | NH—signal peptide sequence—2 transmembrane helices |
| c672-5* | 2255–2813 | 98 | ND | — | — |
| c672-6 | 2814–3438> | — | Orfc672-6/605> | 33.6 | NH |
| p789 insert | |||||
| c789-1 | 1974–2679 | 116 | Orfc789-1/513 | 29.5 | 40% identity to the V. choleraeVCA0405 60 N-terminal amino acids and 35% identity to the Lactococcus lactis prophage pi2 protein 2 (AE006335_3) 75 C-terminal amino acids—signal peptide sequence—2 transmembrane helices |
| c789-2** | 2680–3028> | — | Orfc789-2/327> | 33.7 | NH (identical to c669-6) |
Cassettes (c) have been named according to their plasmid number (see Table 1) and their position in the insert; the two families of repeated cassettes are indicated by * and **, respectively.
Sequences missing 5′ or 3′ in incomplete cassettes are indicated by < and >, respectively.
The given attC site length is from the last Y of the inverse core site (RYYYAAC) to the G located upstream of the recombination point in the core site of the integrated cassette (GTTRRRY).
ORFs are in classical positive orientation that is in the same direction as their associated attC site. ORFs in the opposite orientations are underlined; ND, no ORF > 150-bp detected.
NH, no homologous protein detected by BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/). When related to a V. cholerae SI cassette, the corresponding VCAxxx name is underlined.
Number of equal scoring matches expected by chance, results with value ≤10−2 have been considered.
Demonstrated activities are in bold characters. Motifs have been evidenced through the CDD search option in BLAST analysis and by using the signal peptide and transmembrane segment prediction programs SignalP and TMHMM (Center for Biological Sequence Analysis; http://www.cbs.dtu.dk/services/).
The p667 and p641 inserts overlapping each other, we have annotated and deposited to GenBank the corresponding contig.
Irrelevant, as the sequence only corresponds to the 3′ part of the attC site.
This 147-bp ORF is preceded by a canonical ribosome-binding site and consequently has been considered as a potential gene.
The V. fischeri ccdAB Cassette Encodes a Cytotoxic Protein and Its Antidote
The V. fischeri cassette c669-2 was found to carry two ORFs related to the control of cell death systems that form the gyrase-inhibiting family of proteins, CcdB, and their antidotes, CcdA. Interestingly, the closest relatives of the cassette encoded Ccd proteins were neither functionally nor structurally associated. The closest relative of CcdBVfi was found to be the CcdBF of the plasmid addiction system from the F plasmid (42% amino acid identity), whereas CcdAVfi was only 19% identical to the associated antidote, CcdAF (Miki et al. 1984). Furthermore, CcdAVfi was found to be 42% identical to the antidote protein, CcdAO157, found on the chromosome of Escherichia coli O157:H7 (accession no. NP_285744; Perna et al. 2001), whereas CcdBVfi shared only 35% identity with the toxin, CcdBO157 (accession no. NP_2857445.1). To functionally characterize this putative ccdAB system, the 739-bp NsiI fragment from p669 encompassing the two ORFs of V. fischeri cassette c669-2, hereafter called ccdAB, was cloned into pNOT218 that had been digested with PstI (Table 1). This plasmid, p1357, contained the ccdAB genes in the opposite orientation relative to the lacZ promoter. To ensure that the ccdAB operon would be expressed in E. coli, the p1357 insert was recloned in pTZ19R, placing the ccdAB genes in the same relative orientation as the inducible lac promoter (p1400, Table 1). The toxigenic activity associated with ccdB expression was demonstrated by subcloning the AclI–EcoRI internal fragment of p1400, which contained only the 3′ end of ccdA and an intact ccdB, into pSU19 (CmR) digested by AccI and EcoRI to put ccdB under the control of Plac. The ligation product was then used to transform DH5α and ω106, a DH5α strain containing the plasmid p1400. Whereas transformation of ω106 gave rise to thousands of CmR clones, only a single CmR clone was obtained in transformations with DH5α. Furthermore, characterization of the single DH5α CmR clone by sequence analysis of the corresponding plasmid (p1446) revealed a C → T transition in the ccdB coding region. This transition resulted in the conversion of codon 35 from an Arg (CGA) to a stop codon (TGA), leading to early termination of ccdB translation. These results demonstrated that expression of ccdB in the absence of ccdA coexpression was lethal in E. coli.
Analysis and Comparison of the VXRs
We developed a program, XXR, that could detect the attC sites of integron gene cassette arrays. We used XXR to extract the attC sites from the cassette array of the V. cholerae N16961 SI and recovered 176 complete VCRs. We also used the program to extract the VMeRs (V. metschnikovii) and VFRs (V. fischeri) from the various contigs that we built. Dendograms of the VXRs associated with the cassettes from the different contigs and V. cholerae N16961 were compiled using CLUSTALX and TreeView (Fig. 3). As can be seen, 149 of the 176 VCRs showed an overall similarity of >90%. The other 27 VCRs partitioned into three more remote subclades. All but one VMeR, VMeR 253-5, grouped together to form a distinct clade from the group gathering the 149 VCRs. Interestingly, most of the 27 remote VCRs appeared to be more closely related to the VMeRs as they branched among them. The VFRs also formed a coherent clade distantly related to the VCRs and VMeRs. The attC sites carried by the cassettes found in multiple copies inside the V. metschnikovii and V. fischeri SIs formed subclades within the respective VXR families (Fig. 3), indicating that they all descended from a common ancestor.
Figure 3.
Phylogenetic relationships of the VXRs. The unrooted dendrogram was compiled following extraction of the repeat sequences from the SIs of V. cholerae (VCRs in black ), V. metschnikovii (VMeRs in blue), and V. fischeri (VFRs in red) using the XXR program. The VXRs of the other species are omitted for clarity. The significance of the red- and green-framed VXRs are discussed in the text. Filled boxes denote VXRs that are descended from a common ancestor.
Comparison of the Related Cassettes Found in the V. metschnikovii and V. cholerae SIs
An important issue to be addressed is the degree of cassette exchange among Vibrio species. Among the 32 V. metschnikovii cassettes examined, 6 were found to have counterparts in the V. cholerae SI. However, only two of these, c253-3 and c253-5, were complete, and subsequent analysis focused on their phylogenetic relationship to their V. cholerae counterparts. Interestingly, these two families of cassettes showed different types of relationships. Cassette c253-3 of V. metschnikovii was related to two nearly identical V. cholerae cassettes, VCA0338 and VCA0415. Cassette c253-3 was 84.5% and 83% identical to VCA0338 and VCA0415, respectively. However, this overall homology was in fact partitioned within the cassettes; that is, the ORFs of the cassettes shared 91%–92% identity, but the VXR components shared only 62%–63% identity. In addition, the VMeR carried by cassette c253-3 was closely related to the majority of the VMeRs, whereas the VCRs carried by the V. cholerae cassette homologs, VCA0338 and VCA0415, branched with the majority of the VCRs (see Fig. 3).
Conversely, cassette c253-5, which is unique in our V. metschnikovii cassette sample, is related to a V. cholerae cassette that appears twice in the N16961 SI, cassettes VCA0414 and VCA0425. Cassette c253-5 shares 65%–66% identity with VCA0414 and VCA0425. Here, however, we observed that the divergences between the ORFs and the VXRs were in the same range, 63%–64% and 73%–75% identical, respectively. Noticeably, the repeat sequences of the VCA0414 and VCA0425 cassettes in V. cholerae have close homologs to unrelated V. cholerae cassettes, whereas the repeat sequence from its V. metschnikovii counterpart was less related to any of the VMeRs from our cassette sample (best score 70% identity) and clearly branch out of the VMeRs (Fig. 3).
Structural Characteristics of the VXRs
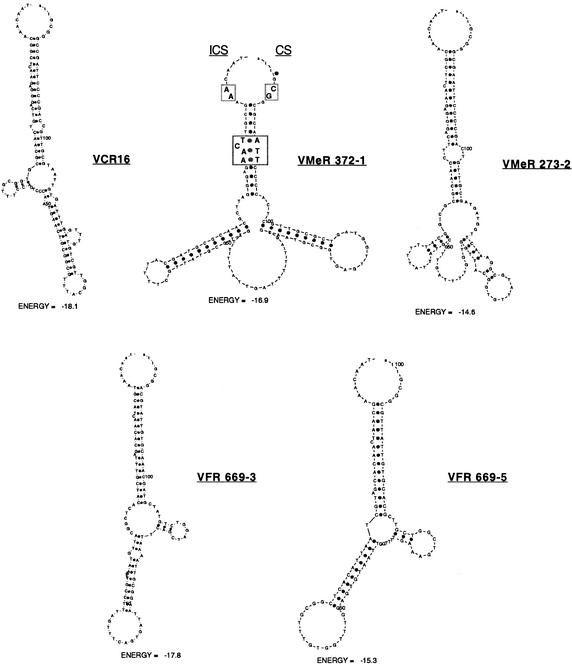
We have analyzed the different families of Vibrio attC sites for the presence of specific structural features similar to those described for the VCRs by Manning and colleagues (Barker et al. 1994). Like the VCRs, the VFRs and the VMeRs were also found to show imperfect dyad symmetry with the potential to adopt stable single-strand secondary conformations. Figure 4 shows examples of such potential secondary structures found for different representatives of the VXR families. We noticed that, in all cases, a potential stem structure was formed starting 4 nt downstream of the last “C” of the ICS sequence RYYYTAAC and hybridizing with a sequence ending 4 nt upstream of the first “G” in the CS sequence GTTARRY. More interestingly, we observed that the nucleotides in positions 5 to 8 in the ICS arm of this stem, including a protruding nucleotide (a C in 201 of the 219 VXRs analyzed), as well as the complementary nucleotides in the CS arm of the stem were conserved in all of the 219 VXRs analyzed (see alignment in supplementary figure; available online at http://www.genome.org). We also noticed that these specific characteristics were conserved in the PAR sequences recently described for the Pseudomonas alcaligenes SI cassettes (data not shown; Vaisvila et al. 2001). In each case, this “protruding C” region was found to overlap with the 2L/2R complementary sequences described by Hall and collaborators; however, we could not identify any convincing homology to the 2L and 2R consensus sequences within these regions. As shown in Figure 4, we observed that in addition to the protruding C mentioned above, protruding nucleotides were always found in the ICS arm of the stem and never in the CS arm, regardless of the primary sequence of the VXR.
Figure 4.
Proposed secondary structures of representative VXRs. Secondary structures and free energies were determined using the MFOLD (Walter et al. 1994) online interface at l'Institut Pasteur. The protruding C region of the proposed 2L site within the stem is boxed in VMeR 372-1, as are the nucleotides adjacent to the ICS and CS that are absolutely conserved in all 219 VXRs examined. A:T and G:C base pairs are marked with dark gray dots, respectively. (VCR) V. cholerae repeats; (VMeR) V. metschnikovii repeats; (VFR) V. fischeri repeats.
The VCRs of the V. cholerae SI cassettes were found in most cases to have a stretch of 9–11 consecutive complementary nucleotides. The majority of the VFRs and VMeRs also displayed a complementary stretch in the same length range, 9–11 nt. The CS consensus sequences found for the VCRs, VMeRs, and VFRs were determined to be GTTAKGYN, GTTAtRYK, and GTTAtRYg, respectively (the consensus is written in capital letters if >75% of the VXRs carry the same type of nucleotide at that position and in small letters if the incidence is 50%–75%; [K] G or T; [Y] C or T; [N] A, C, G, or T).
DISCUSSION
The recent evidence exalting the impact of lateral gene transfer (LGT) on bacterial evolution supports the conclusion that LGT is both an ancient and a perpetual phenomenon that has been a major force in shaping the genomes of extant species. The relatively recent discovery of integrons left open the question of the extent, beyond the dissemination of antibiotic-resistance genes, to which this particular system had an impact on bacterial evolution. The discovery of ancestral chromosomal SIs in diverse proteobacteria established this system as a key player in the evolution and plasticity of bacterial genomes (Rowe-Magnus and Mazel 2001). We used several independent approaches to analyze the SIs among remote species in the Vibrionaceae. We examined the SIs and the regions flanking the integron integrases for (1) clusters of orthologous genes, (2) conservation of local gene order, (3) the distribution of orthologs between species, and (4) comparison of phylogenetic trees constructed from 16S RNA and rplT and intIA genes. The data presented here from our extended analysis of known and novel SIs in members of the Vibrionaceae supports the contention of an ancient and a perpetual role for integrons in the engineering of extensive genetic diversity in this and other genera.
Integrons are Ancient Structures
We previously established that the evolutionary history of the superintegron platforms, that is, the intIA genes and their associated attI sites, paralleled that of the Vibrio radiation according to 16S RNA analysis, indicating that integrons are ancient structures (Rowe-Magnus et al. 2001). We have now extended our analysis through the identification and characterization of new SI platforms within the genomes of Vibrio species remote from those previously characterized. Every Vibrio species examined thus far has been found to harbor an SI. In addition, we have now identified the SI integrase and the first cassettes carried in the SIs of L. anguillarum and V. vulnificus, a fish pathogen and an emerging human pathogen, respectively. V. natriegens was also included in our study to validate the recent proposal for reclassification of the L. pelagia species CIP 10276.2 to the V. natriegens group (Macian et al. 2000). We observed that the intIA genes, as well as the chromosomal genes located at the SI boundary, were almost identical in L. pelagia CIP 10276.2 and V. natriegens 103193T, lending strong support to the reassignment of L. pelagia strain CIP 102762 to V. natriegens.
To provide additional support to the aforementioned congruence of the 16S rRNA and intI-based phylogenetic trees, we have characterized the genetic context in the neighborhood of the SIs by examining the chromosomal region flanking the intIA boundary in each Vibrio species. As shown in Figure 2, we observed that the location was in agreement with the grouping drawn from the phylogeny. Indeed, the position of the SI island on the chromosome was conserved within, but not between, the subclades, that is, the intIA genes in the V. cholerae clade were all located downstream of the same ribosomal operon, and all these strains belong to the same phylogenetic clade. Likewise, the intIA genes of the SIs in the V. parahaemolyticus clade were also located within the same genetic context, but their chromosomal location was different from that of the V. cholerae clade. Interestingly, the integrase gene of the V. vulnificus SI, VvuintIA, branched within the V. parahaemolyticus clade but was located in the same genetic context as the intIA genes of the V. cholerae clade. This indicated that the chromosomal rearrangement that led to the change in SI location from the neighborhood of the rplT gene to that of ORF474 likely occurred in the ancestor of V. parahaemolyticus subsequent to its divergence from V. vulnificus. In accordance with the topology of the intI tree, the location of the V. fischeri SI was completely unrelated to the other Vibrios, as its intIA was found adjacent to an ORF most closely related to VCA0034, which is located 250 kb away from the SI on Chromosome 2 of V. cholerae N16961. The conserved chromosomal context within the respective subclades in the intIA phylogenetic tree could be very useful in recovering the SIs from other species within the same subclade.
Informational genes, that is, those involved in numerous protein–protein interactions such as ribosomal genes, are proposed to be refractory to LGT (Jain et al. 1999). For this reason, ribosomal genes have been used as the cornerstone for phylogenetic analysis. Our phylogenetic analysis of the intIA genes revealed that the V. fischeri intIA gene had the deepest branching point among all the integron-integrases, but the V. fischeri branching point derived from 16S rRNA analysis placed this species inside the Vibrio radiation, between V. metschnikovii and V. cholerae (Rowe-Magnus et al. 2001; Vaisvila et al. 2001). Lateral transfer of the integron platform in V. fischeri would provide a simple explanation for this inconsistency. However, although less problematic in the prokaryotic realm, it has been shown that rRNA trees can sometimes be misleading in inferring phylogenies because of differences in base composition and unequal rates of evolution among species (Philippe and Laurent 1998). Hence, phylogenies based on alternative or multiple gene sets are being used more frequently to support the accuracy of rRNA trees.
In a phylogenetic analysis based on the ribosomal L20 genes (rplT), another essential chromosomal gene unlikely to be subject to horizontal transfer, we retrieved a branching order congruent with the intIA gene-based tree for all species examined (Rowe-Magnus et al. 2001). This discrepancy might be attributed to either a specific and congruent evolution of the intIA and rplT genes compared with the 16S rRNA genes in these species, or to an acquisition of the intIA and rplT genes from an unknown and phylogenetically remote bacterial source in V. fischeri. Although this question is still open, two observations in the work described here lead us to favor the first hypothesis. First, if we examine the integrase genes of V. cholerae and V. fischeri we observe 39% identity between them. The sole homolog to the ORF abutting VfiintIA is VCA0034 of V. cholerae, at 34% identity. These values are within the same range and are consistent with the placement of these species in the rplT gene. This indicates that these three genes, intIA, rplT, and the VCA0034 orthologs, coevolved together within these species. Second, if acquired from an exogenous source, the evolutionary congruence among intIA, rplT, and the VCA0034 ortholog in V. fischeri implies the simultaneous and relatively recent capture of all three genes. Because rplT is not in the vicinity of VfiintIA and the VCA0034 ortholog, this would necessitate the large-scale acquisition of a genetic element bearing an essential gene, that is, a chromosome, in a V. fischeri ancestor. Furthermore, the replacement of a single ribosomal component, such as rplT, or a subpopulation of components would give rise to heterogeneous ribosomal complexes that are likely to have a drastic effect on cell viability. Maintenance of such a chromosomal element thus seems highly unlikely. Likewise, coevolution of the three genes following their independent capture within a small evolutionary timeframe is also an improbable scenario. Finally, comparison of the intergenic sequences located between the different intIA and their adjacent chromosomal genes did not reveal any conserved sequences or structures to indicate that SIs are mobile. We favor the notion of a specific and congruent evolution of the intIA and rplT genes compared with the 16S rRNA genes in these species, and believe that the V. fischeri SI integrase branching point is consistent with the ancient and sedentary attributes of the system, and its coevolution with the host genome.
The Gene Cassette Reservoir
A comparison of subsets of cassettes from the SIs of V. metschnikovii, V. fischeri, and V. cholerae indicated that the majority of the cassettes found in the SI of one species were not found in that of another. None of the 19 unique V. fischeri cassettes had a counterpart in the SIs of the other Vibrio species examined, and only 6 out of the 32 V. metschnikovii cassettes examined were found to have counterparts in the V. cholerae SI. Two of these, c253-3 and c253-5, were complete and allowed further comparative study with their V. cholerae counterparts. Interestingly, these two families of cassettes showed different degrees of relatedness between the ORF and attC portions of the cassettes. Our analyses indicated that some common cassettes, such as c253-3 of V metschnikovii and VCA0338 of V. cholerae, were independently assembled within these bacteria or that a conversion mechanism corrected the associated XXR sequence into the signature attC site of that species following acquisition. However, the existence of cassettes carrying remote attC sites within the same SI renders this second alternative less probable. For comparison of other cassettes, such as c253-5 of V. metschnikovii and VCA0414 of V. cholerae, it is tempting to speculate that these specific cassettes both descended from a common ancestral cassette that was constructed in V. cholerae or a closely related Vibrio species and was then subsequently acquired in V. metschnikovii via horizontal transfer. Thus, some cassettes may have been constructed independently in the two species, whereas others were likely descendents of a common ancestral cassette that were acquired via horizontal transfer. The unique cassettes may have important roles in the exploitation of a particular niche by a particular species, and their shear numbers lend credence to the hypothesis of an in vivo cassette assembly process that is independently active within each species.
Analysis of the attC sites carried by the cassettes found in the V. cholerae N16961 SI showed that a large majority (∼85%) of the cassette-associated VCRs shared an overall similarity of >90% (Fig. 3), as previously observed among the first 13 VCRs identified (Barker et al. 1994). The rest of the VCRs showed at most 70% identity to the first group and were more closely related to VXRs of other species, such as the VMeRs of V. metschnikovii (Fig. 3). Because completely unrelated ORFs were associated with almost identical VCRs, this indicated that (1) the VCRs and the ORFs had independent origins, and the cassettes corresponded to the addition of an attC site to a gene through a specific assembly process; and (2) the cassettes carrying an attC site that was poorly related to the VCRs likely had an exogenous origin and have been acquired as complete cassettes through lateral transfer.
Cassette-Encoded Functions
The activity of only a handful of SI cassettes has been demonstrated experimentally. These include pathogenicity factors (van Dongen et al. 1987; Ogawa and Takeda 1993), antibiotic-resistance determinants (Melano et al. 2002; Rowe-Magnus et al. 2002b), metabolic genes (Barker and Manning 1997; Rowe-Magnus et al. 2001), and restriction enzymes (Rowe-Magnus et al. 2001; Vaisvila et al. 2001; R. Vaisvila, R. Morgan, and E. Raleigh, unpubl.). As seen for the V. cholerae and the P. alcaligenes SI cassettes (Heidelberg et al. 2000; Rowe-Magnus et al. 2001; Vaisvila et al. 2001), a large number of the V. metschnikovii and V. fischeri cassette-encoded genes have no counterparts in the databases or the sole homologs are ORFs of unassigned function (Tables 2 and 3). We also noticed that the putative products of a large number of cassettes are likely associated with the cell envelope as they contain signal peptide sequences and/or transmembrane segments (Tables 2 and 3). This might be indicative of a role for these cassettes in membrane integrity, variation, or adaptation to environmental changes.
Potential Secondary Structure of the attC Sites
Perhaps the most striking aspect of the activity of integron-integrases is their ability to recognize remote DNA sequences as targets (Mazel et al. 1998; Hall et al. 1999; Collis et al. 2001; Rowe-Magnus et al. 2001; Drouin et al. 2002; Hansson et al. 2002). Several common structural features among attC sites have been previously identified from the comparison of various antibiotic-resistance cassettes (Francia et al. 1997; Stokes et al. 1997). All attC sites possess (1) an internal imperfect dyad symmetry, (2) a core-site (CS) consensus of sequence GTTAGSC ([S] G or C, originally defined as GTTRRRY) and a perfectly complementary inverse core site (ICS) of consensus GSCTAAC (Hansson et al. 1997; Stokes et al. 1997), and (3) two sets of inversely oriented integrase binding sites, the LH and RH simple sites. We observed that regardless of the primary sequence of the VXR, in all cases a potential stem structure was formed starting 4 nt downstream of the ICS and hybridizing with a sequence ending 4 nt upstream of the CS sequence. More interestingly, we observed that the protruding C region and the nucleotides immediately flanking it in the stem were conserved in all but a few exceptions of the 219 VXRs analyzed. We also noticed that these specific characteristics were conserved in the much shorter attC sites of other SIs that have been recently identified (Vaisvila et al. 2001). In each case, this protruding C region was found to overlap with the 2L/2R complementary sequences described by Hall and collaborators; however, we could not identify any convincing homology to the 2L and 2R consensus sequences in these regions. Despite this, the VCRs have been experimentally shown to be substrates for the integrase of class 1 integrons (Mazel et al. 1998; Rowe-Magnus et al. 2001, 2002b). We observed that, in addition to the protruding C, protruding nucleotides were always found in the ICS arm of the stem and never in the CS arm sequence of the VXR. Along with the lack of identifiable 2L and 2R sites within the VXRs, these results support the hypothesis that, outside of the ICS and CS, the particular structural characteristics that result from the primary sequence in the imperfect inverted repeat regions of attC sites are more important in defining an attC site as a target for the integrase than the specific sequence itself. It would thus seem that symmetry, rather than sequence, is a key feature in the recognition of such a varied collection of target recombination sequences by a single site-specific recombinase. We are presently conducting experiments to test this hypothesis.
Microevolution Versus Macroevolution in SIs
Postsegregational killing (PSK) systems are normally found either on plasmids or within prophages. These systems generally consist of a pair of genes organized in an operon, with the downstream gene specifying a stable toxin and the upstream gene specifying a specific but unstable antitoxin. Once the operon is expressed, the cells are “addicted” to the short-lived antidote polypeptide, because its continued de novo synthesis becomes essential for cell survival (Engelberg-Kulka and Glaser 1999). Because loss of the extrachromosomal elements bearing these modules selectively kills the cured cells, these types of genetic systems have been found to enhance plasmid segregation and phage maintenance. The chromosomal equivalents are known as control of cell death (CCD) systems. We have demonstrated that one of the V. fischeri cassettes encodes an addiction module of the ccdAB family, a gyrase toxin and its antidote. Other chromosomal- or plasmid-encoded PSK systems that inhibit varied essential cellular functions have been characterized (Jensen and Gerdes 1995; Couturier et al. 1998; Engelberg-Kulka and Glaser 1999), but this is the only example, with the exception of a ccdAB operon found on the E. coli O157:H7 chromosome (Perna et al. 2001), of a gyrase-targeted killing system that is not carried by a plasmid (Couturier et al. 1998). Cassettes carrying genes with functions related to other PSK systems can also be identified in other SIs, such as the putative parDE (VCA0359/VCA0360) and higAB (VCA0391/VCA0392) cassette homologs of V. cholerae, and the death on curing (doc) P1 toxin analogs identified in the V. cholerae and V. metschnikovii SIs. Upstream of the doc-related gene (ORFc253-6b) in the V. metschnikovii cassette c253-6, is a small ORF (ORFc253-6a, Table 2), which almost certainly encodes a prevents host death (phd) antidote counterpart, even if its product, apart from a similar small size, shows little homology to the P1 phd gene. Interestingly, even if the annotated V. cholerae N16961 genome did not reveal a bona fide phd-like antidote gene associated with the doc analog, careful analysis of the sequence located upstream of the doc analog (VCA0475) led us to identify the same putative phd gene. However, in V. cholerae, a deletion event occurred inside the upstream phd–doc cassette, resulting in its fusion with an acetyl transferase gene, masking both the gene and the real boundary with the phd–doc cassette.
Several features, including the presence of the phd–doc, higAB, and parDE orthologs within the V. cholerae SI, led Heidelberg and coworkers to suggest that Chromosome 2 of V. cholerae was originally a megaplasmid that was captured by an ancestral Vibrio species (Heidelberg et al. 2000). The subsequent relocation of essential genes from Chromosome 1 to this megaplasmid then created a chromosome, enhancing the stable maintenance of this smaller replicon. The similar nucleotide composition and percentage G + C content between the two chromosomes would support the acquisition event to have occurred hundreds of millions of years ago. However, two features of the PSK systems weaken the argument that Chromosome 2 was once a megaplasmid. First, all three of these PSK systems are structured as gene cassettes and could have been acquired at any time. Second, in addition to being mobile, the GC composition of the aforementioned PSK gene cassettes is quite different (40%–44%) from the rest of the chromosome (47%). This contradicts the megaplasmid hypothesis because, if the phd–doc, higAB, or parDE PSK gene cassettes participated in the initial retention of the megaplasmid in an ancestor Vibrio species, then they would be expected to have a GC composition more in line with the genome. This indicates a more recent origin for the PSK systems and that they were acquired by a preexisting Chromosome 2.
An intriguing question is what is the function of these chromosomal addiction modules? PSK/CCD systems may have varied complex roles in bacterial behavior and evolution. Bacteria are unicellular organisms. Hence, they would not be expected to undergo programmed cell death (PCD). However, Glaser and colleagues recently discovered a chromosomal PCD locus, mazEF, in E. coli. This locus has the same organization as the PSK systems, encoding a stable toxin and an unstable but specific antidote, and functions in a similar manner. Although the target of the MazF toxin is not known, the operon was shown to be subject to regulation by guanosine 3′,5′-bispyrophosphate and antibiotics that are general inhibitors of transcription or translation (Aizenman et al. 1996; Sat et al. 2001). These compounds act as signaling molecules to trigger PCD in E. coli. This discovery has prompted biologists to revisit the view that bacteria may behave in multicellular ways. Furthermore, Orejas and coworkers have shown that the antidote of a second chromosomal ccd loci in E. coli, chpB, can neutralize the plasmid-borne toxin of the parD system (Santos Sierra et al. 1998). These types of functional interactions between systems may play an important role in bacterial evolution by permitting the acquisition of plasmids bearing only the toxin partner of the operon or, to reduce the genetic burden on the bacterium, the loss of plasmids carrying homologous PSK systems.
Clark et al. (2000) examined the global SI organization of 65 different V. cholerae O serotypes by PCR and Southern hybridization. Extensive restriction polymorphism was observed even among closely related isolates, indicating a plasticity in the SI structures and in their microevolution through integrase-mediated events. This is not unexpected because the gene cassettes are independently mobile genetic units that are supposedly subject to episodic selection and we have demonstrated that the integron–integrase randomly excises cassettes from SIs (Rowe-Magnus et al. 2002b). SIs may contain hundreds of attC sites and multiple copies of gene cassettes, insertion sequences, and pseudogenes. Because identical and likely functional copies of the same cassettes are found in the V. cholerae SI (such as the three copies of the glutathione-transferase, VCA0328, VCA0341, and VCA0463; Heidelberg et al. 2000; Rowe-Magnus et al. 2002b), it is unlikely that a mechanism specifically inactivating multicopy cassettes exists in these species to suppress homologous recombination between them. It is also commonly proposed that pseudogenes are rapidly eliminated from bacterial genomes by selection to provide streamlined, compact chromosomes to minimize the genetic burden (Lawrence et al. 2001), and insertion sequences are renowned for their activities in restructuring genomes (Shapiro 1999). Clearly, ample opportunities exist for large-scale deletions and rearrangements to occur between attC sites, IS elements, or multicopy cassettes, so the macroevolution of SIs would also be anticipated. Yet, paradoxically, SIs can remain remarkably stable. The genetic organization of MRIs promotes coexpression of the inserted gene cassettes from a single promoter, PC, that is internal to the integrase gene. Hence, selection for one resistance determinant often coselects for the maintenance of the entire array. However, a similar situation for SIs is difficult to imagine. We propose that the PSK/CCD systems act to stabilize the massive arrays of SI cassettes. A parDE paralogous gene family of seven members has been identified in the V. cholerae genome (gvc family 331, http://www.tigr.org/tigr-scripts/CMR2/ParalogousList.spl?db=gvc). Interestingly, all seven of these paralogs are structured as gene cassettes, bringing the total number for PSK-type systems in the V. cholerae SI to nine. Kobayashi and colleagues have shown that restriction-methylation systems (RMSs) fit all the properties of PSK systems (Kobayashi 1998; Kobayashi et al. 1999). Indeed, once acquired, they become essential for the survival of the bacteria because the long half-life of the nuclease compared with the methylase will eventually cause cell death if the RMS is lost (Kusano et al. 1995). Thus, the bacteria become dependent on the invading RMS. This facet has been demonstrated to enhance plasmid segregation stability in E. coli and Bacillus subtilis (Kulakauskas et al. 1995; Handa et al. 2000). We suggest that the presence of PSK systems in the SIs of the Vibrionaceae or RMS in the SIs of Xanthamonads (Rowe-Magnus et al. 2001) and Pseudomonads (Vaisvila et al. 2001) may act to stabilize these massive arrays of independently mobile genetic units by minimizing large-scale random excisions, because a probability exists to lose or shut off expression of the PSK or RMS cassettes. Such an event would result in cell death. In this scenario, the maintenance of other cassettes within these arrays would not have to arise from a direct link to the PSK or RMS cassettes and the array could be stably maintained in the absence of selection. Thus, the selfish nature of PSK/CCD systems could contribute positively to the fitness of bacteria, and this may help to explain the presence of these types of systems within SIs. Experiments are presently underway to test this hypothesis.
METHODS
Bacterial Strains, PCR, and Sequencing
Bacterial strains were provided by the Collection de l'Institut Pasteur (CIP). All genomic DNA was isolated by using the QIAGEN DNEasy Kit. Genomic DNA from each species was digested with the indicated restriction enzyme (V. metschnikovii, BamHI, EcoRI, or EcoRV; V. fischeri, BamHI, HindIII, or XbaI; L. anguillarum, HindIII), and partial libraries were constructed as previously described (Rowe-Magnus et al. 2001). PCR-amplified genes were cloned using the TA TOPO cloning kit (Invitrogen) and verified by sequencing of the corresponding genomic clones performed by MWG-Biotech. Primers were obtained from Genset.
Sequence and Phylogenetic Analysis
ORFs of at least 150 nt were identified using the ORF Finder at the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Each amino acid sequence was used as a BLAST query (Altschul et al. 1997) to search the GenBank, EMBL, SWISS-PROT, and the NCBI unfinished Microbial Genome BLAST Web site sequence databases. Signal peptides were identified with SignalP program v1.1 at the Center for Biological Sequence Analysis (http://www.cbs.dtu.dk/services/SignalP; Nielsen et al. 1997). Transmembrane proteins were predicted with TMHMM at the Center for Biological Sequence Analysis at the Technical University of Denmark (http://www.cbs.dtu.dk/ services/TMHMM-1.0/). Putative promoter sequences were predicted using the BDGP promoter software program (http://www.fruitfly.org/seq_tools/promoter.html). DNA secondary structures were determined using MFOLD analysis (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). Alignments were generated with the CLUSTALX software program (version 1.8), and dendrograms were compiled by using the neighbor-joining method (computed from 1000 independent trials) of CLUSTALX and PHYLIP or TREEVIEW.
Developing the XXR Program
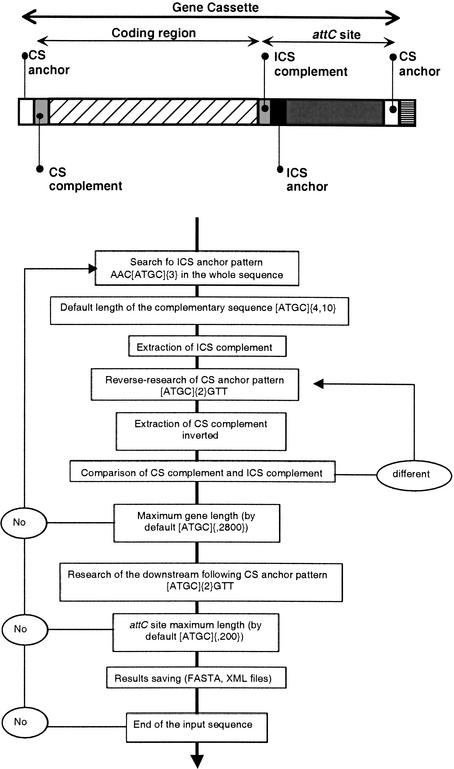
To automatically identify nucleotide sequences that are structurally organized as integron cassette arrays in any nucleotide sequence, we developed the program XXR. XXR is written in Perl (5.6 version) and is functional on any platform that uses this programming language (Linux, Unix, Cygwin). In principle, using any nucleotide sequence file in FASTA format, XXR is able to extract putative cassette structures that fulfill the criteria established from analysis of previously known cassettes from integrons and superintegrons. These criteria are (1) anchoring at an AAC to define the end of the inverse core site (ICS) and a GTT to define the start of the core site (CS); (2) the minimum and maximum length of the complementary sequences between the ICS and the cognate upstream CS in the cassette as well as their identification; and (3) the maximum length of the two building blocks of a cassette, the ORF and its associated attC site, in order to avoid aberrant cassettes. Most parameters can be modified; however, some, like the anchor sequences in the CS (GTT) and in the ICS (AAC) and the minimal length of the complementarity, are fixed. The most important point is the definition of the ICS, as this sequence is the anchoring point for the research of the full cassette sequence. The boundary is then searched in the neighborhood upstream and downstream of the ICS. After analysis of a given sequence, XXR outputs two different file types: a file of data gathering all the extracted cassettes in FASTA format and XML (eXtensible Markup Language) files for each sequence that likely corresponds to a cassette. Figure 5 shows the procedure followed by XXR for the retrieval of cassette sequences from the input sequence file. XXR can be used through our Web site, http://www.pasteur.fr/recherche/unites/ptmg/integ.
Figure 5.
Graphical representation of the search parameters used to retrieve attC sites by the program XXR. See text for details.
Cloning Novel SI Loci and Gene Cassettes in the Vibrionaceae
Gene cassettes from the V. metschnikovii SI were identified by screening of EcoRI and BglII partial libraries with the primers VMeR1 (GTCCCTCTTGAAGCGTTTGTTA) and VMeR2 (GC CCCTTAAGCGGGCGTTA). The same strategy was used to screen V. fischeri BamHI, HindIII, and XbaI partial libraries with the primers VFR1 and VFR2 (Rowe-Magnus et al. 2001). Cloning of the integrase genes from the SI loci carried in V. vulnificus 75.4 and V. natriegens 103193 was performed by PCR using primers LPR1 (CGATCCCTCTTGAAGTTTGTTA) and I456/5 (TCTTGTAC(C/G)GT(A/T)CGTATATCA) or LPR1-2 (GAATCACTCTTAAACAGTTTGTTA) and LPINT8-2 (CCTT TACCTTGCCAGACACG), respectively. LPR1 and LPR1-2 were designed from the comparison of the attC sites of the cassettes found in the L. pelagia SI (LPRs), and correspond to the outer end of the LPR. I456/5 was designed from comparison of known Vibrio SI integrases and corresponds to a highly conserved segment at the 3′ end of the gene. The PCR products were cloned using the TA TOPO cloning kit to give p1184 and p1216. Sequence analysis of the corresponding PCR products allowed us to identify most of the intIA genes, the cognate attI sites, and the first cassettes of both the V. vulnificus and V. natriegens SIs. Southern analysis of L. anguillarum genomic DNA probed with the V. fischeri intIA gene showed that the LanintIA gene was carried on a 2.5-kb HindIII fragment. Screening of a L. anguillarum HindIII partial library with the same probe allowed the isolation of p1456, a plasmid carrying the corresponding insert. Sequence analysis revealed that this fragment carried the intIA gene and the first three cassettes of the SI. As the L. anguillarum intIA gene was most closely related to the V. metschnikovii intIA gene, we tested whether the LanintIA was located in the same chromosomal context, that is, downstream of the rplT gene (Fig. 2). This was achieved by PCR using RPLT (ATGCCTCGCGTAAAACGTGGTGTAC), a primer corresponding to the highly conserved 5′ region of the rplT gene, and Lang1 (TCGGCATTGCAGCGAGCAGTTCGG), a primer internal to the LanintIA (p2010, Table 1). The result was a 630-nt product that was similar in size to the corresponding fragment in V. metschnikovii. Sequencing of this fragment confirmed the intIA location downstream of the rplT gene (Fig. 2). Using an analogous PCR strategy with oligonucleotides Vna1 (GATTTCTTTCAGACACCGCTCTCAC) and ORF474 (GACTGAATGTCTTATTTGCCTTTGG), we were able to show that the V. natriegens intIA gene was located in the same genetic context as the L. pelagia and V. parahaemolyticus IntIA genes (p2018, Table 1). Identification of the V. vulnificus intIA chromosomal context was achieved by using the same PCR strategy with oligonucleotides RPLT and Vvu1 (CTG CAGAAACAGGCACTCATCAGGATG; p2019, Table 1).
Cloning the ccd Cassette of V. fischeri
A 739-bp NsiI fragment of p669 that encompassed the two ORFs of the V. fischeri cassette c669-2 was cloned into pNOT218 that had been digested with PstI. This plasmid, p1357, contained the ccdAB genes in the opposite orientation relative to the lacZ promoter. To ensure that the ccdAB operon would be expressed in E. coli, the p1357 insert was recloned into pTZ19R, placing the ccdAB genes in the same relative orientation as the inducible lac promoter (p1400). The toxigenic activity associated with ccdB expression was demonstrated by subcloning the AclI–EcoRI internal fragment of p1400, which contained only the 3′ end of ccdA and an intact ccdB, into pSU19 digested by AccI and EcoRI in order to put ccdB under the control of Plac (p1446). The ligation product was then used to transform either DH5α or ω106, a DH5α strain containing the plasmid p1400.
WEB SITE REFERENCES
http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html; MFOLD analysis, Institut Pasteur.
http://www.cbs.dtu.dk/services/SignalP; SignalP program v1.1, the Center for Biological Sequence Analysis.
http://www.cbs.dtu.dk/services/TMHMM-1.0/; transmembrane protein prediction, the Technical University of Denmark.
http://www.fruitfly.org/seq_tools/promoter.html; BDGP promoter software program, Berkley Drosophila Genome Project.
http://www.ncbi.nlm.nih.gov/gorf/gorf.html; ORF Finder, the National Center for Biotechnology Information.
http://www.pasteur.fr/recherche/unites/pmtg/integ; authors' Web site.
http://www.tigr.org/tigr-scripts/CMR2/ParalogousList.spl?db=gvc; paralogous gene families, The Institute for Genomic Research.
http://www.ncbi.nlm.nih.gov/BLAST/; Similarity search programs, the National Center for Biotechnology Information.
Acknowledgments
We dedicate this paper to the memory of Pr. Maurice Hofnung, former Chef de l'Unité PMTG, for his encouragement, guidance and friendship. D.R-M. is an EMBO and a Fondation de la Recherche Medicale (FRM) Post-doctoral fellow. This work was supported by the Institut Pasteur, the CNRS, and the Programme de Recherche Fondamentale en Microbiologie et Maladies Infectieuses et Parasitaires from the MENRT and the DGA (contract no. 0134020).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
E-MAIL mazel@pasteur.fr; FAX 33 1 45 68 87 90.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.617103.
REFERENCES
- 1.Aizenman E., Engelberg-Kulka, H., and Glaser, G. 1996. An Escherichia coli chromosomal “addiction module” regulated by guanosine [corrected] 3′,5′-bispyrophosphate: A model for programmed bacterial cell death. Proc. Natl. Acad. Sci. 93: 6059-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker A. and Manning, P.A. 1997. VlpA of Vibrio cholerae O1: The first bacterial member of the α 2-microglobulin lipocalin superfamily. Microbiology 143: 1805-1813. [DOI] [PubMed] [Google Scholar]
- 4.Barker A., Clark, C.A., and Manning, P.A. 1994. Identification of VCR, a repeated sequence associated with a locus encoding a hemagglutinin in Vibrio cholerae O1. J. Bacteriol. 176: 5450-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caporale L.H. 1999. Chance favors the prepared genome. Ann. NY Acad. Sci. 870: 1-21. [DOI] [PubMed] [Google Scholar]
- 6.Clark C.A., Purins, L., Kaewrakon, P., Focareta, T., and Manning, P.A. 2000. The Vibrio cholerae O1 chromosomal integron. Microbiology 146: 2605-2612. [DOI] [PubMed] [Google Scholar]
- 7.Collis C.M., Kim, M.J., Stokes, H.W., and Hall, R.M. 1998. Binding of the purified integron DNA integrase Intl1 to integron- and cassette-associated recombination sites. Mol. Microbiol. 29: 477-490. [DOI] [PubMed] [Google Scholar]
- 8.Collis C.M., Recchia, G.D., Kim, M.J., Stokes, H.W., and Hall, R.M. 2001. Efficiency of recombination reactions catalyzed by class 1 integron integrase IntI1. J. Bacteriol. 183: 2535-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Couturier M., Bahassi, el-M., and Van Melderen, L. 1998. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 6: 269-275. [DOI] [PubMed] [Google Scholar]
- 10.Drouin F., Melancon, J., and Roy, P.H. 2002. The IntI-like tyrosine recombinase of Shewanella oneidensis is active as an integron integrase. J. Bacteriol. 184: 1811-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Engelberg-Kulka H. and Glaser, G. 1999. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu. Rev. Microbiol. 53: 43-70. [DOI] [PubMed] [Google Scholar]
- 12.Francia M.V., Avila, P., de la Cruz, F., and Garcia Lobo, J.M. 1997. A hot spot in plasmid F for site-specific recombination mediated by Tn21 integron integrase. J. Bacteriol. 179: 4419-4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall R.M. and Stokes, H.W. 1993. Integrons: Novel DNA elements which capture genes by site-specific recombination. Genetica 90: 115-132. [DOI] [PubMed] [Google Scholar]
- 14.Hall R.M., Collis, C.M., Kim, M.J., Partridge, S.R., Recchia, G.D., and Stokes, H.W. 1999. Mobile gene cassettes and integrons in evolution. Ann. NY Acad. Sci. 870: 68-80. [DOI] [PubMed] [Google Scholar]
- 15.Handa N., Ichige, A., Kusano, K., and Kobayashi, I. 2000. Cellular responses to postsegregational killing by restriction-modification genes. J. Bacteriol. 182: 2218-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansson K., Skold, O., and Sundstrom, L. 1997. Non-palindromic attl sites of integrons are capable of site-specific recombination with one another and with secondary targets. Mol. Microbiol. 26: 441-453. [DOI] [PubMed] [Google Scholar]
- 17.Hansson K., Sundstrom, L., Pelletier, A., and Roy, P.H. 2002. IntI2 integron integrase in Tn7. J. Bacteriol. 184: 1712-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heidelberg J.F., Eisen, J.A., Nelson, W.C., Clayton, R.A., Gwinn, M.L., Dodson, R.J., Haft, D.H., Hickey, E.K., Peterson, J.D., Umayam, L., et al. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406: 477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hochhut B., Lotfi, Y., Mazel, D., Faruque, S.M., Woodgate, R., and Waldor, M.K. 2001. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob. Agents Chemother. 45: 2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain R., Rivera, M.C., and Lake, J.A. 1999. Horizontal gene transfer among genomes: The complexity hypothesis. Proc. Natl. Acad. Sci. 96: 3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen R.B. and Gerdes, K. 1995. Programmed cell death in bacteria: Proteic plasmid stabilization systems. Mol. Microbiol. 17: 205-210. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi I. 1998. Selfishness and death: Raison d'etre of restriction, recombination and mitochondria. Trends Genet. 14: 368-374. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi I., Nobusato, A., Kobayashi-Takahashi, N., and Uchiyama, I. 1999. Shaping the genome—Restriction-modification systems as mobile genetic elements. Curr. Opin. Genet. Dev. 9: 649-656. [DOI] [PubMed] [Google Scholar]
- 24.Kulakauskas S., Lubys, A., and Ehrlich, S.D. 1995. DNA restriction-modification systems mediate plasmid maintenance. J. Bacteriol. 177: 3451-3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kusano K., Naito, T., Handa, N., and Kobayashi, I. 1995. Restriction-modification systems as genomic parasites in competition for specific sequences. Proc. Natl. Acad. Sci. 92: 11095-11099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence J.G., Hendrix, R.W., and Casjens, S. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9: 535-540. [DOI] [PubMed] [Google Scholar]
- 27.Levesque C., Brassard, S., Lapointe, J., and Roy, P.H. 1994. Diversity and relative strength of tandem promoters for the antibiotic-resistance genes of several integrons. Gene 142: 49-54. [DOI] [PubMed] [Google Scholar]
- 28.Liebert C.A., Hall, R.M., and Summers, A.O. 1999. Transposon Tn21, flagship of the floating genome. Microbiol. Mol. Biol. Rev. 63: 507-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macian M.C., Ludwig, W., Schleifer, K.H., Garay, E., and Pujalte, M.J. 2000. Vibrio pelagius: Differences of the type strain deposited at various culture collections. Syst. Appl. Microbiol. 23: 373-375. [DOI] [PubMed] [Google Scholar]
- 30.Mazel D., Dychinco, B., Webb, V.A., and Davies, J. 1998. A distinctive class of integron in the Vibrio cholerae genome. Science 280: 605-608. [DOI] [PubMed] [Google Scholar]
- 31.Melano R., Petroni, A., Garutti, A., Saka, H.A., Mange, L., Pasteran, F., Rapoport, M., Rossi, A., and Galas, M. 2002. New carbenicillin-hydrolyzing β-lactamase (CARB-7) from Vibrio cholerae non-O1, non-O139 strains encoded by the VCR region of the V. cholerae genome. Antimicrob. Agents Chemother. 46: 2162-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miki T., Yoshioka, K., and Horiuchi, T. 1984. Control of cell division by sex factor F in Escherichia coli. I. The 42.84–43.6 F segment couples cell division of the host bacteria with replication of plasmid DNA. J. Mol. Biol. 174: 605-625. [DOI] [PubMed] [Google Scholar]
- 33.Naas T., Mikami, Y., Imai, T., Poirel, L., and Nordmann, P. 2001. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J. Bacteriol. 183: 235-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nield B.S., Holmes, A.J., Gillings, M.R., Recchia, G.D., Mabbutt, B.C., Nevalainen, K.M., and Stokes, H.W. 2001. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 195: 59-65. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen H., Engelbrecht, J., Brunak, S., and von Heijne, G. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10: 1-6. [DOI] [PubMed] [Google Scholar]
- 36.Ochman H., Lawrence, J.G., and Groisman, E.A. 2000. Lateral gene transfer and the nature of bacterial evolution. Nature 405: 299-304. [DOI] [PubMed] [Google Scholar]
- 37.Ogawa A. and Takeda, T. 1993. The gene encoding the heat-stable enterotoxin of Vibrio cholerae is flanked by 123-base pair direct repeats. Microbiol. Immunol. 37: 607-616. [DOI] [PubMed] [Google Scholar]
- 38.Perna N.T., Plunkett, G., III, Burland, V., Mau, B., Glasner, J.D., Rose, D.J., Mayhew, G.F., Evans, P.S., Gregor, J., Kirkpatrick, H.A., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409: 529-533. [DOI] [PubMed] [Google Scholar]
- 39.Philippe H. and Laurent, J. 1998. How good are deep phylogenetic trees? Curr. Opin. Genet. Dev. 8: 616-623. [DOI] [PubMed] [Google Scholar]
- 40.Recchia G.D. and Hall, R.M. 1995. Gene cassettes: A new class of mobile element. Microbiology 141: 3015-3027. [DOI] [PubMed] [Google Scholar]
- 41.Rowe-Magnus D.A. and Mazel, D. 1999. Resistance gene capture. Curr. Opin. Microbiol. 2: 483-488. [DOI] [PubMed] [Google Scholar]
- 42.___, 2001. Integrons: Natural tools for bacterial genome evolution. Curr. Opin. Microbiol. 4: 565-569. [DOI] [PubMed] [Google Scholar]
- 43.Rowe-Magnus D.A., Guerout, A.-M., and Mazel, D. 1999. Super-integrons. Res. Microbiol. 150: 641-651. [DOI] [PubMed] [Google Scholar]
- 44.Rowe-Magnus D.A., Guerout, A.-M., Ploncard, P., Dychinco, B., Davies, J., and Mazel, D. 2001. The evolutionary history of chromosomal super-integrons provides an ancestry for multiresistant integrons. Proc. Natl. Acad. Sci. 98: 652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rowe-Magnus D.A., Davies, J., and Mazel, D. 2002a. Impact of integrons and transposons on the evolution of resistance and virulence. Curr. Top Microbiol. Immunol. 264: 167-188. [PubMed] [Google Scholar]
- 46.Rowe-Magnus D.A., Guerout, A.M., and Mazel, D. 2002b. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 43: 1657-1669. [DOI] [PubMed] [Google Scholar]
- 47.Santos Sierra S., Giraldo, R., and Diaz Orejas, R. 1998. Functional interactions between chpB and parD, two homologous conditional killer systems found in the Escherichia coli chromosome and in plasmid R1. FEMS Microbiol. Lett. 168: 51-58. [DOI] [PubMed] [Google Scholar]
- 48.Sat B., Hazan, R., Fisher, T., Khaner, H., Glaser, G., and Engelberg-Kulka, H. 2001. Programmed cell death in Escherichia coli: Some antibiotics can trigger mazEF lethality. J. Bacteriol. 183: 2041-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shapiro J.A. 1999. Transposable elements as the key to a 21st century view of evolution. Genetica 107: 171-179. [PubMed] [Google Scholar]
- 50.Stokes H.W., O'Gorman, D.B., Recchia, G.D., Parsekhian, M., and Hall, R.M. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26: 731-745. [DOI] [PubMed] [Google Scholar]
- 51.Sundstrom L. 1998. The potential of integrons and connected programmed rearrangements for mediating horizontal gene transfer. APMIS Supplementum 84: 37-42. [DOI] [PubMed] [Google Scholar]
- 52.Sundstrom L., Roy, P.H., and Skold, O. 1991. Site-specific insertion of three structural gene cassettes in transposon Tn7. J. Bacteriol. 173: 3025-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaisvila R., Morgan, R.D., Posfai, J., and Raleigh, E.A. 2001. Discovery and distribution of super-integrons among Pseudomonads. Mol. Microbiol. 42: 587-601. [DOI] [PubMed] [Google Scholar]
- 54.van Dongen W.M.A.M., van Vlerken, M.M.A., and De Graaf, F.K. 1987. Nucleotide sequence of a DNA fragment encoding a Vibrio cholerae haemagglutinin. Mol. Gen. (Life Sci. Adv.) 6: 85-91. [Google Scholar]
- 55.Walter A.E., Turner, D.H., Kim, J., Lyttle, M.H., Muller, P., Mathews, D.H., and Zuker, M. 1994. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. 91: 9218-9222. [DOI] [PMC free article] [PubMed] [Google Scholar]